Abstract
Protein homeostasis is critical for cellular survival and its dysregulation has been implicated in Alzheimer's disease (AD) and other neurodegenerative disorders. Despite the growing appreciation of the pathogenic mechanisms involved in familial forms of AD, much less is known about the sporadic cases. Aggregates found in both familial and sporadic AD often include proteins other than those typically associated with the disease. One such protein is a mutant form of ubiquitin, UBB+1, a frameshift product generated by molecular misreading of a wild-type ubiquitin gene. UBB+1 has been associated with multiple disorders. UBB+1 cannot function as a ubiquitin molecule, and it is itself a substrate for degradation by the ubiquitin/proteasome system (UPS). Accumulation of UBB+1 impairs the proteasome system and enhances toxic protein aggregation, ultimately resulting in cell death. Here, we describe a novel model system to investigate how UBB+1 impairs UPS function and whether it plays a causal role in protein aggregation. We expressed a protein analogous to UBB+1 in yeast (Ubext) and demonstrated that it caused UPS impairment. Blocking ubiquitination of Ubext or weakening its interactions with other ubiquitin-processing proteins reduced the UPS impairment. Expression of Ubext altered the conjugation of wild-type ubiquitin to a UPS substrate. The expression of Ubext markedly enhanced cellular susceptibility to toxic protein aggregates but, surprisingly, did not induce or alter nontoxic protein aggregates in yeast. Taken together, these results suggest that Ubext interacts with more than one protein to elicit impairment of the UPS and affect protein aggregate toxicity. Furthermore, we suggest a model whereby chronic UPS impairment could inflict deleterious consequences on proper protein aggregate sequestration.
Author Summary
The accumulation of cytotoxic protein aggregates occurs in many neurodegenerative diseases. It is difficult to determine if the protein aggregates found in these diseases represent a cause or consequence of the disorder. Degradation pathways, such as the ubiquitin/proteasome system (UPS), remove misfolded proteins that are prone to aggregate. The UPS involves many players that work in concert to target proteins for degradation by the proteasome. A mutant form of ubiquitin has been associated with many diseases, including Alzheimer's disease. We developed a yeast model of the mutant ubiquitin protein in order to investigate its effect on UPS function and protein aggregation. We demonstrate that this mutant ubiquitin causes impairment of the UPS and suggest that it does so by interacting with multiple components of the pathway. Using this model, we evaluated the effects of the mutant ubiquitin on nontoxic protein aggregates and found that they were unaltered by its presence. We demonstrate that the mutant ubiquitin acts as a modifier, which increases cellular susceptibility to the phenotypic effects of deleterious protein aggregates by altering UPS functionality and substrate ubiquitination. Furthermore, the system we developed can be utilized to further understand the complex interplay of proteasomal impairment and protein aggregate toxicity.
Introduction
As technology and medicine further extend the human lifespan, age-related diseases will become more prevalent. Alzheimer's disease (AD) is a neurodegenerative disorder that affects 20 million people worldwide and is the most common form of late-onset dementia [1]. The study of genetic mutations that cause early onset AD has provided insight into some of the factors involved, but most cases of AD are sporadic and of unknown origin. Uncovering the risk factors involved in any multi-factorial disease is challenging but vital for disease treatment and prevention. Many fundamental pathways, including the ubiquitin proteasome system (UPS), have been suggested to play a role in AD. Therefore, investigating the relationship between AD and the UPS could lead to new therapeutic targets.
The UPS is an evolutionarily conserved pathway that selectively eliminates short-lived and damaged proteins. A number of cellular processes, including the cell cycle, stress response, and DNA repair, require the UPS [2]. Protein degradation by the UPS involves a series of enzymes that ultimately attach ubiquitin, a small well-conserved protein, to an internal lysine residue in the target protein [3]–[5]. Multiple ubiquitin proteins can be connected to form a polyubiquitin chain which serves as a degradation signal recognized by the 26S proteasome. A series of events involving E1, E2 and E3 enzymes are required to attach ubiquitin via its C-terminal glycine residue to the target protein. The formation of polyubiquitin chains and the process of ubiquitin conjugation to protein targets displays exquisite specificity, in part by the multitude of E2 and E3 enzymes. Despite intensive study, the roles of many components of the UPS remain to be elucidated.
