Abstract
For efficient transcription, RNA PolII must overcome the presence of nucleosomes. The p38-related MAPK Hog1 is an important regulator of transcription upon osmostress in yeast and thereby it is involved in initiation and elongation. However, the role of this protein kinase in elongation has remained unclear. Here, we show that during stress there is a dramatic change in the nucleosome organization of stress-responsive loci that depends on Hog1 and the RSC chromatin remodelling complex. Upon stress, the MAPK Hog1 physically interacts with RSC to direct its association with the ORF of osmo-responsive genes. In RSC mutants, PolII accumulates on stress promoters but not in coding regions. RSC mutants also display reduced stress gene expression and enhanced sensitivity to osmostress. Cell survival under acute osmostress might thus depend on a burst of transcription that in turn could occur only with efficient nucleosome eviction. Our results suggest that the selective targeting of the RSC complex by Hog1 provides the necessary mechanistic basis for this event.
Keywords: chromatin remodelling, gene expression, osmostress, RSC, SAPK Hog1
Introduction
Adaptation to environmental stress requires changes in many aspects of cellular physiology. In eukaryotic cells, stress-activated protein kinases (SAPKs) have an essential function for proper cell adaptation to extracellular stimuli (Kyriakis and Avruch, 2001). Exposure of cells to high osmolarity results in rapid activation of a conserved family of SAPKs, which include mammalian p38 and yeast Hog1 (de Nadal et al, 2002; Sheikh-Hamad and Gustin, 2004). The SAPKs are signalling molecules that can modulate gene expression in response to specific environmental stimuli. Until recently, the prevailing view on how SAPKs, and kinases in general, modulate gene expression has been through direct phosphorylation of transcription factors or co-regulatory proteins. However, the observation that the SAPK Hog1 is recruited to chromatin has lead to a new role for signalling kinases as integral components of transcription complexes, influencing gene expression in an unexpected way (Alepuz et al, 2001; Proft and Struhl, 2002).
Recruitment of Hog1 to promoters through its association with transcription factors bound to target promoters was shown to be important to stimulate the recruitment of PolII, the Rpd3 histone deacetylase and SAGA complexes allowing for proper transcription initiation (Alepuz et al, 2003; de Nadal et al, 2004; Zapater et al, 2007). Thus, binding of Hog1 to promoters is critical to induce gene expression upon stress and maximize cell survival. A recent report suggests that nuclear localization of the MAPK and regulation of gene expression might not be critical for cell viability at high osmolarity (Westfall et al, 2008); however, a number of mutants on transcription elements required for proper gene expression upon stress display a phenotype of osmosensitivity.
Recent reports have shown that binding of the Hog1 MAPK to chromatin is not only restricted to promoters but it also extends to coding regions of stress-responsive genes (Pokholok et al, 2006; Proft et al, 2006). Activated Hog1 associates with elongating PolII and components of the elongation complex and is found selectively recruited to the entire coding region of osmotic stress genes. Hog1 increases levels of PolII elongation complex and mRNA. Thus, in addition to its various functions during transcriptional initiation, Hog1 behaves as a transcriptional elongation factor that is selective for genes induced upon osmotic stress (Proft et al, 2006).
Global in vivo binding analyses (ChIP-on-chip) have revealed that in addition to Hog1, other signalling kinases such as Tpk1 (the catalytic subunit of the PKA) or the Fus3 and Kss1 MAP kinases are also found in coding regions of specific genes (Pokholok et al, 2006). Thus, protein signalling kinases may have a more general role as chromatin-associated enzymes than previously anticipated. At this point, however, the role of these kinases in the coding regions remains unclear, which opens a new dimension on transcription regulation by signalling kinases.
Chromatin structure imposes significant obstacles on all aspects of transcription, from initiation to elongation. Modifications at the surface of nucleosomes are critical for transcription as they can disrupt chromatin contacts or affect the recruitment of non-histone proteins to chromatin (Kouzarides, 2007; Li et al, 2007). The dynamics of chromatin structure are tightly regulated through multiple mechanisms, including chromatin remodelling, histone variant incorporation, histone eviction and histone modification. Chromatin remodellers are specialized multi-protein complexes that enable access to nucleosomal DNA by altering the structure, composition and positioning of nucleosomes (Saha et al, 2006). The family of SWI/SNF remodellers is known to participate in many aspects of gene expression, in general to promote transcription. The RSC complex is a member of the SWI/SNF family and is characterized for modifying nucleosome structure through ATP hydrolysis. Interestingly, it has been shown that in vitro stimulation of transcription through a chromatin template requires RSC for efficient PolII elongation through DNA loop formation to make nucleosomal DNA accessible and making possible the process of nucleosome mobilization and histone eviction (Sengupta et al, 2001; Carey et al, 2006; Lorch et al, 2006; Zhang et al, 2006). In addition, chromatin alterations have been shown to occur at RSC-occupied genes in rsc mutants (Moreira and Holmberg, 1999; Soutourina et al, 2006). The RSC complex is known to be targeted to specific genes by binding to activators or other components of the transcriptional machinery (e.g. Govind et al, 2005).
Global binding assays showed that Rsc1 and Rsc2 associate with about 700 target promoters that include RNA PolIII- and RNA PolII-dependent genes. RSC displays distinct modes of association with promoters but, it is worth noting that regulated association of RSC is correlated with transcriptional activation of genes involved in carbohydrate metabolism and occurs prior to RNA PolII recruitment. In addition, genome-wide localization of Rsc9 indicated a relationship between genes targeted by RSC and genes regulated by stress (i.e. hydrogen peroxide or rapamycin). Although these studies did not shed light on how RSC was targeted to chromatin and the specific function of RSC on the transcription process, they clearly showed that external stimuli induced changes in RSC genome-wide localization that correlated with induction or repression of specific families of genes (Damelin et al, 2002; Ng et al, 2002).
