Abstract
Hepatocyte growth factor (HGF) and its receptor c-Met are involved in liver regeneration. The role of HGF and c-Met in liver regeneration in rat following two-thirds partial hepatectomy (PHx) was investigated using RNA interference to silence HGF and c-Met in separate experiments. A mixture of 2 c-Met-specific short hairpin RNA (ShRNA) sequences, ShM1 and ShM2, and 3 HGF-specific ShRNA, ShH1, ShH3, and ShH4, were complexed with linear polyethylenimine. Rats were injected with the ShRNA/PEI complex 24 hours before and at the time of PHx. A mismatch and a scrambled ShRNA served as negative controls. ShRNA treatment resulted in suppression of c-Met and HGF mRNA and protein compared with that in controls. The regenerative response was assessed by PCNA, mitotic index, and BrdU labeling. Treatment with the ShHGF mixture resulted in moderate suppression of hepatocyte proliferation. Immunohistochemical analysis revealed severe suppression of incorporation of BrdU and complete absence of mitosis in rats treated with ShMet 24 hours after PHx compared with that in controls. Gene array analyses indicated abnormal expression patterns in many cell-cycle- and apoptosis-related genes. The active form of caspase 3 was seen to increase in ShMet-treated rats. The TUNEL assay indicated a slight increase in apoptosis in ShMet-treated rats compared with that in controls.
Conclusion
The data indicated that in vivo silencing of c-Met and HGF mRNA by RNA interference in normal rats results in suppression of mRNA and protein, which had a measurable effect on proliferation kinetics associated with liver regeneration.
Hepatocyte growth factor (HGF, also known as scatter factor, SF) is a pleiotropic growth factor initially identified as the hepatocyte mitogen rising in the plasma of hepatectomized rats.1–4 The HGF/ c-Met pathway is among the earliest signaling pathways activated after partial hepatectomy (PHx).5 HGF protein levels in the plasma increase more than 20-fold within 1 hour,1 followed by a dramatic increase in HGF mRNA 3–6 hours after a two-thirds PHx.6 The rapid rise of HGF and its binding to its receptor c-Met, occurring within 30 minutes after PHx, is a very early signal of hepatocyte proliferation during liver regeneration. The importance of c-Met and HGF in liver development was previously demonstrated utilizing c-Met and HGF knockout mice. Elimination of HGF/Met results in a lethal embryonic phenotype.7–9 Studies by 2 laboratories found that c-met was specifically eliminated in the liver.10,11 Both studies demonstrated that hepatic-targeted elimination of c-Met prevents regenerative response after two-thirds PHx. c- Met was essential for completion of regeneration, and its elimination blocked the process in the early stages of the S phase in hepatocytes. However, mice in both studies had developed significant histopathologic alterations in their livers when the regenerative capacity was assessed, thus complicating interpretation of the results.12
siRNA has been used to knock out genes in vitro 13 and in vivo 14 by transfection of synthetic 21- to 23-nucleotide SiRNA, plasmid-based expression of short hairpin RNA (ShRNA) by RNA polymerase III or RNA plymerase II promoter,15 and viral vectors.16 In view of the in vivo stability issues regarding synthetic RNA and the induction of the immune response and the associated toxicity of viral vectors, we decided to use a ShRNA plasmid complexed with a nonviral vector, polyethylenimine (PEI), to deliver ShRNA in vivo. Among the nonviral vectors, PEI has proved to be an efficient nonviral gene transfer vector for hepatocytes in vivo and in culture.17 PEI functions as a proton trap and protects the complexed DNA from degradation, resulting in higher transfection efficiencies in vivo and in vitro.18
We therefore decided to use RNA interference to silenceHGFand c-Met in vivo in normal animals (rats) and to examine the impact of their elimination on liver regeneration in a setting not complicated by pathologic alterations of the liver. We addressed the role of HGF and its receptor c-Met in liver regeneration in rats by using ShRNA targeting c-Met and HGF to directly inhibit these genes in vivo. We present evidence that silencing HGF and c-Met by RNA interference had a strong interfering effect at the RNA and protein levels for both HGF and Met. Interference with HGF had measurable but moderate effects on the proliferation kinetics of hepatocytes. However, interference with Met abolished all mitosis, decreased incorporation of BrdU, and increased apoptosis 24 hours after PHx. Microarray analyses also indicated that suppression of Met caused profound alterations in expression of many cell-cycle- and apoptosis-related genes.
Materials and Methods
See Materials and Methods on the HEPATOLOGY website (http://interscience.wiley.com/jpages/0270-9139/ suppmat/index.html).
Results
Cellular Distribution of the Vector in the Liver
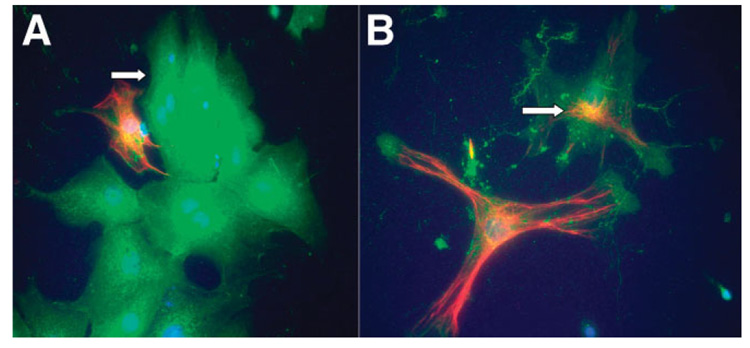
The ShRNA vector employed encoded a red-shifted variant of the jellyfish GFP. We used the plasmid containing ShRNA targeted against HGF (expressed by stellate cells) to investigate the cellular distribution of the plasmid after injection. Stellate cells and endothelial cells are difficult to assess by immunohistochemistry on frozen or paraffin sections. To better identify the cellular types involved in plasmid uptake, we used mixed primary cultures of cells from liver treated with ShRNA. Cells were isolated 4 hours after the second ShRNA injection and were cultured for 12 hours. The cultures contained hepatocytes and a sufficient number of contaminant stellate cells, endothelial cells, and Kupffer cells. As shown in Fig. 1A,B, most of the hepatocytes stained positive for GFP immunofluorescence. Stellate cells also stained positive for desmin (red) and GFP (green), generating focal yellow coloration based on the proximity of the chromophores. More than 90% of the hepatocytes and approximately 60% of the stellate cells stained positive for GFP. However, we could not detect a significant number of GFP-positive endothelial cells (identified by CD31, data not shown).
