Abstract
The ESCRT pathway functions in vesicle formation at the multivesicular body, the budding of enveloped RNA viruses such as HIV-1, and the final abscission stage of cytokinesis. As the only known enzyme in the ESCRT pathway, the AAA ATPase Vps4 provides the energy required for multiple rounds of vesicle formation. Like other Vps4 proteins, yeast Vps4 cycles through two states: a catalytically inactive disassembled state that we show here is a dimer, and a catalytically active higher order assembly that we have modeled as a dodecamer composed of two stacked hexameric rings. We also report crystal structures of yeast Vps4 proteins in the apo- and ATPγS-bound states. In both cases, Vps4 subunits assembled into continuous helices with six-fold screw axes that are analogous to helices seen previously in other Vps4 crystal forms. The helices are stabilized by extensive interactions between the large and small AAA ATPase domains of adjacent Vps4 subunits, suggesting that these contact surfaces may be used to build both the catalytically active dodecamer and catalytically inactive dimer. Consistent with this model, we have identified interface mutants that specifically inhibit Vps4 dimerization, dodecamerization, or both. Thus, the Vps4 dimer and dodecamer likely form distinct but overlapping interfaces. Finally, our structural studies have allowed us to model the conformation of a conserved loop (Pore Loop 2) that is predicted to form an arginine-rich pore at the center of one of the Vps4 hexameric rings. Our mutational analyses demonstrate that Pore Loop 2 residues Arg241 and Arg251 are required for efficient HIV-1 budding, thereby supporting a role for this “arginine collar” in Vps4 function.
Keywords: Vps4, AAA ATPase, Oligomerization, MVB pathway, X-ray Crystallography
Introduction
The processes of vesicle budding into multivesicular bodies (MVBs), retrovirus release, and cytokinesis share a common requirement for an overlapping subset of cellular machinery, termed the Endosomal Sorting Complexes Required for Transport (ESCRT)1; 2; 3. The ESCRT machinery may perform analogous functions at the final stages of all three processes, where a membrane fission event is required to resolve a thin, cytosol-containing membrane tubule. At steady state, ESCRT factors bind weakly to membranes throughout the cell4, but they can be recruited to function at specific sites of membrane remodeling including the late endosome, the plasma membrane, and the midbody. Most ESCRT factors function as subunits of one of four different complexes, termed ESCRT-0, -I, -II, and -III. Although their functions are not yet fully defined, ESCRT-0, -I, and -II appear to act as adaptors, binding directly to both membranes and protein cargoes as they are sorted into vesicles or virions. ESCRT-III subunits, in contrast, appear to co-assemble into membrane-associated filaments that play a more direct role in membrane remodeling, possibly mediating membrane extrusion and/or fission5.
Once assembled on membranes, the ESCRT machinery is released by the action of the Vps4 ATPases, the only known enzymes in the ESCRT pathway. A direct role for Vps4 function in the ESCRT pathway is indicated by a number of observations, including: 1) Vps4 localizes to endosomal membranes6 and to the midbody during cytokinesis7; 8, 2) MVB protein sorting and intralumenal vesicle formation are inhibited in the absence of Vps4 or upon expression of dominant negative Vps4 that cannot bind or hydrolyze ATP6; 9; 10; 11; 12, and 3) Depletion or dominant inhibition of Vps4 blocks late stages of both cytokinesis7; 8 and enveloped virus budding (reviewed in 13; 14; 15). At a minimum, Vps4 ATPase activity appears to be required to release and recycle the assembled ESCRT machinery, because the other ESCRT factors assemble but are not released in the absence of Vps4 activity16. Vps4 activity may also be mechanistically coupled to membrane deformation and/or fission, though such coupling has not been demonstrated experimentally.
Vps4 proteins belong to the large and diverse family of AAA ATPases (ATPases Associated with diverse cellular Activities) (reviewed in 17; 18; 19; 20; 21; 22; 23). AAA ATPases can contain either one (class I) or two (class II) ATPase cassettes, and Vps4 is a class I ATPase. Yeasts express a single Vps4 protein, whereas humans and other mammals express two closely related Vps4 proteins, termed VPS4A and VPS4B/SKD124. As summarized in Fig. 1a, Vps4 proteins contain an N-terminal MIT domain that binds ESCRT-III protein substrates, a central ATPase cassette composed of large and small domains, and a three-stranded antiparallel sheet (the β domain) inserted within the small ATPase domain 25; 26; 27; 28. On its own, the linker between the MIT and ATPase regions lacks a persistent structure26, and can be hyper-susceptible to proteolysis in the context of the full length protein. This linker undergoes nucleotide-dependent changes in proteolytic susceptibility28, however, suggesting that it serves as a semi-flexible tether that can adopt different conformations.
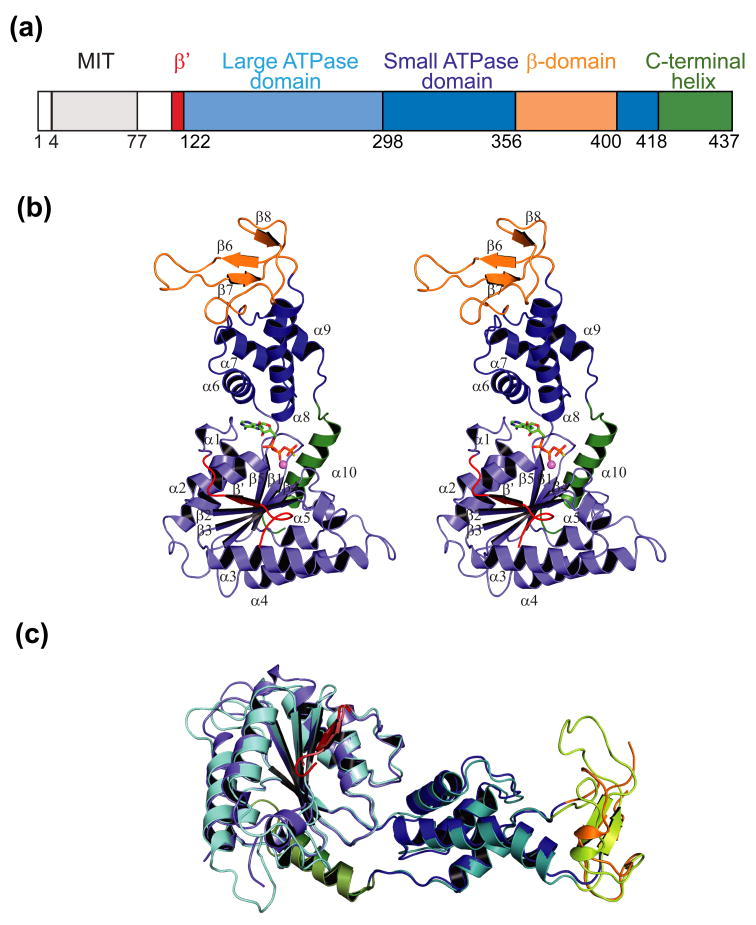
Figure 1.
Structure of yeast Vps4.
(a) Domain organization of the human and yeast Vps4 proteins. Numbering scheme corresponds to the yeast Vps4 protein.
(b) Stereoview of the Vps4ΔMIT structure. This structure corresponds to molecule A in the ATPγS bound state in crystal form 2. Domains are color coded as in part (a). Note that the small helix spanning residues 199–206 is not numbered in order to maintain consistency with previous conventions.
(c) Superposition of yeast Vps4ΔMIT (in crystal form 1) and human VPS4BΔMIT (lighter shades) in their sulfate-bound states. The two proteins overlay with an RMSD of 1.2 Å over 225 Cα positions within the AAA ATPase cassette.
Most AAA ATPases function as closed hexameric rings, although other arrangements are known29. Ring formation is mediated by the AAA ATPase cassette, and ATP binding typically promotes ring assembly because nucleotides bind between and contact adjacent subunits18. Vps4 is typical in this regard because ATP binding promotes enzyme assembly16; 25, and Vps4 appears to have a canonical ATP binding site (although high resolution structures of ATP-bound Vps4 proteins have not previously been reported). Upon ATP binding, Type I ATPases generally assemble into single ring structures, whereas Type II ATPases generally form double ring structures in which the two ATPase cassettes form separate, stacked rings (e.g.,21; 30). The Type I Vps4 enzymes are atypical in this regard in that they appear to form double stacked rings16; 25; 27; 31. Crystal structures of double ring Vps4 complexes are not yet available, and current models for the stoichiometry and structures of this assembly have therefore been deduced from gel filtration 16; 25, protein crosslinking16; 31, mutagenesis25, single particle cryo-EM reconstructions27; 31, and modeling studies25; 27. Vps4 proteins also bind an activator, Vta1/SBP1/LIP5, which contacts the β domain and promotes assembly and ATPase activity25; 31; 32; 33; 34; 35; 36; 37; 38. Recent biochemical and EM studies of the yeast Vps4-Vta1 supercomplex suggest that Vta1 may bind just one of the two rings in the double-ring assembly31.
Although there is general agreement that Vps4 enzymes drive the ESCRT pathway by cycling between an inactive, low molecular weight state and an active, high molecular weight state, uncertainties surround the structures and stoichiometries of both the active and inactive conformations. For example, the inactive state of Vps4 has been described as both a monomer28 and a dimer16; 25; 27, and the nature of the putative dimer is not clear. Similarly, two different single particle reconstructions of the fully assembled core enzyme at moderate resolution have been interpreted as showing a dodecamer31 or a tetradecamer27, and these studies also differ on other aspects of the global architecture. In an effort to characterize the architectures and oligomeric states of Vps4 further, we have determined two new crystal structures of the S. cerevisiae Vps4 core enzyme, performed analytical ultracentrifugation analyses that define the oligomeric state of the inactive Vps4 complex, and carried out extensive mutagenesis aimed at discriminating between different Vps4 assembly models and identifying residues that perform key functions in the active and inactive states.
