Abstract
Cancer stem cells (CSC) are remarkably similar to normal stem cells: both self-renew, are multipotent and express common surface markers, e.g., PROMININ-1 (PROM1, CD133)1. What remains unclear is whether CSC are the direct progeny of mutated stem cells, or more mature cells that reacquire stem cell properties during tumor formation. Answering this important question will require knowledge of whether normal stem cells are susceptible to cancer causing mutations; however, this has proved difficult to test since the identity of most adult tissue stem cells is not known. Here, using an inducible-Cre-nuclear(n)LacZ reporter allele knocked into the Prom1 locus (Prom1C-L), we show that Prom1 is expressed in a variety of developing and adult tissues. Lineage-tracing studies of adult Prom1+/C-L mice containing the Rosa26YFP reporter allele showed that Prom1+ cells are located at the base of crypts in the small intestine, co-express Lgr52, generate the entire intestinal epithelium, and are therefore likely to be the small intestinal stem cell. Prom1 was reported recently to mark CSC of human intestinal tumors that arise frequently as a consequence of aberrant Wingless (WNT) signaling3-5. Activation of endogenous Wnt signaling in Prom1+/C-L mice containing a Cre-dependent mutant allele of Beta-catenin (Ctnnb1lox(ex3)) resulted first in a gross disruption of crypt architecture and a disproportionate expansion of Prom1+ cells at the crypt base. Lineage-tracing demonstrated that the progeny of these cells replaced the mucosa of the entire small intestine with neoplastic tissue that was characterized by focal high-grade intraepithelial neoplasia and crypt adenoma formation. Although all neoplastic cells arose from Prom1+ cells in these mice, only 7% of tumor cells retained Prom1 expression. Our data indicate that Prom1 marks stem cells in the adult small intestine, which are susceptible to transformation into tumors retaining a fraction of mutant-Prom1+ tumor cells.
The hypothesis that cancers are generated by rare populations of CSC that are more tumorigenic than other cancer cells has gained increasing credence1. This has followed consistent observations that some solid tumors and leukemias contain small numbers of self-renewing cells that propagate the disease when transplanted in mice3,4,6-8. CSC also express markers of normal stem cells, and in the brain, exist in microenvironments that mimic normal stem cell niches9,10.
Although there is an expanding literature to support the existence of CSC, important caveats of these studies continue to provoke controversy and debate. The current definitive test of a CSC is the capacity to propagate tumors as xenografts in immunocompromised mice11; however, it has been argued that species differences alone might account for the selective growth of subpopulations of cells in these assays. Indeed, the great majority of cells in a mouse lymphoma were shown recently to possess tumor initiating capacity when allografted into syngenic mice12. Further controversy has dogged the use of stem cell surface markers to isolate CSC. Notable among these is PROM1, a five-transmembrane domain containing glycoprotein that is expressed on the surface of a variety of normal stem cells13-15. CSC of brain7,16, gut3,4 and pancreatic17 tumors have been isolated using PROM1 antibodies; however, most PROM1 antibodies recognize glycosylation-dependent epitopes that vary with the differentiation and transformation status of the cell18, complicating the use of these reagents to fractionate stem cells. Without better understanding of normal tissue stem cells and their susceptibility to neoplastic transformation, it will be difficult to conduct definitive studies of the existence and origins of CSC.
To understand better the identity of normal tissue stem cells and their role in the cancer process, we generated a knock-in allele in which we integrated a creERT2-IRES-nLacZ cassette at the first ATG codon of Prom1 (Prom1C-L); thereby creating a null allele (Figure 1a and Supplemental Figure 1). Prom1C-L/C-L mice were born and aged normally, indicating that Prom1 is dispensable for gross embryonic and postnatal development. nLacZ expression in Prom1+/C-L embryos that retained one copy of the wild-type allele, was restricted to the central nervous system, kidney, intestine and developing skeletal system (Figure 1b). The tissue distribution of Prom1 expression increased with subsequent development and was detected ultimately in several organs of adult Prom1+/C-L mice (Figure 1c, Supplemental Figure 2 and Supplemental Table 1). Analysis of nLacZ and Prom1 protein expression from the modified and wild-type alleles, respectively, as well as tissue specific markers, detected Prom1 in Nestin+ cells of the ependymal layer and subventricular zone of the adult brain; Clara Cell Specific Protein (CCSP)+ cells at the bronchoalveloar junction in the lung; and cells lining the pancreatic ducts (Figure 1c). Each of these cell populations has been reported to include adult stem cells19-22. nLacZ and Prom1 expression were also observed in numerous differentiated cells in Prom1+/C-L mice, including proximal renal tubule cells, photoreceptors in the retina (both Figure 1c), neurons in the adult brain, acinar and islet cells in the pancreas, and goblet and columnar epithelial cells lining the colon (Supplemental Figure 2). This pattern of Prom1 expression is in agreement with recent studies of the Prom1 locus in mouse tissues23, and indicates that Prom1 expression is not confined to stem cells, but is expressed also in multiple types of differentiated cells.
