Abstract
Simulations of a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine lipid bilayer interacting with a solid surface of hydroxylated nanoporous amorphous silica have been carried out over a range of lipid-solid substrate distances. The porous solid surface allowed the water layer to dynamically adjust is thickness, maintaining equal pressures above and below the membrane bilayer. Qualitative estimates of the force between the surfaces leads to an estimated lipid-silicon distance in very good agreement with the results of neutron scattering experiments. Detailed analysis of the simulation at the separation suggested by experiment shows that for this type of solid support the water layer between surfaces is very narrow, consisting only of bound waters hydrating the lipid headgroups and hydrophilic silica surface. The reduced hydration, however, has only minor effects on the headgroup hydration, the orientation of water molecules at the interface, and the membrane dipole potential. While these structural properties were not sensitive to the presence of the solid substrate, the calculated diffusion coefficient for translation of the lipid molecules was altered significantly by the silica surface.
Keywords: molecular dynamics, diffusion, lipid bilayer, solid support
INTRODUCTION
Solid supported lipid bilayer membranes have found wide use as an experimental system to study membrane properties 1 and show promise in the design of devices with practical applications such as chemical sensing 2. A particularly promising class of solid supported membranes are those employing nanoporous silica as the solid substrate 3. The nanoporous silica films allow encapsulation of molecules and reservoirs of fluid having controlled compositions differing from that outside the membrane, potentially generating architectures with properties of cellular membranes. Despite the interest in supported lipid bilayers, both as a research tool and as a technological development, few detailed molecular models of the systems have been developed.
Molecular dynamics (MD) computer simulation has emerged in the past decade as a powerful tool for probing the structure and dynamics of phospholipid membranes at the atomic scale. The technique is especially useful for understanding the biologically relevant fluid phase of the bilayer that is characterized not by a single molecular conformation but by a large ensemble of states. Previously we have used MD simulation to examine a number of lipid systems, including bilayers differing in their headgroup and acyl chains composition 4, 5, transmembrane proteins 6, membrane bound peptides 7, phospholipid sterol mixtures 8, and bilayers with small molecules 9 or detergents 10 incorporated. While simulations of lipid bilayers and complicated bilayer membranes are now relatively common, only a few such computational studies of supported bilayers have been undertaken. An early study described an MD simulation of a hybrid bilayer formed from a phospholipid monolayer and an alkanethiol monolayer covalently attached to a solid (gold) surface. 11 Very recently a coarse grained model was applied to model a phospatidylcholine bilayer in contact with a solid, hydrophilic support.12 An atomically detailed model, similar in form to the one described here, has been used to study a lipid bilayer supported by a quartz substrate.13
In the following we describe a series of MD simulations at varying lipid-substrate distances that have been analyzed with a focus on changes in the structure and dynamics of the lipid bilayer and the water layer between the membrane and the substrate. By comparing structural features observed in the simulation to experimental neutron scattering results we obtain an atomically detailed model that matches laboratory observation. The simulation trajectory is then used to predict other properties of the solid supported membrane. We conclude with some observations from the simulation that may be useful in understanding the properties of supported bilayer systems.
PROCEDURE
MD simulations were carried out using the program CHARMM. The lipid and water force field were standard CHARMM versions14, 15 while the silica parameters were those employed by Leung et al.16 The CHARMM force field includes internal energy terms describing bond stretching, bond angle bending, and torsional rotation. Intermolecular interactions (and intra-molecular interactions between atoms separated by more than two chemical bonds) are given by Coulombic interactions between partial atomic charges and a Lennard-Jones 6–12 potential between atomic centers. Standard CHARMM combining rules were used for interactions between different atom types, including lipid-silica interactions. The 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer consisted of 46 lipids in each monolayer. The silica slab was ~30 Å thick with lateral dimensions of ~51 and ~56 Å and was spanned by a pore ~ 2 nm in diameter. The construction of the initial coordinates of the silica substrate is described by Leung et al.16 who use a combination of quantum mechanical and force-field based Monte Carlo calculations to produce the slab of amorphous silica with the desired pore radius. The lipid and silica were hydrated with 7325 water molecules, giving a total system size of ~40k atoms. The simulation was carried out under conditions of constant volume and temperature of (298K). Three dimensional periodic boundary conditions were applied and long range electrostatics computed with the particle mesh Ewald summation.17 Bonds involving hydrogen were held rigid with the SHAKE algorithm18 allowing a 2 fs time step.