Results
Structure Determinations
Two different Saccharomyces cerevisiae Vps4 constructs that spanned residues 104–437 and 122–437 were expressed in E. coli, crystallized, and structurally characterized by X-ray crystallography using the human VPS4B structure as an initial search model25. Both constructs lacked N-terminal MIT domains and segments of the ensuing linker (see Fig. 1), and both also contained the E233Q mutation, which allows ATP binding but blocks ATP hydrolysis16. Vps4122–437 crystallized in space group P6522, with one protein molecule per asymmetric unit and a sulfate ion bound at the active site. This structure was refined at a resolution of 2.7 Å and an Rfree value of 28.7% (Table 1). Vps4104–437 crystallized in space group P212121 with three molecules per asymmetric unit and was refined at a resolution of 3.25 Å to an Rfree value of 28.9%. Molecules A and B showed good density for ATPγS at the active site (Fig. 2), whereas molecule C also had active site electron density but clearly lacked density for the adenine nucleoside. The density could be adequately modeled as an ethylene glycol molecule in complex with a magnesium ion, and this is therefore our tentative assignment for the bound ligand in this case. Crystallographic statistics are summarized in Table 1, and both structures are generally well ordered, albeit at modest resolution. As detailed in Table 2, the ordered regions of all constructs begin at approximately residue 122, and we therefore refer to the proteins interchangeably as Vps4ΔMIT, with their bound ligands designated as “SO4” (crystal form 1), “ATPγS” (molecules A and B in crystal form 2), and “ethylene glycol-Mg2+” (molecule C in crystal form 2).
Table 1.
Data collection and refinement statistics
| Vps4122–437, E233Q Crystal form 1 | Vps4104–437, E233Q Crystal form 2 | |
| Data collection | ||
| Space group | P6522 | P212121 |
| Cell parameters (Å) | a = 110.8 | a = 77.0 |
| b = 110.8 | b = 119.8 | |
| c = 169.1 | c = 156.8 | |
| Wavelength (Å) | 1.00 | 1.00 |
| Resolution (Å) | 30-2.7 | 30-3.25 |
| Outer Shell (Å) | 2.77-2.7 | 3.37-3.25 |
| Number of Reflections | ||
| Total observed | 117,584 | 121,837 |
| Unique | 17,628 | 23,550 |
| Completeness (%) | 99.8 (97.5) | 99.8 (100) |
| Rsym (%)a | 11.1 (52.6) | 12.0 (62.9) |
| Mean I/σ (I) | 9.8 (2.3) | 8.0 (1.9) |
| Refinement statistics | ||
| Rcryst/Rfree (%)b,c | 24.1/28.7 | 21.6/28.9 |
| RMSD from ideal geometryd | ||
| Bond lengths (Å) | 0.012 | 0.017 |
| Bond angles (°) | 1.489 | 1.970 |
| Average B-factor (Å2) | 51.5 | 86.6 |
| Phi/psi angles (non Gly/Pro) | ||
| Most favorable region (%) | 86.7 | 77.4 |
| Additional allowed region (%) | 10.7 | 20.5 |
| Generous allowed region (%) | 2.7 | 1.6 |
| Disallowed region (%) | 0.4 | 0.7 |
Values in parenthesis are for the highest resolution shell.
Rsym = (|(ΣI − <I>)|)/(ΣI), where <I> is the average intensity of multiple measurements.
Rcryst = Σhkl||Fobs(hkl)|| − Fcalc(hkl)||/Σhkl|Fobs(hkl)|
Rfree = the crossvalidation R factor for 5% of reflections against which the model was not refined
Geometry was analyzed in PROCHECK67.
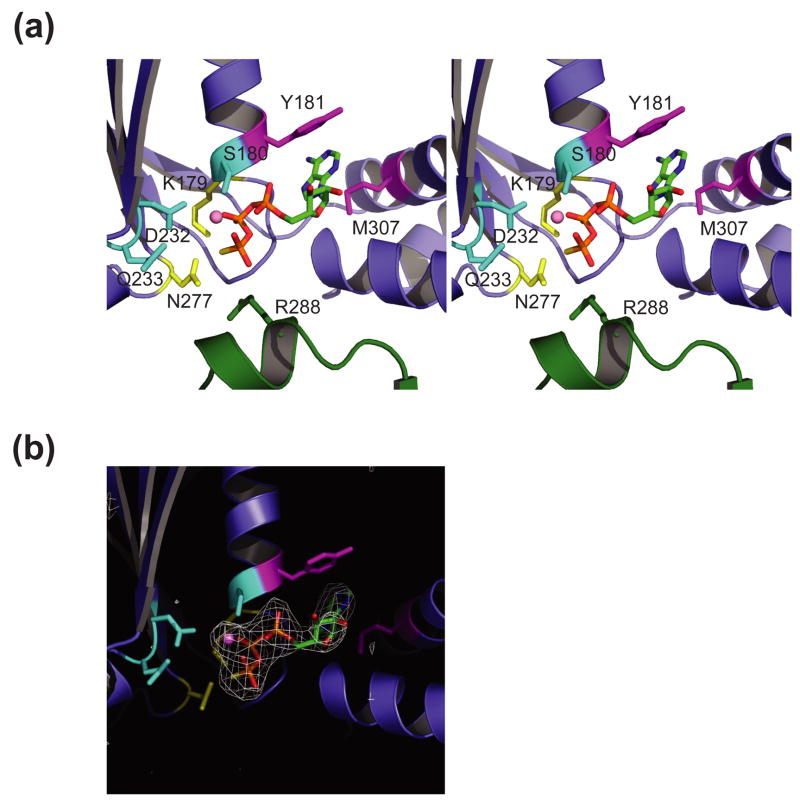
Figure 2.
Vps4 nucleotide binding pockets.
(a) Stereoview of the Vps4-ATPγS nucleotide binding site of molecule A in crystal form 2. Active site Vps4 residues are color coded according to their functional roles in Mg2+ coordination/ATP hydrolysis (cyan, S180, D232 and E/Q233), adenine ring stacking (magenta, Y181 and M307) and phosphate sensing (yellow, K179 and N277) with the “arginine finger” residue R288 from an adjacent molecule in the modeled hexamer shown in green. Note that the E233Q mutant was used here and throughout to allow ATP/ATPγS binding while inhibiting hydrolysis.
(b) Electron density for the ATPγS nucleotides in molecule A of Vps4ΔMIT crystal form 2. The densities show (F0 – FC) omit maps contoured at 2.5 σ.
Table 2.
The intermolecular lattice contacts in all known Vps4ΔMIT structures
| Structure | PDB | Mol/AU | Ligand bound | Hinge angle I | Hinge angle II | Total buried area per monomer [Å2] |
|---|---|---|---|---|---|---|
| Vps4 (126–433) | 2RKO | 1 | apo | 116 | 157 | Interface 1 – 637
Interface 2 – 290 Interface 3 – 156 Interface 6 – 722 |
| Vps4 (129–433) | 2QP9 | 1 | SO4 | 116 | 161 | Interface 1 – 740
Interface 2 – 431 Interface 3 – 284 Interface 6 - 740 |
| Vps4 (122–433) | This work | 1 | SO4 | 118 | 156 | Interface 1 – 477
Interface 2 – 333 Interface 3 – 409 Interface 6 – 767 |
| Vps4
Molecule A (119–435) Molecule B (119–435) Molecule C (123–432) |
2QPA | 3 | ADP
PO4 PO4 |
116
119 124 |
147
160 153 |
Interface 2(AB) - 457
Interface 4(BC) - 216 Interface 5(AA) - 280 Interface 5(BC) - 240 Interface 6(AB) - 701 Interface 6(AC) - 673 Interface 6(BC) - 721 |
| Vps4
Molecule A (119–437) Molecule B (120–435) Molecule C (123–437) |
This work | 3 | ATPγS
ATPγS Ethylene glycol-Mg2+ |
115
118 134 |
145
155 150 |
Interface 2(AB) - 676
Interface 4(BC) - 426 Interface 5(AA) - 480 Interface 5(BC) - 451 Interface 6(AB) - 895 Interface 6(AC) - 772 Interface 6(BC) - 720 |
Vps4 Structures
As expected, the Vps4 structures reported here resemble published structures of human VPS4BΔMIT-SO425, yeast (His)6Vps4ΔMIT27, and yeast Vps4ΔMIT-ADP28. For example, the ATPase cassettes of our yeast Vps4ΔMIT-SO4 and human VPS4BΔMIT-SO4 structures overlap with a Cα root mean square deviation (RMSD) between ordered residues of just 1.2 Å (Fig. 1c). Nevertheless, our two new structures have several ordered loops that have not been seen in previous structures, and the new structures extend the existing ranges of bound ligands, interdomain angles, and intermolecular packing interactions.
As in other AAA ATPases, the ATPase cassette of Vps4 is composed of two domains. The larger amino-terminal domain has six α-helices (helices 1–5 and 10) that pack on either side of a six-stranded β sheet. The first strand of the sheet (β′), which is found in only a subset of AAA ATPases, runs antiparallel to the remaining five strands (strands 1–5). The smaller carboxyl terminal domain is an antiparallel four-helix bundle (helices 6–9) (Fig. 1b). The two domains are structurally distinct, although the final helix extends away from the smaller domain to pack along one edge of the large domain sheet. Vps4 is unique amongst known AAA ATPases in that a three-stranded β-sheet, called the β domain, is inserted between helices 8 and 9 of the four-helix bundle. The β domain contributes to Vta1 binding25 and enzyme assembly39.
Both of our Vps4 crystal forms were grown in the presence of ATPγS, and two of the three molecules in crystal form 2 (A and B) show clear electron density for bound ATPγS nucleotides (Figures 2a, b). In these two molecules, the nucleotide binds in a site between the large and small domains, and makes all of the expected contacts with the Walker A/P Loop (173GPPGTGKSY181), Walker B (230FIDEVD235), and sensor 1 (275ATN277) elements18 (Figure 2a). In particular, the adenine ring adopts the anti conformation, and stacks between Tyr181 (Walker A) and Met307 (helix 6) 40; 41; 42; 43; 44. The ATPγS triphosphate hydrogen bonds with backbone amide NH groups in the P Loop (173GPPGTGKSY181, underlined residues make H bonds), and the phosphates also contact the conserved Lys179 (Walker A/P Loop, β-phosphate salt bridge), Asn277 (sensor 1, γ-sulfate) and Pro175 (P Loop, γ-sulfate). A bound Mg2+ ion is coordinated by oxygen atoms from the β and γ phosphates, and by the conserved side chains of Ser180 (P Loop) and Asp232 (Walker B). The structure is consistent with the observation that the K179A mutation inhibits ATP binding (and higher order assembly)16. The Walker B element also contains another highly conserved acidic residue (Glu233), which is thought to function as the catalytic base that activates a nucleophilic water16; 25. This residue was mutated to Gln in the Vps4ΔMIT constructs to minimize ATP hydrolysis, and the Gln233 side chain projects out of the active site where it contacts Ser198 and Ser200. We have not attempted to compare the positions of the active site nucleotides and side chains with those of the ADP-bound Vps4 structure because both complexes were solved at modest resolution and because the bound ADP ligand has a very high B factor (179 Å2)28.
