Abstract
Recent advances in far-field fluorescence microscopy have led to substantial improvements in image resolution, achieving a near-molecular resolution of 20 - 30 nm in the two lateral dimensions. Three-dimensional (3D) nanoscale-resolution imaging, however, remains a challenge. Here, we demonstrate 3D stochastic optical reconstruction microscopy (STORM) by determining both axial and lateral positions of individual fluorophores with nanometer accuracy using optical astigmatism. Iterative, stochastic activation of photo-switchable probes enables high-precision 3D localization of each probe and thus the construction of a 3D image without scanning the sample. Using this approach, we achieved an image resolution of 20 - 30 nm in the lateral dimensions and 50 - 60 nm in the axial dimension. This development allowed us to resolve the 3D morphology of nanoscopic cellular structures.
Due to its non-invasive nature and multi-color capability, far-field optical microscopy offers three-dimensional (3D) imaging of biological specimens with minimal perturbation and biomolecular specificity when combined with fluorescent labeling. These advantages make fluorescence microscopy one of the most widely used imaging methods in biology. The diffraction barrier, however, limits the imaging resolution of conventional light microscopy to 200 - 300 nm in the lateral dimensions, leaving many intracellular organelles and molecular structures unresolvable. Recently, the diffraction limit has been surpassed and lateral imaging resolutions of 20 - 50 nm have been achieved by several “super-resolution” far-field microscopy techniques, including stimulated emission depletion (STED) and its related RESOLFT microscopy (1, 2), saturated structured illumination microscopy (SSIM) (3), STORM (4, 5), photoactivated localization microscopy (PALM) (6, 7) and other methods using similar principles (8-10).
While these techniques have improved 2D image resolution, resolving most organelles and cellular structures requires high-resolution imaging in all three dimensions. Three-dimensional fluorescence imaging is most commonly performed using confocal or multi-photon microscopy, the axial resolution of which is typically in the range of 500 - 800 nm, two to three times worse than the lateral resolution (11, 12). The axial imaging resolution can be improved to roughly 100 nm by 4Pi and I5M microscopy (13-15). Furthermore, an axial resolution as high as 30 - 50 nm has been obtained by employing stimulated emission depletion along the axial direction using the 4Pi illumination geometry, but the same scheme does not provide super resolution in the lateral dimensions (1).
Here we demonstrate 3D STORM imaging with a spatial resolution that is 10 times better than the diffraction limit in all three dimensions without invoking sample or optical beam scanning. STORM and PALM rely on single-molecule detection (16) and exploit the photo-switchable nature of certain fluorophores to temporally separate the otherwise spatially overlapping images of numerous molecules, thereby allowing the high-precision localization of individual molecules (4, 6, 7, 9). Limited only by the number of photons detected (17), localization accuracies as high as 1 nm can be achieved in the lateral dimensions for a single fluorescent dye at ambient conditions (18). While the lateral position of a particle can be determined from the centroid of its image (19, 20), the shape of the image contains information about the particle’s axial (z) position. Nanoscale localization accuracy has been previously achieved in the z dimension by introducing defocusing (21-24) or astigmatism (25, 26) into the image, without significantly compromising the lateral positioning capability.