The initial configuration of our bilayer-substrate system placed the POPC bilayer ~ 30Å from the silica substrate. Additional systems were constructed sequentially by applying a planar harmonic restraint (force constant 100 kcal/mol/Å2) to the center of mass of the bilayer at distances nearer to the silica substrate, allowing each simulation to equilibrate the solvent density before decreasing the silica-lipid distance. The nanopore in the silica substrate allows water to flow through the substrate, correcting the water density as the bilayer moves closer. The spring position was moved ~2 Å in each cycle with a total of 13 simulations carried out. After equilibration the force constant was reduced to 1 kcal/mol/Å2 and each of the 13 simulations was continued for 5 nanoseconds with some simulations carried out to greater times as detailed below.
RESULTS
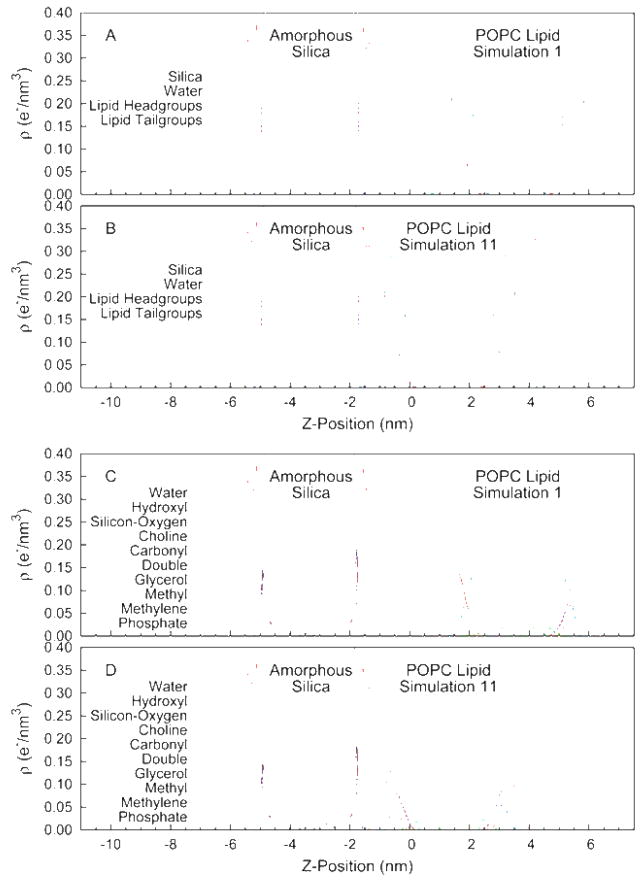
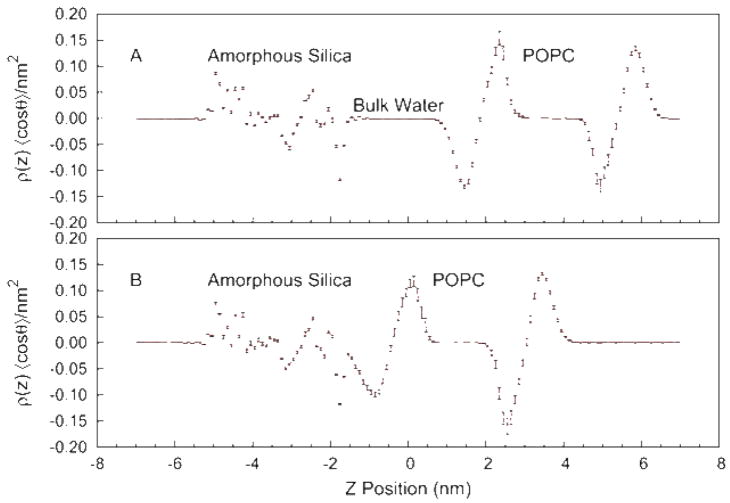
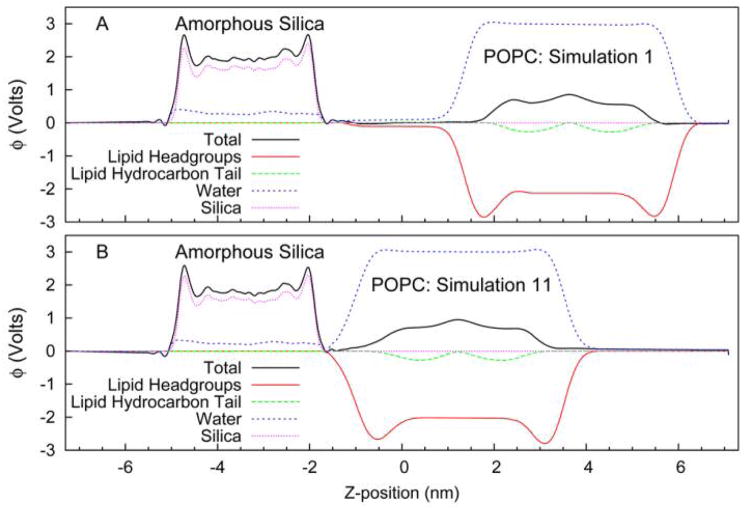
The structure of the fluid phase lipid bilayer is best described in terms of probability distributions due to the high degree of disorder and the significant thermal motions of the phospholipid molecules. In Figure 1 we plot the distribution of silica, water, and lipid as a function of position along the direction perpendicular to the surface. For this calculation the system was divided into 144 slabs (~1 Å thick) perpendicular to the bilayer normal. The occurrences for each molecular fragment were measured for each coordinate set and averaged over the final 2 ns of trajectory. The upper panel (Figure 1 A) shows the result at a large silica-lipid separation, corresponding to non-interacting surfaces. The distribution of lipid headgroups and acyl chains is consistent with previous simulations of phospholipid bilayers, showing broad peaks and a complex mixture of water and lipid components at the membrane/water interface. At this large separation there is a significant layer, 10–15 Å in thickness, of bulk water unperturbed by the silica or lipid surfaces. To quantify the influence of the surfaces on the structure of water we have computed the average orientation of the water molecules, 〈cos(θ)〉, where θ is the angle between the dipole moment vector of the water molecule and the vector normal to the surface. The upper panel (A) of Figure 2 gives the mean water orientation weighted by the water density distribution, showing regions of water molecules strongly ordered by the phosphatidylcholine and carbonyl groups of the lipid (with opposite orientation) and by the hydrophilic hydroxyl groups on the silica surface. The decay length of the orientational distribution is ~ 6 Å in the direction of the center of the water lamella. In summary, the results at large lipid-substrate distance suggest no perturbation of the bilayer structure and indicate a significant layer of water molecules having bulk density and no orientational preference.