Our remaining two Vps4 structures do not exhibit clear density for bound ATPγS nucleotides. Specifically, Molecule C in crystal form 2 has density in the phosphate binding sites that likely corresponds to an ethylene glycol molecule (which presumably came from the 48 % ethylene glycol crystallization solution) in complex with a magnesium ion, which is coordinated by the conserved Asp232 and Ser180 side chains and a hydroxyl group of the ethylene glycol. The single unique molecule in the VPS4ΔMIT-SO4 crystal form also lacks nucleotide density, but has density within the β and γ phosphate binding sites that likely corresponds to a sulfate ion from the 1.6 M ammonium sulfate crystallization solution. A sulfate ion is also seen in the same position in the structure of human VPS4BΔMIT-SO425.
The two different crystal forms reported here extend the range of interdomain angles seen for Vps4, and further show how the different domains can move relative to one another (Fig. 3 and Table 2). As has been noted previously28, relative domain motions are centered about two different hinges. The first hinge is Pro297, which is located within the linker between the two domains of the ATPase cassette. Molecule A in crystal form 2 has the most closed hinge I angle seen to date (115°), whereas molecule C has the most open angle (134°). The second hinge is centered at Pro350 in the center of helix 8 in the small domain. This intradomain hinge is evident in overlays of the core of the small domain (Fig. 3 and data not shown). This hinge angle can vary by up to 16°, and this variation allows the β domains to project in different directions from the small ATPase domain, which may be required to accommodate different β domain packing arrangements in the two rings of the dodecamer31.
Figure 3.
Interdomain flexibility in different Vps4ΔMIT crystal structures.
Superposition of six independent structures of Vps4ΔMIT in complex with: no ligand (2RKO, white), sulfate (crystal form 1, green), phosphate (2QPA, molecule B, blue), ADP (2QPA, molecule A, red), ATPγS (crystal form 2, molecule A, pink), and ATPγS (crystal form 2, molecule C, purple). Superposition on the large ATPase domains reveals that the small ATPase domain can rotate by up to 19° about a hinge angle located in the linker between the two domains and centered at residue Pro297. A second hinge within the small ATPase domain is also evident in the middle of the extended helix α8, centered about residue Pro350. These two hinge angles were defined by first finding the centers of masses of the large ATPase domain (residues 122–298 and 418–433), the core of the small domain (300–350; 402–414), and the β domain (358–399) using the program 6d_moleman2. Hinge angle I was then calculated as the angle between the large and small domains, with Pro297 Cα at the apex, and hinge angle II was calculated as the angle between the small domain and the β domain, with Pro350 Cα at the apex.
As in all previously reported Vps4 crystal structures, the subunits in our two crystal forms pack as continuous helices that repeat every six subunits. In crystal form 1, the helix follows the 65 crystallographic screw axis, and every subunit is therefore in an equivalent position. In crystal form 2, the two ATPγS-bound Vps4 molecules are nearly equivalent, whereas the more open third molecule sits in a slightly different position within the spiral. Analogous 6-fold (or pseudo 6-fold) screw arrangements have been seen in the crystal structures of other AAA ATPases, which presumably reflects their propensity to form six membered rings45; 46. As discussed below, adjacent helices are connected by different types of two-fold symmetric packing interactions in the different Vps4 crystal forms.
Nucleotide-free Vps4 Dimerizes in Solution
The oligomeric state of the inactive, low molecular weight Vps4 complex has been controversial, and we therefore performed gel filtration and analytical ultracentrifugation analyses to examine the oligomerization of nucleotide-free Vps4 proteins. Pure recombinant Vps4 (MW=48,803 g/mol) and Vps4ΔMIT (MW=37,700 g/mol) proteins eluted from a Superdex S200 gel filtration column as single species with apparent molecular weights of 127 kDa and 88 kDa, respectively. Thus, the Stokes radii of both constructs were most consistent with dimers, but could possibly be explained by extended monomers. Equilibrium sedimentation analyses were used to distinguish between these two possibilities because this approach provides shape-independent measures of protein solution mass.
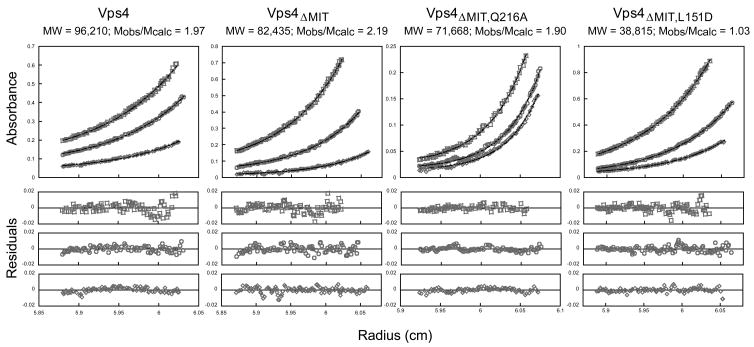
As shown in Fig. 4 and Supplemental Fig. 2, the radial distributions of full length Vps4 (panel 1) and Vps4ΔMIT (panel 2) fit well to single-species dimer models, but not to monomer or trimer models. In each case, sedimentation data were collected at three different protein concentrations and two rotor speeds, yielding a total of six data sets for each protein. The six data sets were globally fit to single species models in which the molecular weight was allowed to float, and also to monomer, dimer, or trimer models with fixed molecular weights. The distribution of Vps4 fit a protein with a predicted mass of 96,210 g/mol, which matched the mass expected for a Vps4 dimer (Mobs/Mmonomer = 1.97). Data for three initial concentrations of Vps4 centrifuged at 12,000 RPM are shown together with the global fits (Fig. 4, panel 1), and the small, random residuals indicate that the data were satisfactorily fit by a simple single species model (lower panels). Global fits to the three data sets collected at 16,000 RPM were also satisfactory (data not shown). Similarly, when fixed monomer, dimer, or trimer molecular weights were used during the global fitting procedure, the dimer model was clearly the best fit to the data (Supplemental Fig. 2). Likewise, sedimentation data for the Vps4ΔMIT protein estimated a solution mass of 82,435 g/mol (Mobs/Mmonomer = 2.19, Fig. 4, panel 2), and the dimer model was also the best fit of the different possible fixed molecular weight models (Supplemental Fig. 2).
Figure 4.
Vps4 proteins dimerize in solution
Representative equilibrium sedimentation profiles for full length Vps4 (panel 1), Vps4ΔMIT (panel 2), Vps4ΔMIT, Q216A (panel 3, Interface 1 mutant), and Vps4ΔMIT, L151D (panel 4, Interface 2 mutant). Sedimentation data are plotted as absorbance versus the distance from the center of the axis of rotation (radius). To simplify the plot, the radius was normalized so that the data from all three sectors (i.e. three different concentrations) overlap. In each case, data from three protein concentrations and one speed are displayed (open symbols) along with the best single ideal species fit (solid lines). The best fits for each protein were derived from global fits to data collected at three concentrations and two speeds. The residuals (differences between the raw absorbance data and the fit are shown below each panel.
As in most of our other experiments, these analyses utilized Vps4 proteins with E233Q mutations because the constructs were also used to examine the effects of ATP binding in the absence of hydrolysis. To ensure that the E233Q mutation did not alter the oligomerization state, gel filtration analyses were also performed on wild type Vps4ΔMIT proteins (both Vps4101–437 and Vps4124–437). In both cases, these proteins eluted with the same retention times as their E233Q analogues, indicating that the E233Q mutation did not affect the oligomerization state (not shown). We therefore conclude that in the absence of nucleotide, Vps4 and Vps4ΔMIT proteins form stable dimers in solution.
The Vps4 Dimer Interface
We reasoned that recurring crystal packing interactions might recapitulate aspects of Vps4 dimerization in solution, and therefore examined all of the intermolecular lattice contacts in the different known Vps4ΔMIT crystal forms. The different interfaces are summarized in Table 2 and Supplemental Figure 1, and subsets of the interfaces are shown in Fig. 5a. The various yeast Vps4 crystal lattices show six different classes of dimer interfaces, five of which exhibit two-fold rotational symmetry (Interfaces 1–5) and one of which lacks symmetry (Interface 6). Interface 1, the largest of the two-fold symmetric interfaces, involves contacts between residues in helix 3 and strand 2 in adjacent large domains. This interface is seen in three different crystal forms, and buries an average of 620±130Å2/subunit. Variants of Interface 1 were seen in our Vps4ΔMIT-SO4 crystals (480Å2) and also in two other Vps4ΔMIT crystal forms, one with a bound sulfate (2QP9) and one without a bound ligand (2RKO) (740Å2 and 640Å2, respectively). We note that although similar contact surfaces are used to create Interface 1 in these three crystal forms, the interactions differ significantly in detail (see Supplemental Figure 1).
Figure 5.
Mutational analyses of crystallographic Vps4 dimer interfaces.
(a) Crystallographic Vps4 dimer interfaces. Interface 1 is a symmetric interface between two large ATPase domains (residue Q216 is shown in cyan), Interface 2 is a symmetric interface between two small ATPase domains (residue L407 is shown in blue), and Interface 6 is an asymmetric interface between the large and small ATPase domains (residues L151 and W388 are shown in orange and green, respectively).
(b) Gel filtration chromatograms of Vps4ΔMIT proteins with the following mutations: Q216A (Interface 1), L407D (Interface 2), L151D (Interface 6), and W388A (Interface 6). Vps4ΔMIT proteins used here and elsewhere contained the E233Q mutation, which allowed ATP binding but inhibited hydrolysis. For reference, the elution profile of the “wild type” Vps4ΔMIT, E233Q protein is shown in red in each panel, elution positions for monomeric (1) and dimeric (2) proteins are shown as dotted vertical lines, and the elution positions of molecular weight standards are shown below the chromatograms. Vps4 protein concentrations were 150 μM in all cases. Note that at low micromolar concentrations, the dimeric proteins exhibited concentration-dependent mobilities (not shown), indicating that appreciable concentrations of monomers could accumulate under these low protein and non-equilibrium conditions.
