Abstract
Multiplexed nanobarcodes have been prepared with quantum dots (QDs) and alternating amphiphilic copolymers consisted of hydrocarbons and maleic anhydride groups. In homogenous solution, the QD-polymer complexes grow epitaxially into nanobeads of narrow size dispersity, which has been previously achieved only for micrometer-sized beads in the presence of solid supports. As a result of this new nanostructure formation mechanism, more than 250 QDs can be loaded into a nanobead of 100 nm in diameter. A model assay for sensitive detection of human prostate specific antigen (PSA) has also been demonstrated using the QD-nanobeads as fluorescent reporters. This nanoparticle-polymer self-assembly technology is capable of producing a variety of nanostructures and is expected to open new opportunities in nanoparticle-based ultrasensitive detection and imaging.
Introduction
The development of fluorescent probes that are stable, compact, and significantly brighter than traditional fluorophores (e.g. organic dyes and fluorescent proteins) is of considerable interests to many research areas including DNA sequencing, gene expression profiling, molecular imaging, fundamental biophysics, as well as clinical diagnostics.1–5 Despite recent success with semiconductor QDs, which are 20–50 times brighter than single dye molecules, fluorescent probes with improved brightness and multiplexing capability are highly desirable for analysis of low-abundance targets in bioassays including single molecule detection, immunoassays, and fluorescence in situ hybridization, and for understanding of complex human diseases involving a large number of genes and proteins (e.g. cancer and atherosclerosis).
In this context, optical encoding technologies by multiplexing colors and fluorescence intensities of fluorophores have become an attractive strategy because a large number of high-brightness probes can be readily produced.6–10 Compared with organic fluorophores, semiconductor QDs are of particular importance to this application because of their favorable optical properties such as simultaneous excitation of multiple colors with a single light source, minimal spectral overlap between adjacent colors, and remarkable photostability.11–13 For example, we have previously reported the preparation of QD-tagged optical barcodes by incorporating multicolor QDs into mesoporous microspheres at predefined intensity ratios.14 The use of 10 intensity levels of 6 colors has a theoretical coding capacity of one million (106). The encoded microspheres are highly fluorescent and uniform, because they typically contains 5,000 – 10 million QDs per bead depending on the microbead size and doping level. This remarkable nanoparticle doping capacity has also captivated scientists to develop alternative preparation methods (e.g., nanoparticle layer-by-layer deposition) and improved readout apparatus.15–18 Unfortunately, because of the large size (typically 1–15 µm), these QD-doped microspheres are not suitable for applications such as gene, protein and cell labeling.
Toward the development of uniform and bright QD-encoded beads in the nanometer regime, a few attempts have been made. Most common reaction schemes include encapsulation of nanoparticles in silica beads,19–23 hydrogels,24 and block-copolymer micelles.25–27 However, these existing methods are limited by low nanoparticle loading capacity, fluorescence quenching or broad size distribution. For example, QD-doped silica beads can be made with low size distribution, but only a small number of QDs can be incorporated,19, 20 and their quantum efficiency is often significantly reduced (75% decrease). 21 This is because the optical properties of QDs are highly sensitive to the environment in contrast to embedded metallic and magnetic nanoparticles.28–31 In this regard, the block copolymer based micelle formation should be more attractive because high-quality QDs (prepared in organic solvents at elevated temperatures) are generally clustered in the hydrophobic core of block copolymer micelle.27, 32 On the other hand, the micelle size distribution are much broader than silica nanobeads,27 and the number of QDs can be encapsulated is still limited. Therefore, new methods are needed to simultaneously achieve the high brightness and narrow size dispersity for wide-spread application of the nanobeads in fluorescence based imaging and detection. In addition, multicolor doping with tunable fluorescence intensity ratios and low level of fluorophore self-quenching in the nanometer regime has not been achieved.
Results and Discussion
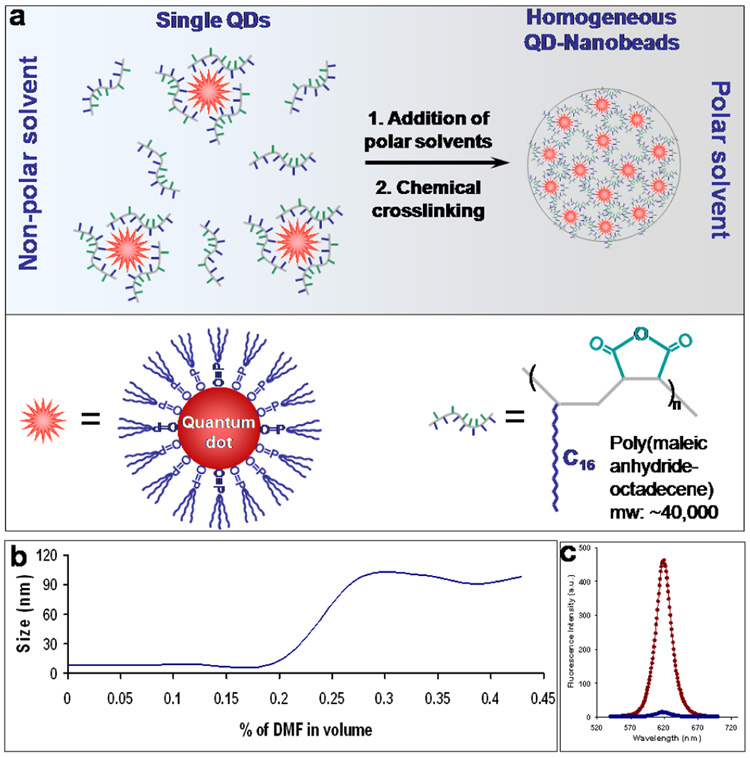
Here, we report a new approach for preparation of QD-tagged nanobeads based on epitaxial growth of nanoparticle-amphiphilic polymer complexes in homogeneous solution. This new generation of fluorescent probe is uniform in size, thousands of times brighter than single organic dyes, stable against photobleaching, and free of ‘blinking’ effect. The key finding was an amphiphilic alternating copolymer that is not only capable of coating multicolor QDs but also capable of preventing them from forming irregular agglomerates. In contrast to nanoparticles clustered inside block copolymer micelles, the QDs are pre-protected by the amphiphilic alternating polymers and thus preventing them from ‘touching’ to each other. The encapsulated QD-polymer complexes self-assemble epitaxially into nanobeads with QDs distributed inside homogeneously. This process is similar to the growth of nanocrystals except that the building blocks are not ions or small molecules but are nanoparticles. As schematically illustrated in Figure 1a, QDs and poly(maleic anhydride-octadecene) (PMAO) bond to each other via multivalent hydrophobic interactions. The QD-PMAO conjugates are highly soluble in tetrahydrofuran (THF) but form aggregates in polar solvents such as dimethylformamide (DMF). Interestingly, a solvent gradient created by slow addition of DMF into THF leads to epitaxial growth of highly fluorescent nanobeads with narrow size dispersity. In this process, the PMAO polymer plays a critical role in controlling the nanobead size and size distribution.