Figure 1.
Transverse distribution of lipid, silica, and water segments in the simulation unit cell. The segment densities are given in terms of the number of electrons per unit volume. (A&C) Results from simulation 1 (negligible lipid-substrate interactions). (B&D) Results from simulation 11 (strong lipid-substrate interactions).
Figure 2.
Mean orientation of water molecule dipole moment as a function of transverse position in the simulation unit cell, weighted by the density of water molecules. (A) Results from simulation 1 (negligible lipid-substrate interactions). (B) Results from simulation 11 (strong lipid-substrate interactions). The error bars are calculated as the standard error, computed from 1 ns block averages.
Given the disordered nature of the membrane interface it is difficult to develop a single precise meaning of lipid-substrate distance, therefore we have chosen to define a metric based on a structural feature that has been measured experimentally and which may be useful in characterizing the lipid/silica interface. We compute the hydrophilic layer thickness (HT) as the distance between the silica interface, measured as the location where the silica density reaches 50% of its bulk value, and the hydrophobic interface of the bilayer, measured as the location where the acyl chain density reaches 50% of its bulk value. These two interfaces have the advantage of being among the sharpest structural features of the system and defining important parameters describing the electrostatics of the environment. Additionally, this distance has been measured experimentally for the silica-POPC system using neutron scattering techniques 19, giving a distance of 14.6 Å. The simulation at large lipid-substrate distance (described in the preceding paragraph and hereafter referred to as simulation 1) gives HT = 37.4 Å, however our simulations covered a range of distances including one (simulation 11) with HT = 15.2 Å. The transverse distribution functions for simulation 11 are given in the lower panel (B) of Figure 1. The most striking feature of a comparison between Figures 1A and 1B is the complete loss of that portion of the water layer having bulk-like density. Interestingly the silica surface and lipid headgroups maintain nearly identical levels of hydration, the equilibrium spacing between the membrane and solid support is the minimum value that allows each interface to remain fully hydrated. This is supported by Figure 2B that shows the region of unoriented water has disappeared at this spacing yet the orientation of the surface bound waters, to both the lipid and to the silica, is little changed.