Hartmann et al. have argued that the crystallographic Vps4 Interface 1 also forms in solution based upon their report that the Q216A interface mutation blocks Vps4ΔMIT dimerization in solution27. In our hands, however, the Q216A mutation did not alter the gel filtration mobility of Vps4ΔMIT, indicating that this mutation did not inhibit dimer formation in solution (Fig. 5b, panel 1, compare red and black chromatograms). This conclusion was confirmed in analytical ultracentrifugation experiments, which showed that the Vps4ΔMIT, Q216A protein remained dimeric over the entire concentration range tested (Fig. 4, panel 3, MWobs = 71,668 g/mol, MWobs/MWmonomer = 1.90). The Vps4ΔMIT mobility was similarly unaffected by two other Vps4 Interface 1 mutations (N224A and M220D, data not shown), and we therefore conclude that Interface 1 does not mediate Vps4 dimerization in solution.
Four other classes of two-fold symmetric subunit contacts were seen in at least two different Vps4ΔMIT crystal forms (Interfaces 2–5). However, these interfaces were approximately half the size of Interface 1, with average buried surface areas of: 350±70, 280±130 Å2, 320±150 Å2, and 360±120 Å2, respectively. None of these interfaces occurred in all of the reported crystal forms, and the residues within Interfaces 2, 4, and 5 are not well conserved across species. Interface 3 residues are well conserved, but this is likely explained by the fact that they form the “Rossmann fold” element of the nucleotide binding site. These observations imply that Interfaces 2–5 are also unlikely to mediate Vps4 dimerization in solution.
Interface 2 connects the two small domains of Vps4 and primarily involves contacts between residues in helices 7 and 9 in adjacent small domains. This interface is present in our Vps4ΔMIT-SO4 crystal structure (buried surface area, 330Å2), and also in Vps4ΔMIT crystals with a bound sulfate (2QP9) and without a bound ligand (2RKO) (430Å2, and 290Å2, respectively). Xiao et al. reported that two mutations within this interface (M330D and L407D) did not alter the gel filtration mobility of Vps4ΔMIT28. In good agreement with this report, we also found that the L407D mutation had no effect on the gel filtration mobility of Vps4ΔMIT (Fig. 4b, panel 2). These data confirm that Vps4 Interface 2, like Interface 1, does not mediate Vps4 dimerization in solution.
These observations indicate that none of the symmetric dimer interfaces seen in more than one Vps4 crystal form are likely to mediate Vps4 dimerization in solution. We therefore considered the possibility that Vps4 might form an asymmetric dimer in solution. In particular, all known Vps4 crystal forms contain an asymmetric sixth interface that is larger than any of the two-fold symmetric interfaces (760±80 Å2). Interface 6 connects the small domain of one subunit with the large domain of an adjacent subunit, and is used to create the continuous helix of Vps4 molecules. The interface is complex, and involves contacts between helix 1 and the helix 1/strand 1 loop on one side of the interface, and helices 7–9 and the C-terminal edge of the β domain on the other. Additional contacts between adjacent large domains are also made between the two molecules that bind ATP-γS in our crystal form 2 (between helix 5 on one side of the interface and the strand 2-helix 3 loop on the other). These additional contacts result from closure of the interdomain angle upon ATPγS binding (see Fig. 3).
The requirement for Interface 6 in Vps4 dimerization was tested using four different point mutations (L151D, I351A, I354D, and W388A). We initially changed the highly buried L151, I351 and I354 residues to aspartate in order to robustly disrupt the interface. However, the I351D protein was only partially soluble, and we therefore replaced this residue with alanine. W388 was changed to alanine rather than aspartate because this dramatic reduction in side chain volume was expected to yield a sufficient change in interface stability. Strikingly, every one of these mutations reduced the gel filtration mobility of Vps4ΔMIT to that expected for a monomer (compare red and black curves in Fig. 5, panels 3 (L151D) and 4 (W388A), and similar data not shown for the I351A, I354D mutants). Analytical ultracentrifugation experiments confirmed that Vps4ΔMIT, L151D was indeed monomeric in solution (Fig. 4, panel 4, MWobs = 38,815 g/mol, MWobs/MWmonomer = 1.03)). Thus, residues used to create the asymmetric Interface 6 seen in all known Vps4ΔMIT crystal forms are also required for solution dimerization. As discussed below, we propose that the Vps4 solution dimer interface is likely to be a variant of Interface 6 in which the subunit conformations are altered so that the assembly does not propagate into a polymer.
Requirements for Vps4 Dodecamerization
Mutant Vps4ΔMIT constructs that can bind but not hydrolyze ATP (e.g., VPS4ΔMIT, E233Q) form stable higher order assemblies that have been modeled as double rings, with six31 or seven27 subunits per ring. Conversion of Vps4ΔMIT dimers into higher order structures (here termed dodecamers) can also be analyzed by gel filtration chromatography (Fig. 6, panel 1), which provides a convenient assay for mutations that inhibit Vps4 ring formation.
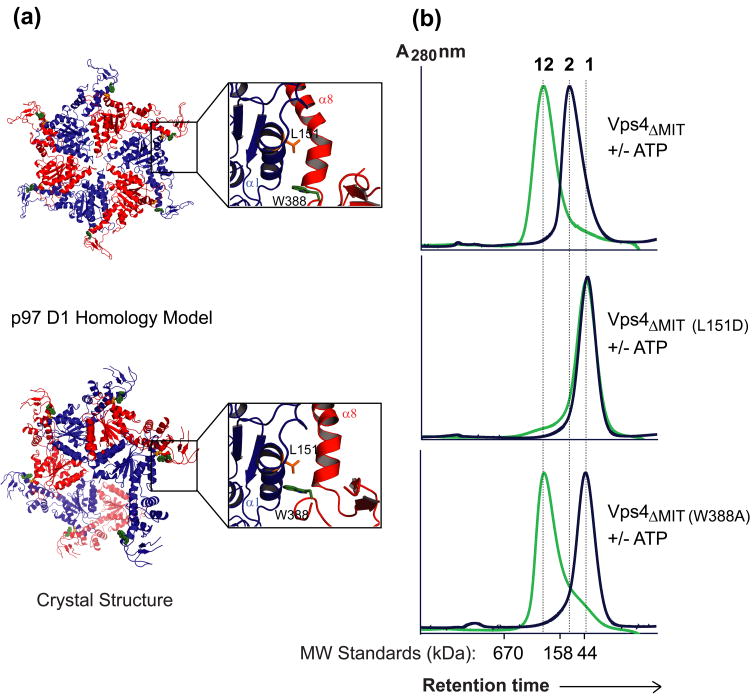
Figure 6.
Mutagenesis test of the Vps4ΔMIT hexamer homology model.
(a) The “p97 D1 Homology” model for a Vps4ΔMIT hexameric ring within the dodecameric enzyme (upper) is shown together with the arrangement of Vps4 along the crystallographic six-fold screw axis (lower). The p97 D1 Homology Model was created by superimposing Vps4ΔMIT subunits onto the crystal structure of the hexameric ring of the homologous p97 D1 protein44. The Trp388 (green) and Leu151 (orange) residues are shown explicitly in homology model and crystal structure to illustrate how the Leu151 residue contributes to the interface in both models whereas the Trp388 residue contributes to the interface in the crystal structure but not in the p97 D1 Homology Model (see insets).
(b) Gel filtration chromatograms of wild type and mutant Vps4ΔMIT proteins in the absence (black) or presence (green) of ATP. For reference, elution positions for monomeric (1), dimeric (2), and dodecameric (12) proteins are shown as dotted vertical lines, and the elution positions of molecular weight standards are shown below the chromatograms. Note that upon addition of ATP, the wild type Vps4ΔMIT protein converts from a dimer to a dodecamer, Vps4ΔMIT, L151 remains a monomer, and Vps4ΔMIT, W388A converts from a monomer to a dodecamer.
We previously proposed a homology model for one of the two rings of the Vps4ΔMIT dodecamer. This model was created by substituting Vps4 subunits in place of the crystallographically characterized p97 D1 subunits in their hexameric ring conformation25; 47. This model for the Vps4 ring generates subunit interfaces that are similar, but not identical to the crystallographically defined Vps4 Interface 6. Differences between the crystallographic interface and the homology model are illustrated in Fig. 6a, where the upper model shows the p97-based homology model and the lower model shows the spiraling “hexamer” of crystal form 1 (crystal form 2 is very similar). The p97 D1 homology model predicts that ring formation will be dependent upon residues located at the center of crystallographic Interface 6 (e.g., Leu151), but not upon residues located at the exterior of Interface 6 that are separated when the subunit orientation is adjusted to match the p97 D1 ring (e.g., Trp388, see insets). We therefore probed the validity of the p97 D1 homology model by testing the assembly properties of VPS4ΔMIT proteins with L151D and W388A point mutations.
As expected, the Vps4ΔMIT, L151D mutant remained monomeric in both the presence and absence of ATP, indicating that this mutation blocks both dimerization and dodecamerization (compare panels 1 and 2 in Fig. 6b). We note that the inability of this mutant to assemble into any higher order structure implies either that formation of the second ring in the Vps4 dodecamer depends upon forming the first ring (our preferred explanation) or, alternatively, that Leu151 makes important contacts in both rings of the dodecamer. In contrast, the Vps4ΔMIT, W388A mutant was monomeric in the absence of ATP, but assembled into stable dodecamers upon addition of ATP (panel 3). This result supports the idea that one of the two rings in the Vps4ΔMIT dodecamer resembles the hexameric ring formed by the homologous p97 D1 domain. This experiment also indicates that the Vps4ΔMIT dimer is not an obligate on-pathway intermediate for forming the Vps4 dodecamer, because the W388A mutation inhibits dimerization but not dodecamerization. Conversely, we previously showed that the Vps4R352A mutant dimerized normally, but failed to form higher order assemblies in the presence of ATP25. Arg352 sits within the subunit interface of our p97 D1 homology model, but is exposed at the edge of Interface 6 in the different Vps4 crystal structures. Thus, W388A and R352A are examples of mutations that allow formation of one Vps4 state but inhibit formation of the other, and reflect the use of overlapping but distinct interfaces for dimer and dodecamer formation.