Fig. 1.
Cellular localization of ShRNA in rat liver tissue by fluorescence detection of GFP and desmin. (A) Hepatocytes expressing GPF are green (arrow); stellate cells expressing desmin appear red. (B) Higher magnification of a stellate cell expressing GFP plasmid. Colocalization of GFP and desmin in some regions of the stellate cell cytoplasm resulted in yellow coloration (indicated by arrow).
Suppression of HGF and c-Met mRNA by ShRNA
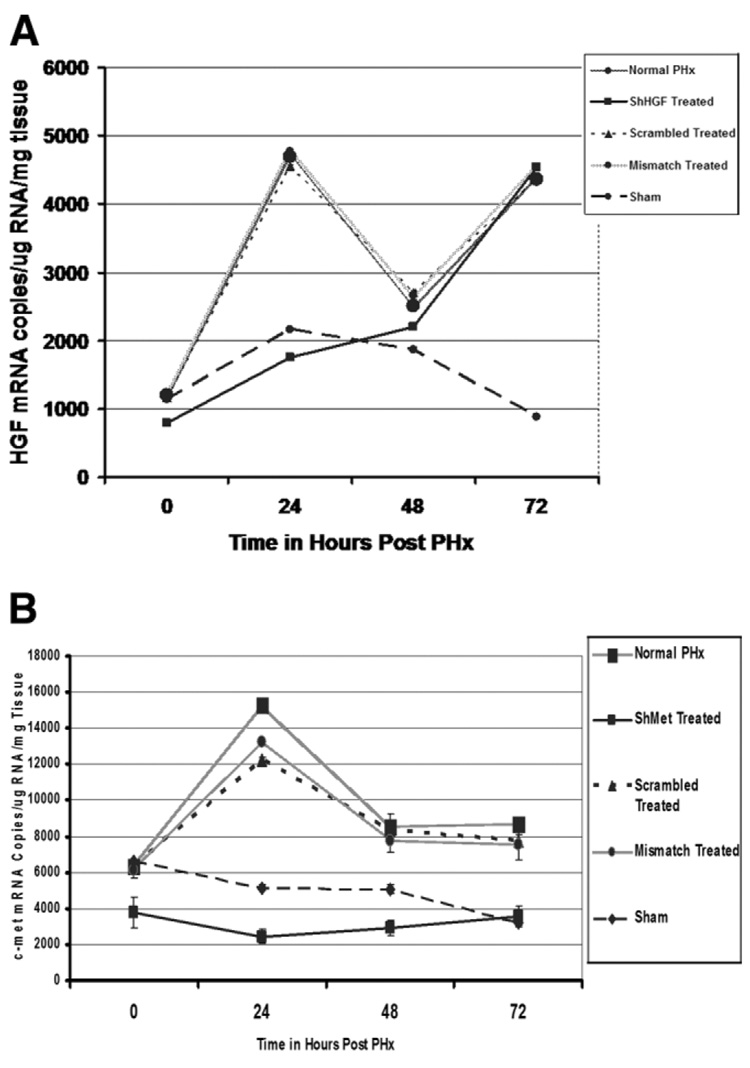
We investigated the suppression of HGF and c-Met mRNA by injection of their respective silencing constructs. Plasmids were injected via the superior mesenteric vein 24 hours before and at the time of the PHx. Absolute quantitative RT-PCR was used to quantify mRNA expression by constructing a standard curve using in vitro synthesized cRNA standards (see Materials and Methods section in the supplemental text). As seen in Fig. 2A, 24 hours after the first injection (equal to the time of PHx), there was a 1.5-fold reduction in HGF mRNA in ShHGF-treated rats compared with that in the normal untreated control. By 48 hours after the first injection (24 hours after the second injection), there was 2.6-fold suppression in HGF mRNA. By day 3, however, the expression of HGF mRNA was almost equal to that of the untreated control. Treatment with mismatch and scrambled had no effect on HGF mRNA.
Fig. 2.
Real-time PCR analysis of HGF and c-Met expression in regenerating rat livers injected with a mixture of ShHGF and ShMet vector (n = 3). HGF and c-Met mRNA levels in ShRNA-treated rats and controls were determined by absolute quantitation. (A) Significant suppression of HGF mRNA was evident (P < 0.005) 24 hours after PHx compared with that in mismatch and scrambled injected controls. Levels returned to normal level 48 hours after PHx. (B) ShMet treatment resulted in significant suppression of c-Met mRNA (P < 0.005) 24 hours after PHx that was maintained for 72 hours after PHx. Injection of scrambled and mismatch RNA had no effect on c-Met mRNA expression.