Figure 1. QD encapsulation with alternating amphiphilic polymers and subsequent formation of nanobeads.
(a) Schematic illustration of the nanobead formation mechanism. Single QDs are pre-coated with PMAO polymers interaction. The QD-polymer complexes form uniform nanobeads as a result of increasing solvent polarity. The nanobeads were further stabilized through hydrophobic by crosslinking of polymer chains with diamines. (b) Nanobead formation monitored by DLS measurements. QD-polymer complexes are dispersed when DMF concentration is under 20% in volume. Increasing DMF concentration from 20% to 30% leads to quick formation of QD-nanobeads as indicated by the size increase from approximately 10 nm to 100 nm. In comparison, QDs alone start to form irregular nanoparticle aggregates when DMF reaches a concentration of ~5%. The delayed aggregation process of QD and PMOA mixture indicates that the polymers interact with QDs leading to formation of QD-PMAO hybrid structure with reduced hydrophobicity compared to original QDs. (c) Fluorescence measurement showing the QD incorporation efficiency. Nanobeads were removed by centrifugation, and the supernatants were measured on a fluorometer (blue curve). Fluorescence spectra indicate that more than 95% of the QDs are incorporated into the nanobeads.
It is noteworthy that similar QD-polymer hybrid structures have been previously reported by several groups including our own for solublization of single hydrophobic QDs.33–36 For example, Manna, Parak and coworkers developed a simple and versatile procedure for solublization of hydrophobic semiconductor, metallic, and oxide nanoparticles using low-molecular weight poly(maleic anhydride-tetradecene) (Mw ~9,000).34 In contrast, the PMAO copolymers used in the current research are significantly larger (Mw 30,000–50,000) and are insoluble in aqueous buffers. To increase the water solubility of this polymer, Colvin et al has grafted it with multiple polyethylene glycol (PEG) chains for stabilization of single QDs.36
Using single color QDs, we systematically investigated the conditions for nanobead formation. Dynamic light scattering measurements indicate that QDs remain single in THF/DMF solvent mixture when DMF concentration is under 20% in volume (Figure 1b). Increasing DMF concentration from 20% to 30% leads to spontaneous formation of QD-nanobeads as indicated by the size shift from approximately 10 nm to 100 nm. This self-assembly process is highly efficient at encapsulating QDs. Fluorescence measurement of single QDs left in the supernatant after isolation of nanobeads by centrifugation indicates that more than 95% of QDs are incorporated into the nanobeads (Figure 1c). To further enhance the nanobead stability, such as preventing potential QD leaching or release in bioassays, the polymer chains in the nanobeads are crosslinked with small-molecule diamines. Due to the rich anhydride contents in PMAO polymer and the high reactivity between anhydrides and primary amines, no catalytic reagents are needed to crosslink the polymers into a stable network. Similar reaction of crosslinking of neighboring polymer chains has been previously demonstrated with block copolymers for enhanced micelle stability.37, 38 Following the nanobead formation, the resulting fluorescent nanobeads must be made water-soluble for biological applications. We found that the nanobeads isolated by centrifugation cannot be directly suspended in aqueous buffers. This is understandable because majority of the anhydride groups are not hydrolyzed into carboxylic acids, and thus the nanobeads are not sufficiently hydrophilic. We solved this problem by using a slow dialysis procedure against Tris buffer, which contains 20 mM Tris(hydroxymethyl)aminomethane for efficient hydrolysis of the anhydride groups as the solvents gradually change from THF/DMF mixture to aqueous solution. The resulting nanobeads are stable in aqueous buffers for at least several months.