A Functional Role for Pore Loop 2
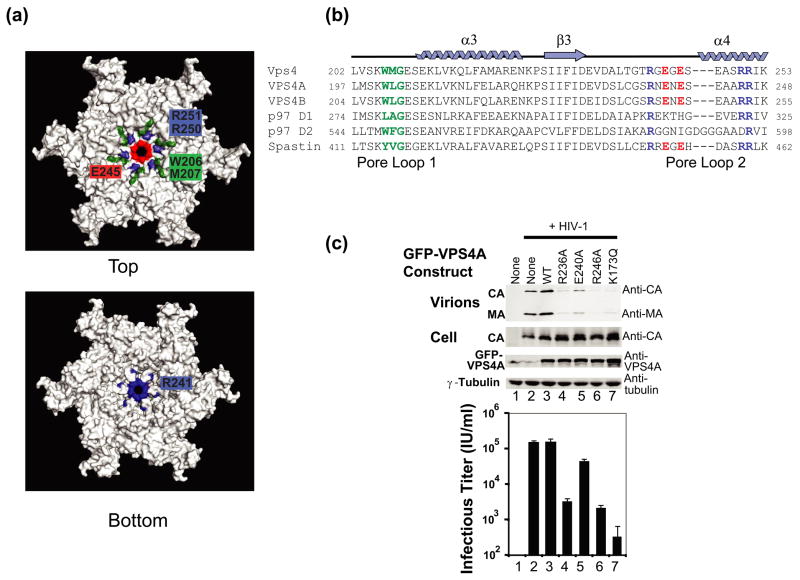
Our model for the p97 D1 homology model for the hexameric ring of Vps4 places two different loops near the center of the narrow pore (Fig. 7). Analogous loops are also present in other AAA ATPases, and they have been termed Pore Loop 1 (206WMG208, green in Figures 7a, b) and Pore Loop 2 (241RGEGESEASRR251, blue) 48; 49; 50. A section of Pore Loop 2 is also known as the “Arginine Collar”, owing to the presence of three conserved arginine residues (Arg241, Arg250, and Arg251 in Vps4)51; 52; 53. Unlike previous Vps4 structures, Pore Loops 1 and 2 are both well ordered in the three crystallographically independent molecules of Vps4 crystal form 2. Pore Loop 1 forms a short two turn helix between strand 2 and helix 3, and the conserved Trp206/Met207 dipeptide forms a hydrophobic patch that sits above the pore in the hexamer model, with Met207 projecting into the central cavity (green in Figs. 7a, b). Pore Loop 2 forms an extended loop between strand 3 and helix 4, and the conserved Pore Loop 2 Arg residues at positions 250 and 251 form a positively charged “collar” that sits immediately beneath the hydrophobic Pore 1 residues (blue in Fig. 7a, upper panel). Charged Pore 2 residues also serve to restrict the diameter of the pore itself to ~10 Å, with Glu243 and 245 (red), forming a negatively charged ring on the Pore Loop 1 side, and Arg241 (blue), forming a positively charged ring on the opposite side (Fig. 7a, lower panel).
Figure 7.
VPS4A Pore Loop 2 residues are required for HIV budding.
(a) Two views of the p97 D1 Homology model for a hexameric ring of yeast Vps4ΔMIT in the ATPγS-bound state. Pore loop 1 residues (206WMG208) are shown in green, and a subset of the charged Pore Loop 2 residues are shown in blue (basic residues) or red (acidic residues).
(b) Sequence alignments highlighting Pore Loop 1 (green) and Pore Loop 2 (blue, basic, and red, acidic) residues from human and yeast VPS4 proteins. Equivalent residues from the related p97 and Spastin AAA ATPase cassettes are highlighted following the same color scheme (except for non-conservative changes).
(c) Vps4 Pore Loop 2 mutations dominantly inhibit HIV-1 release and infectivity. 293T cells were co-transfected with an empty vector control (lane 1) or with a proviral HIV-1 expression vector (lanes 2–7) together with the designated GFP-VPS4A expression constructs. The top two western blots show the levels of virion-associated CA and MA proteins released into the supernatant (Panel 1, Virion) or expressed in the cells (Panel 2, Cell). The bottom two western blots show cellular levels of GFP-VPS4A and of an γ-Tubulin control. The bottom panel shows vector titers in the supernatants under the different conditions (infectious units/ml). Note that expression of the different Pore Loop 2 mutant GFP-VPS4A constructs (lanes 3–6) or of the VPS4A ATPase mutant (lane 7, positive control) dominantly inhibited virion release and titers. Infectivity values are the mean of measurements from quadruplicate repeats and error bars indicate standard deviations.
We have shown previously that point mutations in Pore Loop 1 residues of both human VPS4A and VPS4B inhibit HIV-1 release and infectivity, implying that this element is important for Vps4 protein function25. To examine the functional importance of Pore Loop 2 residues, we assayed the efficiency of HIV-1 release and infectivity from cells expressing exogenous wild type VPS4A or VPS4A proteins with mutations in the two most highly conserved Pore Loop 2 arginines (R236A or R246A in VPS4A), the central Pore Loop 2 glutamate (E240A), or the ATP binding site (K173Q, positive control)16. As shown in Figure 7c, overexpression of VPS4AR236A and VPS4AR246A inhibited the release of an HIV-1 vector, as indicated by reductions in the levels of the virion-associated MA and CA proteins released into the supernatant (upper panel, compare lanes 4 and 6 to lane 3) and in viral titers (lower panel). In both cases, titer reductions were substantial (48- and 73-fold, respectively, note the log scale in Fig. 7c). Virion release and titers were also impaired by overexpression of the VPS4AE240A mutant, but in this case the reduction was modest (4-fold). Control experiments behaved as expected in that overexpression of the VPS4AK173Q ATP binding mutant strongly inhibited virion release and titer (lanes 7, 470-fold titer reduction), overexpression of the wild type VPS4A protein did not impair virion release (compare lanes 2 and 3), and cellular expression levels of the HIV-1 CA and GFP-VPS4A proteins were equivalent in all cases (panels 2 and 3, respectively). We also demonstrated that the analogous Pore Loop 2 point mutations did not affect yeast Vps4E233Q protein dodecamerization in vitro, implying that these residues are not critical for Vps4 assembly (data not shown). We therefore conclude that the conserved arginine residues of Pore Loop 2 play important functional roles in Vps4 enzymes, and that the central Glu residue also contributes, albeit to a lesser extent.
DISCUSSION
Vps4 Oligomerization
At steady state, Vps4 is distributed throughout the cytoplasm in an inactive state, but can be recruited to membrane sites of action where it binds ATP and associates into enzymatically active higher order complexes16. Our studies imply that Vps4 is likely a dimer in its inactive state. Specifically, we found that Vps4 forms stable dimers in solution, as evidenced by the unambiguous fit of equilibrium sedimentation data to a single species dimer model and by the identification of a series of different point mutations that block Vps4ΔMIT dimerization. As is true for any dimeric protein, however, Vps4 will dissociate into monomeric species at sufficiently low protein concentrations. Indeed, we observed both monomeric and dimeric species for the Human VPS4B protein25, presumably because the human VPS4B homodimer is less stable than the yeast Vps4 homodimer. Moreover, we observed that even the yeast Vps4 protein exhibited concentration-dependent mobility in gel filtration experiments performed at low micromolar concentrations, indicating that monomeric yeast Vps4 species can also be detected at low protein concentrations under non-equilibrium conditions (not shown). These observations raise the possibility that monomeric Vps4 species may accumulate in vivo if the protein concentration is sufficiently low. Nevertheless, the dissociation constant for the yeast Vps4 dimer must be less than ~5 μM in order to allow an adequate fit by a single species dimer model over the concentration ranges tested (Fig. 4 and data not shown).
ATP binding promotes formation of higher order Vps4 assemblies, provided that non-hydrolyzable ATP analogs such as ATPγS are used, or that the Vps4 protein is mutated to prevent nucleotide hydrolysis (or both). These higher order assemblies appear to be dodecamers composed of two non-equivalent hexameric rings. The conclusion that VPS4 likely forms hexameric rings is based upon: 1) six-fold symmetry observed in cryo-EM reconstructions of assembled Vps4ΔMIT and Vps4 complexes31, 2) the finding that equilibrium sedimentation data for the Vps4 assembly is best fit by a dodecamer model25, and 3) strong homologies between Vps4 and the related p97 D1 and spastin proteins, both of which form hexameric rings52; 54; 55. The Vps4 dimer/oligomer assembly cycle is therefore reminiscent of NtrC, another AAA ATPase that also forms catalytically inactive dimers, and then assembles into active hexameric rings upon ATP binding56; 57. In both cases, the dimers are “off pathway” with respect to hexameric ring assembly, and therefore appear to represent autoinhibited states that negatively regulate enzymatic activity. There is no indication, that the inhibitory dimers of NtrC and Vps4 proteins are structurally related, however, because the NtrC dimer subunits associate in a two-fold symmetric, head to tail orientation that is completely distinct from the orientations of subunits in the hexamer.
Models for Vps4 Assemblies
All Vps4ΔMIT crystal forms reported to date show a similar packing arrangement in which the subunits form a continuous helix with a six-fold screw axis. These helices, in turn exhibit different types of two-fold symmetric packing interactions in different crystal forms, but our experiments indicate that none of these symmetric crystallographic interfaces is likely to mediate Vps4 dimerization in solution. In contrast, Vps4 dimerization is inhibited by a series of different mutations within the interface used to form the continuous helix (e.g., I351A, I354D, L151D and W388A). This leads to the surprising conclusion that the Vps4 dimer is likely formed by an asymmetric interface. We suggest that this interface likely resembles, but is not identical to, the asymmetric dimer interface that mediates Vps4 helix formation in all crystal forms reported to date. If correct, this model begs the question why the interaction does not repeat infinitely to form a helical (or linear) polymer in solution. Our preferred explanation is that the two subunits of the asymmetric dimer have different interdomain angles, making the dimer incompatible with repeating interactions. This type of asymmetric interaction is hinted at in the crystal structure of the Vps4-ATPγS complex, where differing interdomain angles in the three different subunits produce distinct dimeric interactions between the A:B, B:C, and C:A molecule pairs. However, a more precise understanding of this structure will clearly require high resolution crystals of a discrete Vps4 dimer.