In this study, we show that the Hog1 MAPK interacts with the RSC chromatin-remodelling complex to mediate its recruitment to osmo-responsive genes. The RSC targeting is required for the massive nucleosome rearrangements found in osmostress genes in response to high osmolarity. Indeed, we show that a defect in RSC function results in distinctly lower gene expression of these genes during acute osmostress, albeit showing accumulation of PolII at the promoters. Therefore, our results define a novel function for Hog1 at the stress loci—that is the targeting of the RSC complex to a specific set of genes to mediate chromatin remodelling for efficient polymerase progression and proper gene expression and maximal cell survival upon stress.
Results
Mutants in the RSC complex are sensitive to osmostress and display reduced osmostress gene expression
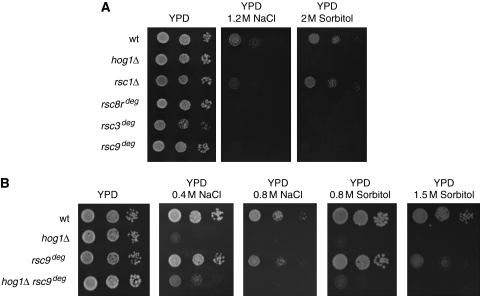
The ability of cells to survive at high osmolarity depends on the HOG signalling pathway and the control of gene expression exerted by the Hog1 MAPK. We performed a genome-wide screen to identify activities required for full expression of the gene programme required for cell survival upon osmostress. This screen identified several complexes such as the Rpd3 histone deacetylase complex, SAGA, Mediator and components of the TEC that are important for gene expression in response to osmostress (Proft et al, 2006; Zapater et al, 2007). In addition, mutations in RSC1 and NPL6 were identified that yielded cells osmosensitive at high osmolarity. Both genes encode non-essential components of the RSC complex. We then analysed whether cells containing mutations on key components of the RSC complex also displayed a similar phenotype. We analysed cell growth in the presence of high osmolarity in strains containing RSC components tagged with a degron mark to stimulate degradation of the proteins (Campsteijn et al, 2007). Cells with reduced amount of Rsc8, Rsc3 or Rsc9 are even more sensitive to high osmolarity than rsc1- or npl6-deficient strains while they still grow in media without osmostress (Figure 1A; Supplementary Figure S5 and not shown). We then asked whether a double mutant hog1 rsc9deg was more osmosensitive than the single mutants were. As shown in Figure 1B, the osmosensitivity of the double mutant is very similar to the single hog1 mutant, suggesting that both genes might fall into the same pathway.
Figure 1.
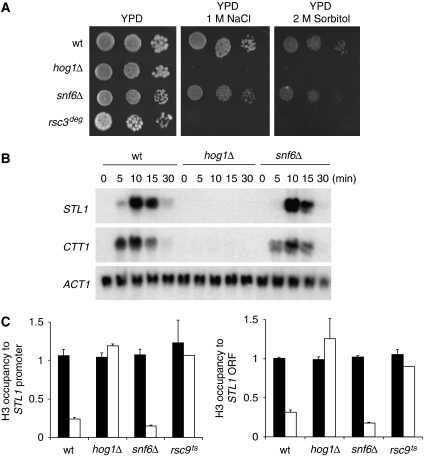
Mutations in the components of the RSC complex affect cell survival at high osmolarity. (A) Wild-type and the indicated mutant strains were spotted on YPD plates without and with 1.2 M NaCl or 2 M sorbitol. Growth was scored after 4 days. (B) The rsc9deg hog1Δ double mutant grows similarly at high osmolarity conditions than the single hog1Δ strain. Wild-type and the indicated mutant strains were spotted on YPD plates without and with osmotic stress conditions. Growth was scored after 4 days.
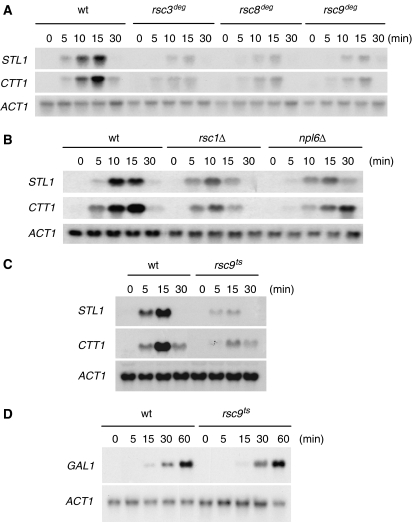
We then tested whether mutations in RSC components resulted in decreased osmostress gene expression. Expression of osmo-responsive genes such as STL1, CTT1 and GRE2 is significantly affected in these mutant strains (Figure 2A and B, and not shown. Quantitative data are shown in Supplementary Figure S1). These three genes are driven by different transcription factors under the control of the Hog1 MAPK (i.e. Hot1, Msn2/Msn4 and Sko1, respectively) and thus indicate a general defect on stress-responsive genes rather than a defect associated with a given transcription factor. Importantly, the RSC mutants, although showing a reduction in gene expression, they display similar Hog1 activation than the wild type in response to osmostress (data not shown). Correspondingly, the analysis of gene expression in a rsc9ts strain under non-permissive temperature showed that a ts mutation in RSC9 also strongly reduces expression of osmo-responsive genes upon stress (Figure 2C). The deletion of rsc1 or npl6 results in cells not as osmosensitive as to inactivation of RSC3, RSC8 or RSC9. Correspondingly, osmostress gene expression is reduced to a lesser degree in npl6 and rsc1 strains (Figure 2B). Under the same non-permissive conditions, induction of GAL1 in the presence of galactose was not affected (Figure 2D). Therefore, as it is the case for promoter-associated factors and elongation factors, the RSC complex is important for gene expression in response to osmotic stress and for the ability of the cells to grow at high osmolarity.
Figure 2.