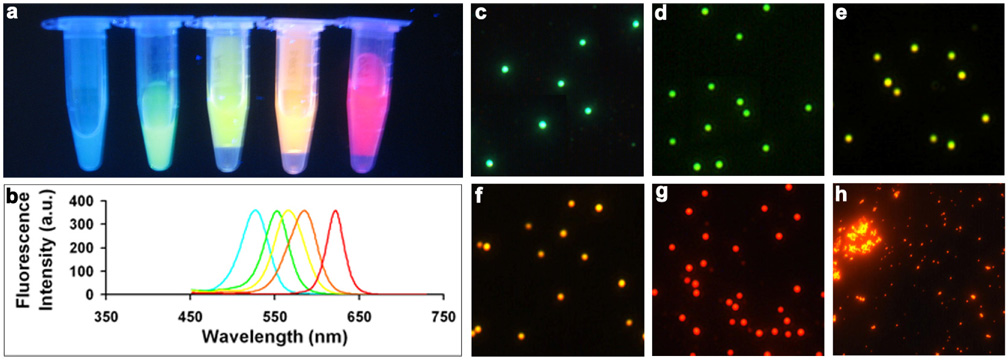
Figure 2 shows a series of water-soluble nanobeads doped with single-color QDs. The nanobeads are uniform in size and highly luminescent. The sizes of these monochrome nanobeads are about 100 nm in diameter, which is suitable for molecular and cellular labeling. Single-particle fluorescence spectroscopy reveals that the nanobeads are significantly brighter than the constituting single QDs. This remarkable brightness can be attributed to the large number of QDs incorporated because the QDs self-assemble into nanobeads in homogeneous solutions without any structural template. In contrast, in our previous reported QD-doped mesoporous microbeads, the QDs only occupy <0.1% of the total bead volume at the highest doping level,39, 40 which resulted in a low density of incorporated nanoparticles. Note that both the multicolor nanobead solutions and microscopic images were obtained using single light sources, a handheld UV lamp and a mercury lamp, respectively. Simultaneous excitation of multiple colors is a unique optical property of QDs and will have significant impact on bioassays such as fluorescence cross-correlation spectroscopy (FCCS), because cofocusing of two or more laser beams is an exceedingly difficult task due to chromatic and spherical aberrations of microscope objective.41 Indeed, this has been a major problem for dual-probe based imaging and detection when organic dyes or organic dye-labeled nanobeads are used.41–43
Figure 2. True-color fluorescence images of monochromatic QD-nanobeads emitting light at 525 nm, 550 nm, 565nm, 585 nm, and 620 nm.
(a) QD-nanobeads in solution. The nanobead solutions in microcentrifuge tubes were placed on a flat surface, illuminated with a handheld UV lamp, and imaged with a standard consumer digital camera. (b) Emission spectra of the nanobeads. (c–g) Fluorescent micrographs of nanobeads dispersed on coverslips. (h) Control experiment showing the importance of the PMAO polymer. Without the polymer, QDs form aggregates of irregular shapes and sizes.
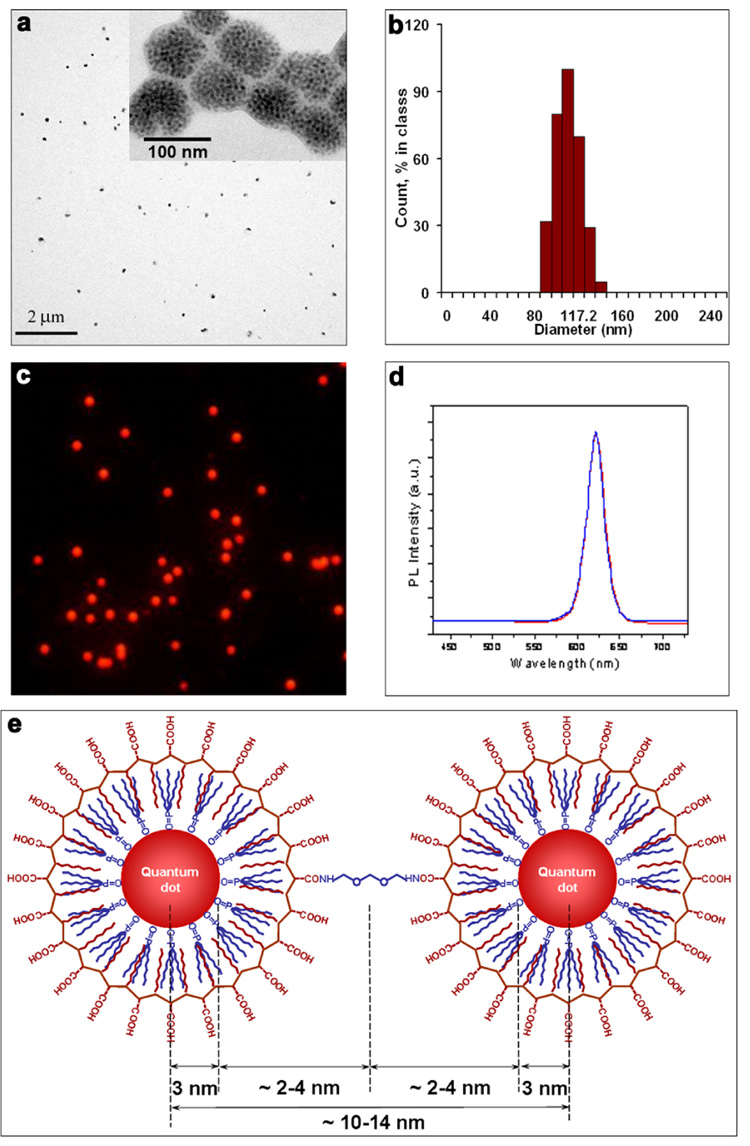
Next, we thoroughly characterized the structural and optical properties of QD-nanobeads. The size of red QD-doped nanobeads was measured by both transmission electron microscopy (TEM) and dynamic light scattering (DLS). The ‘dry’ size of the nanobeads measured by TEM is 92 ± 13 nm (Figure 3a), whereas the DLS measurement indicates that the hydrodynamic size of nanobeads in solution is 112 ± 18 nm (Figure 3b). The discrepancy between the two measurements could be attributed to two effects. First, because the nanobeads are made from polymer-coated QDs that are inter-connected, they are unlikely to be rigid structures in solution. When the solvent is dried (such as the condition of TEM measurement), the polymer-coated QDs could collapse closer to each other, which renders the nanobeads to shrink. Second, the nanobeads are highly negatively charged in aqueous buffer due to the abundant carboxylic acid groups. This negative surface charge not only prevents nanobeads from aggregation, but also creates an electrical double layer in aqueous buffers, thereby slightly increasing the colloidal hydrodynamic radius compared to the actual size.