Treatment with ShMet also suppressed c-Met mRNA (Fig. 2B). Twenty-four hours after the first injection, there was a 1.6-fold reduction in c-Met mRNA compared with that in the untreated normal control. By 48 hours after the first injection (and 24 hours after PHx), there was 6.2-fold reduction in c-Met mRNA. By 72 hours after the first injection, a 3-fold reduction in c-Met mRNA was evident. Injection with scrambled or mismatch ShRNA did not affect c-Met mRNA expression.
Changes in HGF and Met Proteins
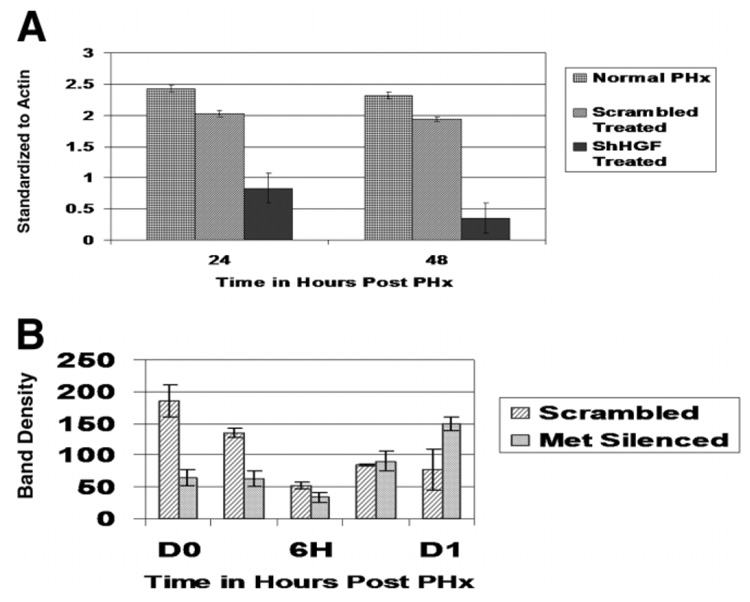
Results shown in Fig. 3A demonstrate amarked decrease in HGFprotein 24 and 48 hours after the last injection of the ShHGF RNA. There was a moderate decrease in the overall expression of Met protein (data not shown), probably reflecting that endothelial cells, known to heavily express Met, did not appreciably take up the ShRNA plasmid. However, relevant changes were noted when we assayed expression of tyrosine-phosphorylated Met during the first 24 hours after PHx. Because activation of Met by tyrosine phosphorylation in the first 24 hours after PHx occurred predominantly in the proliferating cells (hepatocytes, which did take up the ShRNA plasmid), changes in Tyr-phosphorylated Met in the first 24 hours after PHx would tend to reflect the ShRNA in hepatocytes. The results are shown in Fig. 3B. There was a dramatic decrease in Tyr-phosphorylated Met both at the time of PHx (time 0, 24 hours after the first ShMet injection) and up to 6 hours after PHx. This is significant because Tyr phosphorylation of Met occurs predominantly in the first 30–60 minutes after PHx.11 There was a gradual increase in Tyr-phosphorylated Met after 6 hours, with a rebound increase after 24 hours (correlating with the greatly increased hepatocyte proliferation seen in the ShMet-treated animals 48 hours after PHx, discussed later).
Fig. 3.
(A) Decrease in HGF protein induced by injection of ShFGF RNA. HGF protein was assayed in Western blots of whole-liver homogenate. 29 (B) Levels of tyrosine-phosphorylated Met protein during first 24 hours after PHx. Western blots were run using proteins from whole-cell lysates immunoprecipitated with Met antibody (SP260). Blots were probed with antiphosphotyrosine antibody to detect tyrosine-phosphorylated Met. Significant suppression of tyrosine-phosphorylated Met was seen 24 hours after the first injection (time of PHx, P < 0.005) and 1 hour after PHx was observed.
Effect of ShHGF on Proliferation Kinetics after PHx
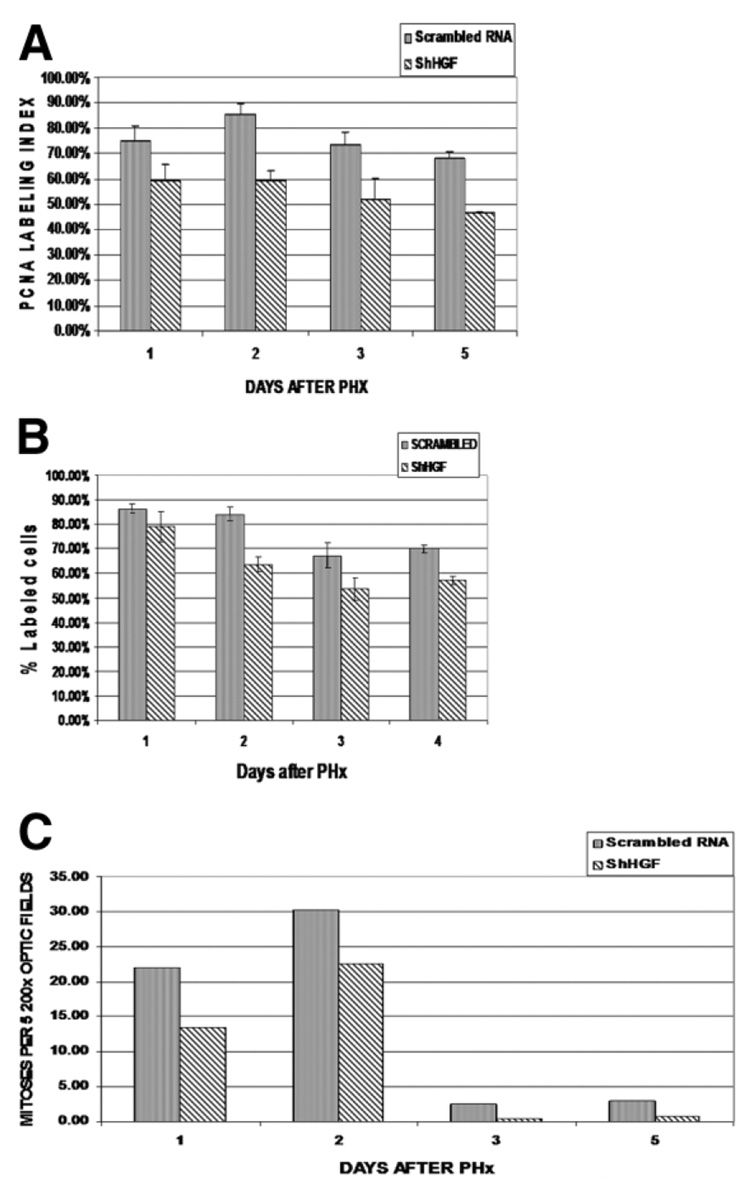
The effects of ShHGF treatment on the proliferative indices of hepatocytes are shown in Fig. 4A–C. A decrease in HGF had a moderate but measurable impact on the proliferative indices, including the indices of PCNA and BrdU labeling and the hepatocyte mitotic index. The decrease in the proliferative indices ranged from 10% to 30%.