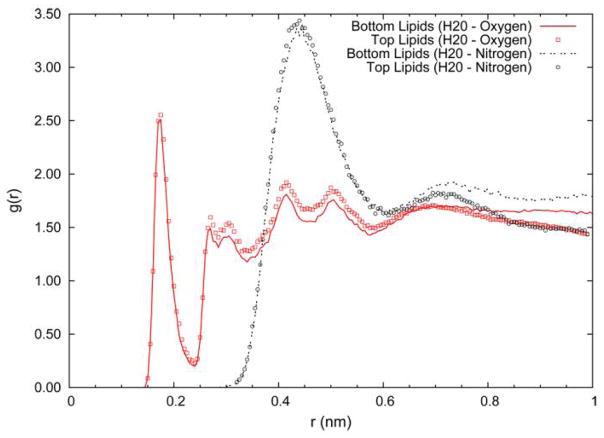
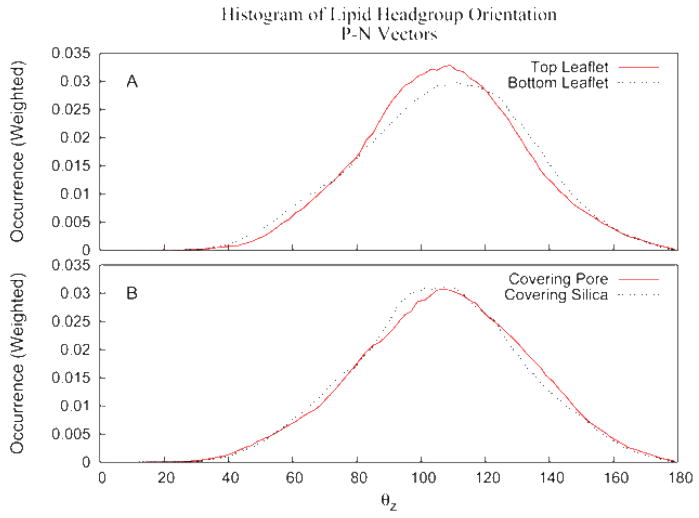
A molecular graphic of a snapshot taken from simulation 11 is given in Figure 3 and shows the crowded hydrophilic region between lipid and substrate. The graphic clearly shows that the concept of a water layer between the surfaces is not applicable for this type of solid supported bilayer. But while the density of water is low, the lipid headgroups remain fully hydrated. Figure 4 shows the radial distribution function between phosphate oxygens and water (solid line and squares) and between the choline nitrogen and water (dashed line and circles). The calculation has been carried out separately for the two bilayer leaflets with the lines showing the results for the inner leaflet (facing silica) and the symbols showing the outer leaflet results. At this lipid-substrate distance the headgroups of the membrane are able to maintain near complete solvation. Even the headgroup orientation is maintained largely unperturbed. To characterize the headgroup structure, the probability distribution for the orientation of the P – N vectors with respect to the surface normal was computed from simulation 11 and the results plotted in Figure 5. As observed in previous MD simulations, the distribution is peaked at values with near parallel orientation of the P – N vector and the bilayer surface. Figure 5A compares the distribution of lipids that are substrate exposed to those that face the aqueous region. The effect of the interaction with the substrate is to broaden the distribution on the inner leaflet with a greater population of conformations both more extended toward the surface and more pointed in towards the hydrocarbon core. This interesting compensation allows the membrane to accommodate the presence of the solid substrate without changing the local electric field. In Figure 5B the distribution of the inner leaflet has been further subdivided into those lipids above the water filled pore and those above the planar silica surface. The lipids above the pore produce a distribution similar to those in the outer layer while those interacting with the silica surface produce the deviations noted above. Figure 6 shows the electrostatic potential profile computed from the trajectories of simulation 1 and simulation 11. The total dipole potential, as well as the magnitude of each of its contributing components, is indistinguishable between the two simulations.