High resolution structural studies will also be required to reveal the precise conformation of the Vps4 dodecamer. In this case, however, moderate resolution cryo-EM reconstruction of dodecameric Vps4 assemblies are available, and these structures reveal that the two hexameric rings adopt very different conformations31. The structure of the more constricted ring appears to be consistent with a homology model based upon the hexameric ring formed by the D1 AAA ATPase cassette of the related p97 ATPase47. Although the resolution of the Vps4ΔMIT reconstruction is modest (~25Å), the homology model matches the reconstructed ring in overall shape and dimensions, and the six Vps4 β-domains appear to protrude from the exterior of reconstructed ring density as predicted by the homology model. Moreover, several mutations in the predicted hexamer interface block higher order Vps4 assembly, providing biochemical support for the homology model. It is noteworthy that some of these mutations also inhibit Vps4 dimerization (L151D), whereas others do not (R352A)25, conversely some mutations block dimerization but not assembly (W388A). Thus, our data indicate that the dimer and hexameric ring interfaces utilize overlapping, but non-identical, surfaces.
Comparisons to Previous Vps4 Models
Our conclusions differ in important ways from two previous analyses of the unassembled Vps4 protein. Specifically, Xiao et al. argued that the low molecular weight Vps4 species formed in the absence of nucleotide was a monomer rather than a dimer28. This conclusion was based in part on their observation that mutations in the symmetric crystallographic Vps4 dimer Interface 2 did not alter the Vps4 gel filtration mobility. Similarly, we also found that the interface mutation L407D did not alter the gel filtration mobility of Vps4ΔMIT (Fig. 5, panel 2) However, Xiao et al. did not consider the alternative possibility that Vps4 might dimerize through a variant of the asymmetric Interface 6 (Figs. 4 and 5), whereas we found that mutations within this interface blocked Vps4 dimerization as analyzed both by gel filtration chromatography and analytical ultracentrifugation. Hence, our data demonstrate that Vps4 is primarily dimeric in its low molecular weight form.
Our analysis also differs from that of Hartmann et al., who argued that the Vps4 dimer consists of two molecules held together by the symmetrical crystallographic Interface 127. This conclusion was based upon their report that the Q216A mutation appeared to inhibit Vps4ΔMIT dimerization, as analyzed by gel filtration chromatography. In our hands, however, Vps4ΔMIT dimerization was not inhibited by the Q216A mutation or by other mutations within Interface 1. Furthermore, we do not believe that the data presented in Fig. 6 of Hartmann et al. constitute compelling evidence that the Q216A mutation inhibits Vps4ΔMIT dimerization in solution.
These differences have important implications for models of higher order Vps4 assembly because Hartmann et al. used the Interface 1 dimer as a constraint in creating their model for the fully assembled Vps4 double ring27. This constraint compelled a model in which the two Vps4 rings pack in a head-to-head orientation. However, our data indicate that the crystallographic dimer Interface 1 does not actually form in solution, as we find that Interface 1 mutants such as Q216A dimerize in solution (Fig. 5b). Furthermore, we find that the Q216A mutant also dodecamerizes normally, implying that that Gln216 does not make an essential contact in the fully assembled enzyme, as would be required by the head-to-head model (see Supplemental Fig. 3). We also find the head-to-head model unattractive because: 1) it differs from the ring orientations seen in all other well characterized Type II AAA ATPases, 2) it would require that the substrate-binding MIT domains emanate from the center of the double ringed structure (rather than from the end(s), as is seen for substrate binding domains of other AAA ATPases), and 3) it requires the ATPase activities of the two rings to work in opposition, which is difficult to envision if substrates are pumped up and through the rings.
Our cryo-EM model for the Vps4 dodecamer indicates instead that the two hexameric rings adopt very different conformations, the more constricted of which resembles the p97 D1 homology model. In this model, the Vps4 subunits interact through a surface that is similar, but not identical to the crystallographically defined Interface 6, which can explain why only a subset of mutations within Interface 6 inhibit Vps4 assembly. Vps4 residues that make up the intersubunit interaction surface in the p97 homology model are shown in detail in Supplemental Fig. 4. The conservation of many of these interface residues between the Vps4, p97 D1, and spastin proteins, further supports the homology model. This model can also explain why Vps4 becomes an active ATPase upon assembly because hexamerization would position each 288RR289 dipeptide into the active site of an adjacent subunit, in close proximity with the γ-phosphate of the ATP. The use of such “arginine fingers” (sometimes also termed the second region of homology (SRH)) is a recurring feature in AAA ATPase active sites18, and serves an important catalytic role because the arginine(s) interacts with the γ-phosphate, making it a better leaving group. However, in the spiraling hexamers seen in crystal form 2 and related Vps4 crystal forms, the SRH residues of the adjacent subunit are located at least 12Å away from the γ-sulfate and therefore do not make interactions that would promote ATP hydrolysis. In our p97-based Vps4 hexamer homology model, these residues move much closer to the γ-sulfate, with the Arg288 residue brought to within 3.5Å of a γ-sulfate oxygen, where it could contact the terminal phosphate/sulfate and perform its expected role in catalysis.
A Functional Role for Vps4 Pore Loop 2
The central pores of AAA ATPases are typically defined by two adjacent loops that project into the central cavity. We have previously shown that hydrophobic Pore Loop 1 residues are required for VPS4A function, as judged by the ability of Pore Loop 1 mutants to dominantly inhibit HIV-1 release and infectivity25. In the current study, we used the same approach to show that Pore Loop 2 residues that help create the arginine collar are also required for human VPS4A function. Specifically, we found that overexpression of either VPS4AR236A or VPS4AR246A inhibited HIV-1 vector release and reduced viral titers more than 40-fold, showing that these mutant proteins were potent dominant negative inhibitors of HIV budding. As illustrated in Fig. 7, the two arginine residues appear to perform different functions, with VPS4A Arg236 (equivalent to yeast Vps4 Arg241) forming part of the positively charged collar that sits beneath the Pore 1 loop, and VPS4A Arg246 (equivalent to yeast Vps4 Arg251) lining the opposite side of the pore itself. Equivalent arginine residues are also important for the activity of other AAA ATPases, including the two closest relatives of VPS4; p9751, and spastin52; 53. The functional importance of these conserved residues highlights the similarities between Vps4 and the other two related AAA ATPases, and suggests that conserved residues within the arginine collar may perform analogous mechanistic functions, possibly by binding the substrate and/or by assisting in protein denaturation51 as polypeptide substrates are transferred through the central pore.
MATERIALS AND METHODS
Cloning
DNA encoding Vps4 from S. cerevisiae was amplified by PCR from yeast genomic DNA, and directionally cloned into the pET151 E. coli expression vector (Invitrogen). Constructs expressing mutant proteins were created by Quick-change mutagenesis (Stratagene) and verified by DNA sequencing.
Protein Expression and Purification
Vps4122–437, E233Q and Vps4104–437, E233Q were expressed in BL21 Codon+ E. coli cells (Stratagene) in ZY autoinduction media for 6 h at 37°C and then overnight at 21°C. Cells were harvested by centrifugation and lysed by resuspension in 10 mg/ml lysozyme in lysis buffer (10 mM imidazole, 300 mM NaCl, 50 mM Tris pH 7.4, 5% glycerol and protease inhibitors (PMSF, aprotinin, leupeptin, pepstatin), 45 min, 4°C), followed by sonication. The lysate was clarified by centrifugation (45 min, 35 000 × g) and the soluble Vps4 proteins were bound to a Ni2+ sepharose column (Amersham), washed with 10 column volumes of lysis buffer, and eluted in 75 mM imidazole (in lysis buffer). Fractions were assayed for Vps4 by SDS-PAGE, pooled, dialyzed into 100 mM NaCl, 25 mM Tris pH 7.4, and 1 mM DTT, and purified by anion-exchange chromatography (Q-sepharose, Invitrogen, 100–500 mM NaCl gradient in 25 mM Tris pH 7.4, and 1 mM DTT). Vps4 fractions were pooled, and the (His)6 affinity tag was removed by incubation with TEV protease (~1 mg/100mg protein, 20 h at 4°C), followed by dialysis into 20 mM Tris pH 7.4, 100 mM NaCl, and removal of unprocessed protein by Ni2+ sepharose chromatography. The cleaved protein was collected in the flow through, concentrated, and purified to homogeneity by gel filtration chromatography (SD200, Amersham, in 50 mM NaCl, 25 mM Tris pH 7.4, 1mM DTT). Yields were typically 25 mg/l culture.
Gel Filtration Chromatography
The gel filtration chromatography experiments shown in Fig. 5, Fig. 6, and Supplemental Fig. 3 were performed on 150 μM Vps4 proteins using a Superdex 200 column in 100 mM NaCl, 25 mM Tris–HCl (pH 7.5), and 1 mM DTT in the presence or absence of ATP. Both columns were calibrated using molecular weight standards (Biorad).
Analytical Ultracentrifugation
Equilibrium sedimentation experiments on full length Vps4E233Q, Vps4ΔMIT, E233Q and Vps4ΔMIT, E233Q with L151D and Q216A interface mutations, were performed on an XL-A analytical ultracentrifuge (Beckman Coulter). Protein concentrations ranged from 5–30 μm (110-μl) and the corresponding buffer blanks (120 μl; 50 mM NaCl, 25 mM Tris pH 7.5) were run in parallel in sample cells fitted with six-channel equilibrium centerpieces. All data were collected at 4°C and at two rotor speeds for each protein: Vps4 at 12,000 and 16,000 rpm, for Vps4ΔMIT at 14,000 and 18,000 rpm, and for Vps4ΔMIT interface mutants L151D and Q216A at 16,000 and 20,000 rpm. The attainment of equilibrium after 24 h was confirmed by comparing repetitive absorption scans. Theoretical molecular weights, partial specific volumes (vbar), and molar extinction coefficients at 280 nm of the Vps4 variants were calculated using the program SEDNTERP (Version 1.09, available on the World Wide Web)58. All data were analyzed using the program Heteroanalysis59.