Figure 3. Characterization of structural and optical properties of QD-nanobeads.
(a) TEM images showing the size of the nanobeads is 92 ± 13 nm. Individual QDs can be resolved when imaged at high-magnification (inset). (b) DLS measurement indicating the hydrodynamic diameter of the nanobeads in aqueous buffer is 112 ± 18 nm. (c) Fluorescence imaging of QD-nanobeads confirming their low size dispersity and high brightness. (d) Spectroscopy measurement of the QD-nanobeads and original QDs in THF. Identical fluorescence emission spectra were observed for single QDs and QDs embedded inside the nanobeads. When green QDs were used in the same measurement, a slight spectral shift toward red was observed (Supplementary Figure 1). (e) Schematic plot of two adjacent QDs inside nanobeads. See text for details.
Fluorescence imaging confirms the low size dispersity of the nanobeads. As shown in Figure 3c, the red QD doped nanobeads are uniform and highly fluorescent. Wavelength-resolved spectroscopy measurement of single nanobeads indicates that the nanobeads are as bright as approximately 200 single QDs. This number is calculated by dividing the average fluorescence intensity of nanobeads by that of the single QDs. Statistical analysis of a population of more than 100 nanobeads shows that the standard deviation of their fluorescence intensity is 30%. This value is close to the calculated variation of the nanobead volume using its radius (24% variation based on TEM and 28% variation based on DLS), suggesting that the observed fluorescence signal variation is mainly determined by the nanobead size uniformity. Although this intensity distribution is not as tight as the values previously reported for QD-doped microspheres (typically 3–15%), 14, 39, 40 it is substantially improved over the micelle-based QD nanobeads.27 In general, preparation of nanobeads with highly uniform fluorescence intensity is extremely difficult because of the small number of QDs that can be incorporated relative to microspheres. As a consequence, small variations in bead sizes and spectroscopy measurement conditions, in addition to shot noise, will translate into significant variance in single-bead fluorescence intensity.
The parameters that control nanobead size and size distribution are not entirely clear through current studies. Above a critical DMF concentration, QD-PMAO cluster into nanobeads due to their low solubility in DMF. The PMAO molecules distributed at the interface of nanobeads and the solvent mixture help lower the interfacial energy. The nanobeads are unlikely to be equilibrium structures, which favor the formation of large agglomerates. Therefore, the size and tight size distribution might be controlled by kinetics in which high percentage of DMF (in the current experiment ~30%) freezes the growth of nanobeads at a certain size leading to formation of uniform and stable QD clusters. We note that this and other possible mechanisms deserve further systematic studies. Nevertheless, if the nanobead formation is indeed controlled by kinetics, their size and size dispersity will be tunable by changing experiment conditions such as polymer composition, nanoparticle and polymer concentrations, and the rate of polar solvent addition.
The fluorescent micrograph also shows that the QD-nanobeads do not ‘blink’ under continuous excitation. The blinking effect is characteristic of single quantum systems such as single dye molecules and single QDs, 44, 45 and this on-and-off behavior could be problematic for detection and imaging of fast biological processes, such as biomolecular transport and trafficking. Although individual QDs exhibit the fluorescence intermittency, the nanobeads, which contain an ensemble of QDs, collectively have constant fluorescence intensity.
An important question to ask is how many QDs are incorporated into each nanobead. TEM measurements at high magnification can resolve individual QDs (inset of Figure 3a), but the image is a 2-D representation of a 3-D structure, which makes it impossible to count the number of QDs per bead. We, therefore, estimated the number based on the optical properties. Comparison of the fluorescence quantum efficiency (QE) of the nanobeads and the original single QDs in THF indicates that the QE of nanobeads is 25% lower than that of QDs, which is typically around 30–40% (The concentration of QDs in the nanobead samples was first derived from the UV absorption measurement. Fluorescence intensity was then evaluated for QD solution and nanobead solution at the same absorbance value). This QE reduction is likely resulted from concentration-dependent fluorophore self-quenching, a phenomenon first observed in the 1880s.46 When fluorophores are in close proximity to each other, such as under high concentration, the fluorescence intensity does not increase linearly with increasing concentration of fluorophores and may even decrease. Because the fluorescence intensity of single nanobeads is as bright as 200 single QDs (see the single bead spectroscopy measurement discussed above) and because their QE is 25% lower than single QDs, we estimated that each nanobead of 112 nm in diameter is packed with 267 . Despite the variance in QD spacing, this calculation suggests that average distance between two adjacent QDs is approximately 14 nm (center to center), which matches the result of theoretical analysis of the particle packing geometry (schematically illustrated in Figure 3e). Considering the radius of red QDs of 3 nm and the intercalating hydrocarbon layer from QD surface ligands and PMAO polymer of 2–4 nm in thickness, the overall distance between two QDs (center to center) will be approximately 10–15 nm, which is also equivalent to the diameter of the a polymer coated QD. Indeed, recent TEM study of single QDs coated with the same PMAO polymer with an additional layer of poly(ethylene glycol) (PEG) showed an overall size of 17 nm,36 which is slightly larger than our results due to the PEG layer. We also note that additional optimization of this technology is needed to reduce or eliminate the QD self quenching. This could be achieved with increased separation distance between QDs, such as by using amphiphilic polymers grafted with longer side chains.