The importance of the UPS in cellular homeostasis is apparent not only by the redundancy and conservation of the components, but also by its role in disease [5],[6]. The complex interplay between protein aggregation and UPS function is easily appreciated, yet it is often difficult to determine the causal nature of the problem. UPS dysfunction can prevent the degradation of misfolded proteins, which can lead to aggregation. Conversely, protein aggregates can be challenging substrates for the UPS and can thus cause proteasomal impairment [7]. Protein aggregation is a hallmark of many neurodegenerative disorders [6]. In addition, mutations in ubiquitin processing enzymes, such as UCHL1 and Parkin, can lead to inherited forms of neurodegenerative diseases [8],[9]. Furthermore, many protein aggregates associated with disease show ubiquitin deposition [10], suggesting that dysfunctional UPS activity may contribute to pathogenesis. Understanding the interplay between protein aggregation and clearance is an active area of research, but most systems are complicated by cellular toxicity, which alone can have negative consequences on protein homeostasis.
A mutant form of ubiquitin was found associated with AD and other diseases and was proposed to act as a natural proteasome inhibitor [11]. The generation of this mutant ubiquitin protein is unusual - the mutation is found in the messenger RNA, but not in the DNA sequence of the ubiquitin-B gene. The mutant ubiquitin results from a dinucleotide deletion near the 3′ end of the mRNA transcript which shifts the reading frame for translation. The mutant protein has been named UBB+1 [12]. The dinucleotide deletion event in the mRNA has been termed “molecular misreading”, though the mechanism by which the deletion occurs remains elusive [13],[14]. Many human mRNA transcripts, including all copies of ubiquitin, contain potential sites for molecular misreading, since hotspots for these events are hypothesized to occur near simple repeat sequences (e.g. GAGAG) [15]. The best characterized +1 mutant ubiquitin protein has a short C-terminal extension, with the majority of the protein being identical to ubiquitin [12]. As such, the protein is presumably folded and recognized as ubiquitin, but the C-terminal glycine residue essential for conjugation to substrates is absent.
The accumulation of the UBB+1 protein in the neurological hallmarks of AD is curious, since the mutant cannot be conjugated to target proteins [12]. The presence of UBB+1 has been proposed to represent an endogenous readout of proteasomal dysfunction [16],[17]. Due to its association with protein aggregation, it was also suggested that UBB+1 could contribute to disease pathology [18]. UBB+1 protein accumulation has been documented in multiple disorders such as polyglutamine expansion diseases (including Huntington's disease), Pick's disease and even non-neuronal tissue diseases [11],[19]. However, the mechanism of UBB+1 action in these diseases remains unclear.
To evaluate the role of UBB+1 in disease, the effects of ectopic UBB+1 expression have been investigated in cultured mammalian cells. Although UBB+1 cannot be conjugated to target substrates, it can be ubiquitinated by wild type ubiquitin and degraded by the proteasome [20]. However, high levels of UBB+1 expression cause proteasomal impairment [16],[21],[22]. As a natural inhibitor of the UPS, UBB+1 could be another example whereby proteasomal impairment induces protein aggregation. Therefore, UBB+1 might act as a disease modifier. Recently, a UBB+1 transgenic mouse has been characterized [23]. UBB+1 expression resulted in constant UPS impairment that caused a minor learning deficit and caused changes in transcription profiles that mirror those found in brains of humans with AD [23]. The expression of UBB+1 in mammalian cells enhances the toxicity and aggregation of an expanded polyglutamine protein [24]. However, measuring changes in protein aggregation in cells that are dying from toxic protein aggregates is challenging. Hence, it remains to be determined if UBB+1 affects protein aggregation per se, or if it affects the ability of the cells to cope with the aggregates.
We developed a model system using Saccharomyces cerevisiae to evaluate the cellular effects of UBB+1. We expressed a mutant ubiquitin protein (Ubext) analogous to UBB+1 and found that it caused UPS impairment in yeast. Furthermore, we found that Ubext changed the ubiquitination pattern on a UPS substrate. Taking advantage of non-toxic protein aggregates in yeast, we demonstrated that the expression of Ubext neither induced nor changed these aggregates. However, Ubext did make cells more susceptible to toxic protein aggregates. We propose that Ubext does not cause protein aggregation, but rather acts as a phenotypic enhancer of deleterious aggregation. We present a model based on our work and other recent advances in the field to explain how this might occur.