Figure 1. Generation of Prom1C-L mice and reporter analysis.
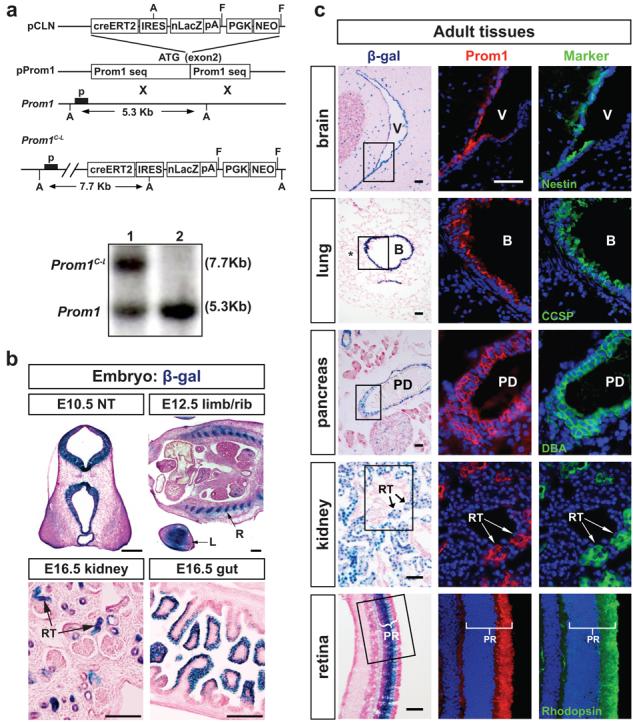
(a) Top to bottom: CreERT2-IRES-nuclear(n)LacZ-PGK-Neo was cloned into the ATG site of Prom1. pProm1 was subject to homologous recombination with wild type Prom1 to generate the Prom1C-L targeted allele. PGK-Neo was excised by flip-recombination. p=Southern blot probe; A=ApaL1 sites; FRT=flip recombinase targets. Southern blot of mouse tissues heterozygous for the targeted allele (lane 1) and homozygous for the wild-type allele (lane 2). (b) Expression of nLacZ from the Prom1C-L allele in embryonic tissues. NT, neural tube; R, rib; L, limb; RT, renal tubule. Scale bars=200μm. (c) Prom1 and nLacZ expression in adult mouse (3 month) tissues. β-galactosidase staining (left panel), Prom1 protein (middle panel, immunofluorescence) and markers (right panel) of neural stem and progenitor cells (Nestin), Clara cells (Clara cell specific protein [CCSP]), pancreatic ductal cells (Dolichos Biflorus Agglutinin [DBA]-fluorescein), proximal renal tubular endothelial cells (Lotus Tetragonolobus Lectin [LTL]-fluorescein), and photoreceptors (rhodopsin). Middle and right panels are adjacent sections of the boxed areas in left panels. V, ventricle; B, bronchiole; * vessel at bronchioalveolar junction; PD, pancreatic duct; RT, renal tubule; PR, photoreceptor. All scale bars=50μm.
In contrast to most tissues, Prom1 expression was relatively restricted in the small intestine of Prom1+/C-L adult mice. Less than three percent of cells covering the villi expressed nLacZ (n=82/3305), versus 26% (n=106/402) of cells at the crypt base (β-galactosidase staining, P<0.0001, Chi-square test, Figure 2a and Supplemental Figure 3). Prom1 mRNA expression was similarly distributed in the small intestine and overlapped closely with that of Lgr5, which was reported recently to mark intestinal stem cells2 (Figure 2b and c, and Supplemental Figure 3). Therefore, as a first step to test if Prom1 might be expressed by Lgr5+ crypt stem cells, we performed concurrent Lgr5 in situ hybridization and nLacZ immunofluorescence analyses of adult Prom1+/C-L mouse small intestine. Over 75% of Lgr5+ crypt cells contained nLacZ+ nuclei (n=390/500 cells counted), suggesting that Prom1+ crypt cells are predominantly stem cells (Figure 2d). Further, of the 4.1 LacZ positive nuclei (average, range 0 to 8 cells in 210 crypts counted) observed in each Prom1+/C-L crypt, 23%, 30% and 17% were located at the 0, 1′ and 2′ cell positions, respectively, that are the sites most frequently occupied by Lgr5+ cells2 (Figure 2e). Together, these data suggest strongly that Lgr5+ small intestinal stem cells express Prom1.
Figure 2. Prom1 marks small intestinal crypt stem cells.