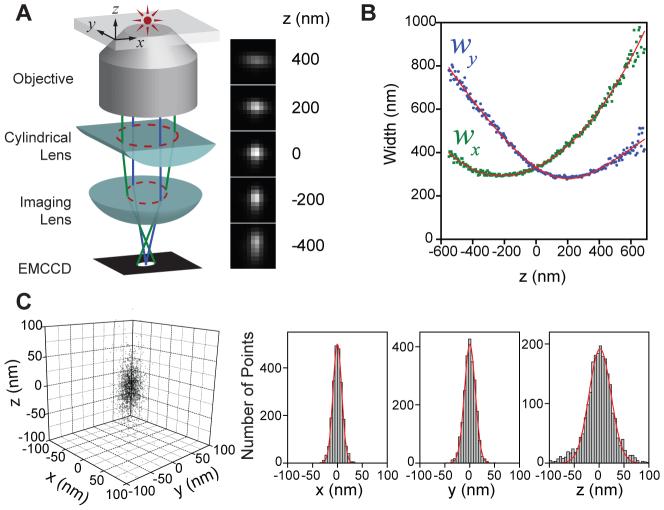
In this work, we used the astigmatism imaging method to achieve 3D STORM imaging. To this end, a weak cylindrical lens was introduced into the imaging path to create two slightly different focal planes for the x and y directions (Fig. 1A) (25, 26). As a result, the ellipticity and orientation of a fluorophore’s image varied as its position changed in z (Fig. 1A): When the fluorophore was in the average focal plane (approximately half-way between the x and y focal planes where the point-spread-function (PSF) has equal widths in the x and y directions), the image appeared round; when the fluorophore was above the average focal plane, its image was more focused in the y direction than in the x direction and thus appeared ellipsoidal with its long axis along x; conversely when the fluorophore was below the focal plane, the image appeared ellipsoidal with its long axis along y. By fitting the image with a 2D elliptical Gaussian function, we obtained the x and y coordinates of peak position as well as the peak widths wx and wy, which in turn allowed the z coordinate of the fluorophore to be unambiguously determined. To experimentally generate a calibration curve of wx and wy as a function of z, we immobilized Alexa 647-labeled streptavidin molecules or quantum dots on a glass surface and imaged individual molecules to determine the wx and wy values as the sample was scanned in z (Fig. 1B). In 3D STORM analysis, the z coordinate of each photo-activated fluorophore was then determined by comparing the measured wx and wy values of its image with the calibration curves. In addition, for samples immersed in aqueous solution on a glass substrate, All z localizations were rescaled by a factor of 0.79 to account for the refractive index mismatch between glass and water (see Supporting Online Material for a detailed description of the analysis procedures) (27).
Fig. 1.
The scheme of 3D STORM. (A) Three-dimensional localization of individual fluorophores. The simplified optical diagram illustrates the principle of determining the z-coordinate of a fluorescent object from the ellipticity of its image by introducing a cylindrical lens into the imaging path. The right panel shows the images of a fluorophore at various z positions. (B) The calibration curve of the image widths wx and wy as a function of z obtained from single Alexa 647 molecules. Each data point represents the average value obtained from 6 molecules. The data were fit to a defocusing function (red curve) as described in the Supporting Online Materials (27). (C) Three-dimensional localization distribution of single molecules. Each molecule gives a cluster of localizations due to repetitive activation of the same molecule. Localizations from 145 clusters were aligned by their center-of-mass to generate the overall 3D presentation of the localization distribution (left panel). Histograms of the distribution in x, y and z (right panels) were fit to a Gaussian function, yielding the standard deviation of 9 nm in x, 11 nm in y, and 22 nm in z.
The 3D resolution of STORM is limited by the accuracy with which individual photo-activated fluorophores can be localized in all three dimensions during a switching cycle. We recently discovered a family of photo-switchable cyanine dyes (Cy5, Cy5.5, Cy7 and Alexa Fluor 647) that can be reversibly cycled between a fluorescent and a dark state by light of different wavelengths. The reactivation efficiency of these photo-switchable “reporters” depends critically on the proximity of an “activator” dye, which can be any one of a variety of dye molecules (e.g. Cy3, Cy2, Alexa Fluor 405) (5, 28). Here we used Cy3 and Alexa 647 as the activator and reporter pair to perform 3D STORM imaging. A red laser (657 nm) was used to image Alexa 647 molecules and deactivate them to the dark state, whereas a green laser (532 nm) was used to reactivate the fluorophores. Each activator-reporter pair could be cycled on and off hundreds of times before permanent photobleaching occurred and an average of 6000 photons were detected per switching cycle using objective-type total-internal-reflection fluorescence (TIRF) or epi-fluorescence imaging geometry (5, 28). This reversible switching behavior provided an internal control to measure the localization accuracy. To this end, we immobilized streptavidin molecules doubly labeled with Cy3 and Alexa 647 on a glass surface (27). The molecules were then switched on and off for multiple cycles, and their x, y, and z coordinates were determined for each switching cycle (27). This procedure resulted in a cluster of localizations for each molecule (Fig. 1C). The standard deviations of the localization distribution obtained within 100 nm of the average focal plane were 8 - 9 nm in x, 10 - 11 nm in y, and 18 - 22 nm in z, and the corresponding full widths at half maximum (FWHM) were 18 - 21 nm, 24 - 26 nm, and 42 - 52 nm, providing a quantitative measure of the localization accuracy in 3D (Fig. 1C). The localization accuracies in the two lateral dimensions were similar to our previous 2D STORM resolution obtained without the cylindrical lens (5). The localization accuracy in z was, as expected, approximately twice the localization accuracy measured in x and y. Because the image width increases as the fluorophore moves away from the focal plane, the localization accuracy decreases with increasing z values, especially in the lateral dimensions. Therefore, we typically chose a z imaging depth of about 600 nm near the focal plane, within which the lateral and axial localization accuracies changed by less than 1.6 fold and 1.3 fold, respectively, in comparison with the values obtained at the average focal plane. The imaging depth may, however, be increased by employing z scanning in future experiments.