Results
Ubext Expression in Yeast Cannot Functionally Rescue a Decrease in Wild Type Ubiquitin
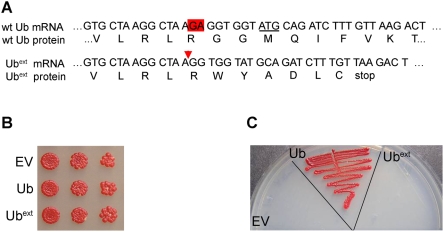
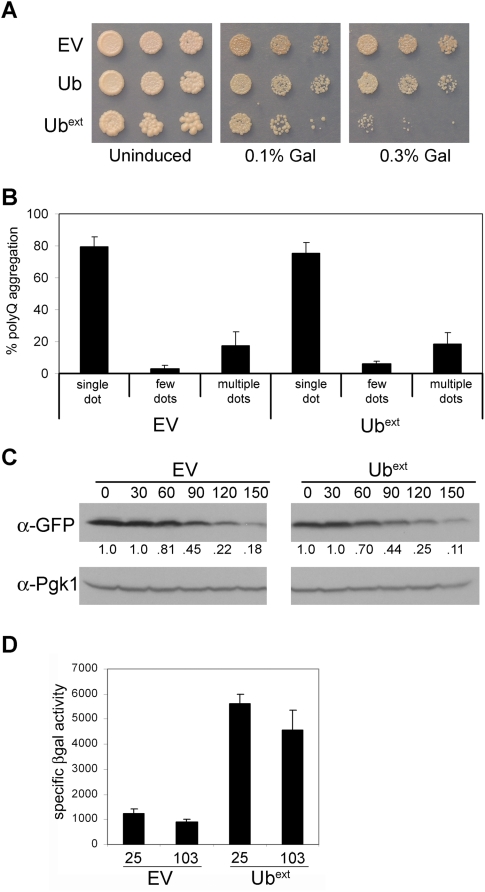
The mechanism by which +1 proteins, such as UBB+1, are produced is currently unknown. To create a yeast model of UBB+1, we generated an expression vector containing the sequence of the first ubiquitin-coding region of the yeast tandem ubiquitin gene, UBI4, such that a dinucleotide deletion occurred near the carboxy terminus (Figure 1A). The deletion caused a frameshift in the coding sequence of ubiquitin and extended the open reading frame to the next stop codon (termed extended ubiquitin or Ubext). This construct mimics the generation of UBB+1 from the human tandem ubiquitin gene (ubiquitin-B). Constitutive expression of Ubext in log-phase yeast did not cause a growth defect when assessed in either liquid medium (data not shown) or on solid medium (Figure 1B). Wild type cells expressing Ubext did show a reduced growth rate after recovery from stationary phase (data not shown).
Figure 1. Ubext does not function as ubiquitin.
(A) A schematic diagram depicting the wild type and mutant ubiquitin (Ubext) mRNA and protein sequences beginning at nucleotide 207 of UBI4. The underlined ATG denotes the beginning of the next ubiquitin open reading frame in the tandem array. The red triangle signifies the site of the dinucleotide GA deletion. (B) Ubext expression does not affect logarithmically growing yeast. Serial dilutions of wild type yeast ectopically expressing Ubext, excess wild type ubiquitin (Ub), or an empty vector (EV) control were spotted onto selective medium. (C) Ubext does not behave as wild type ubiquitin and cannot compensate for the loss of UBI4. The Δubi4 strain was transformed with EV, Ub, and Ubext. Transformants were plated onto selective medium following growth into stationary phase.
To evaluate the functionality of Ubext, we analyzed its ability to replace wild type ubiquitin. The stress-inducible UBI4 gene encodes a tandem array of five ubiquitin moieties that are separated post-translationally by deubiquitinating enzymes (DUBs) that cleave after the C-terminal glycine residue, G76 [25]. UBI4 is non-essential in vegetatively growing cells but is required for cells to recover from various stress conditions [26],[27]. We utilized a strain lacking UBI4 to evaluate the functionality of Ubext. Δubi4 cells were transformed with expression plasmids that contain wild type ubiquitin, Ubext or empty vector. The transformants were grown for two weeks to allow them to reach stationary phase and then plated again to evaluate their ability to recover. Only cells expressing extra wild type ubiquitin were rescued from the loss of UBI4 and could grow after this stress (Figure 1C). This demonstrates that Ubext is a non-functional ubiquitin, as expected due to the lack of the C-terminal glycine residue required for conjugation to target substrates.