Crystallization
Crystals were grown by sitting drop vapor diffusion at 21°C. Vps4 104–437, E233Q crystallized in drops comprising 2 μl of 13 mg/ml protein, 50 mM NaCl, 25 mM Tris pH 7.4, 1 mM DTT, 4 mM MgCl2, and 2 mM ATPγS, mixed with 1 μl of reservoir solution (48% ethylene glycol (vol/vol), Bis-Tris pH 5.0). Crystals were cryo-cooled by plunging into liquid nitrogen directly from the crystallization solution. Vps4122–437, E233Q was crystallized in drops comprising 2 μl of 12 mg/ml protein, 100 mM NaCl, 25 mM Tris pH 7.4, 1 mM DTT, 4 mM MgCl2, and 1 mM ATPγS, mixed with 2 μl reservoir solution (1.6 M ammonium sulfate, 0.1 M Bis-Tris pH 5.5). Crystals were briefly transferred to cryoprotectant (mother liquor made up with 20% glycerol), suspended in a nylon loop, and cryo-cooled by plunging into liquid nitrogen.
Structure Determination and Refinement
Diffraction data from Vps4104–437 and Vps4122–437 crystals were collected at Beamline X25 of the National Synchrotron Light Source, Brookhaven National Laboratory, and scaled and integrated by using the HKL2000 suite60. Crystal structures were determined by molecular replacement using PHASER61. A polyalanine/homology search based on the human VPS4B structure (pdb code 1XWI)25 was used to determine Vps4122–437, E233Q, and this refined structure was used to determine the structure of Vps4104–437, E233Q. Model building was performed with COOT62. Structures were refined using REFMAC563 with TLS refinement using TLSMD64 and TLSANL65 in the CCP4 suite66. Structures were analyzed with PROCHECK67 and figures were generated using PYMOL68.
Infectivity Assays
293T cells were seeded in 6-well plates at 8×105 cells per well and co-transfected with a pEGFP-VPS4A expression vector and an HIV-1 vector system (packaging and transfer plasmids kindly provided by D. Trono, and envelope plasmid kindly provided by J. Burns). Briefly, 12 μl Fugene 6 Transfection Reagent (Roche Diagnostics) was combined with 85.14 μl Optimem (no additives) and incubated at room temperature for 5 minutes. 1 μg of pCMVDR8.2, 1 μg of pWPTS-nlsLacZ, 0.36 μg phCMV-VSVG, and 0.5 μg of wild type or mutant pEGFP-VPS4A was combined with the transfection mixture (final volume of 100 μl) and incubated at room temperature for 20 minutes. Virions were harvested ~48 h post-transfection and used to transduce the reporter HeLa-M cell line as described previously38 or used in Western blotting experiments to analyze virion release.
Virion Release Assays
Virion-containing supernatants and proteins from transfected cells were prepared for Western blotting as described69; 70. Primary antibodies used were: rabbit anti-HIV-1 MA (1:1000, D. Trono), rabbit anti-HIV-1 CA UT415 (1:1000, Covance), rabbit anti-VPS4A UT289 (1:000, Covance), and mouse anti-γ-Tubulin GTU-88 (1:5000, Abcam). Secondary antibodies used were: goat anti-rabbit ALEXA680nm (1:20,000, Molecular Probes) and donkey anti-mouse IR800nm (1:10,000, Rockland). Proteins were detected by imaging fluorescent secondary antibodies on an Odyssey scanner (LiCor Inc.).
Protein Data Bank Accession Numbers
Atomic coordinates and structure factors for yeast Vps4ΔMIT have been deposited in the RCSB Protein Data Bank with accession number 3EIE for the SO4-bound form and 3EIH for the ATPγS-bound form.
Supplementary Material
Intermolecular interfaces in different Vps4ΔMIT crystal forms. Note that in some cases similar, but non-identical interfaces have been grouped together.
Comparative models for Vps4 protein oligomerization. Representative equilibrium sedimentation profiles for full length Vps4 (panel 1), Vps4ΔMIT (panel 2), Vps4ΔMIT, Q216A (panel 3, Interface 1 mutant), Vps4ΔMIT, L151D (panel 4, Interface 2 mutant) fit to monomer, dimer and trimer single species models with fixed molecular weights. Sedimentation data are plotted as absorbance versus the distance from the center of the axis of rotation (radius). To simplify the plot, the radius was normalized so the data from all three sectors (i.e. three different concentrations) overlap. In each case, data from three protein concentrations and one speed are displayed (gray) along with the indicated single ideal species fit (black lines). The fits for each protein were globally fit to three concentrations and two speeds. The residuals (differences between the raw absorbance data and the fit) are shown below each panel. A good fit is indicated by random and small residuals.
Vps4ΔMIT, Q216A dimerizes and dodecamerizes normally. Gel filtration chromatograms of the mutant Vps4ΔMIT, Q216A protein in the absence (black) or presence (green) of ATP. For reference, elution positions of dimeric (2) and dodecameric (12) proteins are shown as dotted vertical lines, and the elution positions of molecular weight standards are shown below the chromatograms. Note that upon addition of ATP, the Vps4ΔMIT, Q216A protein converts from a dimer to a dodecamer.
Relationship between the p97 D1 hexamer interface and the modeled Vps4 hexamer interface.
(a) Structures of the crystallographically defined p97 D1 hexamer (left) and the analogous interfaces in the p97 homology model of the Vps4 hexamer (right). The inset shows a detailed view of the major Vps4 interface. Residues that are located within the interface and are identical between Vps4 and p97 D1 are highlighted in red, and conservative changes are highlighted in blue.
(b) Sequence alignments of residues within Vps4 Interface 6 and equivalent regions of the p97 D1 and spastin AAA ATPase cassettes. The aligned regions span both sides of Interface 6 (helix α1-strand β1, upper sequence and helix α8-α9, lower sequences). Colored residues contribute to the interdomain interface between subunits in the p97 D1 hexamer, and the color coding is the same as in part (a). Residues that mediate interdomain contacts in the p97 D1 but are not identical in Vps4 are underlined.
Acknowledgments
We thank Ian Huggins for help with protein purification, Heidi Schubert for advice with crystallography, and a reviewer for suggesting the assignment of electron density as an ethylene glycol molecule. We gratefully acknowledge the DNA Synthesis and Sequencing Core Facilities at the University of Utah. Data for this study were measured at beamline X25 of the National Synchrotron Light Source. Financial support comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health. This work was supported by National Institutes of Health Grants AI51174 (to W.I.S.) and P50GM082545 (to W.I.S. and C.P.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci. 2007;32:561–73. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–68. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 4.Welsch S, Habermann A, Jager S, Muller B, Krijnse-Locker J, Krausslich HG. Ultrastructural analysis of ESCRT proteins suggests a role for endosome-associated tubular-vesicular membranes in ESCRT function. Traffic. 2006;7:1551–66. doi: 10.1111/j.1600-0854.2006.00489.x. [DOI] [PubMed] [Google Scholar]
- 5.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. Embo J. 1997;16:1820–31. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–12. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 8.Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. Embo J. 2007;26:4215–27. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finken-Eigen M, Rohricht RA, Kohrer K. The VPS4 gene is involved in protein transport out of a yeast pre-vacuolar endosome-like compartment. Curr Genet. 1997;31:469–80. doi: 10.1007/s002940050232. [DOI] [PubMed] [Google Scholar]
- 10.Fujita H, Yamanaka M, Imamura K, Tanaka Y, Nara A, Yoshimori T, Yokota S, Himeno M. A dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: class E vps phenotype in mammalian cells. J Cell Sci. 2003;116:401–14. doi: 10.1242/jcs.00213. [DOI] [PubMed] [Google Scholar]
- 11.Yoshimori T, Yamagata F, Yamamoto A, Mizushima N, Kabeya Y, Nara A, Miwako I, Ohashi M, Ohsumi M, Ohsumi Y. The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol Biol Cell. 2000;11:747–63. doi: 10.1091/mbc.11.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bishop N, Woodman P. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol Biol Cell. 2000;11:227–39. doi: 10.1091/mbc.11.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujii K, Hurley JH, Freed EO. Beyond Tsg101: the role of Alix in ‘ESCRTing’ HIV-1. Nat Rev Microbiol. 2007;5:912–6. doi: 10.1038/nrmicro1790. [DOI] [PubMed] [Google Scholar]
- 14.Welsch S, Muller B, Krausslich HG. More than one door - Budding of enveloped viruses through cellular membranes. FEBS Lett. 2007;581:2089–97. doi: 10.1016/j.febslet.2007.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 16.Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. Embo J. 1998;17:2982–93. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunger AT, DeLaBarre B. NSF and p97/VCP: similar at first, different at last. FEBS Lett. 2003;555:126–33. doi: 10.1016/s0014-5793(03)01107-4. [DOI] [PubMed] [Google Scholar]
- 18.Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure--diverse function. Genes Cells. 2001;6:575–97. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 19.Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, Neher SB, Oakes ES, Siddiqui SM, Wah DA, Baker TA. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–29. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 21.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 22.Frickey T, Lupas AN. Phylogenetic analysis of AAA proteins. J Struct Biol. 2004;146:2–10. doi: 10.1016/j.jsb.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 23.Licht S, Lee I. Resolving Individual Steps in the Operation of ATP-Dependent Proteolytic Molecular Machines: From Conformational Changes to Substrate Translocation and Processivity. Biochemistry. 2008;47:3595–605. doi: 10.1021/bi800025g. [DOI] [PubMed] [Google Scholar]
- 24.Scheuring S, Rohricht RA, Schoning-Burkhardt B, Beyer A, Muller S, Abts HF, Kohrer K. Mammalian cells express two VPS4 proteins both of which are involved in intracellular protein trafficking. J Mol Biol. 2001;312:469–80. doi: 10.1006/jmbi.2001.4917. [DOI] [PubMed] [Google Scholar]
- 25.Scott A, Chung HY, Gonciarz-Swiatek M, Hill GC, Whitby FG, Gaspar J, Holton JM, Viswanathan R, Ghaffarian S, Hill CP, Sundquist WI. Structural and mechanistic studies of VPS4 proteins. Embo J. 2005;24:3658–69. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott A, Gaspar J, Stuchell-Brereton MD, Alam SL, Skalicky JJ, Sundquist WI. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc Natl Acad Sci U S A. 2005;102:13813–8. doi: 10.1073/pnas.0502165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartmann C, Chami M, Zachariae U, de Groot BL, Engel A, Grutter MG. Vacuolar protein sorting: two different functional states of the AAA-ATPase Vps4p. J Mol Biol. 2008;377:352–63. doi: 10.1016/j.jmb.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 28.Xiao J, Xia H, Yoshino-Koh K, Zhou J, Xu Z. Structural characterization of the ATPase reaction cycle of endosomal AAA protein Vps4. J Mol Biol. 2007;374:655–70. doi: 10.1016/j.jmb.2007.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lupas AN, Martin J. AAA proteins. Curr Opin Struct Biol. 2002;12:746–53. doi: 10.1016/s0959-440x(02)00388-3. [DOI] [PubMed] [Google Scholar]