As an initial test of 3D STORM, we imaged a model bead sample prepared by immobilizing 200 nm biotinylated polystyrene beads on a glass surface and then incubating the sample with Cy3-Alexa 647 labeled streptavidin to coat the beads with photo-switchable probes (27). Three-dimensional STORM images of the beads were obtained by iterative, stochastic activation of sparse subsets of optically resolvable Alexa 647 molecules, allowing the x, y and z coordinates of individual molecules to be determined. Over the course of multiple activation cycles, the positions of numerous fluorophores were determined and used to construct a full 3D image (27). The projections of the bead images appeared approximately spherical when viewed along all three directions with average diameters of 210 ± 16, 225 ± 25 and 228 ± 25 nm in x, y and z respectively (fig. S1) (27), indicating accurate localization in all three dimensions. As the image of each fluorophore simultaneously encodes its x, y and z coordinates, no additional time was required to localize each molecule in 3D STORM as compared with 2D STORM imaging.
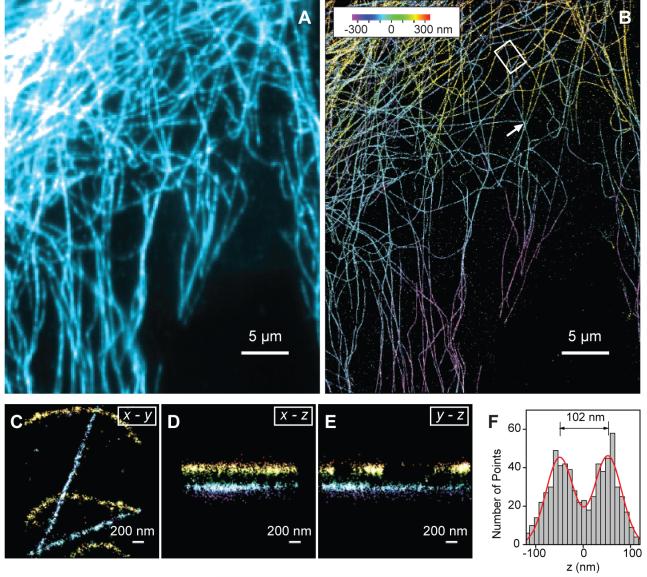
Applying 3D STORM to cell imaging, we next performed indirect immunofluorescence imaging of the microtubule network in green monkey kidney epithelial (BS-C-1) cells. Cells were immunostained with primary antibodies and then with Cy3 and Alexa 647 doubly labeled secondary antibodies (27). The 3D STORM image not only showed a substantial improvement in resolution as compared to the conventional wide-field fluorescence image (Figs. 2A,B), but also provided the z-dimension information (color-coded in Fig. 2B) that was not available in the conventional image. Multiple layers of microtubule filaments were clearly visible in the x-y, x-z and y-z cross sections of the cell (Figs. 2C - E and Movie S1) (27). To characterize our cell imaging resolution more quantitatively, we identified point-like objects in the cell that appeared as small clusters of localizations away from any discernable microtubule filaments. These clusters likely represent individual antibodies nonspecifically attached to the cell. The FWHM of these clusters, which were randomly chosen over the entire measured z-range of the cell, were 22 nm in x, 28 nm in y and 55 nm in z (fig. S2) (27), similar to those determined for individual molecules immobilized on a glass surface (compare fig. S2 with Fig. 1C). Two microtubule filaments separated by 100 nm in z appeared well separated in the 3D STORM image (Fig. 2F). The apparent width of the microtubule filaments in the z dimension was 66 nm, slightly larger than our intrinsic imaging resolution in z and in quantitative agreement with the convolution of the imaging resolution and the independently measured width of antibody-coated microtubule (Fig. 2F). Since the effective resolution is determined by a combination of the intrinsic imaging resolution as characterized above and the size of the labels (such as the antibodies), improved resolution may be achieved by using direct immunofluorescence to remove one layer of antibody labeling, as we show in the next example, or by using Fab fragments or genetically encoded peptide tags (29, 30) in place of antibodies.
Fig. 2.