Ubext Expression Causes UPS Impairment
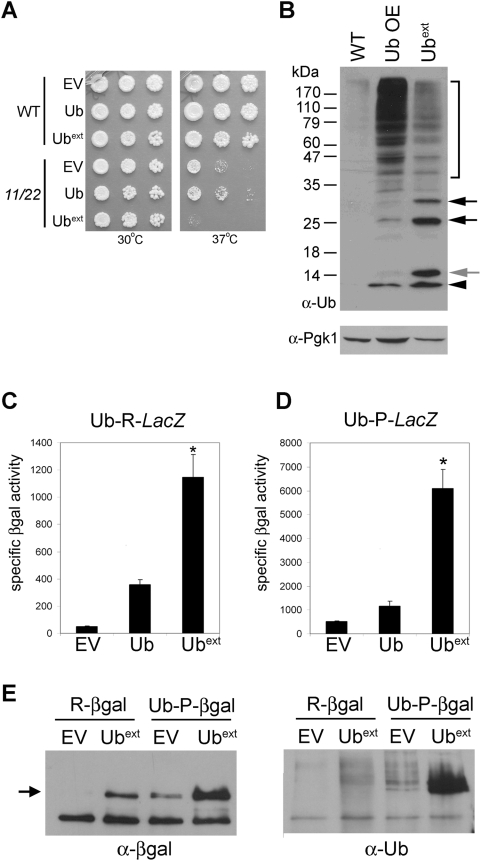
If Ubext affects UPS functionality in yeast as UBB+1 does in mammals, then we hypothesized that Ubext would display synthetic lethality with a proteasome mutant. We evaluated the cellular viability of a temperature-sensitive catalytic proteasome mutant strain (pre1-1 pre2-2) [28] expressing Ubext. As predicted, Ubext-expressing pre1-1 pre2-2 cells were inviable at the restrictive temperature (Figure 2A). Wild type cells expressing Ubext grown at the restrictive temperature did not show a growth defect (Figure 2A). Next we evaluated another ubiquitination-dependent process to determine if Ubext effects are more widespread. We challenged Ubext-expressing cells to DNA damage induced by UV irradiation and found that they survived as well as the control cells (data not shown).
Figure 2. Expression of Ubext causes proteasomal impairment.
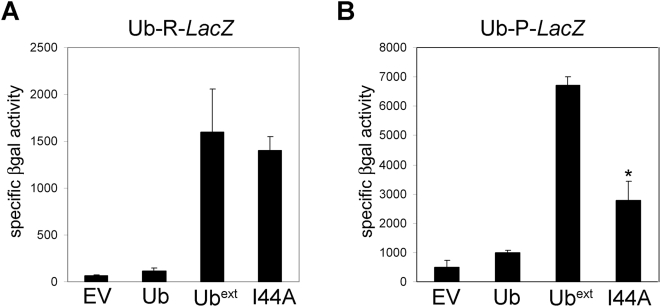
(A) Ubext displays synthetic lethality with proteasome mutants. Wild type (WT) and temperature-sensitive proteasome mutant cells, pre1-1 pre2-2 (11/22), were transformed with plasmids containing empty vector (EV), ubiquitin (Ub) and Ubext. Serial dilutions of cells were spotted onto selective medium and grown at 30°C and 37°C. (B) Cells expressing Ubext show a distinct pattern of ubiquitin conjugation. Protein lysate from wild type yeast cells containing an empty vector (WT), extra ubiquitin (Ub OE), or Ubext were analyzed by SDS-PAGE and western blot using an anti-ubiquitin antibody. Ubext causes an increase in ubiquitin-conjugated proteins (bracket) as compared to WT. The black arrowhead indicates ubiquitin monomer. The grey arrow points to Ubext. Black arrows represent conjugated Ubext. Pgk1p expression was probed to assess protein loading on the membrane. (C) Ubext-expressing cells impair the degradation of the N-end rule substrate R-β-galactosidase (βgal). Cells containing EV, Ub, or Ubext were transformed with pGal-Ub-R-LacZ. The stability of R-βgal was measured by specific activity (luminescence units/µg protein). The asterisk (*) indicates statistical significance between wild type Ub and Ubext (p = 0.0013). (D) Ubext-expression prevents the efficient proteasomal degradation of a ubiquitin fusion degradation substrate. The stability of Ub-P-LacZ was evaluated as in B. The asterisk (*) indicates statistical significance between wild type Ub and Ubext (p = 0.0005). (E) Ubiquitinated reporter substrates are present in Ubext-expressing cells. Wild type cells containing the Ub-X-LacZ reporter constructs and expressing Ubext or the control (EV) were analyzed for ubiquitinated βgal protein. βgal protein was immunoprecipitated with an anti-βgal antibody (left) and the bound fractions were blotted with an anti-ubiquitin antibody (right). The arrow indicates full length βgal protein.