Mutations in the components of the RSC complex display impaired osmostress gene expression. (A) RNA levels in wild-type and mutant strains (rsc3deg, rsc8deg and rsc9deg strains) grown in YPGal medium up to mid-log phase, subjected to 2 h at 37°C and then subjected to osmotic shock (0.4 M NaCl) for the indicated times are shown. (B) RNA levels in wild-type and RSC mutant strains (rsc1Δ and npl6Δ) grown in YPD medium up to mid-log phase and subjected to a mild osmotic shock (0.4 M NaCl) for the indicated times are shown. (C) rsc9ts mutation affects osmostress gene expression. Indicated strains were subjected to osmotic stress (0.4 M NaCl) for the indicated times. Total RNA was assayed by northern blot for STL1, CTT1 and ACT1 (as a loading control). (D) Induction of the GAL1 gene is not affected by mutation in rsc9. Wild-type and rsc9ts strains were grown at 25°C in YPRaf, shifted to 37°C for 1 h 30 min until OD660 0.7–0.9 and induced with galactose 2% for the indicated times. Total RNA was assayed by northern blot for GAL1 expression and ACT1 as a loading control.
Hog1 interacts with the RSC complex
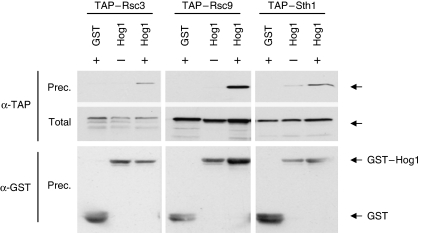
We previously showed that Hog1 interacted with several complexes (e.g. SAGA and Rpd3) to recruit them at the osmo-responsive promoters. We therefore tested whether Hog1 is able to interact with the RSC chromatin remodelling complex by performing GST pull-down experiments in extracts from osmotically stressed cells expressing GST–Hog1 and TAP-tagged versions of Rsc3, Sth1 and Rsc9 (all bona fide components of the RSC complex). In all cases, GST–Hog1, but not the GST control, co-precipitates the TAP-tagged RSC components and not with a control protein such as TAP–Bdf1 (Figure 3; Supplementary Figure S2C). Binding of RSC subunits with Hog1 might be stress dependent. Correspondingly, GST–Rsc9 was also able to pull down HA–Hog1 from osmotically stressed cells (Supplementary Figure S2A). Also, interaction was observed between Rsc3 and Hog1 both chromosomally tagged (Supplementary Figure S2B). Similar results were obtained using extracts treated with DNAse (not shown). Thus, the pull-down experiments indicate that Hog1 physically associates with the RSC complex. In addition, they strongly support the idea that Hog1 could be targeting RSC to the stress-responsive genes.
Figure 3.
Hog1 physically associates with the RSC complex. TAP-tagged Rsc3, Rsc9 and Sth1 strains that express GST or GST–Hog1 were (+) or were not (−) subjected to a brief osmotic shock (10 min, 0.4 M NaCl). GST proteins were pulled down by glutathione Sepharose 4B beads, and the presence of TAP proteins was probed by immunoblotting using anti-TAP antibody (PAP; Sigma) (top). Total extracts represent <20% of total input protein (middle). The amount of precipitated GST proteins was detected using anti-GST (bottom).
RSC associates with osmo-responsive genes upon stress
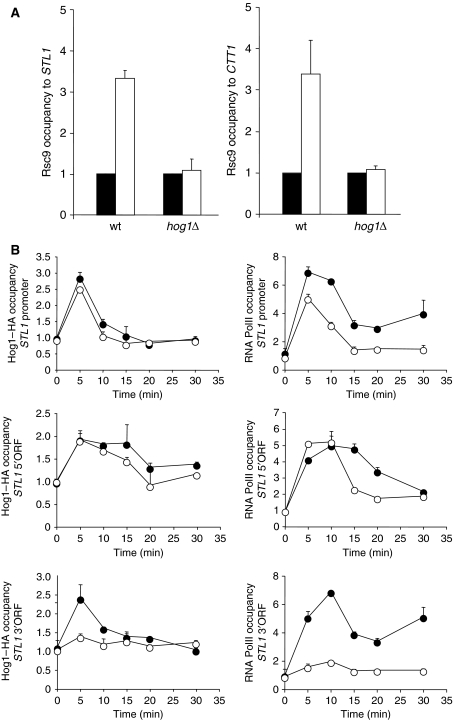
Hog1 associates with osmo-responsive gene promoters and throughout the entire transcribed region of target genes in response to stress (Alepuz et al, 2001; Pokholok et al, 2006; Proft et al, 2006). Considering that Hog1 associates with the RSC complex, we asked whether RSC was recruited to osmo-responsive genes in response to stress by Hog1. We utilized chromatin immunoprecipitation to follow the binding of Rsc1 and Rsc9 components of the RSC complex to various regions of osmostress-activated genes (STL1 and CTT1) before and after the addition of NaCl. Chromatin from wild-type and hog1Δ cells expressing functional epitope-tagged Rsc1 and Rsc9 from their natural locus was immunoprecipitated and analysed by PCR. As shown in Figure 4A (and data not shown), both subunits of the RSC complex associate specifically with osmo-responsive genes only in response to osmostress and its recruitment is completely dependent on HOG1. Results are shown as the fold induction of treated against the untreated cultures (see Materials and methods). It is worth noting that we could only detect binding of RSC to the coding regions and not to the promoters of stress-responsive genes (see Discussion). The binding kinetics of Rsc9 to stress coding regions is similar to Hog1 (data not shown). Therefore, our data suggested a possible role for RSC in a transcriptional process that occurs after the initiation step.
Figure 4.