Three-dimensional STORM imaging of microtubules in a cell. (A) Conventional indirect immunofluorescence image of microtubules in a large area of a BS-C-1 cell. (B) The 3D STORM image of the same area with the z-position information color-coded according to the colored scale bar. Each localization is depicted in the STORM image as a Gaussian peak, the width of which is determined by the number of photons detected (5). (C-E) The x-y, x-z and y-z cross-sections of a small region of the cell outlined by the white box in (B), showing 5 microtubule filaments. Movie S1 shows the 3D representation of this region, with the viewing angle rotated to show different perspectives (27). (F) The z profile of two microtubules crossing in the x-y projection but separated by 102 nm in z, from a region indicated by the arrow in (B). The histogram shows the distribution of z-coordinates of the localizations, fit to two Gaussians with identical widths (FWHM = 66 nm) and a separation of 102 nm (red curve). The apparent width of 66 nm agrees quantitatively with the convolution of our imaging resolution in z (represented by a Gaussian function with FWHM of 55 nm) and the previously measured width of antibody-coated microtubules (represented by a uniform distribution with a width of 56 nm) (5).
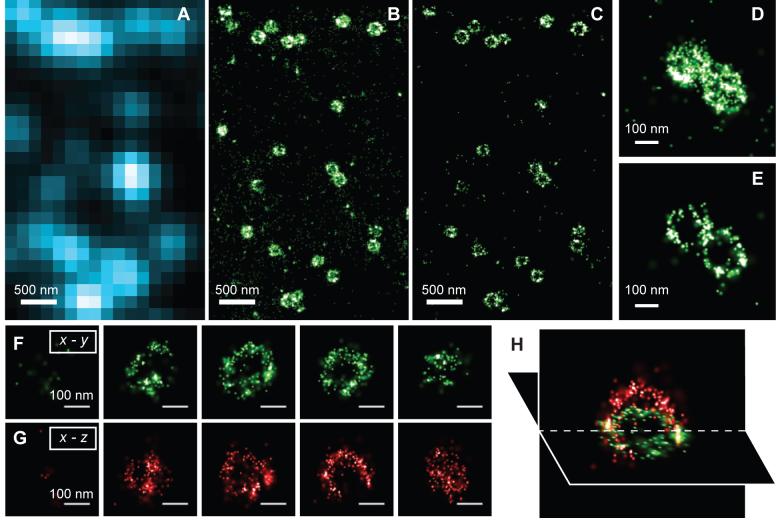
Finally, to demonstrate that 3D STORM can resolve the 3D morphology of nanoscopic structures in cells, we imaged clathrin-coated pits (CCP) in BS-C-1 cells. CCPs are spherical cage-like structures, approximately 150 - 200 nm in size, assembled from clathrin and cofactors on the cytoplasmic side of the cell membrane to facilitate endocytosis (31). To image CCPs, we adopted a direct immunofluorescence scheme using Cy3 and Alexa 647 doubly labeled primary antibodies against clathrin (27). When imaged by conventional fluorescence microscopy, all CCPs appeared as nearly diffraction-limited spots with no discernable structure (Fig. 3A). In 2D STORM images in which the z-dimension information was discarded, the round shape of CCPs was clearly seen (Figs. 3B, D). The size distribution of CCPs measured from the 2D projection image, 180 ± 40 nm, agrees quantitatively with the size distribution determined using electron microscopy (32). Including the z-dimension information allows us to clearly visualize the 3D structure of the pits (Figs. 3C, E - H). Figures 3C and 3E show the x-y cross-sections of the image, taken from a region near the opening of the pits at the cell surface. The circular ring-like structure of the pit periphery was unambiguously resolved. Consecutive x-y and x-z cross-sections of the pits (Figs. 3F-H) clearly revealed the three-dimensional half-spherical-cage like morphology of these nanoscopic structures that was not observable in the 2D STORM images. In summary, we have demonstrated 3D super-resolution imaging with 20 - 30 nm lateral and 50 - 60 nm axial resolution by STORM microscopy. This imaging capability allowed nanoscale features of cellular structures to be resolved optically under ambient conditions, at a resolution previously only seen with electron microscopy.
Fig. 3.
Three-dimensional STORM imaging of clathrin-coated pits in a cell. (A) Conventional direct immunofluorescence image of clathrin in a region of a BS-C-1 cell. (B) The 2D STORM image of the same area with all localizations at different z positions included. (C) A x-y cross-section (50 nm thick in z) of the same area showing the ring-like structure of the periphery of the CCPs at the plasma membrane. (D, E) Magnified view of two nearby CCPs in 2D STORM (D) and their 100 nm thick x-y cross-section in the 3D image (E). (F - H) Serial x-y cross-sections (each 50 nm thick in z) (F) and x-z cross-sections (each 50 nm thick in y) (G) of a CCP, and an x-y and x-z cross section presented in 3D perspective (H), showing the cage like structure of the pit.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institute of Health (GM 068518). X.Z. is a Howard Hughes Medical Institute Investigator.