Ubext cannot be conjugated to target protein substrates, but can be recognized as a UPS substrate. Therefore, we assessed its ubiquitination. Protein lysate from Ubext-expressing cells and control cells were evaluated by SDS-PAGE and western blot. Cells expressing Ubext exhibited a unique band which represents the extended mutant ubiquitin protein (Figure 2B, grey arrow) which is larger than wild type ubiquitin (Figure 2B, arrowhead). Cells expressing Ubext also displayed a distinctive laddering pattern which suggests that Ubext is conjugated by wild type ubiquitin moieties (Figure 2B, black arrows). A similar laddering pattern was previously observed in cells expressing UbΔGG [29], a mutant ubiquitin protein lacking only the two C-terminal glycine residues, and we observed the same pattern when we expressed UbΔGG in yeast (data not shown). Additionally, a strain lacking the ubiquitin recycling DUB (Δubp14) accumulates free ubiquitin chains [29] and we also observed that Δubp14 cells show the same ubiquitin laddering pattern as cells expressing Ubext (data not shown).
The expression of Ubext also caused an increase in the level of unconjugated wild type ubiquitin, which was evident by the accumulation of the mono-ubiquitin band in the Ubext lane in comparison to the empty vector control lane (Figure 2B, black arrowhead). Further analysis by quantitative western blot showed approximately a 10-fold increase in wild type mono-ubiquitin in the presence of Ubext (data not shown). Transcriptional activity from the UBI4 promoter using a UBI4promoter-LacZ reporter in Ubext-expressing cells demonstrated a modest two-fold increase (data not shown), suggesting that UBI4-induced transcription may be one, but perhaps not the only source for the increased ubiquitin. Cells expressing Ubext also displayed an increase in the abundance of high molecular weight ubiquitin-conjugated proteins in comparison to the empty vector control (Figure 2B, compare left lane WT to right lane Ubext ). The fact that Ubext caused lethality in the proteasome mutant strain and Ubext-expressing cells accumulated ubiquitinated-protein conjugates, suggests that it is affecting protein degradation. An accumulation of high molecular weight ubiquitinated proteins also occurred with the over expression of wild type ubiquitin (Figure 2B, middle lane). Most likely this occurs because of more ubiquitination of endogenous proteins due to an excess of functional ubiquitin provided by the over expression construct.
We tested the functionality of the UPS in cells expressing Ubext using two different proteasome reporters constructs: an N-end rule substrate and a ubiquitin fusion degradation (UFD) substrate [30]. These substrates are processed by the UPS using distinct enzymes [3],[31],[32]. The N-end rule substrate is a Ub-R-LacZ fusion. The ubiquitin moiety is efficiently cleaved by endogenous DUBs to expose the N-terminal amino acid (arginine) of β-galactosidase (βgal). According to the N-end rule, R-βgal is an unstable protein that is polyubiquitinated and rapidly degraded by the 26S proteasome [33]. The UFD reporter substrate is Ub-P-LacZ. In yeast, no DUB can cleave ubiquitin from βgal if the first amino acid after ubiquitin is proline. Because of the ubiquitin fusion, Ub-P-βgal is unstable and is rapidly degraded by the proteasome. These constructs, along with a stable LacZ control (Ub-M-LacZ), were transformed into cells expressing Ubext to assess UPS function by βgal activity assays. Cells expressing Ubext and either of the unstable proteasome reporters displayed higher levels of specific βgal activity (Figure 2C and 2D). Cells expressing extra wild type ubiquitin showed a slight increase in the stabilization of the reporter constructs. The expression of extra wild type ubiquitin also generated a large steady state population of ubiquitin-conjugated proteins (Figure 2B, middle lane), which could be taxing the degradation capacity of the proteasome. To evaluate if LacZ fusion expression was affected by Ubext, stable M-βgal activity was measured and showed no difference (data not shown). These results demonstrate that the expression of Ubext in yeast inhibits the degradation of two different UPS reporter substrates.