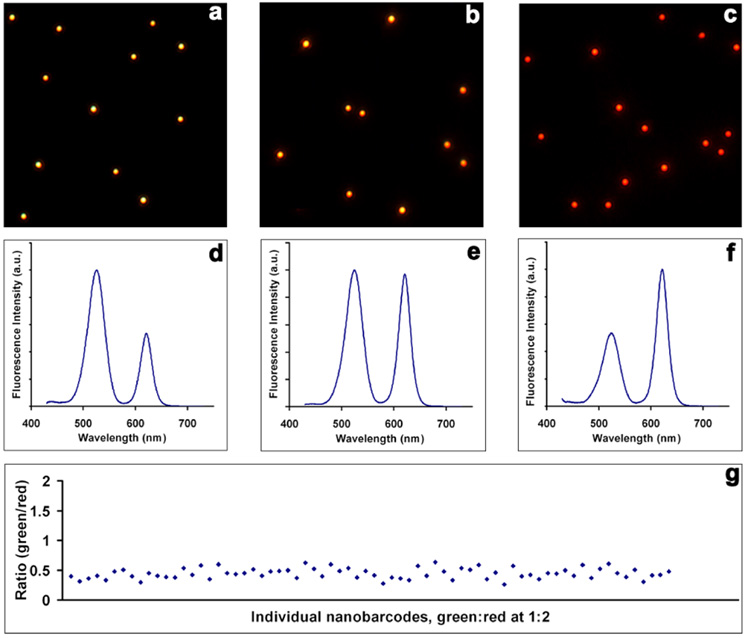
Following the study of single color nanobeads, we proceeded to a more challenging task, making homogeneous dual-color QD-nanobarcodes. Optical barcoding based on fluorescence intensity – color multiplexing can produce more fluorescence probes using a limited number of colors. To demonstrate the feasibility, monodisperse QDs with fluorescence emission maxima at 520 nm (green) and 615 nm (red) were premixed at various fluorescence intensity ratios and used in the incorporation experiment. Figure 4 shows quantitative doping results obtained from the dual-color encoded beads. Using two intensities, there are three unique intensity ratios (green/red 1:2, 1:1, and 2:1). The nanobarcodes with ratios of 2/1, 1/1, and 1/2 appear yellow, orange, and red in fluorescence imaging. Spectral measurement of nanobeads in solution confirms the three fluorescence intensity ratios as shown in Figure 4 d–f. Despite these encouraging results, our initial concern was whether the fluorescence intensity at the ensemble-average level represents those of individual nanobarcodes. To address this question, we exploited a novel hyperspectral imaging approach47, 48 that is capable of examining the spectral features of a large number of nanobeads under exactly the same conditions. A standard hyperspectral imaging setup includes two major components, a passband controlling device (such as liquid crystal tunable filter and diffraction grating) and a scientific-grade monochrome charge-coupled device CCD. Controlled by data acquisition software, the filter or grating automatically steps in a certain wavelength while the camera captures a series of images (image cube) of the sample at each wavelength with constant exposure. This process is repeated for each pure spectral component to generate a spectrum library, which is then used to quantitatively deconvolute mixed colors into separate channels. In our experiments, a tunable filter was set to automatically step in 2 nm (tunable at 1 nm precision). The spectrum at every pixel (or binned as region-of-interest (ROI)) are extracted from the image stack.
Figure 4. Fluorescence imaging and spectroscopy measurements of dual-color QD encoded nanobeads.
(a–c) Fluorescence micrograph of three nanobarcodes with green-to-red intensity ratios of 2:1, 1:1, and 1:2. (d–f) Corresponding fluorescence emission spectra of the three nanobarcodes. (g) Scatter plot of the 1:2 intensity ratio of individual nanobarcodes measured by hyperspectral imaging technique. The values are tightly clustered around the average at 0.44.
More than 100 nanobeads with green/red intensity ratio of ½ were analyzed as an example. As can be seen from Figure 4g, the fluorescence intensity ratios are remarkably robust, although the absolute intensities could vary considerably from bead to bead (because of variations in bead size). As reported by Weiss and co-workers for biomolecular imaging, ratiometric measurements are much more reliable than absolute intensities because the ratio values are not affected by simultaneous drifts or fluctuations of the individual signals.49 The average fluorescence intensity ratio is 0.44 in contrast to the value (0.5) measured by a fluorometer. This is because the differential spectral response of the detectors in fluorometer and hyperspectral imaging system, which are based on a photomultiplier tube and a monochrome silicon CCD, respectively.