- 30.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 31.Yu Z, Gonciarz MD, Sundquist WI, Hill CP, Jensen GJ. Cryo-EM structure of dodecameric Vps4p and its 2:1 complex with Vta1p. J Mol Biol. 2008;377:364–77. doi: 10.1016/j.jmb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujita H, Umezuki Y, Imamura K, Ishikawa D, Uchimura S, Nara A, Yoshimori T, Hayashizaki Y, Kawai J, Ishidoh K, Tanaka Y, Himeno M. Mammalian class E Vps proteins, SBP1 and mVps2/CHMP2A, interact with and regulate the function of an AAA-ATPase SKD1/Vps4B. J Cell Sci. 2004;117:2997–3009. doi: 10.1242/jcs.01170. [DOI] [PubMed] [Google Scholar]
- 33.Azmi I, Davies B, Dimaano C, Payne J, Eckert D, Babst M, Katzmann DJ. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J Cell Biol. 2006;172:705–17. doi: 10.1083/jcb.200508166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeo SC, Xu L, Ren J, Boulton VJ, Wagle MD, Liu C, Ren G, Wong P, Zahn R, Sasajala P, Yang H, Piper RC, Munn AL. Vps20p and Vta1p interact with Vps4p and function in multivesicular body sorting and endosomal transport in Saccharomyces cerevisiae. J Cell Sci. 2003;116:3957–70. doi: 10.1242/jcs.00751. [DOI] [PubMed] [Google Scholar]
- 35.Shiflett SL, Ward DM, Huynh D, Vaughn MB, Simmons JC, Kaplan J. Characterization of Vta1p, a class E Vps protein in Saccharomyces cerevisiae. J Biol Chem. 2004;279:10982–90. doi: 10.1074/jbc.M312669200. [DOI] [PubMed] [Google Scholar]
- 36.Lottridge JM, Flannery AR, Vincelli JL, Stevens TH. Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc Natl Acad Sci U S A. 2006;103:6202–7. doi: 10.1073/pnas.0601712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowers K, Lottridge J, Helliwell SB, Goldthwaite LM, Luzio JP, Stevens TH. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 2004;5:194–210. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- 38.Ward DM, Vaughn MB, Shiflett SL, White PL, Pollock AL, Hill J, Schnegelberger R, Sundquist WI, Kaplan J. The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J Biol Chem. 2005;280:10548–55. doi: 10.1074/jbc.M413734200. [DOI] [PubMed] [Google Scholar]
- 39.Vajjhala PR, Wong JS, To HY, Munn AL. The beta domain is required for Vps4p oligomerization into a functionally active ATPase. Febs J. 2006;273:2357–73. doi: 10.1111/j.1742-4658.2006.05238.x. [DOI] [PubMed] [Google Scholar]
- 40.Huyton T, Pye VE, Briggs LC, Flynn TC, Beuron F, Kondo H, Ma J, Zhang X, Freemont PS. The crystal structure of murine p97/VCP at 3.6A. J Struct Biol. 2003;144:337–48. doi: 10.1016/j.jsb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Sousa MC, Trame CB, Tsuruta H, Wilbanks SM, Reddy VS, McKay DB. Crystal and solution structures of an HslUV protease-chaperone complex. Cell. 2000;103:633–43. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- 42.Suno R, Niwa H, Tsuchiya D, Zhang X, Yoshida M, Morikawa K. Structure of the whole cytosolic region of ATP-dependent protease FtsH. Mol Cell. 2006;22:575–85. doi: 10.1016/j.molcel.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 43.Kim DY, Kim KK. Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J Biol Chem. 2003;278:50664–70. doi: 10.1074/jbc.M305882200. [DOI] [PubMed] [Google Scholar]
- 44.Dreveny I, Kondo H, Uchiyama K, Shaw A, Zhang X, Freemont PS. Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47. Embo J. 2004;23:1030–9. doi: 10.1038/sj.emboj.7600139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo F, Maurizi MR, Esser L, Xia D. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J Biol Chem. 2002;277:46743–52. doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- 46.Lee S, Sowa ME, Watanabe YH, Sigler PB, Chiu W, Yoshida M, Tsai FT. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–40. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, Shaw A, Bates PA, Newman RH, Gowen B, Orlova E, Gorman MA, Kondo H, Dokurno P, Lally J, Leonard G, Meyer H, van Heel M, Freemont PS. Structure of the AAA ATPase p97. Mol Cell. 2000;6:1473–84. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 48.Wang J, Song JJ, Franklin MC, Kamtekar S, Im YJ, Rho SH, Seong IS, Lee CS, Chung CH, Eom SH. Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure. 2001;9:177–84. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- 49.Yamada-Inagawa T, Okuno T, Karata K, Yamanaka K, Ogura T. Conserved pore residues in the AAA protease FtsH are important for proteolysis and its coupling to ATP hydrolysis. J Biol Chem. 2003;278:50182–7. doi: 10.1074/jbc.M308327200. [DOI] [PubMed] [Google Scholar]
- 50.Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, Mogk A, Bukau B. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–65. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 51.DeLaBarre B, Christianson JC, Kopito RR, Brunger AT. Central pore residues mediate the p97/VCP activity required for ERAD. Mol Cell. 2006;22:451–62. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 52.Roll-Mecak A, Vale RD. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 2008;451:363–7. doi: 10.1038/nature06482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White SR, Evans KJ, Lary J, Cole JL, Lauring B. Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J Cell Biol. 2007;176:995–1005. doi: 10.1083/jcb.200610072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DeLaBarre B, Brunger AT. Nucleotide dependent motion and mechanism of action of p97/VCP. J Mol Biol. 2005;347:437–52. doi: 10.1016/j.jmb.2005.01.060. [DOI] [PubMed] [Google Scholar]
- 55.DeLaBarre B, Brunger AT. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat Struct Biol. 2003 doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- 56.Lee SY, De La Torre A, Yan D, Kustu S, Nixon BT, Wemmer DE. Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains. Genes Dev. 2003;17:2552–63. doi: 10.1101/gad.1125603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Carlo S, Chen B, Hoover TR, Kondrashkina E, Nogales E, Nixon BT. The structural basis for regulated assembly and function of the transcriptional activator NtrC. Genes Dev. 2006;20:1485–95. doi: 10.1101/gad.1418306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laue T, Shah B, Ridgeway T, Pelletier S. Computer-aided interpretation of analytical sedimentation data for proteins. In: Rowe A, Horton J, editors. Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge, England: 1992. pp. 90–125. [Google Scholar]
- 59.Cole JL. Analysis of heterogeneous interactions. Methods Enzymol. 2004;384:212–32. doi: 10.1016/S0076-6879(04)84013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 61.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–64. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 62.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 63.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr. 1999;55 (Pt 1):247–55. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 64.Merritt EA, Painter J. TLSMD web server for the generation of multi-group TLS models. J Appl Crystallogr. 2006;39 [Google Scholar]
- 65.Howlin B, Butler SA, Moss DS, Harris GW, Driessen HPC. TLSANL: TLS parameter analysis program for segmented anisotropic refinement of macromolecular structures. J Appl Crystallogr. 1993;26:622–624. [Google Scholar]
- 66.Group C. C. P. The CCP4 Suite:Programs for Protein Crystallography. Acta Crystallogr D50. 1994 November 4;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 67.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 68.DeLano WL. The PyMOL Molecular Graphics System and Users Manual. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 69.von Schwedler UK, Stemmler TL, Klishko VY, Li S, Albertine KH, Davis DR, Sundquist WI. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. Embo J. 1998;17:1555–68. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Langelier C, von Schwedler UK, Fisher RD, De Domenico I, White PL, Hill CP, Kaplan J, Ward D, Sundquist WI. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J Virol. 2006;80:9465–80. doi: 10.1128/JVI.01049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Intermolecular interfaces in different Vps4ΔMIT crystal forms. Note that in some cases similar, but non-identical interfaces have been grouped together.
Comparative models for Vps4 protein oligomerization. Representative equilibrium sedimentation profiles for full length Vps4 (panel 1), Vps4ΔMIT (panel 2), Vps4ΔMIT, Q216A (panel 3, Interface 1 mutant), Vps4ΔMIT, L151D (panel 4, Interface 2 mutant) fit to monomer, dimer and trimer single species models with fixed molecular weights. Sedimentation data are plotted as absorbance versus the distance from the center of the axis of rotation (radius). To simplify the plot, the radius was normalized so the data from all three sectors (i.e. three different concentrations) overlap. In each case, data from three protein concentrations and one speed are displayed (gray) along with the indicated single ideal species fit (black lines). The fits for each protein were globally fit to three concentrations and two speeds. The residuals (differences between the raw absorbance data and the fit) are shown below each panel. A good fit is indicated by random and small residuals.
Vps4ΔMIT, Q216A dimerizes and dodecamerizes normally. Gel filtration chromatograms of the mutant Vps4ΔMIT, Q216A protein in the absence (black) or presence (green) of ATP. For reference, elution positions of dimeric (2) and dodecameric (12) proteins are shown as dotted vertical lines, and the elution positions of molecular weight standards are shown below the chromatograms. Note that upon addition of ATP, the Vps4ΔMIT, Q216A protein converts from a dimer to a dodecamer.
Relationship between the p97 D1 hexamer interface and the modeled Vps4 hexamer interface.
(a) Structures of the crystallographically defined p97 D1 hexamer (left) and the analogous interfaces in the p97 homology model of the Vps4 hexamer (right). The inset shows a detailed view of the major Vps4 interface. Residues that are located within the interface and are identical between Vps4 and p97 D1 are highlighted in red, and conservative changes are highlighted in blue.
(b) Sequence alignments of residues within Vps4 Interface 6 and equivalent regions of the p97 D1 and spastin AAA ATPase cassettes. The aligned regions span both sides of Interface 6 (helix α1-strand β1, upper sequence and helix α8-α9, lower sequences). Colored residues contribute to the interdomain interface between subunits in the p97 D1 hexamer, and the color coding is the same as in part (a). Residues that mediate interdomain contacts in the p97 D1 but are not identical in Vps4 are underlined.