Such stabilization of the proteasome reporter constructs could be due to a lack of ubiquitination of the reporter, since the expression of Ubext also causes accumulation of unconjugated wild type ubiquitin. The reporter substrates (βgal protein) were immunoprecipitated from cells with and without the co-expression of Ubext. Western blot with an anti-βgal antibody revealed that more β-gal protein was precipitated in Ubext-expressing cells (Figure 2E, left). This result correlates with the higher levels of βgal activity measured in Ubext-expressing cells (Figure 2C and D). Analysis with an anti-ubiquitin antibody showed ubiquitin-conjugated R-βgal and Ub-P-βgal in cells expressing Ubext (Figure 2E, right). This data demonstrates that Ubext is not stabilizing these UPS substrates by blocking their ubiquitination.
Expression of Ubext Does Not Directly Block Proteasome Function
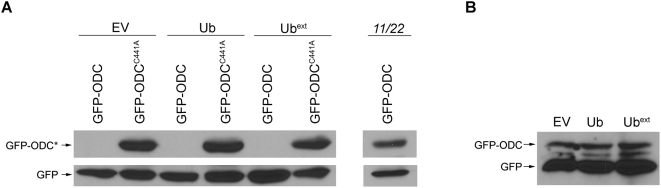
Another plausible explanation for the UPS inhibition could be that Ubext binds to the proteasome and this interaction precludes other proteasome substrates from being efficiently degraded. Alternatively, Ubext could interact with other component(s) of the UPS and inhibit their function. To examine whether Ubext is clogging the proteasome, we took advantage of a ubiquitin-independent proteasome substrate. Ornithine decarboxylase (ODC) is an enzyme involved in polyamine biosynthesis [34],[35] and a short peptide from this protein serves as a ubiquitin-independent degradation signal (i.e. degron) [36]. Measuring the degradation of ODC reflects the functionality of the proteasome in a manner independent of the non-proteasomal components of the UPS cascade. A fusion of GFP with the degron of ODC (GFP-ODC) serves to target GFP to the proteasome where it is rapidly degraded [37]. A point mutation in the ODC degron (C441A) stabilizes the fusion protein by lowering its affinity for the proteasome [38],[39]. GFP-ODC fusions were transformed into cells expressing Ubext and the steady state level of GFP-ODC was evaluated by western blot (Figure 3A). Cells expressing Ubext were able to degrade the GFP-ODC protein while the stable GFP-ODCC441A protein accumulated (Figure 3A). Even prolonged exposure showed that the steady state level of GFP-ODC was approximately equal with or without Ubext expression (Figure 3B). Thus, Ubext permits the degradation of a ubiquitin-independent proteasome substrate, suggesting that the proteasomal degradation capacity is not significantly impaired in cells expressing Ubext.
Figure 3. Ubext-expressing cells can degrade a ubiquitin-independent substrate.
(A) Expression of Ubext does not impair the degradation of GFP-ODC. Cells containing a stable GFP construct were transformed with empty vector (EV), Ub, or Ubext. These cells were then transformed with plasmids expressing either GFP-ODC or GFP-ODCC441A. A proteasome mutant strain (pre1-1 pre2-2, abbreviated 11/22) was transformed with both the GFP and GFP-ODC constructs and as expected, both were stable. Protein lysates were separated by SDS-PAGE and analyzed by western blot using an anti-GFP antibody. GFP-ODC* denotes either GFP-ODC or GFP-ODCC441A. (B) Ubext-expressing cells accumulate approximately an equal amount GFP-ODC in comparison to controls. Protein lysates from cells containing EV, Ub or Ubext co-expressing GFP-ODC were analyzed by SDS-PAGE and western blot with an anti-GFP antibody and visualized after prolonged exposure (1 hour).
Simple Modifications Do Not Alleviate the UPS Impairment Caused by Ubext
We sought to determine how Ubext exerts its negative effects on the UPS pathway. We asked whether Ubext was sequestrating wild type ubiquitin proteins. Ubiquitinated-Ubext could be refractory to DUBs, thereby tying up ubiquitin, as suggested for UBB+1 [20]. To test this hypothesis, we expressed extra ubiquitin in the presence of Ubext and found that the UPS test substrates were still stabilized (data not shown). This result was not surprising since monomeric ubiquitin appears to be abundant in cells expressing Ubext (Figure 2B, arrowhead). This suggests that a lack of wild type ubiquitin is not the cause of the UPS impairment elicited by Ubext.