Fig. 4.
(A) PCNA labeling and (B) BrdU labeling of nuclei of hepatocytes in ShHGF and scrambled ShRNA-treated rats. ShHGF treatment resulted in significant suppression of incorporation of BrdU 48 hours after PHx (P < 0.005) compared with that in controls. (C) Progression of hepatocytes through mitosis was estimated by using mitotic index (number of labeled cells per 5 200× optic fields). Modest suppression of the mitotic index was observed in ShHGF-treated rats compared with that in scrambled ShRNA-treated control group.
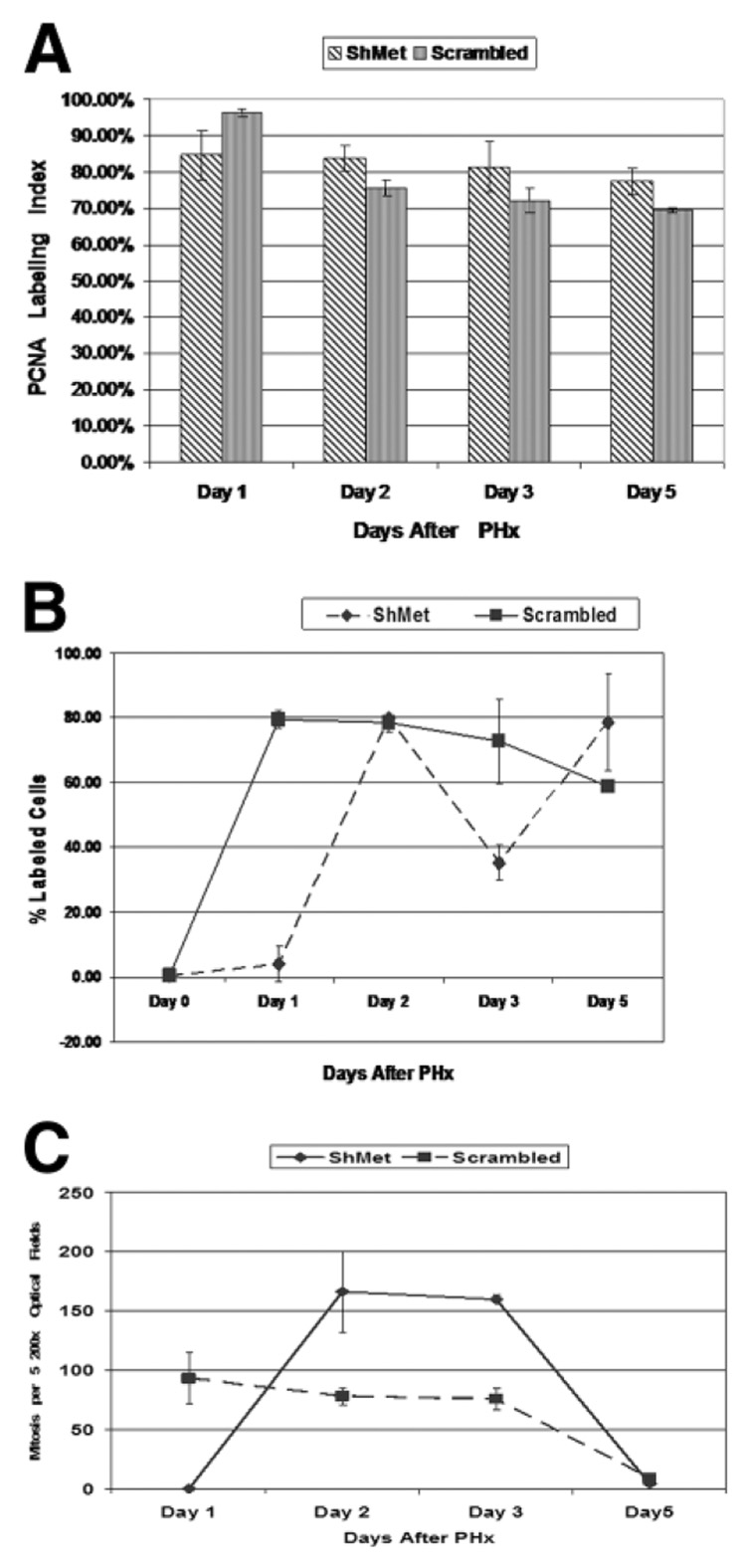
Effect of ShMet on Proliferation Kinetics after PHx
The effects of ShMet treatment on the proliferative indices of hepatocytes are shown in Fig. 5A–C. There was a moderate decrease in the PCNA labeling index of hepatocytes for all days. The effect on the BrdU nuclear labeling index was more dramatic 24 hours after PHx (ShRNA: 4%; scrambled RNA: 80%). The BrdU labeling index of ShRNA-treated animals after 48 hours returned to that of the animals treated with scrambled RNA. There was complete suppression of mitosis in hepatocytes 24 hours after PHx following ShMet treatment (ShMet, 0 cells; scrambled RNA, 92 mitoses per 10 200× optic fields). However, the mitotic rate of the ShMet-treated animals 48 and 72 hours after PHx surpassed the rate of the scrambled RNA–treated animals (mitoses per 10 200× optic fields after 48 hours: scrambled RNA, 78; ShMet RNA, 166; after 72 hours: scrambled RNA, 75; ShMet RNA, 160).