Hog1 mediates the recruitment of RSC to stress genes, whereas binding of Hog1 is independent of RSC. (A) Hog1 mediates the recruitment of the RSC complex to stress-responsive genes in response to osmotic shock. Rsc9 association in vivo by ChIP with the ORF regions of STL1 and CTT1 loci in wild-type and hog1Δ strains (containing HA-tagged Rsc9) that were (open bars) or were not (filled bars) subjected to osmostress (0.4 M NaCl, 10 min) is shown. Results are shown as the fold induction of treated against the untreated cultures normalized to a telomere internal control. Data represent the mean and standard deviation of three independent experiments. (B) The RSC complex is required for proper RNA PolII occupancy at the 3′ end of the coding region of STL1 after osmostress. Wild-type (filled circles) and rsc9ts (open circles) strains expressing Hog1–HA tagged were grown in YPD at 25°C until OD660 0.5, shifted to 37°C for 1 h 30 min and samples were taken after treatment with 0.4 M NaCl for the indicated times. ChIP samples were immunoprecipitated using anti-HA antibody or anti-Rpb1 antibody (8WG16; Covance). Real-time PCR was performed to determine relative occupancy of Hog1–HA (left panels) and RNA PolII (right panels) to STL1 promoter and two different regions within the coding region (5′ORF and 3′ORF) normalized to the telomere control. Results are shown as the fold induction of treated against the non-treated (time 0). Data represent the mean and standard deviation of three independent experiments.
Our results suggest that RSC is recruited by Hog1 to stress genes and thus, binding of Hog1 should be independent of RSC. To determine whether binding of Hog1 was independent of RSC, we utilized chromatin immunoprecipitation to follow the binding of Hog1 to STL1 before and after the addition of NaCl. Chromatin from wild-type and rsc9ts cells expressing functional epitope-tagged Hog1 from their natural locus was immunoprecipitated and analysed by PCR. Results are shown as the fold induction of treated against the untreated cultures (time 0; see Materials and methods). As shown in Figure 4B (left panels), Hog1 binds to the promoter of STL1 either in wild-type and rsc9-deficient strains upon stress. Therefore, in contrast to RSC binding, that is dependent on Hog1, binding of Hog1 does not depend on RSC. It is worth noting that Hog1 recruitment is reduced at the 3′ terminus of the coding region in rsc9-deficient cells. The lack of Hog1 in this region correlates with the lack of polymerase on the same region (Figure 4B, right panels).
Defects on specific promoter-associated factors, which are important in the regulation of osmo-responsive genes, result in reduced accumulation of PolII at stress genes. Thus, we assessed by ChIP the presence of polymerase on the promoter and coding region of STL1 in wild-type and rsc9ts strains in response to osmostress. As shown in Figure 4B (right panels), PolII is recruited on the promoter of STL1 in response to stress in the rsc9ts strain as efficiently as in the wild type. However, when PolII binding was analysed by ChIP on other regions along STL1, PolII recruitment was clearly defective on the 3′ end of the coding region in rsc9ts mutant cells. Therefore, although the lack of RSC does not seem to affect the recruitment of PolII at stress promoters, it strongly affects PolII progression throughout the gene.
Hog1 mediates nucleosome reorganization at stress-responsive loci
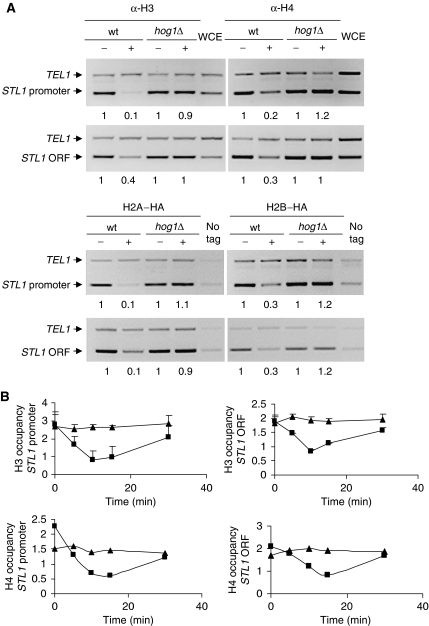
It has been shown that RSC can promote nucleosome remodelling when associates with chromatin (Saha et al, 2006). The targeting of RSC by Hog1 to osmostress genes prompted us to analyse whether Hog1 was promoting nucleosome rearrangements at osmo-responsive genes upon stress. Chromatin from HA-tagged H2A or H2B in wild-type and hog1Δ cells subjected to osmostress was immunoprecipitated using specific antibodies against anti-HA epitope or anti-H3 and H4 and analysed by PCR. As shown in Figure 5A, a dramatic reduction of all histones is observed at both promoter and coding regions of STL1 gene only in response to osmostress. Interestingly, no reduction in binding of any of the histones is observed in stressed hog1Δ cells. Similar effects were observed at GRE2 and CTT1 (Supplementary Figure S3A), whereas no changes in histone composition could be observed in a non-stress-responsive gene such as ACT1 (Supplementary Figure S3B). Kinetic analyses of the in vivo binding of histone H3 and H4 at STL1 in response to osmostress showed that histone eviction is transient and correlates with Hog1 activation (Figure 5B; Supplementary Figure S4). Thus, Hog1 is promoting massive histone eviction at osmo-responsive genes in response to osmostress.
Figure 5.
Hog1 is required for nucleosome reorganization in response to osmostress. (A) Association of histones H3, H4, HA–H2A and HA–H2B with the STL1 promoter and STL1 ORF in wild-type and hog1Δ strains that were (+) or were not (−) exposed to hyperosmotic stress (0.4 M NaCl) is shown. Histones were immunoprecipitated with antibodies against H3 (Abcam), H4 (Abcam) and anti-HA. Binding to STL1 promoter and open-reading frame (ORF) was determined by ChIP. As an internal loading control, PCR samples were amplified with a telomere region (upper band). Control lanes show DNA amplified from extracts prior to immunoprecipitation (WCE) or without tagged protein (no tag). Quantification is depicted as fold binding over TEL and results are shown as the fold induction of treated against the untreated cultures. (B) Kinetics of histone H3 and H4 binding under osmostress conditions. In vivo binding of histone H3 and H4 to STL1 promoter and coding region is shown in wild-type (squares) and hog1Δ (triangles) strains after osmostress. Quantitative data were obtained by real-time PCR and TEL was used as a reference control. Results are shown as the fold induction of treated against the untreated (time 0) cultures. Data represent the mean and standard deviation of three independent experiments.