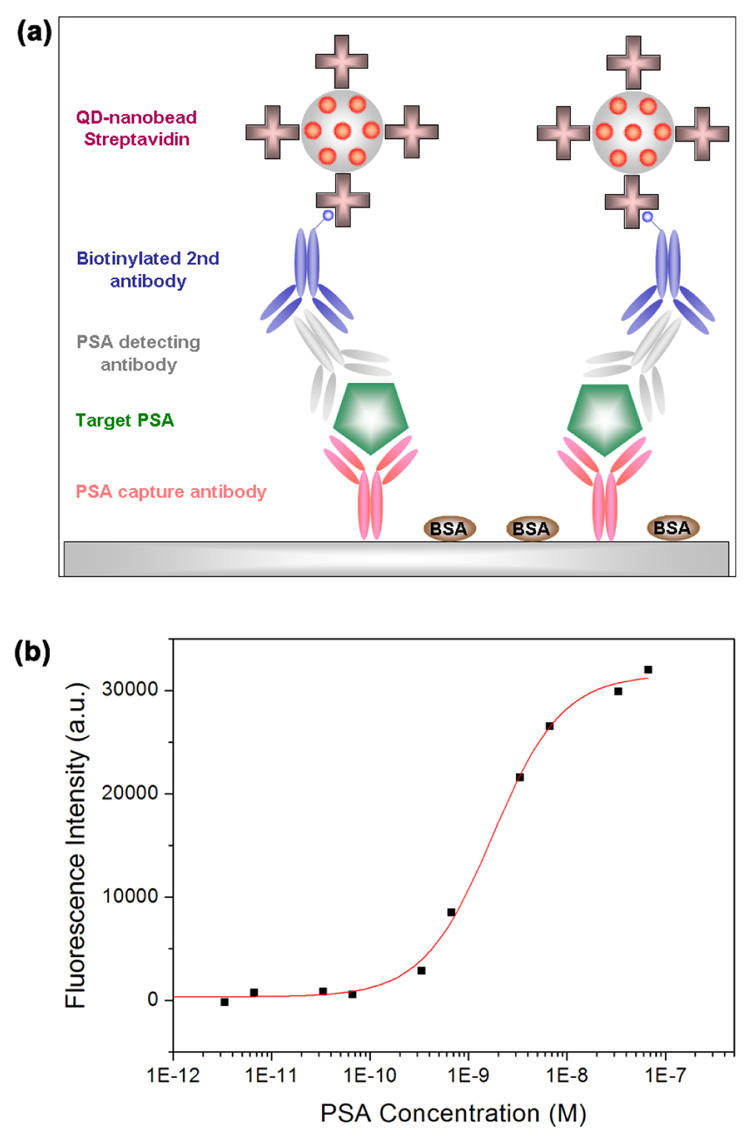
To demonstrate the application of QD-nanobeads in biomolecular imaging and detection, we functionalized the nanobeads with streptavidin using standard carbodiimide crosslinking chemistry and designed a model immunosandwich assay for PSA (prostate specific antigen) detection. As schematically illustrated in Figure 5a, rabbit-anti-human PSA polyclonal antibodies were coated on a 96-well microplate as the capture probe. Serial dilutions of human PSA (target molecules) were added into the microplate followed by incubation with monoclonal mouse anti-human PSA antibodies. Biotinylated anti-mouse IgG and the nanobead-streptavidin bioconjugates were used to generate a fluorescent signal for detection and quantification with a microplate reader. Although the off-the-shelf plate reader is not optimized to measure the fluorescence of a monolayer of nanobeads on microplate surface, Figure 5b shows that PSA molecules still can be readily detected with detection sensitivity in the sub-nanomolar range. If spectrometers are used in conjunction with fluorescence microscopes, the perfectly focused QD-nanobeads are detectable on single bead level. Further development and optimization of the QD-nanobead technology (such as using poly(ethylene glycols)50 as spacers between nanobeads and streptavidin) in combination with exquisite assay design and advanced instrumentation should significantly enhance the detection limit.
Figure 5. Fluorescence immunoassay for PSA detection using QD-nanobeads.
(a) Schematic illustration of the immunoassay. Bioconjugates of QD-nanobeads and streptavidin are used as the reporter probe. (b) Fluorescence emission of QD-nanobeads depending on PSA concentration. Autofluorescence of the microplate imaged under the same condition was subtracted from the fluorescence intensity values. Sub-nanomolar PSA can be readily detected using a standard optical plate reader.
Conclusion
In summary, we have developed a new generation of encoded nanobeads using QD-alternating copolymer complexes. In comparison with QDs embedded in traditional block-copolymer micelles, a key feature of this new technology is that the QDs are pre-coated with amphiphilic polymers prior to formation of nanobeads, which prevents physical contact between QDs. A solvent mixture of THF and DMF leads to epitaxial growth of uniform and highly fluorescent nanobeads from QDs in homogeneous solution without the need for structural templates. As a result of this new bead formation mechanism, a large number of multicolor QDs can be loaded into a nanobead of narrow size dispersity. A model sandwich immunoassay for PSA detection was also demonstrated by conjugating QD-nanobeads with streptavidin molecules. Further development and optimization of this technology should offer new opportunities to ultrasensitive detection of genes, proteins, and cells in fundamental biophysics as well as clinical diagnostics.
Experimental Methods
Reagents and instruments
Poly(maleic anhydride-alt-octadecene), 2,2’-9(ethylenedioxyl) bis(ethylamine), Sulfo-NHS, EDC, BSA, and Tween-20 were purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification. PSA antigen and antibodies targeting PSA were obtained from Fitzgerald Industries International, Inc. (Concord, MA) and Biodesign International (Saco, ME). 96-well microplates with high-binding surface and clear bottom were a gift from Corning Incorporated Inc. (Corning, NY). A UV-2450 spectrophotometer (Shimadzu, Columbia, MD) and a Fluoromax4 fluorometer (Horiba Jobin Yvon, Edison, NJ) were used to characterize the absorption and emission spectra of QDs and QD-nanobeads in solution. The dry and hydrodynamic radii of QDs and QD-nanobeads were measured on a CM100 transmission electron microscope (Philips EO, Netherlands) and a Zetasizer NanoZS size analyzer (Malvern, Worcestershire, UK). True-color fluorescence images were obtained with an IX-71 inverted microscope (Olympus, San Diego, CA) and a Q-color5 digital color camera (Olympus). Broad-band excitation in the near-UV range (330–385 nm) was provided by a mercury lamp. A longpass dichroic filter (400 nm) and emission filter (420 nm, Chroma Technologies, Brattleboro, VT) were used to reject the scattered light and to pass the Stokes-shifted fluorescence signals. Wavelength-resolved fluorescence spectroscopy was accomplished by coupling the fluorescence microscope with a USB4000 single-stage spectrometer (Ocean Optics, Dunedin, FL). For single particle fluorescence measurement, a pinhole of 200 µm diameter is placed at the objective focal plane between the spectrograph and microscope to reject the out-of-focus lights. The QDs and QD-beads were manually positioned, and spectra were recorded. The average fluorescence intensity of single nanobeads and QDs were used to calculate the number of dots per nanobead. For hyperspectral imaging, the images were obtained with a Nuance hyperspectral imaging machine that response to a spectral window from 500 to 950 nm (Cambridge Research and Instrumentation, Inc. Woburn, MA). The immunoassay experiments were carried out on a TECAN Safire™ plate reader (Switzerland).