Fig. 5.
(A) PCNA labeling of hepatocytes in ShMet and scrambled treated rats. A moderate decrease in the PCNA labeling index of hepatocytes was evident until 120 hours after PHx compared with that in scrambled treated animals. (B) BrdU labeling of hepatocyte nuclei. ShMet treatment resulted in significant suppression of the incorporation of BrdU (P < 0.005) compared with that in controls 24 hours after PHx. By day 2, levels in ShMet were comparable to those in scrambled ShRNA control. (C) Progression of hepatocytes through mitosis was estimated by using mitotic index. No mitosis was seen in ShMet-treated rats 1 day after PHx. However, 48 and 72 hours after PHx, the mitotic rate in ShMet-treated rats surpassed that observed in scrambled ShRNA treated rats.
Active Form of Caspase 3 Was Increased After ShMet Treatment
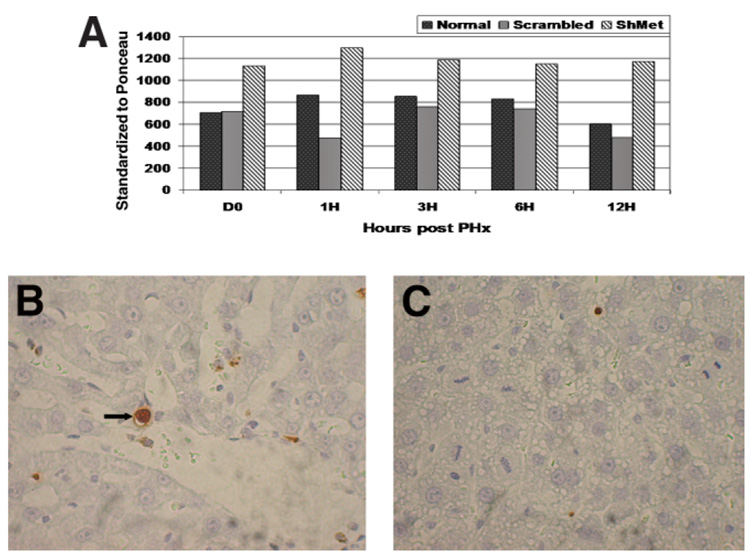
As mentioned in the Supplement, many apoptosis-related genes were up-regulated in animals treated with ShMet. (Supplemental Table 1). We therefore investigated changes in the active form of caspase 3 by Western blot analyses. As seen in Fig. 6A, the active form of caspase 3 (p11 subunit) was increased in ShMet-treated rats compared with that in normal and scrambled-RNA-treated rats. Twenty-four hours after the first injection, the active form showed 1.6-fold up-regulation compared with that in normal PHx-treated rats. Caspase expression remained higher until 12 hours after PHx.
Fig. 6.
(A) Change in active caspase-3 protein during the first 24 hours after PHx. A Western blot was run using whole-cell lysates. Blots were probed with anti-caspase-3 antibody (H-277) to detect active caspase-3. Band density was standardized to total protein from Ponceau stain of the Western blot. Each data point represents a pool of 3 rats per time point. Up-regulation of the p11 subunit of the active form of caspase 3 was observed in ShMet-treated rats compared with that in untreated and scrambled-RNA-treated rats. (B,C) TUNEL assay analysis in histologic sections 24 hours after PHx. An increase was observed in ShMet RNA treated rats (B) compared with that in control rats treated with scrambled ShRNA (C). However, the percentage of apoptotic hepatocytes never exceeded 0.5% of the total in any of the animals.
TUNEL Assay
In view of the increase in the active form of caspase 3, we carried out a TUNEL assay for detection of apoptotic cells in liver tissue by in situ labeling of genomic DNA using terminal deoxynucleotidyl transferase (TdT) as described in the Material and Methods section of the Supplement. As seen in Fig. 6B, in livers from the ShMet-treated group there was slight increase (2.5-fold on average) in TUNEL-positive cells 24 hours after PHx, whereas the livers from the scrambled-treated and normal rats showed few apoptotic nuclei, as seen in Fig. 6C.However, at all times the percentage of apoptotic TUNEL-positive hepatocytes was low and did not exceed 0.5% of the total hepatocyte population. The caspase 3 and apoptosis data also indicate that injection of ShMet was directly responsible for the induction of caspase 3 and that it was not a result of some side effect of PEI or the vector used, as up-regulation of the active caspase form and an increase in the number of TUNEL-positive cells were not observed in the scrambled RNA–treated rats.