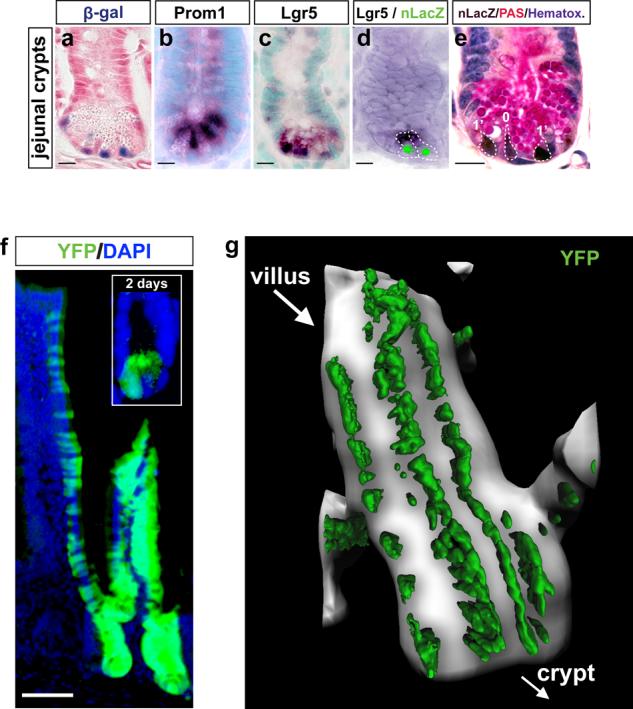
Expression of (a) Prom1C-L (β-galactosidase) and (b) Prom1 wild-type (Prom1 in situ hybridization) alleles is confined to the small intestinal crypt base in Prom1+/C-L mice and overlaps with (c) Lgr5 expression (in situ hybridization). (d) Lgr5 (in situ hybridization, cyoplasmic dark stain) and Prom1 (green, nLacZ immunofluorescence) are co-expressed in the same cells (broken lines indicate cell boundaries). (e) nLacZ immunohistochemistry (brown nuclear immunostain) counterstained with Periodic Acidic Schiff (PAS [pink], to detect Paneth cells) and hematoxylin (blue) reveals the position of Prom1+ cells (numbers denote crypt cell positions) in Prom1+/C-L mice. Scale bar=10μm. (f) Inset shows YFP+ cells in the crypt base of Prom1+/C-L ; RosaYFP mice two days following tamoxifen-induction. Main panel, YFP-labeling of the crypts and epithelial surface of the villi, 60 days post tamoxifen-induction. Scale bar=50μm. (g) Two-photon laser-scanning microscopy (YFP, green) through 464μm of adult Prom1+/C-L mouse small intestinal villus 10 days following tamoxifen-induction.
To test more directly if Prom1 marks stem cells in the small intestine, we used the Cre-dependent Rosa26-Yellow Fluorescence Protein (RosaYFP) reporter allele to trace the lineage of Prom1+ crypt cells. Two month old Prom1+/C-L ; RosaYFP mice were treated with tamoxifen to activate CreERT2 expressed from the Prom1C-L locus; thereby irreversibly activating YFP expression in Prom1+ cells and their progeny. Mice were sacrificed 2, 10 and 60 days following induction. Two days following induction, an average of 2.5 YFP-labelled cells was seen in 25% of small intestinal crypts (>50 labelled crypts counted; Figure 2f inset). By 10 days, trains of contiguous YFP+ cells were observed emanating in single file from the base of crypts in a manner identical to that reported for the progeny of Lgr5+ stem cells2 (Figure 2g). YFP cells remained present at 60 days, extended to the tip of the villi, and included the four differentiated cell types of the intestinal lining: enterocytes, Paneth, goblet, and enteroendocrine cells (Figure 2f and Supplemental Figure 4). Thus, we conclude that the stem cells of the mouse small intestine are Prom1+. In contrast, immediately following induction, YFP-labelling in the colon of these mice displayed the same broad distribution as Prom1 expression, and gradually disappeared within 60 days. Therefore, Lgr5+/Prom1+ stem cells of the small intestine are distinct from Lgr5+/Prom1− stem cells of the large intestine2,23. YFP labelling of the brain, kidney, lung, and pancreas remained relatively static between 2 and 60 days post tamoxifen induction (Supplemental Figure 5 and data not shown). Since these organs have low rates of cell turn-over, it remains possible that some Prom1+ cells in these tissues are quiescent adult stem cells.