Figure 3.
Molecular graphic of a snapshot from simulation 11 (strong lipid-substrate interactions) with coloring: red – lipid and silica oxygen, gold – silicon, white – hydrogen, blue – water oxygen, orange – lipid headgroups, teal – lipid hydrocarbon tails.
Figure 4.
Radial distribution function between water oxygen atoms and lipid phosphate oxygens (squares and solid line) and between water oxygen atoms and lipid choline nitrogen atom (circles and dashed line). All results are from simulation 11 (strong lipid-substrate interactions). The lines give the results for the inner lipid leaflet (facing silica), the symbols give the results for the outer lipid leaflet (facing bulk water).
Figure 5.
Probability distribution function for the orientation of the P – N vector of the lipid headgroups with respect to the surface normal. The dashed line gives the results for the inner lipid leaflet (facing silica), the solid line gives the results for the outer lipid leaflet (facing bulk water).
Figure 6.
Electrostatic potential as a function of transverse position in the simulation unit cell. The potential was computed from Poisson’s equation by double integration of the charge density in the system. (A) Results from simulation 1 (negligible lipid-substrate interactions). (B) Results from simulation 11 (strong lipid-substrate interactions).
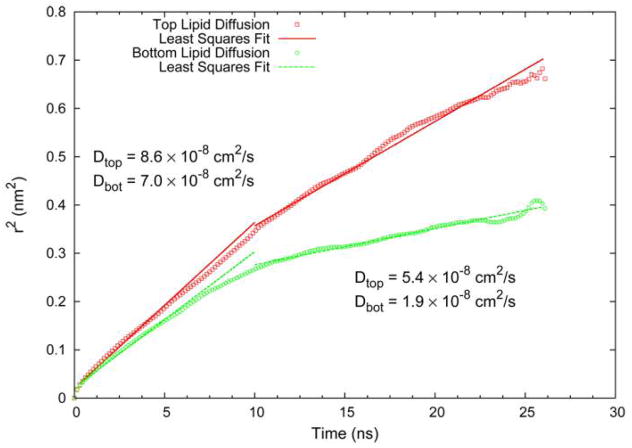
Having examined a number of average properties of the lipids and water, we now turn to quantities probing the fluctuations and dynamics of the membrane. The magnitude of fluctuations in lipid acyl chains is often described in terms of an order parameter. The deuterium order parameter, measured experimentally by the quadrapolar splitting in the 2H NMR spectrum, is a measure of the fluctuations of the C-H vectors in the lipid acyl chains, , where θ is the angle between the C-H vector and the membrane normal. For saturated chains in a fluid phase lipid bilayer the order parameter takes on values near −0.2 in the upper half of the acyl chain, decreasing to near zero at the methyl terminus. The SCD values, computed separately for upper and lower leaflets, showed no statistically significant changes as a function of lipid-substrate distance, in agreement with experimental studies of fluid phase bilayers on a glass substrate.20 It is however possible that small changes in order could be detected by much longer simulations due to the difficulty in adequately sampling enough conformations to quantify small differences in fluctuations. Even more challenging to sample that order fluctuations, that depend on isomerization rates and molecular rotations, are translational motions of the lipid molecules. Translational motions of lipids are typically described by the lateral diffusion coefficient (D) that is related to the long time slope of the displacement correlation function, 〈{x(t) − x(0)}2 + {y(t) −y(0)}2〉 = 4Dt. From the simulation, the change in position of the center of mass in the x-y plane for each lipid was measured over a 14.6 ns trajectory with distinctions made between inner and outer leaflet lipids. Also computed was the change in the center of mass position for each monolayer, inner and outer. At each time step the small movement of the monolayer was subtracted from the each individual lipid to correct for drift. The resulting data was used to compute the displacement correlation functions plotted in Figure 7. The displacements at short times, up to ~10 ns, are similar indicating that ‘cage rattling’ motions are very similar for the inner and outer monolayers. The slope of this short time region of the graph gives “apparent” diffusion coefficients that differ by ~18% with the inner leaflet (silica facing) exhibiting slower motion. Computing the long time slope of the displacement correlation function is challenging due to the fact that the variance among lipids grows rapidly for the points at large times. The slope was thus computed by weighting each point in the least squares analysis by the inverse of the standard deviation. Though sampling of longer time displacements, on the order of a molecular diameter, is incomplete, the results show a substantially lower rate of diffusion in the inner leaflet. This is consistent with NMR measurements of phospholipid diffusion in supported bilayers on silica substrates.21
Figure 7.