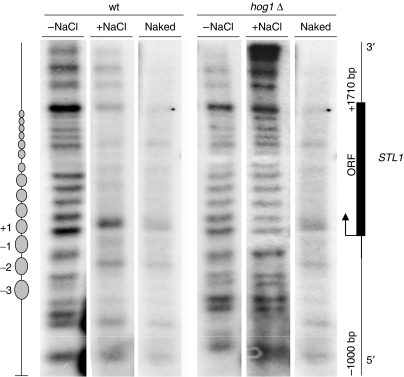
To support the data from ChIP analyses and shed some light on chromatin organization, we analysed the nucleosome pattern in the STL1 gene by performing micrococcal nuclease (MN) digestion of chromatin before and after stress. Wild-type and hog1 strains were subjected with or without osmostress (same conditions used for ChIP analyses) and cells were fixed before chromatin and nuclease digestion to prevent Hog1 activation during spheroplast preparation (see Materials and methods). We found that the digestion pattern of STL1 is consistent with the fact that without stress, STL1 exhibits strongly positioned nucleosomes at the promoter, coding and 3′ regions. Interestingly, in response to osmostress the patterning for nucleosome positioning dramatically changes and chromatin displays a similar digestion pattern as naked DNA (Figure 6). In accordance with the ChIP analyses, MNase digestion showed that exposure to osmostress of hog1Δ cells did not affect nucleosome organization when compared with non-stressed cells (Figure 6). Nucleosome positioning did not change in the non-stress-responsive genes such as ACT1 or GAL1 upon stress as observed by ChIP (data not shown). Therefore, data from both of our chromatin assays support the idea that activated Hog1 induces nucleosome re-positioning at osmo-responsive genes.
Figure 6.
Hog1 mediates major changes in nucleosome positioning in response to high osmolarity. Chromatin and naked DNA from wild-type and hog1Δ cells treated with (+NaCl) or without (−NaCl) were digested with micrococcal nuclease (MN), digested with EcoRI restriction enzyme, resolved in agarose gels and hybridized with a DNA probe to map nucleosomes at chromosomal STL1. The depicted scheme shows the location of nucleosomes (ovals) at the STL1 gene in the wild-type strain without stress.
RSC function is required for nucleosome reorganization upon osmostress
It was reported that the SWI/SNF chromatin remodeller associates with osmo-responsive promoters upon stress (Proft and Struhl, 2002). Thus, we studied whether this complex or the RSC complex could be involved in the dynamic changes in chromatin structure. In contrast to RSC, deletion of SNF6 did not affect cell survival or gene expression of STL1 and CTT1 upon osmostress (Figure 7A and B). Correspondingly, in snf6 cells, histone eviction upon stress was similar to wild type (Figure 7C).
Figure 7.
Mutations in the SWI/SNF complex do not significantly affect cell viability, gene expression and histone eviction in stress-responsive genes upon high osmolarity. (A) Wild-type (BY4741) and the indicated mutant strains were spotted on YPD plates without and with 1 M NaCl or 2 M sorbitol. Growth was scored after 4 days. (B) RNA levels in wild-type and mutant strains (hog1Δ and snf6Δ strains) were grown in YPD medium up to mid-log phase and then subjected to osmotic shock (0.4 M NaCl) for the indicated times. Total RNA was assayed by northern blot for STL1, CTT1 and ACT1 (as a loading control). (C) Association of histone H3 in wild-type, hog1Δ, snf6Δ and rsc9ts strains. Cells were grown at 25°C until OD660 0.5, shifted to 37°C for 1 h 30 min and treated (open bars) or untreated (filled bars) with 0.4 M NaCl for 10 min. Histone H3 was immunoprecipitated with an anti-H3 antibody (Abcam). Quantitative data were obtained by real-time PCR, and TEL was used as a reference control. Results are shown as the fold induction of treated against the untreated cultures. Data represent the mean and standard deviation of three independent experiments.
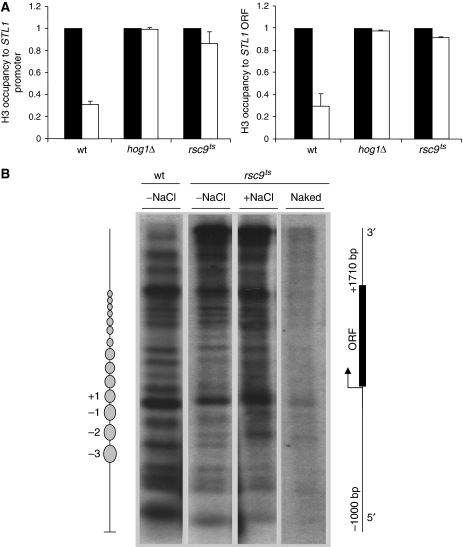
The association of Hog1 with the RSC complex suggested that the RSC complex might be one effector of Hog1 that mediates chromatin remodelling. We therefore tested whether the RSC complex is indeed important for histone eviction in stress-responsive genes. Chromatin from wild-type and rsc9ts cells subjected to osmostress was immunoprecipitated using specific antibodies against H3 and analysed by PCR. As shown in Figure 8A, a decrease in histone H3 binding was observed upon osmostress at the STL1 loci in wild-type cells, which was observed neither in rsc9ts cells nor in hog1-deficient cells. Similar results were obtained when histone H3 eviction was analysed at the CTT1 locus (not shown). Moreover, identical results were obtained when the rsc9 degron strain was tested (not shown). Therefore, in contrast to Snf6, RSC mutation had a clear difference on chromatin dynamics. We then analysed the nucleosome pattern in the STL1 gene by MN digestion of chromatin before and after stress in wild-type and rsc9ts strains as before. We found that although the digestion pattern of STL1 in -type and rsc9ts strains without stress is slightly different, in response to stress the patterning for nucleosome positioning almost do not change in a rsc9ts strain (Figure 8B). Correspondingly, gene expression in response to osmostress was reduced in the rsc9ts strain (Figure 2C). Therefore, both ChIP and MNase analyses have shown that RSC function is required for chromatin remodelling at osmo-responsive genes upon stress. Taken together, although SWI/SNF is recruited to stress genes, RSC is the key remodeller to promote chromatin reorganization upon stress.