Microarray Data Analysis
Analysis of global gene expression in rats treated with ShMet on the first day after PHx by microarray (Supplemental Table 1) indicated dysregulation of many genes involved in the cell cycle and in growth arrest, including p21 and p53, whereas IGFBP1 and CE/BP-beta were down-regulated. Given the dramatic suppression of BrdU incorporation and the absence of mitosis 24 hours after PHx in the ShMet-treated animals, it is significant that expression of cyclin E1 (associated with progression from the G1 to the S phase19) was suppressed and that expression of cyclin B (associated with completion of mitosis20) was dramatically increased. Also of interest was that expression of the TGF-beta receptor II was 27 times higher in the livers receiving ShMet RNA
Absence of Off-Target Effects and of Interferon Response After ShRNA Treatment
A major concern of using ShRNA is the possibility of unexpected silencing of unrelated genes and induction of interferon response.21,22 We used U95 Affymetrix oligo arrays to analyze and compare the pattern of global gene expression 24 hours after PHx in ShRNA-treated rats with that of untreated, sham, and ShRNA- and scrambled RNA–treated rats. There was no detectable difference in the global gene expression pattern between hepatectomized livers and those treated with ShRNA. There were also no detectable alterations in expression of interferon-regulated genes (for details, see Supplemental Figs. 1 and 2).
Discussion
Liver regeneration after PHx is an extremely complex process requiring interaction and cooperation of many growth factors and cytokines and crosstalk between multiple pathways. Of the various agents implicated in liver regeneration, HGF and ligands of the EGF receptor are the only ones that stimulate DNA synthesis in hepatocyte cultures maintained in chemically defined media.23 They are also the only ones that stimulate DNA synthesis in the livers of normal mice and rats.24–28 As already noted, homozygous deletion of HGFor its receptor c-Met results in embryonic lethality.7,9 Thus, it has not been possible to study liver regeneration in such genetically modified mice. There have been 2 recent studies in which c-Met deletion was targeted only to the liver.10,11 This condition resulted in significant histopathology. Because of this, 70% hepatectomies in these mice were associated with high mortality, whereas 30% hepatectomies were tolerated. Both studies concluded that c-Met provides essential signaling for liver regeneration, not compensated by EGFR or receptors of other signaling cytokines. However, interpretation of these studies was complicated by the long-term deletion of hepatocyte Met causing significant hepatic histopathology, including fatty liver and fibrosis. Thus, it was not possible to determine if the effects on liver regeneration were a result of deletion of Met per se or were associated in a secondary manner with the histopathology associated with Met deletion.
The purpose of the present study was to analyze the role of the HGF/Met signaling pathway in liver regeneration by partially or completely eliminating either HGF or c-Met acutely in normal rats without inducing any of the histopathologic alterations associated with long-term Met deletion. The histology of the ShMet- and ShHGF-treated rats was entirely comparable to the control (scrambled RNA treated, mismatch RNA treated, or not RNA treated). Thus, the results obtained should be ascribed only to the interference of expression of the specific gene.
Suppression of HGF
In a previous study we showed that HGF mRNA starts to increase 3 hours after PHx.6 HGF protein (precursor and active heterodimers) decreases in the first 3 hours after PHx, as it is consumed from preexisting stores in the hepatic extracellular matrix. 29 However, both the precursor and the active forms of HGF dramatically increase in whole-liver homogenates after expression of new HGF mRNA beginning 3 hours after PHx and reaching a peak 24–48 hours after PHx.29 In this study ShHGF treatment suppressed both protein and HGF mRNA. Yet the effects of this “knockdown” on hepatocyte DNA synthesis were measurable but modest. Our results suggest that the effects of HGF on hepatocytes are determined by the preexisting HGF protein stores consumed in the first 3 hours after PHx and that the further increase in HGF protein after 3 hours affects primarily nonhepatocyte populations, such as stellate cells, endothelial cells, and biliary epithelium. A more prolonged HGF suppression regimen14 would be required to properly assess the effect of newly synthesized HGF on nonhepatic cell populations.
Suppression of c-Met
The effects of suppression of c-Met need to be examined from 2 perspectives: (1) tyrosine phosphorylation of Met occurs within 30–60 minutes of PHx5; and (2) there is a great overlap in signal transduction pathways between HGF and EGF receptors, except when associated with Met and Fas.30
The results of the present study show that despite the great similarity in signaling pathways of Met and EGFR, Met makes unique signaling contributions that are not compensated for by EGFR or other cytokines related to liver regeneration (e.g., TNF-alpha,31 norepinephrine32). HGF/Met signaling contributions, even though initiated 30–60 minutes after PHx, probably affect downstream signaling cascades, manifesting themselves when hepatocytes are about to enter from theG1 into the S phase (6–12 hours after PHx in the rat). These signals affect both growth-related genes (e.g., cyclins E1 and B, genes p53 and p21) as well as apoptosis-related genes (caspase 3, BclX, etc.). It is possible that the changes seen in apoptosis- related genes are a result of the increased availability of Fas because of decreased association with c-Met following ShMet treatment.30 The overall effects of ShMet RNA observed 24 hours after PHx likely are a result of decreased tyrosine phospho-Met, shown in Fig. 3B. The decrease in Tyr-phospho-Met seen at time 0 (24 hours after the first ShRNA injection, at the time of PHx) and 1 hour after PHx are the ones likely to affect cell-cycle events seen after 24 hours (little BrdU and no mitosis). Tyr-phospho-Met, however, increased more than in the control and far surpass it after 24 hours. This correlates with the increased hepatocyte mitosis in the ShMet above that in the scrambled RNA–treated animals 48 hours after PHx. These results, incidentally, delineate a 24-hour signaling interval between Met activation and hepatocyte mitosis. There is an apparent discrepancy between expression of Tyr-phospho-Met (minimum suppression at ≈40% at time 0 and ≈60% 1 hour after PHx) and the degree of suppression of hepatocyte-related events 24 hours after PHx (no mitosis and very little incorporation of BrdU). This probably reflects the Tyr-phospho-Met in endothelial cells, which do not take up the ShRNA plasmid (data not shown). The data on in vivo stability for the persistence of the plasmid also indicate that the plasmid is lost by the first day after PHx (Supplemental Fig. 1). The loss of plasmid DNA could have occurred for a number of reasons including degradation by nucleases, inactivation of the promoter by methylation, and diluting out because of cellular replication. The dramatic loss of the plasmid probably resulted in the transient silencing observed.