Mutations that activate aberrant WNT signaling, including mutations in Beta-Catenin (CTNNB1), have been identified in over 80% of human colonic cancers5,24 and this tumor type was reported recently to contain Prom1+ CSC3,4. Intestinal tumorigenesis caused by aberrant Wnt signaling has been modeled successfully in genetically engineered mice25. Although tumors in these animals develop predominantly in the small intestine, the disease process mimics much of the cellular and molecular characteristics of human colonic polyposis25-29. Therefore, to determine if intestinal tumors might arise directly from Prom1+ stem cells, we activated endogenous WNT signaling in two month old Prom1+/C-L mice using a Cre-dependent mutant allele of Ctnnb1 (Ctnnb1lox(ex3))29 and traced the lineage of these cells using the Rosa26YFP allele. Two days following tamoxifen-induction, we observed a marked increase in the number of YFP+ cells at the base of small intestinal crypts, but at no other site above crypt cell position 4, in Prom1+/C-L ; RosaYFP ; Ctnnb1+/ lox(ex3) mice relative to Prom1+/C-L ; RosaYFP ; Ctnnb1+/+ control mice (Figure 3a). Two-photon laser scanning microscopy demonstrated that these cells had a slim and elongated morphology compatible with them being crypt stem cells2 (Supplemental Movie 1). Counting of cells isolated from the entire small intestinal mucosae of mice two days following tamoxifen-induction confirmed the marked expansion in YFP+ cells in mice containing the Ctnnb1lox(ex3) allele (P<0.0001, Wilcoxon test, Figure 3b). Remarkably, cultures of these cells yielded four times as many, and much larger, YFP+ clonogenic colonies when isolated from Prom1+/C-L ; RosaYFP ; Ctnnb1+/ lox(ex3) mice than those from control animals (P<0.0001, Wilcoxon test, Figure 3c and d). Thus, activation of Wnt signalling in small intestinal Prom1+ stem cells induces first a marked expansion of these cells in the crypt base.
Figure 3. Tumorigenesis in the mouse small intestine is initiated in Prom1+ crypt stem cells.
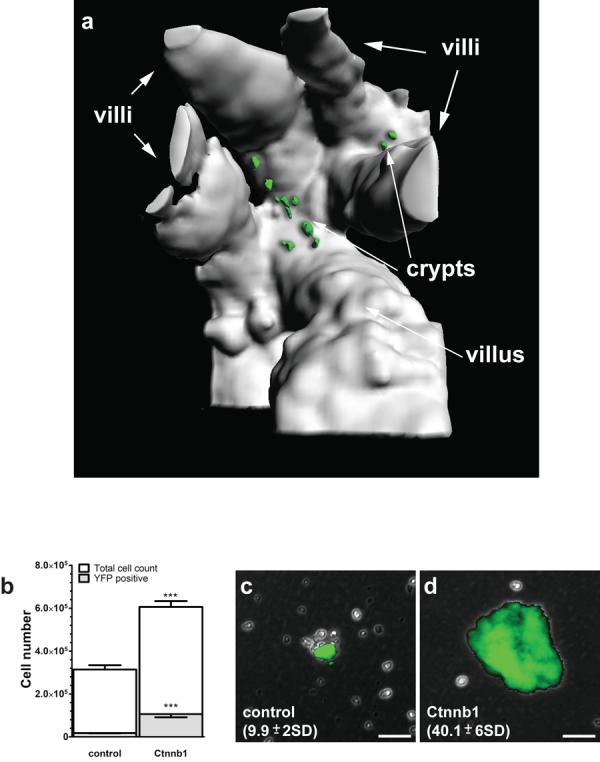
(a) View (394μm depth) into the base of a small intestinal crypt in a Prom1+/C-L ; RosaYFP ; Ctnnb1+/lox(ex3) mouse, two days following tamoxifen-induction. Mice had 8.4 YFP+ cells per crypt (range 2 to 16 >210 crypts counted). (b) Graph reporting the mean (±s.e.m., n=3) number of total and YFP+ cells isolated 2 days following tamoxifen induction, from Prom1+/C-L ; RosaYFP ; Ctnnb1+/+ (control) and Prom1+/C-L ; RosaYFP ; Ctnnb1+/lox(ex3) (Ctnnb1) small intestinal mucosae (***=P<0.0001, Wilcoxon test). Colonies formed following four weeks in stem cell culture medium by single cells isolated 2 days post tamoxifen induction from Prom1+/C-L ; RosaYFP ; Ctnnb1+/+ (c) and Prom1+/C-L ; RosaYFP ; Ctnnb1+/lox(ex3) (d) small intestinal mucosae, numbers=total colonies per small intestinal culture (±s.e.m., n=10, P<0.0001, Wilcoxon test). Scale bars=50μm.