Ubext lacks the essential C-terminal glycine residues (G75 and G76) required for ubiquitin conjugation and these glycine residues are vital for many proteins to interact with ubiquitin [40]. We tested whether adding back two glycine residues to the C-terminal extension of Ubext (Ubext+GG) could restore these interactions and alleviate the proteasomal impairment. Cells expressing Ubext+GG still displayed proteasomal impairment (data not shown), indicating that the C-terminal extension plays a mechanistic role in the phenotype observed.
Ubext-Ubiquitin Conjugation Is Required for N-End Rule Substrate Stabilization but not for UFD Substrate Stabilization
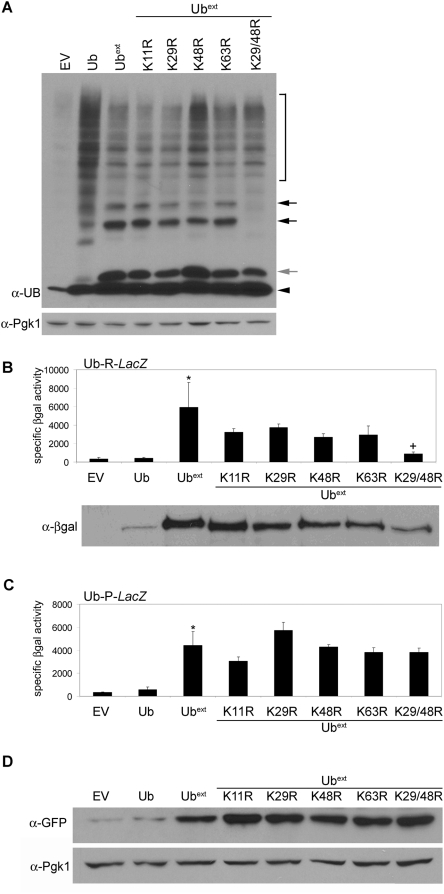
UPS-mediated protein degradation is a selective process and polyubiquitination is the signal which targets proteins to the proteasome for degradation [41],[42]. Therefore, we asked whether blocking the ubiquitination of Ubext would alleviate the associated UPS inhibition. Polyubiquitination can occur on multiple lysine residues of ubiquitin [43]. We mutated four of the lysine residues typically utilized for polyubiquitination by changing them to arginine (referred to as UbextKxR). Ubiquitin conjugation of Ubext was visualized by a distinct laddering pattern on a western blot (Figure 2B, black arrows). While none of the single point mutations prevented ubiquitination of Ubext, the double lysine mutant, UbextK29/48R, did prevent the conjugation (Figure 4A, black arrows).
Figure 4. Ubiquitin conjugation of Ubext is required for stabilization of N-end rule but not UFD substrates.
(A) Lysines 29 and 48 are required for ubiquitin conjugation to Ubext. Protein lysate from cells containing empty vector (EV), Ub, Ubext, or UbextKxR were analyzed by SDS-PAGE and western blot using an anti-ubiquitin antibody. The black arrowhead indicates mono-ubiquitin. The grey arrow points to Ubext. Black arrows represent conjugated Ubext. Pgk1p was probed to assess protein loading (lower). (B) The conjugation of Ubext is necessary for the impaired degradation of the N-end rule substrate R-βgal. Cells containing EV, Ub, Ubext, or UbextKxR mutants were transformed with pGalUb-R-LacZ and analyzed by βgal activity assay. The asterisk (*) denotes statistical significance between Ubext and EV (p = 0.0239). The cross (+) indicates statistical significance between Ubext and UbextK29/48R (p = 0.032). There is no statistically significant difference between Ubext and UbextK11R (p = 0.1602). Lower: Corresponding βgal protein levels from the lysates used in the βgal activity assay were detected by SDS-PAGE and western blot using an anti-βgal antibody. (C) Ubiquitin conjugation of Ubext is not necessary for the impaired degradation of the UFD substrate Ub-P-βgal. Cells containing EV, Ub, Ubext, or UbextKxR mutants were transformed with pGalUb-P-LacZ and analyzed by βgal activity assay. The asterisk (*) denotes statistical significance between Ubext and EV (p = 0.0055). There is no statistically significant difference between Ubext and UbextK48/29R (p = 0.4558). (D) Ubext-ubiquitin conjugation is not necessary to impair the degradation of a second UFD substrate, UbG76V-GFP. Cells containing EV, Ub, Ubext, or UbextKxR mutants were transformed with UbG76V-GFP and analyzed by SDS-PAGE and western blot using an anti-GFP antibody. The blot was reprobed for Pgk1p as a loading control.