Also of interest is that ShMet had no effect on the PCNA nuclear labeling index of the hepatocytes. PCNA nuclear labeling occurs in all cells entering into the G1 phase from the G0 phase. Signaling pathways in addition to the HGF/Met signaling pathway make significant contributions, leading hepatocytes to enter into the cell cycle. These include TNF-alpha,31,33 norepinephrine,32,34 and ligands of the EGFR (EGF,35 TGF-alpha,36 HB-EGF,37 Amphiregulin38). Our studies suggest that other contributing signals are sufficient to stimulate hepatocytes to enter into G1. EGFR is always phosphorylated in tyrosine residues in the rat liver, and its phosphorylation also dramatically increases 60 minutes after PHx.5 However, HGF/Met signaling contributions are essential for hepatocytes to proceed through the G1 phase and enter into the S phase and DNA synthesis.
Acknowledgments
Supported by NIH (grant numbers CA35373 and CA103958) and Rangos Fund for Enhancement of Research in Pathology.
Abbreviations
- HGF
hepatocyte growth factor
- PHx
partial hepatectomy
- ShRNA
short hairpin RNA
- PEI
polyethylenimine
Footnotes
Potential conflict of interest: Nothing to report.
Supplementary materials for this article can be found on the HEPATOLOGY website (http://interscience.wiley.com/jpages/0270-9139/suppmat/index.html).
References
- 1.Lindroos PM, Zarnegar R, Michalopoulos GK. Hepatocyte growth factor (hepatopoietin A) rapidly increases in plasma before DNA synthesis and liver regeneration stimulated by partial hepatectomy and carbon tetrachloride administration. HEPATOLOGY. 1991;13:743–750. [PubMed] [Google Scholar]
- 2.Zarnegar R, Michalopoulos G. Purification and biological characterization of human hepatopoietin A, a polypeptide growth factor for hepatocytes. Cancer Res. 1989;49:3314–3320. [PubMed] [Google Scholar]
- 3.Zarnegar R, Muga S, Enghild J, Michalopoulos G. NH2-terminal amino acid sequence of rabbit hepatopoietin A, a heparin-binding polypeptide growth factor for hepatocytes. Biochem Biophys Res Commun. 1989;163:1370–1376. doi: 10.1016/0006-291x(89)91130-3. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura T, Nishizawa T, Hagiya M, Seki T, Shimonishi M, Sugimura A, et al. Molecular cloning and expression of human hepatocyte growth factor. Nature. 1989;342:440–443. doi: 10.1038/342440a0. [DOI] [PubMed] [Google Scholar]
- 5.Stolz DB, Mars WM, Petersen BE, Kim TH, Michalopoulos GK. Growth factor signal transduction immediately after two-thirds partial hepatectomy in the rat. Cancer Res. 1999;59:3954–3960. [PubMed] [Google Scholar]
- 6.Zarnegar R, DeFrances MC, Kost DP, Lindroos P, Michalopoulos GK. Expression of hepatocyte growth factor mRNA in regenerating rat liver after partial hepatectomy. Biochem Biophys Res Commun. 1991;177:559–565. doi: 10.1016/0006-291x(91)92020-k. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt C, Bladt F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, et al. Scatter factor/hepatocyte growth factor is essential for liver development. Nature. 1995;373:699–702. doi: 10.1038/373699a0. [DOI] [PubMed] [Google Scholar]
- 8.Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud [see comments] Nature. 1995;376:768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- 9.Uehara Y, Minowa O, Mori C, Shiota K, Kuno J, Noda T, et al. Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/ scatter factor. Nature. 1995;373:702–705. doi: 10.1038/373702a0. [DOI] [PubMed] [Google Scholar]
- 10.Borowiak M, Garratt AN, Wustefeld T, Strehle M, Trautwein C, Birchmeier C. Met provides essential signals for liver regeneration. Proc Natl Acad Sci U S A. 2004;101:10608–10613. doi: 10.1073/pnas.0403412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huh CG, Factor VM, Sanchez A, Uchida K, Conner EA, Thorgeirsson SS. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc Natl Acad Sci U S A. 2004;101:4477–4482. doi: 10.1073/pnas.0306068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. HEPATOLOGY. 2006;43 Suppl:S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 13.Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 15.Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 16.Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 17.Chemin I, Moradpour D, Wieland S, Offensperger WB, Walter E, Behr JP, Blum HE. Liver-directed gene transfer: a linear polyethlenimine derivative mediates highly efficient DNA delivery to primary hepatocytes in vitro and in vivo. J Viral Hepat. 1998;5:369–375. doi: 10.1046/j.1365-2893.1998.00126.x. [DOI] [PubMed] [Google Scholar]
- 18.Park TG, Jeong JH, Kim SW. Current status of polymeric gene delivery systems. Adv Drug Deliv Rev. 2006;58:467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Zschemisch NH, Liedtke C, Dierssen U, Nevzorova YA, Wustefeld T, Borlak J, et al. Expression of a cyclin E1 isoform in mice is correlated with the quiescent cell cycle status of hepatocytes in vivo. HEPATOLOGY. 2006;44:164–173. doi: 10.1002/hep.21224. [DOI] [PubMed] [Google Scholar]