Ten days following tamoxifen-induction, Prom1+/C-L ; RosaYFP ; Ctnnb1+/lox(ex3) crypts were markedly disorganized and contained cells that were uniformly nuclear Ctnnb1 immunoreactive, highly proliferative and disproportionately Prom1+ (nLacZ immunopositive) relative to controls (Figure 4a and b and Supplemental Figure 6a to c). Contiguous streams of YFP+ cells were now seen emanating from the crypt and migrating up the surface of the villi (Figure 4c). In contrast to Prom1+/C-L ; RosaYFP ; Ctnnb1+/+ control mice, cells emanating from the crypts of Prom1+/C-L ; RosaYFP ; Ctnnb1+/lox(ex3) mice were both hyperplastic and grossly dysplastic, forming a carpet of contiguous YFP+ cells from the base of the crypt to the tip of the villus (compare Figures 2g, and 4c). All Prom1+/C-L ; RosaYFP ; Ctnnb1+/lox(ex3) mice allowed to age following tamoxifen induction succumbed to their disease within 90 days (n=15/15). Sixty days following tamoxifen induction, the small intestine of Prom1+/C-L ; RosaYFP ; Ctnnb1+/lox(ex3) mice was twice the width of that of control animals (Supplemental Figure 6d). Gross inspection of the duodenal, jejunal and ileal lining revealed a thickened, rugous and YFP-fluorescent mucosa; no regions of normal tissue were identified (Supplemental Figure 6e and f). Microscopic analysis revealed loss of the normal villus architecture across the entire small intestine that was replaced by dysplastic tissue, characterized by focal high-grade intraepithelial neoplasia and crypt adenoma formation (Figure 4d and Supplemental Figure 6f). In contrast, no gross or microscopic abnormalities were seen in the colon which lacks Prom1+ stem cells (Supplemental Figure 6g). The absence of tumors in the colon in which Ctnnb1 was activated in Prom1+ differentiated cells, points to progenitor cells as the source of tumors in this tissue. All cells in the neoplastic small intestine of Prom1+/C-L ; RosaYFP ; Ctnnb1+/lox(ex3) mice expressed YFP and nuclear Ctnnb1 and therefore arose from within the Prom1-lineage; however, only 7%±15.5 (n=36/502 YFP+ cells counted from five separate regions) of tumor cells retained expression of Prom1 (Figure 4 e to g). Dual Ki67 and nLacZ co-immunoflourescence analysis of tumors demonstrated that approximately 10% of Prom1+ tumor cells were proliferating (Figure 4h). These data are compatible with the hypothesis that tumors in these animals contain a fraction of transformed stem cells.
Figure 4. Ctnnb1-mutant Prom1+ crypt stem cells produce diffuse small intestinal tumors in adult Prom1+/C-L ; RosaYFP ; Ctnnb1+/lox(ex3) mice.
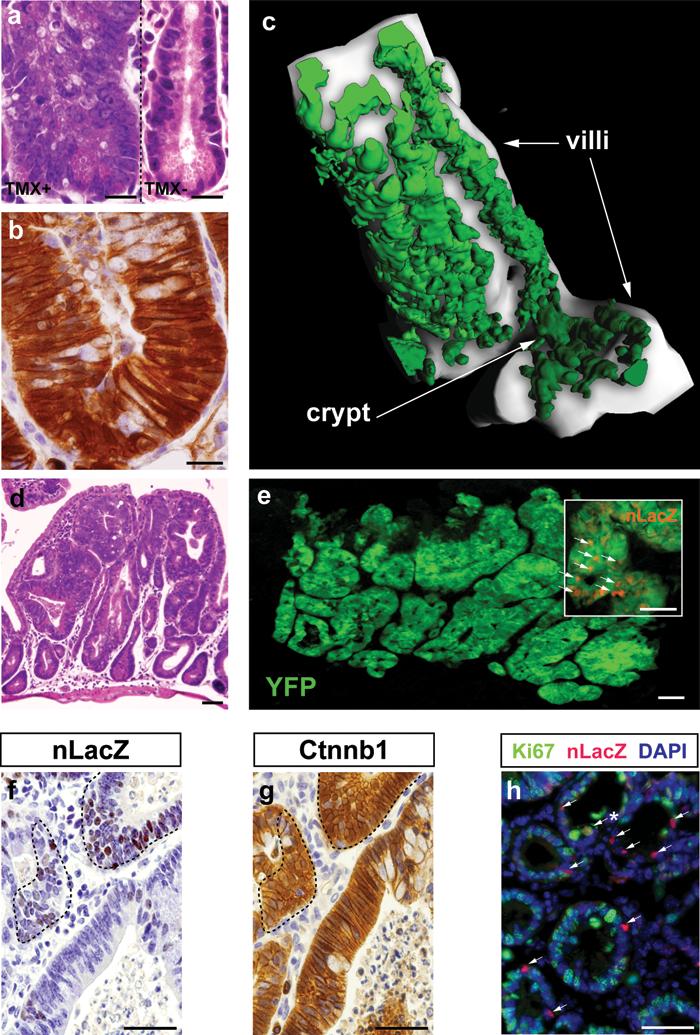
(a) Hematoxylin and eosin (H & E) staining of a dysplastic small intestinal crypt 10 days following tamoxifen-induction (left). Normal crypt in a no tamoxifen control mouse, right. (b) Ctnnb1 immunostaining of crypt cells 10 days following tamoxifen-induction. Scale bars=10μm. (c) Two-photon laser-scanning microscopy (578μm depth; YFP, green) of cells emerging from a small intestinal crypt, 10 days following tamoxifen-induction. (d) H & E stained section of tumorous small intestine 60 days following tamoxifen-induction. (e) YFP (green), nLacZ (red) co-immunofluorscence of neoplastic small intestine. Inset, arrows, nLacZ+ nuclei. nLacZ (f) and Ctnnb1 (g) immunostaining of adjacent sections of tumor. Note, Prom1+ (nLacZ+) cells enclosed by broken lines express nuclear Ctnnb1. (h) Ki67 (green) and nLacZ (red) co-immunofluorscence of tumor. Note the small fraction of proliferating (co-express Ki67+, arrow*) Prom1+ tumor cells (arrows). (d) to (h) scale bars=50μm.