Footnotes
One sentence summary
Fluorescence imaging of cellular structures with 20 - 30 nm lateral and 50 nm axial resolution is achieved in fixed cells at ambient conditions without scanning the sample.
References
- 1.Hell SW. Nat. Biotech. 2003;21:1347. doi: 10.1038/nbt895. [DOI] [PubMed] [Google Scholar]
- 2.Hell SW. Science. 2007;316:1153. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 3.Gustafsson MGL. Proc. Natl. Acad. Sci., USA. 2005;102:13081. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rust MJ, Bates M, Zhuang X. Nat. Meth. 2006;3:793. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates M, Huang B, Dempsey GT, Zhuang X. Science. 2007;317:1749. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betzig E, et al. Science. 2006;313:1642. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 7.Hess ST, Girirajan TPK, Mason MD. Biophys. J. 2006;91:4258. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharonov A, Hochstrasser RM. Proc. Natl. Acad. Sci., USA. 2006;103:18911. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egner A, et al. Biophys. J. 2007;93:3285. doi: 10.1529/biophysj.107.112201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bock H, et al. Appl. Phys. B. 2007;88:161. [Google Scholar]
- 11.Torok P, Wilson T. Opt. Commun. 1997;137:1270135. [Google Scholar]
- 12.Zipfel WR, Williams RM, Webb WW. Nat. Biotechnol. 2003;21:1368. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 13.Nagorni M, Hell SW. J. Struct. Biol. 1998;123:236. doi: 10.1006/jsbi.1998.4037. [DOI] [PubMed] [Google Scholar]
- 14.Gustafsson MGL, Agard DA, Sedat JW. J. Micro. 1999;195:10. doi: 10.1046/j.1365-2818.1999.00576.x. [DOI] [PubMed] [Google Scholar]
- 15.Egner A, Hell SW. Trends Cell Biol. 2005;15:207. doi: 10.1016/j.tcb.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Moerner WE, Orrit M. Science. 1999;283:1670. doi: 10.1126/science.283.5408.1670. [DOI] [PubMed] [Google Scholar]
- 17.Thompson RE, Larson DR, Webb WW. Biophys. J. 2002;82:2775. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yildiz A, et al. Science. 2003;300:2061. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 19.Barak LS, Webb WW. J. Cell Biol. 1981;90:595. doi: 10.1083/jcb.90.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gelles J, Schnapp BJ, Sheetz MP. Nature. 1988;331:450. doi: 10.1038/331450a0. [DOI] [PubMed] [Google Scholar]
- 21.van Oijen AM, Kohler J, Schmidt J, Muller M, Brakenhoff GJ. Chem. Phys. Lett. 1998;292:183. [Google Scholar]
- 22.Speidel M, Jonas A, Florin EL. Opt. Lett. 2003;28:69. doi: 10.1364/ol.28.000069. [DOI] [PubMed] [Google Scholar]
- 23.Prabhat P, Ram S, Ward ES, Ober RJ. Proc. SPIE. 2006;6090:60900. [Google Scholar]
- 24.Toprak E, Balci H, Behm BH, Selvin PR. Nano Lett. 2007;7:2043. doi: 10.1021/nl0709120. [DOI] [PubMed] [Google Scholar]
- 25.Kao HP, Verkman AS. Biophys. J. 1994;67:1291. doi: 10.1016/S0006-3495(94)80601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holtzer L, Meckel T, Schmidt T. Appl. Phys. Lett. 2007;90:053902. [Google Scholar]
- 27.See supporting material on Science online.
- 28.Bates M, Blosser TR, Zhuang X. Phys. Rev. Lett. 2005;94:108101. doi: 10.1103/PhysRevLett.94.108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen I, Ting AY. Curr. Opin. Biotechnol. 2005;16:35. doi: 10.1016/j.copbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 31.Slepnev VI, De Camilli P. Nat. Rev. Neurobiol. 2000;1:161. doi: 10.1038/35044540. [DOI] [PubMed] [Google Scholar]
- 32.Heuser JE, Anderson RGW. J. Cell Biol. 1989;108:389. doi: 10.1083/jcb.108.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.