Displacement correlation function for the lateral diffusion of lipid centers of mass. The dashed line gives the results for the inner lipid leaflet (facing silica), the solid line gives the results for the outer lipid leaflet (facing bulk water).
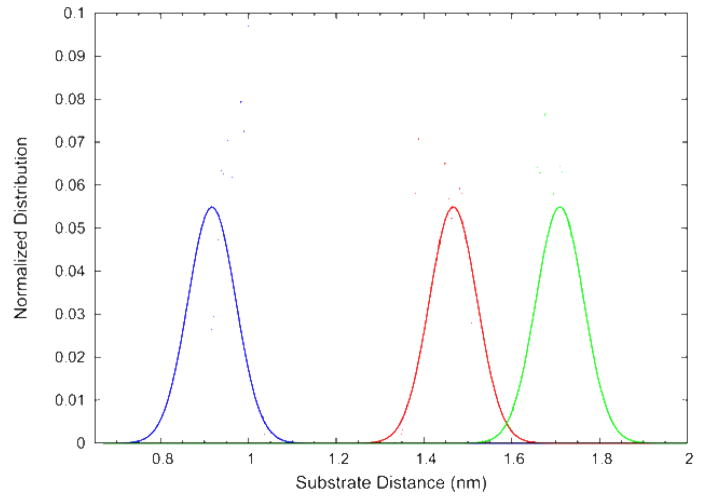
The present calculations are able to provide details on the average structure and fluctuations of the supported bilayer system as well as information on dynamic properties. While it would be desirable to compute thermodynamic properties, in particular the free energy as a function of lipid-substrate distance, such calculations are currently intractable. We can however obtain a qualitative picture of lipid-silica interactions by examining the fluctuations in membrane position during the series of simulations. As described in Methods, the bilayer center of mass is restrained with a harmonic potential during the equilibration phase of each simulation to achieve the desired membrane-substrate distance. Upon reaching the restraint position the force constant for the harmonic potential (kf) was reduced but not eliminated. If the potential of mean force between the bilayer and the silica is zero, one expects the center of mass to fluctuate symmetrically about the restraint position (z0) with a probability distribution function where kB is the Boltzmann constant and T the absolute temperature. In Figure 8, the distribution of bilayer center of mass positions is plotted for three simulations at small lipid-silica separations along with the exponential function expected if there were no lipid-silica interactions. The deviations between the actual and expected distribution functions give a qualitative picture of the potential of mean force between membrane and substrate. The curves on the left show that the observed distribution (symbols) is centered to the right, i.e. at greater separation, than the origin of the restraint potential (line). This indicates a repulsion between the bilayer and the silica surface at this separation. In contrast, the curves in the center describing an intermediate lipid-silica distance is skewed to the left, i.e. to shorter separations, indicating that the restraint is acting to stop the lipid bilayer from moving in the direction of the attraction to the solid substrate. Finally, at the largest separation shown in the figure the observed distribution is approximately centered near the restraint indicating only weak interaction between the surfaces. While the present results are merely qualitative, they suggest that longer simulations with more closely spaced restraints positions could potentially map out the entire force-separation curve for a lipid-solid interaction. An issue not addressed by the present simulations, but that should also be included in a quantitative model of lipid-silica interactions, is the change in surface area per molecule that would likely accompany the dehydration of the lipid surface brought on by decreasing the bilayer-substrate spacing.
Figure 8.
Probability distribution functions as a function of lipid-substrate distance from simulations at three different locations for the restraining potential.