Our data demonstrate that Prom1 marks stem cells in the small intestine and that activation of endogenous Wnt signaling in these cells disrupts normal tissue maintenance that begins first in the crypt, expanding aberrantly the Prom1+ stem cell population, resulting ultimately in neoplastic transformation of the small intestinal mucosa. Therefore, we show for the first time that Prom1 marks an adult solid tissue stem cell that is susceptible to neoplastic transformation, forming a model of a human tumor that contains Prom1+ CSC. Further analysis will determine if the fraction of Prom1+ cells observed in the neoplastic small intestine of Prom1+/C-L ; RosaYFP ; Ctnnb1+/lox(ex3) mice represents a remnant of mutated stem cells that may function as CSC. Our Prom1C-L knock-in provides the research community with an extremely useful tool to explore further the relationship between normal and malignant stem cells in the lung, kidney, brain, pancreas and other tissues.
Methods Summary
Mice
Prom1C-L mice were generated by homologous recombination in 129SvEv mouse embryonic stem cells targeting a CreERT2-IRES-nLacZ-PGK-Neo cassette to the ATG of Prom1. PGK-Neo flanked by flip-recombinase target (FRT) sites was subsequently excised by flip-recombination by breeding with Rosa26-FLPe mice. Rosa26YFP reporter and Rosa26FLPe mice were obtained from the Jackson Laboratory. Ctnnb1lox(ex3)/lox(ex3) mice were kindly provided by Dr. Makoto29. Mice aged 2 months were treated by oral gavage with 250 μl tamoxifen in corn oil at 40 mg/ml for two consecutive days.
Histology and microscopy
For nLacZ analysis, YFP microscopy and immunofluoresence, tissues were perfused and fixed in 2% paraformaldehyde (PFA) overnight, cryo-protected and frozen in OCT. For immunohistochemistry, tissues were fixed in 4% PFA and paraffin embedded. In situ probes for Lgr5 and Prom1 were generated from cDNA clones and BC156649 and BC028286, respectively. Alcian blue and PAS staining were performed on frozen sections following standard protocols. See supplemental methods for details.
Explants and two-photon microscopy
Intestine from tamoxifen treated mice was removed surgically and immediately submerged in Kreb's solution and superfused with a gas mixture of 95% O2/CO2. One millimetre slices were placed on glass coverslips and stabilized with overlying mechanical support and maintained at 37°C within the microscope chamber. Two-photon laser-scanning microscopy was performed using an Ultima imaging system (Prairie Technologies, Middletown, WI), a Ti:sapphire Chameleon Ultra femtosecond-pulsed laser (910 nm) (Coherent, Santa Clara, CA). Images were analyzed using Imaris software (Bitplane, Switzerland). 3-dimensional images were constructed from Z-series stacks taken in 1 μm steps.
Culture of intestinal crypt stem cells
Intestinal epithelial cells were grown in RPMI supplemented with 2mM L-glutamine, 100 units/ml Penicillin, 100ug/ml Streptomicin and 2.5% FBS. YFP+ cells and colonies (formed after 4 weeks in culture) were visualized and counted using fluorescence microscopy. See supplemental methods for details.
Methods
Generation of Prom1+/C-L mice
The 8.0 kb 5′ end of the mouse Prom1 gene (4.8 kb upstream of the ATG start codon and 3.2 kb downstream of ATG) was cloned into pBluescript KS+ plasmid by recombineering from the Ensembl 129S7-derived genomic BAC clone bMQ446F18. The ATG start codon was then replaced by the CreERT2-IRES-LacZ-PgkNeo cassette using the same recombineering-base cloning strategy. The Prom1-CreERT2-IRES-LacZ expression construct (25μg) was linearized and transfected into 129SvEv mouse embryonic stem cells (Millipore, CMTI-1). Neomycin-resistant recombinant ES clones were selected in medium containing 350 μg/ml G418 for day 2-3 and 150 μg/ml for day 4-7. 192 recombinant ES clones were picked into two 96-well plates and cultured to 70% confluence. Plates were then duplicated, one cryo-preserved and one continued cultured to 100% confluence for DNA isolation. DNA from all clones was screened for the correct targeting by long PCR (Platinum® Taq polymerase high fidelity, Invitogen, 11304-011) with primers flanking the 5′ and 3′ homology arms. All positive clones and randomly selected negative clones were then confirmed by Southern blotting using the 5′ probe indicated in Figure 1a. Positive clones were thawed, expanded and injected into C57BL/6 blastocysts following the standard protocol. Chimaeras were mated with C57BL/6 mice and the germline transmission in agouti offspring were confirmed by genotyping PCR. The neomycin selection cassette was later excised in vivo by crossing with Rosa26-FLPe mice (The Jackson Laboratory).