Figure 8.
RSC mediates major changes in nucleosome organization in response to high osmolarity. (A) Rsc9 is required for histone eviction at STL1 loci in response to high osmolarity. Association of histone H3 in wild-type, hog1Δ and rsc9ts strains is shown. Strains were grown at 25°C until OD660 0.5, shifted to 37°C for 1 h 30 min and samples were either untreated (filled bars) or treated (open bars) with 0.4 M NaCl for 10 min. Histone H3 was immunoprecipitated with an anti-H3 antibody (Abcam). Occupancy to STL1 promoter and ORF regions was normalized to TEL as an internal control. Results are shown as the fold induction of treated against the untreated cultures. Data represent the mean and standard deviation of three independent experiments. (B) Chromatin and naked DNA from wild-type and rsc9ts cells were grown at 25°C, shifted to 37°C for 1 h 30 min and treated with (+NaCl) or without (−NaCl) 0.4 M NaCl. Chromatin and DNA were digested with micrococcal nuclease (MN), digested with EcoRI restriction enzyme, resolved in agarose gels and hybridized with a DNA probe to map nucleosomes at chromosomal STL1. The scheme shows the location of nucleosomes (ovals) at the STL1 gene in the wild-type strain without stress.
Discussion
The SAPKs regulate the expression of specific classes of genes that permit the adaptation to environmental stress. Often, SAPKs trigger the necessary response by directly regulating transcriptional initiation through phosphorylation of transcriptional regulatory proteins. However, at least in yeast, the SAPK Hog1 engages in the transcription programme in general terms by taking up a role as an integral component of several transcription complexes. In its active form, Hog1 directs the recruitment of the PolII holoenzyme and the Rpd3 and SAGA complexes to stress-responsive promoters (Alepuz et al, 2003; de Nadal et al, 2004; Zapater et al, 2007). A similar scenario seems to be observed on mammalian genes. During skeletal myogenesis, the p38 MAPK (the mammalian homologue of the Hog1 MAPK) is recruited to muscle-specific promoters and there it participates in the recruitment of chromatin-remodelling activities such as SWI/SNF (Simone et al, 2004). Moreover, recent reports have shown that the role of Hog1 may not be just limited to transcription initiation but rather extends to the process of transcriptional elongation. Here, Hog1 interacts with elongating PolII and components of the TEC to stimulate mRNA production, an observation that defines Hog1 as a bona fide elongation factor, with the novel feature that its role in elongation is restricted to osmo-responsive genes (Pokholok et al, 2006; Proft et al, 2006). This contention is underlined by the fact that the active protein kinase can be crosslinked specifically to DNA sequences that span the open reading frame of osmo-responsive genes. In addition to Hog1, other signalling kinases are also present at the coding regions of target genes (Pokholok et al, 2006). Although the mechanism by which these signalling kinases regulate transcription is still unknown, it opens novel roles for these group of kinases in chromatin (Chow and Davis, 2006; Edmunds and Mahadevan, 2006). The study described here clearly provides one important example towards this possibility by establishing a surprising connection among a prototypical signalling kinase, a chromatin remodelling factor (RSC) and a specific state of chromatin.
Recruitment of Hog1 to ORFs of stress genes is initially identical to that of the PolII. However, the presence of SAPK is temporally more restricted, suggesting a role for the SAPK only at the initial stages of elongation. The nucleosomes occupying the stress-responsive genes provide an obvious candidate structure to serve as the target of the Hog1 (Proft et al, 2006). This speculation seems to be correct, as we found that in response to Hog1 activation there is a transient but quite dramatic change in the nucleosome positioning and the overall occupation levels upon stress at the osmo-responsive loci. Hog1, however, mediates nucleosome rearrangements at stress genes not by direct modification of histones but by physically associating and targeting the chromatin remodelling RSC complex to the coding regions. Indeed, mutations that affect RSC activity display severe defects on gene expression of stress genes and mostly abolish nucleosome remodelling at the stress loci. Interestingly, although PolII is recruited efficiently on the promoters of strains deficient on RSC, PolII is not present at the 3′ region of the ORFs. Thus, chromatin reorganization is critical for polymerase progression and proper gene expression upon stress. Recruitment of Hog1 to osmo-responsive genes occurs at the promoters through binding to specific transcription factors and independently at the coding regions of the genes through the 3′ UTRs (Proft et al, 2006). We have observed that RSC is important to mediate nucleosome reorganization at both promoter and coding region of osmo-responsive genes; however, we detect only the recruitment of RSC at the coding regions of stress genes. Although we cannot exclude that binding of RSC also occurs at the promoters with less affinity than in the coding regions, it might be that Hog1 mediates the recruitment of RSC once the TEC is formed after an initial round of transcription. Actually, maximal histone displacement is observed 10 min into the stress response and importantly 5 min after PolII is already maximally recruited at the promoters (Proft et al, 2006). Thus, together these results should define a role for the SAPK at the elongation phase of transcription. It has been shown that other remodellers are associated with stress genes such as SWI/SNF (Proft and Struhl, 2002); however, in contrast to mutations on RSC, mutations on SWI/SNF do not affect gene expression, histone eviction or osmosensitivity.