Preparation of QD-nanobeads
Purified QDs (0.2µM) and PMAO polymers (2.5 µM) were mixed in 0.2ml THF in a glass vial, followed by slow addition of 0.8 ml DMF under vigorous stirring. The concentration of QDs were determined by UV absorption using the molar extinction coefficients for CdSe QDs previously determined by Peng and co-workers.51 2,2’-(ethylenedioxyl) bis(ethylamine) in DMF (10 mM, 2.85 µl) was added into the solution to crosslink the neighboring polymer chains. The solution was stirred at room temperature for 1 h before the dialysis against Tris buffer (20mM, pH10). The nanobeads were isolated by centrifugation and washed multiple times using borate buffer (10 mM, pH8.1) to remove free polymers and diamines in the solution. For multiplexed nanobarcode preparation, procedure similar to the single color nanobead preparation was used except a mixture of green and red QDs (fluorescence emission maxima at 520 and 615 nm) at desired intensity ratios was used.
Conjugation of streptavidin to QD-nanobeads
Red QD-nanobeads suspended in 1 ml of borate buffer (10mM, pH8.1) were incubated with 50 µl of EDC (1 wt%) and 100 µl of Sulfo-NHS (1 wt%) for 15 mins. 10 µl of streptavidin at a concentration of 5 mg/ml was then added and incubated with QD-nanobeads for 2 hrs. The bioconjugates were spun down to remove the unbound streptavidin and this process was repeated twice. The purified bioconjugates were dispersed in borate buffer with 1 wt% BSA.
PSA sandwich immunoassay
Standard sandwich immunoassays were performed for PSA detection using QD-nanobeads. To immobilize PSA capture antibody, 96-well microplate was incubated with polyclonal rabbit-anti-human PSA antibodies (100 µl, 4µg/ml) at 4°C overnight. The microplate was washed with PBS buffer (10mM, pH7.4) with 0.05% Tween-20 (PBST). BSA molecules (100 µl, 2 wt% in PBS buffer) were added to block any un-coated regions. The microplate was again washed with PBST before a series of dilutions of human PSA were introduced into the microplate and incubated for 1h. After removing un-bound PSA, monoclonal mouse-anti-human PSA antibodies (100 µl, 4 µg/ml) were added to form the sandwich. The microplate was subject to incubation with biotinylated anti-mouse IgG and streptavidin labeled with QD-nanobeads or FITC for fluorescence-based detection.
Supplementary Material
Structural and optical properties of green QD-nanobeads, and negative control experiments of the ELISA assay of PSA detection. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgements
This work was supported in part by NIH, NSF, the Seattle Foundation, and the Department of Bioengineering at the University of Washington. X.G. thanks the NSF for a Faculty Early Career Development award (CAREER), and S.R.D. acknowledges the NSF for generous fellowship support. We are especially grateful to Profs. Patrick Stayton, Allan Hoffman, and Miqin Zhang for uses of their particle sizer and plate reader, and Dr. Andrew Y. Wang (OceanNanotech LLC) for help on quantum dot synthesis.
References
- 1.Xie XS, Yu J, Yang WY. Science. 2006;312:228–230. doi: 10.1126/science.1127566. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, Xiao J, Ren X, Lao K, Xie XS. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 3.NCI. Alliance for Nanotechnology in Cancer. 2006 May [Google Scholar]
- 4.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 5.Rosi NL, Mirkin CA. Chem. Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 6.Battersby BJ, Bryant D, Meutermans W, Matthews D, Smythe ML, Trau M. J. Am. Chem. Soc. 2000;122:2138–2139. [Google Scholar]
- 7.Fulton RJ, McDade RL, Smith PL, Kienker LJ, Kettman JR., Jr Clin. Chem. 1997;43:1749–1756. [PubMed] [Google Scholar]
- 8.Steemers FJ, Ferguson JA, Walt DR. Nat. Biotechnol. 2000;18:91–94. doi: 10.1038/72006. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson JA, Boles TC, Adams CP, Walt DR. Nat. Biotechnol. 1996;14:1681–1684. doi: 10.1038/nbt1296-1681. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Tseng1 YD, Kwon SY, d'Espaux L, Bunch JS, McEuen PL, Luo D. Nat. Mater. 2004;3:38–42. doi: 10.1038/nmat1045. [DOI] [PubMed] [Google Scholar]
- 11.Gao XH, Yang L, Petros JA, Marshall FF, Simons JW, Nie S. Curr. Opin. Biotechnol. 2005;16:63–72. doi: 10.1016/j.copbio.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nat. Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 14.Han M, Gao X, Su JZ, Nie S. Nat. Biotechnol. 2001;19:631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 15.Lee JA, Mardyani S, Hung A, Rhee A, Klostranec J, Mu Y, Li D, Chan WCW. Adv. Mater. 2007;19:3113–3118. [Google Scholar]