β-galactosidase (β-gal) staining
To prepare sections of all organs except the retina, mice were perfused and fixed with 2% paraformaldehyde (PFA) overnight at 4°C, cryo-protected in 20 % sucrose overnight at 4°C and frozen in OCT. Frozen sections were washed in PBS for 5 min and incubated in the dark in standard β-gal substrate (5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 1 mg/ml X-gal, 2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% NP-40 in PBS) overnight at 37°C. To stain the retina, mouse eyes were removed and immersed immediately in cold PBS. The retina was isolated from the eyes and fixed at room temperature in 0.5% gluteraldhyde/PBS for 10 minutes, washed briefly and incubated in the rinse buffer (2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% NP-40 in PBS) for 30 minutes. The retina was subsequently stained in the same β-gal substrate for 30 minutes at 37°C, rinsed in PBS and post-fixed in 4% PFA overnight. The retina was cryo-protected in 20 % sucrose for two hours before frozen in OCT.
Microscopy
Standard immunofluorescence (IF) staining was performed on 16 μm frozen sections and immunohistochemistry on 5 μm paraffin sections. Antibodies included rat anti-Prom1 (eBioscience, 14-1331, 1:100), mouse anti-nestin (Millipore, MAB353, 1:50), rabbit anti-uteroglobin (or CCSP) (Abcam, ab40873, 1:1000), mouse anti-rhodopsin (gift from Dr. Michael Dyer, ref. 30, 1:500), rabbit anti-Ki67 (Vector, VP-RM04, 1:2000), mouse anti-β-catenin (BD, 610154, 1:200), chicken anti-β-gal (Abcam, ab9361, 1:1000), rabbit anti-β-gal (MP, 55976, 1:1000), rabbit anti-synaptophysin (Dako, A 0010, 1:150), rabbit anti-amylase (Sigma, A8273, 1:1000), guinea pig anti-insulin (Dako, A 0564, 1:250). Two lectins: fluorescein-labeled Dolichos Biflorus Agglutinin (DBA) (Vector, FL-1031, 1:200), fluorescein-labeled Lotus Tetragonolobus Lectin (LTL) (Vector, FL-1321, 1:200). To obtain the kidney images in Figure 1c, the same section was sequentially stained with X-gal followed by Prom1/LTL co-IF. The same sequential X-gal/IF staining was performed for Supplemental Figure 2b. All cell counts in tissue sections were performed blind to genotype and tamoxifen treatment.
In situ hybridization (ISH)
To obtain Lgr5 probe, T3 promoter sequence (CATTCACTTTTGGCCGTTTT) was added to Lgr5 sense oligo and T7 promoter sequence (AAGTCATGGGGTAAGCTGGTG) to the antisense oligo. Lgr5 template was amplified from its cDNA clone (BC156649) and the 828 bp PCR product was used as the template to generate Dig-cRNA probe (Roche). Probes to the mouse Prom1 mRNA was similarly generated from its cDNA clone (BC028286). 4% PFA fixed frozen sections were pre-treated in graded alcohols from 70% to 100% to facilitate penetration of Dig-labeled cRNA probes. Sections were then incubated in the hybridization buffer (500 ng/ml Dig-labeled cRNA probe, 50% formaldehyde, 4X SSC, 250 μg/ml tRNA, 200 μg/ml ssDNA and 10% dextran sulfate) at 55°C overnight. Following incubation sections were washed in degraded SSC buffers and incubated with anti-Dig antibody (Roche, 1:600) for 1.5 hours at room temperature. Sections were subsequently washed and colorized with NCBI/NBT (Roche) for 3-16 hours, counterstained with methyl green and mounted with 2% gelatin.
Culture of intestinal crypt stem cells
Small intestines were excised from each of two Prom1+/C-L; RosaYFP and two Prom1+/C-L ; RosaYFP ; Ctnnb1+/lox(ex3) mice induced with tamoxifen 2 days prior to sacrifice, and rinsed repeatedly with sterile PBS. Crypts were isolated by incubating minced intestines with 3mM EDTA and 0.5 mM dithiothreitol in PBS without Ca2+ and Mg2+ for 90 minutes at room temperature followed by vigorous shaking for 30 seconds to release large sheets of cells from their calcium- and magnesium-dependent interactions with the basement membrane and stromal cells. These were collected by centrifugation at 500 rpm for 5 minutes at room temperature. To obtain a single-cell suspension, pelleted cells were incubated in 0.3% pancreatin (Sigma) in PBS for 90 min. at room temperature. Cells were collected by centrifugation at 1000 rpm for 5 minutes at room temperature, washed with 3 mM EDTA and 0.5 mM dithiothreitol in PBS without Ca2+ and Mg2+ and grown in RPMI supplemented with 2 mM L-glutamine, 100 units/ml Penicillin, 100 μg/ml Streptomicin and 2.5% FBS. Colonies formed after approximately 4 weeks in culture.