CONCLUSIONS
The simulations reported here constitute an initial step toward developing models with sufficient accuracy and detail to support laboratory investigations involving this scientifically and technologically important system. While our model of a supported bilayer clearly lacks certain elements of the physical system, e.g. surface roughness of the silica and long wavelength undulations of the lipid bilayer membrane, the extremely high spatial and temporal resolution of the simulation provides important insight into the structure and dynamics of the system. However, further calculations are clearly needed to provide greater sampling, especially with regards to lipid diffusion which is frustratingly slow on the MD timescale. Our most important finding is that at the experimentally determined lipid-substrate distance, this system lacks a layer of water having bulk like density or orientation. Perhaps surprisingly, this reduced hydration maintains full solvation of the lipid headgroups, producing minimal changes in headgroup orientation, acyl chain order parameters, and the membrane dipole potential.
An interesting observation from the simulation is that translational diffusion of lipids in the monolayer having direct contact with the silica surface is substantially reduced even under conditions where little to no structural change is observed. Though much longer simulations are needed to confirm this result it does support the interpretation of NMR diffusion measurements of lipids on a silica substrate.21 Other diffusion studies, utilizing fluorescence methods, however have come to the opposite conclusion, i.e. that diffusion in the inner and outer leaflet is the same to within experimental error.22 It would be interesting to examine the effect of variables such as lipid-substrate distance, surface charge, and lipid type with much longer simulations (of the order 100 ns) that could reliably detect changes in the rate of diffusion. A second area where this type of simulation could make connection to experiment is the computation of the free energy of association between membrane and substrate. A set of longer simulations than presented here, at various lipid-substrate distances, could potentially determine force vs. separation curves using the weighted histogram method.23 The potential to examine the effect of changing various surface and/or lipid properties, especially where these variables are difficult to control experimentally, could lead to substantial insight into details of the underlying physics driving organization.
Acknowledgments
We thank Susan Rempe and Chris Lorenz for providing coordinates of the nanoporous silica and thank Larry Scott for helpful discussions. This work was funded by the National Institutes of Health through the NIH Roadmap for Medical Research (PHS 2 PN2 EY016570B).
References
- 1.Boxer SG. Molecular transport and organization in supported lipid membranes. Current Opinion in Chemical Biology. 2000;4(6):704. doi: 10.1016/s1367-5931(00)00139-3. [DOI] [PubMed] [Google Scholar]
- 2.Song X, Nolan J, Swanson BI. Optical Signal Transduction Triggered by Protein-Ligand Binding: Detection of Toxins Using Multivalent Binding. J Am Chem Soc. 1998;120(19):4873–4874. [Google Scholar]
- 3.Baca HK, Ashley C, Carnes E, Lopez D, Flemming J, Dunphy D, Singh S, Chen Z, Liu N, Fan H, Lopez GP, Brozik SM, Werner-Washburne M, Brinker CJ. Cell-Directed Assembly of Lipid-Silica Nanostructures Providing Extended Cell Viability. Science. 2006;313(5785):337–341. doi: 10.1126/science.1126590. [DOI] [PubMed] [Google Scholar]
- 4.Feller SE, Gawrisch K, MacKerell AD., Jr Polyunsaturated fatty acids in lipid bilayers: intrinsic and environmental contributions to their unique physical properties. J Am Chem Soc. 2002;124(2):318–26. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 5.Pitman MC, Suits F, Gawrisch K, Feller SE. Molecular dynamics investigation of dynamical properties of phosphatidylethanolamine lipid bilayers. The Journal of Chemical Physics. 2005;122(24):244715. doi: 10.1063/1.1899153. [DOI] [PubMed] [Google Scholar]