It has been shown that RSC is able to stimulate chromatin remodelling and to mediate active transcription from a paused polymerase within a gene (Carey et al, 2006). However, the relevance in vivo for the remodelling of a specific subset of genes has not been described. Genome-wide binding analyses showed that strong changes in the localization of RSC components occur upon different type of stresses, indicating that external stimuli promote changes in RSC activity that can be mediated by signalling molecules (Damelin et al, 2002; Ng et al, 2002). Interestingly, the specificity for the RSC to stress genes might be determined by the direct targeting of the MAPK to stress-responsive genes. We have also tested whether Hog1 can modify RSC by direct phosphorylation. So far, we have only identified Npl6 as a direct substrate of the MAPK; however, mutation on the phosphorylation site, albeit completely eliminated Hog1 phosphorylation, did not affect gene expression. Thus, this could indicate that selective tethering by the MAPK is critical for the remodelling. Therefore, our results define a novel function for Hog1 at the stress loci, that is the targeting of the RSC complex to a specific set of genes to mediate chromatin remodelling and enabling efficient gene expression and maximal cell survival.
Materials and methods
Yeast strains and plasmid DNAs
Yeast strains used in this study are derived form BY4741 (MATa his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0): chromosomally integrated strains that express tag-TAP proteins under its own promoter RSC3-TAP, RSC9-TAP, DBF1-TAP and STH1-TAP (from the yeast TAP collection; Open Biosystems) and deletion strains YGM61 (MATa hog1∷kanMx4), rsc1∷kanMx4, npl6∷kanMx4 and snf6∷kanMx4. Chromosomally integrated strains YGM164 (MATa HTA1-HA6-HIS3) and YGM177 (MATa HTA1-HA6-HIS3 hog1∷kanMx4) were genomically tagged at the C terminus of the HTA1 locus with a cassette encoding 6-HA epitope tag and HIS3 as selectable marker. YGM165 (MATa HTB1-HA6-HIS3) and YGM181 (MATa HTB1-HA6-HIS3 hog1∷kanMx4) were genomically tagged at the C terminus of the HTB1 locus by means of the described HA6-HIS3 cassette. The degron strains (MATa ADE2; his3-11,15; leu2-3,112; trp1-1; ura3-1; Δlys2∷rKWD50N, PGal1–10-myc∷UBR1∷HIS3) were rsc3 degron-URA3 (PCUP1-degron∷rsc3∷URA3), rsc8 degron-URA3 (PCUP1-degron∷rsc8∷URA3) and rsc9 degron-URA3 (PCUP1-degron∷rsc9∷URA3). The rsc9 degron strain was used to delete the HOG1 gene by the LEU2 selectable marker (YGM264). Derivatives from W303-1A (leu2-3,112 ura3-1 his-11 trp1-1 can1-100) was the rsc9ts strain (MATa fae1-2∷TRP1) from Dr G Ammerer. W303 wild-type and hog1∷kanMx4 strains were tagged in the genomic locus of RSC9 and RSC1 at the C terminus with a sequence encoding 6-HA epitope tag followed by the Schizosaccharomyces pombe HIS3 as a selectable marker (YGM17, YGM23, YGM68 and YGM79). W303 wild-type, a rsc9ts or TAP–Rsc3 strains were genomically tagged at the C terminus of the HOG1 locus with a sequence encoding 6-HA epitope tag followed by the S. pombe HIS3 as a selectable marker (YEN173, YGM248 and YGM285). The strains containing tagged Hog1 or Rsc9 were fully functional. The plasmids used in this study are derived from pRS426TEG1 (PTEF1-GST, URA3+, 2 μm) vector. The GST-Hog1 plasmid was able to suppress the osmosensitivity observed in a hog1 strain. The GST-Rsc9 plasmid was able to complement the temperature sensitivity defect of a rsc9ts strain. Therefore, both plasmid expressed fully functional proteins.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as described previously (Kuras and Struhl, 1999; Alepuz et al, 2001). Yeast cultures were grown to early log phase (OD600=0.6–1.0) before aliquots of the culture were exposed to osmotic stress treatment (0.4 M NaCl) for the time specified in the figure legends. For crosslinking, yeast cells were treated with 1% formaldehyde for 20 min at room temperature. Antibodies used in this study were rabbit polyclonal to histone H3 and H4 (Abcam antibodies ab1791 and ab10158, respectively), and anti-Rpb1 (8WG16; Covance). Monoclonal anti-HA and anti-MYC antibodies were also used to immunoprecipitate HA- and Myc-tagged proteins. Primer mixes were adjusted for balanced signals. Conventional and real-time PCR analysis of stress genes and constitutively expressed genes utilized the following primers with locations indicated by the distance from the respect ATG initiation codon: STL1 (−372/−112 for promoter; +402/+630 for 5′ coding region; +1550/+1793 for 3′ coding region), CTT1 (−452/−160 for promoter; +422/+669 for coding region), GRE2 (−300/+30 for promoter; +340/+620 for coding region), ACT1 (−389/+30), GAL10 (+150/+316) and TEL1 (region 490 bp right arm of chromosome VI). Experiments were performed on three independent chromatin preparations, and quantitative PCR analysis was performed in real time using an Applied Biosystems 7700 sequence detector. Immunoprecipitation efficiencies were calculated in triplicate by dividing the amount of PCR product in the immunoprecipitated sample by the amount of PCR product in the input sample. Data are presented as fold immunoprecipitation over TEL sequence control.
GST pull-down experiments
To analyse the association of Hog1 with components of the RSC complex, 1 mg of yeast extract from cells expressing specific TAP- or HA-tagged protein was incubated with 50 μl of glutathione Sepharose 4B beads overnight at 4°C in buffer A (50 mM Tris–HCl pH 7.5, 15 mM EDTA, 15 mM EGTA, 0.1% Triton X-100, 150 mM NaCl, 2 mM DTT plus antiproteases and phosphatase inhibitors). The beads were washed extensively with buffer A, resuspended in loading buffer and resolved by SDS–PAGE. The antibody used to detect the TAP-/HA-tagged proteins was the PAP antibody from Sigma or specific anti-HA antibody.
Northern blot analysis
Yeast strains were grown to mid-log phase in rich medium and then subjected to osmotic shock (0.4 M NaCl) for the indicated times. Total RNA and expression of specific genes were probed using labelled PCR fragments containing the entire open reading frame of STL1 (1.7 kb), CTT1 (1.7 kb), GAL1 (0.87 kb) and ACT1 (1.4 kb).