Reverse transcription PCR
Total RNA was isolated from the kidney of adult Prom1+/C-L and Prom1C-L/C-L mice following standard Trizol/chloroform/isopropanol protocol. 1 μg RNA was reverse transcribed using the iScript™ reverse transcription kit (Bio-Rad, 170-8890). cDNA was then amplified using Taq polymerase and gene specific primers for the 5′end of the Prom1 transcript (5′- GAGTCCTTATCTGCGCCATC -3′, 5′- CTGCTCCCCAGACTGCTTAG-3′), 3′end of the Prom1 transcript (5′-GGATTGCAAAAGAGGTCGAG-3′, 5′-AATTCAGAGGGTCCGCAAC-3′), and GAPDH (5′-TGTTCCAGTATGACTCCACTCACG-3′, 5′-GCCCTTCCACAATGCCAAAG-3′). Target sequences were amplified within linear ranges using 95°C/30 seconds, 60°C/30 seconds and 72°C/50 seconds conditions. All primer pairs amplified regions crossing intron borders.
Supplementary Material
Acknowledgements
RJG holds the Sydney Schlobohm Leadership Chair of Research from the Brain Tumor Society and the Howard C. Schott Research Chair from the Malia's Cord Foundation, and is supported by grants from the National Institutes of Health (R01CA129541, P01CA96832 and P30CA021765), the Collaborative Ependymoma Research Network (CERN) and by the American Lebanese Syrian Associated Charities (ALSAC). We are grateful to the staff of the ARC for excellent technical assistance.
Footnotes
Supplementary Information
Supplemental Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124:1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 3.Ricci-Vitiani L, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 4.O'Brien CA, et al. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 5.Sparks AB, et al. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res. 1998;58:1130–1134. [PubMed] [Google Scholar]
- 6.Lapidot T, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 7.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 8.Al-Hajj M, et al. From the Cover: Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calabrese C, et al. A Perivascular Niche for Brain Tumor Stem Cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 10.Hambardzumyan D, et al. PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes Dev. 2008;22:436–448. doi: 10.1101/gad.1627008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke MF, et al. Cancer Stem Cells--Perspectives on Current Status and Future Directions: AACR Workshop on Cancer Stem Cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 12.Kelly PN, et al. Tumor growth need not be driven by rare cancer stem cells. Science. 2007;317:337. doi: 10.1126/science.1142596. [DOI] [PubMed] [Google Scholar]
- 13.Yin AH, et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002–5012. [PubMed] [Google Scholar]
- 14.Miraglia S, et al. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood. 1997;90:5013–5021. [PubMed] [Google Scholar]
- 15.Mizrak D, Brittan M, Alison MR. CD133: molecule of the moment. J Pathol. 2008;214:3–9. doi: 10.1002/path.2283. [DOI] [PubMed] [Google Scholar]
- 16.Taylor MD, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8:323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Hermann PC, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Corbeil D, et al. The human AC133 hematopoietic stem cell antigen is also expressed in epithelial cells and targeted to plasma membrane protrusions. J Biol Chem. 2000;275:5512–5520. doi: 10.1074/jbc.275.8.5512. [DOI] [PubMed] [Google Scholar]
- 19.Kim CF, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 20.Xu X, et al. Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell. 2008;132:197–207. doi: 10.1016/j.cell.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 21.Lois C, Alvarez-Buylla A. Proliferating subventricular zone cells in the adult mammalian forebrain can differentiate into neurons and glia. Proc Natl Acad Sci U S A. 1993;90:2074–2977. doi: 10.1073/pnas.90.5.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uchida N, et al. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci U S A. 2000;97:14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shmelkov SV, et al. CD133 expression is not restricted to stem cells, and both CD133 and CD133 metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;22:22–30. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 25.Taketo MM. Wnt signaling and gastrointestinal tumorigenesis in mouse models. Oncogene. 2006;25:7522–7530. doi: 10.1038/sj.onc.1210058. [DOI] [PubMed] [Google Scholar]
- 26.Gaspar C, et al. Cross-species comparison of human and mouse intestinal polyps reveals conserved mechanisms in adenomatous polyposis coli (APC)-driven tumorigenesis. Am J Pathol. 2008;172:1363–1368. doi: 10.2353/ajpath.2008.070851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 28.Su LK, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 29.Harada N, et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. Embo J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, et al. Rb regulates proliferation and rod photoreceptor development in the mouse retina. Nat Genet. 2004;36:351–60. doi: 10.1038/ng1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