- 16.Wang DY, Rogach AL, Caruso F. Nano Lett. 2002;2:857–861. [Google Scholar]
- 17.Wang QB, Liu Y, Lin CX, Yan H. Nanotechnology. 2007;18:405604. [Google Scholar]
- 18.Klostranec JM, Xiang Q, Farcas GA, Lee JA, Rhee A, Lafferty EI, Perrault SD, Kain KC, Chan WCW. Nano Lett. 2007;7:2812–2818. doi: 10.1021/nl071415m. [DOI] [PubMed] [Google Scholar]
- 19.Yi DK, Selvan ST, Lee SS, Papaefthymiou GC, Kundaliya D, Ying JY. J. Am. Chem. Soc. 2005;127:4990–4991. doi: 10.1021/ja0428863. [DOI] [PubMed] [Google Scholar]
- 20.Selvan ST, Tan TT, Ying JY. Adv. Mater. 2005;17:1620–1625. [Google Scholar]
- 21.Yang Y, Gao MY. Adv. Mater. 2005;17:2354–2357. [Google Scholar]
- 22.Rogach AL, Nagesha D, Ostrander JW, Giersig M, Kotov NA. Chem. Mater. 2000;12:2676–2685. [Google Scholar]
- 23.Mokari T, Sertchook H, Aharoni A, Ebenstein Y, Avnir D, Banin U. Chem. Mater. 2005;17:258–263. [Google Scholar]
- 24.Kuang M, Wang DY, Bao HB, Gao MY, Möhwald H, Jiang M. Adv. Mater. 2005;17:267–270. [Google Scholar]
- 25.Moffitt M, Vali H, Eisenberg A. Chem. Mater. 1998;10:1021–1028. [Google Scholar]
- 26.Yusuf H, Kim WG, Lee DH, Guo Y, Moffitt MG. Langmuir. 2007;23:868–878. doi: 10.1021/la0623634. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Rosenzweig Z. Nano Lett. 2002;2:1299–1302. [Google Scholar]
- 28.Mulvaney P, Giersig M, Ung T, Liz-Marzán LM. Adv. Mater. 1997;9:570–575. [Google Scholar]
- 29.Lu Y, Yin Y, Li ZY, Xia Y. Nano Lett. 2002;2:785–788. [Google Scholar]
- 30.Zhao W, Gu J, Zhang L, Chen H, Shi J. J. Am. Chem. Soc. 2005;127:8916–8917. doi: 10.1021/ja051113r. [DOI] [PubMed] [Google Scholar]
- 31.Kim BS, Qiu JM, Wang JP, Taton TA. Nano Lett. 2005;5:1987–1991. doi: 10.1021/nl0513939. [DOI] [PubMed] [Google Scholar]
- 32.Kim BS, Taton TA. Langmuir. 2007;23:2198–2202. doi: 10.1021/la062692w. [DOI] [PubMed] [Google Scholar]
- 33.Wu X, Liu H, Liu J, Haley KN, Treadway JA, Larson JP, Ge N, Peale F, Bruchez MP. Nat. Biotechnol. 2003;21:41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 34.Pellegrino T, Manna L, Kudera S, Liedl T, Koktysh D, Rogach AL, Keller S, Rädler J, Natile G, Parak WJ. Nano Lett. 2004;4:703–707. [Google Scholar]
- 35.Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. Nat. Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 36.Yu WW, Chang E, Falkner JC, Zhang J, Al-Somali AM, Sayes CM, Johns J, Drezek R, Colvin VL. J. Am. Chem. Soc. 2007;129:2871–2879. doi: 10.1021/ja067184n. [DOI] [PubMed] [Google Scholar]
- 37.Thurmond KB, Kowalewski T, Wooley KL. J. Am. Chem. Soc. 1996;118:7239–7240. [Google Scholar]
- 38.Butun V, Lowe AB, Billingham NC, Armes SP. J. Am. Chem. Soc. 1999;121:4288–4289. [Google Scholar]
- 39.Gao XH, Nie S. J. Phys. Chem. B. 2003;107:11575–11578. [Google Scholar]
- 40.Gao XH, Nie S. Anal. Chem. 2004;76:2406–2410. doi: 10.1021/ac0354600. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Zhou D, Browne H, Balasubramanian S, Klenerman D. Anal. Chem. 2004;76:4446–4451. doi: 10.1021/ac049512c. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Ying L, Green JJ, Balasubramanian S, Klenerman D. Anal. Chem. 2003;75:1664–1670. doi: 10.1021/ac026367z. [DOI] [PubMed] [Google Scholar]
- 43.Lacoste TD, Michalet X, Pinaud F, Chemla DS, Alivisatos AP, Weiss S. Proc. Natl. Acad. Sci. USA. 2000;97:9461–9466. doi: 10.1073/pnas.170286097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nirmal M, Dabbousi BO, Bawendi MG, Macklin JJ, Trautman JK, Harris TD, Brus LE. Nature. 1996;383:802–804. [Google Scholar]
- 45.Empedocles SA, Bawendi MG. Science. 1997;278:2114–2117. doi: 10.1126/science.278.5346.2114. [DOI] [PubMed] [Google Scholar]
- 46.Walter B. Ann. Phys. 1888;34:31–326. [Google Scholar]
- 47.True LD, Gao XH. J. Mol. Diagn. 2007;9:7–11. doi: 10.2353/jmoldx.2007.060186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levenson RM. Lab Med. 2004;35:244–251. [Google Scholar]
- 49.Deniz AA, Laurence TA, Dahan M, Chemla DS, Schultz PG, Weiss S. Annu. Rev. Phys. Chem. 2001;52:233–253. doi: 10.1146/annurev.physchem.52.1.233. [DOI] [PubMed] [Google Scholar]
- 50.Matsuya T, Tashiro S, Hoshino N, Shibata N, Nagasaki Y, Kataoka K. Anal. Chem. 2003;75:6124–6132. doi: 10.1021/ac034346e. [DOI] [PubMed] [Google Scholar]
- 51.Yu WW, Qu L, Guo W, Peng X. Chem. Mater. 2003;15:2854–2860. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structural and optical properties of green QD-nanobeads, and negative control experiments of the ELISA assay of PSA detection. This material is available free of charge via the Internet at http://pubs.acs.org.