MNase nucleosome mapping
Yeast spheroplasts and MN digestions were performed as described previously with modifications (Chávez et al, 1995). Spheroplasts were prepared from mid-log phase cultures grown in SC-Ura with 2% glucose and either untreated or treated with salt (0.4 M NaCl, 10 min), following 1% formaldehyde crosslinking for 20 min, treated with 125 mM glycine for 15 min and washed four times with TBS. Cells were then lysed and immediately digested with 7.5–125 mU of MN. For naked DNA controls, genomic DNA was extracted as previously described and digested with 0.003–0.2 mU of MN under the same conditions. MN-cleaved genomic DNA was digested with EcoRI (for STL1) and resolved in 1.5% agarose gel without ethidium bromide. For the analysis of STL1, the probe used was the 200-bp PCR fragment immediately upstream of the EcoRI site present in STL1 (at −1052 from the transcription start site).
Supplementary Material
Supplementary Figures S1–S5
Supplementary Figures Legend
Acknowledgments
We are grateful to Drs C Schüller and R Shiekhattar for helpful discussions. To J Perez and L Subirana for their excellent technical assistance. GM was recipient of PhD fellowships from the Ministerio de Educación y Ciencia. This study was supported by the ‘Ramón y Cajal' programme and grant BFU2005-00202 to EdeN, grant BFU2006-15446-C03-01 to SC, and grants from Ministerio de Educación y Ciencia BFU2006 and Consolider Ingenio 2010 programme (grant CSD2007-0015), from the European Science Foundation (ESF) under the EUROCORES Program EuroDYNA and through contract no. ERAS-CT-2003-980409 of the European Commission, DG Research, FP6 as part of a EURYI scheme award (www.esf.org/euryi) to FP.
References
- Alepuz PM, de Nadal E, Zapater M, Ammerer G, Posas F (2003) Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA Pol II. EMBO J 22: 2433–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alepuz PM, Jovanovic A, Reiser V, Ammerer G (2001) Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol Cell 7: 767–777 [DOI] [PubMed] [Google Scholar]
- Campsteijn C, Wijnands-Collin AM, Logie C (2007) Reverse genetic analysis of the yeast RSC chromatin remodeler reveals a role for RSC3 and SNF5 homolog 1 in ploidy maintenance. PLoS Genet 3: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M, Li B, Workman JL (2006) RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell 24: 481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez S, Candau R, Truss M, Beato M (1995) Constitutive repression and nuclear factor I-dependent hormone activation of the mouse mammary tumor virus promoter in Saccharomyces cerevisiae. Mol Cell Biol 15: 6987–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CW, Davis RJ (2006) Proteins kinases: chromatin-associated enzymes? Cell 127: 887–890 [DOI] [PubMed] [Google Scholar]
- Damelin M, Simon I, Moy TI, Wilson B, Komili S, Tempst P, Roth FP, Young RA, Cairns BR, Silver PA (2002) The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol Cell 9: 563–573 [DOI] [PubMed] [Google Scholar]
- de Nadal E, Alepuz PM, Posas F (2002) Dealing with osmostress through MAP kinase activation. EMBO Rep 3: 735–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E, Zapater M, Alepuz PM, Sumoy L, Mas G, Posas F (2004) The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427: 370–374 [DOI] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC (2006) Cell signaling. Protein kinases seek close encounters with active genes. Science 313: 449–451 [DOI] [PubMed] [Google Scholar]
- Govind CK, Yoon S, Qiu H, Govind S, Hinnebusch AG (2005) Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol Cell Biol 25: 5626–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Kuras L, Struhl K (1999) Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399: 609–613 [DOI] [PubMed] [Google Scholar]
- Kyriakis JM, Avruch J (2001) Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev 81: 807–869 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, Kornberg RD (2006) Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci USA 103: 3090–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira JMA, Holmberg S (1999) Transcriptional repression of the yeast CHA1 gene requires the chromatin-remodeling complex RSC. EMBO J 18: 2836–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2002) Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev 16: 806–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA (2006) Activated signal transduction kinases frequently occupy target genes. Science 313: 533–536 [DOI] [PubMed] [Google Scholar]
- Proft M, Mas G, de Nadal E, Vendrell A, Noriega N, Struhl K, Posas F (2006) The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol Cell 23: 241–250 [DOI] [PubMed] [Google Scholar]
- Proft M, Struhl K (2002) Hog1 kinase converts the Sko1–Cyc8–Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell 9: 1307–1317 [DOI] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR (2006) Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol 7: 437–447 [DOI] [PubMed] [Google Scholar]
- Sengupta SM, VanKanegan M, Persinger J, Logie C, Cairns BR, Peterson CL, Bartholomew B (2001) The interactions of yeast SWI/SNF and RSC with the nucleosome before and after chromatin remodelling. J Biol Chem 276: 12636–12644 [DOI] [PubMed] [Google Scholar]
- Sheikh-Hamad D, Gustin MC (2004) MAP kinases and the adaptive response to hypertonicity: functional preservation from yeast to mammals. Am J Physiol Renal Physiol 287: F1102–F1110 [DOI] [PubMed] [Google Scholar]
- Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL (2004) p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet 36: 738–743 [DOI] [PubMed] [Google Scholar]
- Soutourina J, Bordas-Le Floch V, Gendrel G, Flores A, Ducrot C, Dumay-Odelot H, Soularue P, Navarro F, Cairns BR, Lefebvre O, Werner M (2006) Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol Cell Biol 26: 4920–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PJ, Patterson JC, Chen RE, Thorner J (2008) Stress resistance and signal fidelity independent of nuclear MAPK function. Proc Natl Acad Sci USA 105: 12212–12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapater M, Sohrmann M, Peter M, Posas F, de Nadal E (2007) Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol Cell Biol 27: 3900–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C (2006) DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol Cell 24: 559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S5
Supplementary Figures Legend