- 20.Wolf F, Wandke C, Isenberg N, Geley S. Dose-dependent effects of stable cyclin B1 on progression through mitosis in human cells. EMBO J. 2006;25:2802–2813. doi: 10.1038/sj.emboj.7601163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW. Specificity of short interfering RNA determined through gene expression signatures. Proc Natl Acad Sci U S A. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 23.Block GD, Locker J, Bowen WC, Petersen BE, Katyal S, Strom SC, et al. Population expansion, clonal growth, and specific differentiation patterns in primary cultures of hepatocytes induced by HGF/SF, EGF and TGF alpha in a chemically defined (HGM) medium. J Cell Biol. 1996;132:1133–1149. doi: 10.1083/jcb.132.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patijn GA, Lieber A, Schowalter DB, Schwall R, Kay MA. Hepatocyte growth factor induces hepatocyte proliferation in vivo and allows for efficient retroviral-mediated gene transfer in mice. HEPATOLOGY. 1998;28:707–716. doi: 10.1002/hep.510280317. [DOI] [PubMed] [Google Scholar]
- 25.Gao C, Jokerst R, Gondipalli P, Cai SR, Kennedy S, Ponder KP. Intramuscular injection of an adenoviral vector expressing hepatocyte growth factor facilitates hepatic transduction with a retroviral vector in mice. Hum Gene Ther. 1999;10:911–922. doi: 10.1089/10430349950018319. [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Chen S, Huang L, Michalopoulos GK, Liu Y. Sustained expression of naked plasmid DNA encoding hepatocyte growth factor in mice promotes liver and overall body growth. HEPATOLOGY. 2001;33:848–859. doi: 10.1053/jhep.2001.23438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bucher NL. Liver regeneration: an overview. J Gastroenterol Hepatol. 1991;6:615–624. doi: 10.1111/j.1440-1746.1991.tb00921.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu ML, Mars WM, Zarnegar R, Michalopoulos GK. Collagenase pretreatment and the mitogenic effects of hepatocyte growth factor and transforming growth factor-alpha in adult rat liver. HEPATOLOGY. 1994;19:1521–1527. [PubMed] [Google Scholar]
- 29.Pediaditakis P, Lopez-Talavera JC, Petersen B, Monga SP, Michalopoulos GK. The processing and utilization of hepatocyte growth factor/scatter factor following partial hepatectomy in the rat. HEPATOLOGY. 2001;34:688–693. doi: 10.1053/jhep.2001.27811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, DeFrances MC, Dai Y, Pediaditakis P, Johnson C, Bell A, et al. A mechanism of cell survival: sequestration of Fas by the HGF receptor Met. Mol Cell. 2002;9:411–421. doi: 10.1016/s1097-2765(02)00439-2. [DOI] [PubMed] [Google Scholar]
- 31.Webber EM, Bruix J, Pierce RH, Fausto N. Tumor necrosis factor primes hepatocytes for DNA replication in the rat. HEPATOLOGY. 1998;28:1226–1234. doi: 10.1002/hep.510280509. [DOI] [PubMed] [Google Scholar]
- 32.Cruise JL, Knechtle SJ, Bollinger RR, Kuhn C, Michalopoulos G. Alpha 1-adrenergic effects and liver regeneration. HEPATOLOGY. 1987;7:1189–1194. doi: 10.1002/hep.1840070604. [DOI] [PubMed] [Google Scholar]
- 33.Yamada Y, Kirillova I, Peschon JJ, Fausto N. Initiation of liver growth by tumor necrosis factor: deficient liver regeneration in mice lacking type I tumor necrosis factor receptor. Proc Natl Acad Sci U S A. 1997;94:1441–1446. doi: 10.1073/pnas.94.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Houck KA, Michalopoulos GK. Altered responses of regenerating hepatocytes to norepinephrine and transforming growth factor type beta. J Cell Physiol. 1989;141:503–509. doi: 10.1002/jcp.1041410308. [DOI] [PubMed] [Google Scholar]
- 35.Bucher NL, Patel U, Cohen S. Hormonal factors and liver growth. Adv Enzyme Regul. 1977;16:205–213. doi: 10.1016/0065-2571(78)90074-2. [DOI] [PubMed] [Google Scholar]
- 36.Webber EM, FitzGerald MJ, Brown PI, Bartlett MH, Fausto N. Transforming growth factor-alpha expression during liver regeneration after partial hepatectomy and toxic injury, and potential interactions between transforming growth factor-alpha and hepatocyte growth factor. HEPATOLOGY. 1993;18:1422–1431. [PubMed] [Google Scholar]
- 37.Mitchell C, Nivison M, Jackson LF, Fox R, Lee DC, Campbell JS, et al. Heparin-binding epidermal growth factor-like growth factor links hepatocyte priming with cell cycle progression during liver regeneration. J Biol Chem. 2005;280:2562–2568. doi: 10.1074/jbc.M412372200. [DOI] [PubMed] [Google Scholar]
- 38.Berasain C, Garcia-Trevijano ER, Castillo J, Erroba E, Lee DC, Prieto J, Avila MA. Amphiregulin: an early trigger of liver regeneration in mice [see comment] Gastroenterology. 2005;128:424–432. doi: 10.1053/j.gastro.2004.11.006. [DOI] [PubMed] [Google Scholar]