- 6.Feller SE, Gawrisch K, Woolf TB. Rhodopsin exhibits a preference for solvation by polyunsaturated docosohexaenoic acid. J Am Chem Soc. 2003;125(15):4434–5. doi: 10.1021/ja0345874. [DOI] [PubMed] [Google Scholar]
- 7.Vogel A, Tan KT, Waldmann H, Feller SE, Brown MF, Huster D. Flexibility of Ras Lipid Modifications Studied by 2H Solid-State NMR and Molecular Dynamics Simulations. Biophys J. 2007;93(8):2697–2712. doi: 10.1529/biophysj.107.104562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pitman MC, Suits F, Mackerell AD, Jr, Feller SE. Molecular-level organization of saturated and polyunsaturated fatty acids in a phosphatidylcholine bilayer containing cholesterol. Biochemistry. 2004;43(49):15318–28. doi: 10.1021/bi048231w. [DOI] [PubMed] [Google Scholar]
- 9.Feller SE, Brown CA, Nizza DT, Gawrisch K. Nuclear Overhauser enhancement spectroscopy cross-relaxation rates and ethanol distribution across membranes. Biophys J. 2002;82(3):1396–404. doi: 10.1016/S0006-3495(02)75494-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider MJ, Feller SE. Molecular Dynamics Simulations of a Phospholipid-Detergent Mixture. Journal of Physical Chemistry B. 2001;105(7):1331–1337. [Google Scholar]
- 11.Tarek M, Tu K, Klein ML, Tobias DJ. Molecular Dynamics Simulations of Supported Phospholipid/Alkanethiol Bilayers on a Gold(111) Surface. Biophys J. 1999;77(2):964–972. doi: 10.1016/S0006-3495(99)76947-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xing C, Faller R. Interactions of Lipid Bilayers with Supports: A Coarse-Grained Molecular Simulation Study. J Phys Chem B. 2008;112(23):7086–7094. doi: 10.1021/jp077305l. [DOI] [PubMed] [Google Scholar]
- 13.Heine DR, Rammohan AR, Balakrishnan J. Atomistic simulations of the interaction between lipid bilayers and substrates. Molecular Simulation. 2007;33(4):391–397. [Google Scholar]
- 14.Klauda JB, Brooks BR, MacKerell AD, Jr, Venable RM, Pastor RW. An ab Initio Study on the Torsional Surface of Alkanes and Its Effect on Molecular Simulations of Alkanes and a DPPC Bilayer. Journal of Physical Chemistry B. 2005;109(11):5300–5311. doi: 10.1021/jp0468096. [DOI] [PubMed] [Google Scholar]
- 15.Schlenkrich M, Brickmann J, MacKerell AD, Jr, Karplus M. Empirical potential energy function for phospholipids: criteria for parameter optimization and applications. In: Merz K, Roux B, editors. Biological Membranes: A Molecular Perspective from Computation and Experiment. Birkhauser; Boston: 1996. pp. 31–81. [Google Scholar]
- 16.Leung K, Rempe SB, Lorenz CD. Salt Permeation and Exclusion in Hydroxylated and Functionalized Silica Pores. Physical Review Letters. 2006;96(9):095504. doi: 10.1103/PhysRevLett.96.095504. [DOI] [PubMed] [Google Scholar]
- 17.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. The Journal of Chemical Physics. 1995;103(19):8577. [Google Scholar]
- 18.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 19.Doshi DA, Dattelbaum AM, Watkins EB, Brinker CJ, Swanson BI, Shreve AP, Parikh AN, Majewski J. Neutron Reflectivity Study of Lipid Membranes Assembled on Ordered Nanocomposite and Nanoporous Silica Thin Films. Langmuir. 2005;21(7):2865–2870. doi: 10.1021/la0471240. [DOI] [PubMed] [Google Scholar]
- 20.Bayerl TM, Bloom M. Physical properties of single phospholipid bilayers adsorbed to micro glass beads. A new vesicular model system studied by 2H-nuclear magnetic resonance. Biophys J. 1990;58(2):357–362. doi: 10.1016/S0006-3495(90)82382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hetzer M, Heinz S, Grage S, Bayerl TM. Asymmetric Molecular Friction in Supported Phospholipid Bilayers Revealed by NMR Measurements of Lipid Diffusion. Langmuir. 1998;14(5):982–984. [Google Scholar]
- 22.Zhang L, Granick S. Lipid diffusion compared in outer and inner leaflets of planar supported bilayers. The Journal of Chemical Physics. 2005;123(21):211104. doi: 10.1063/1.2138699. [DOI] [PubMed] [Google Scholar]
- 23.Shankar Kumar JMRDBRHSPAK. THE weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. Journal of Computational Chemistry. 1992;13(8):1011–1021. [Google Scholar]