We evaluated the degradation of the UPS substrates in the presence of the UbextKxR mutants. The expression of each single UbextKxR mutant stabilized the N-end rule substrate, R-βgal (Figure 4B). However, the expression of the UbextK29/48R double mutant allowed for better degradation of the reporter protein, suggesting that the ubiquitination of Ubext is necessary to impair the degradation of the N-end rule substrate. The steady state levels of βgal protein were detected by western blot and corroborated the result of the βgal activity assay (Figure 4B, lower).
Next, we evaluated the degradation of the UFD substrate in the presence of the UbextKxR mutants. Each UbextKxR mutant, including the double mutant (UbextK29/48R), impaired the degradation of the UFD reporter protein Ub-P-βgal (Figure 4C). Since these data contradict the effects of UbextK29/48R on N-end rule substrate stability (Figure 4B) and previously published results with UBB+1 [22], we evaluated another UFD substrate, a ubiquitin-GFP fusion (UbG76V-GFP). Western blot analysis revealed that this UFD substrate was also stabilized by Ubext as well as each UbextKxR mutant, including the double mutant (Figure 4D). Taken together, these data demonstrate that the conjugation of Ubext is necessary to cause impaired degradation of an N-end rule substrate, but mono-Ubext (i.e. UbextK29/48R) can still impair the degradation of UFD substrates. Based on these data, we suggest that ubiquitin conjugation to N-end rule substrates and UFD substrates is different. The degradation pathways utilized for these two reporters are distinct [3],[31],[32], however they typically report on the same degradation competence of the proteasome, although differences have been cited under certain circumstances [29],[44],[45]. The observed differences here could be explained if different proteins interact with the substrates to perform the ubiquitin conjugation. Perhaps preformed ubiquitin chains are conjugated en masse to N-end rule substrates but ubiquitin is added sequentially to UFD substrates. Thus, in the presence of UbextK29/48R the substrates would be affected differently. Furthermore, this emphasizes that the mode of ubiquitin conjugation, which remains somewhat of a mystery [46], may be an important factor in the differential ability of the cells to cope with one UPS substrate versus another.
Disruption of the Hydrophobic Patch of Ubext Modulates Proteasomal Impairment of a UFD Substrate
Our data suggest that Ubext might be interacting with multiple components of the ubiquitin processing pathway, sequestering proteins required for efficient degradation of proteasome target substrates. Ubiquitin contains a hydrophobic patch (L8, I44 and V70) that is critical for its interaction with many other proteins and the proteasome [47],[48]. The ubiquitin mutation I44A disrupts the hydrophobic patch and this mutant fails to interact with some of its partner proteins [48]. We created a UbextI44A mutant and tested whether its expression caused UPS impairment. Cells expressing UbextI44A still stabilized the N-end rule substrate, R-βgal (Figure 5A). However, expression of UbextI44A resulted in a modest, yet reproducible, increase in the degradation of UFD substrate Ub-P-βgal (Figure 5B). This differential stabilization of the reporters did not occur with different type of mutant ubiquitin, UbΔGG I44A (data not shown). These data suggest that the interaction of Ubext with other proteins is partially disrupted by mutating the hydrophobic patch and further supports that Ubext may have multiple interacting partners to impose the UPS impairment.
Figure 5. Mutation of the Ubext hydrophobic patch (I44A) moderately affects proteasomal impairment.
(A) UbextI44A still inhibits N-end rule substrate degradation. Cells containing pGal-Ub-R-LacZ were transformed with empty vector (EV), Ub, Ubext or UbextI44A (I44A) and the stability of R-βgal was measured by βgal activity assay. (B) UbextI44A moderately enhances the degradation of a UFD substrate. Cells containing pGal-Ub-P-LacZ were transformed with EV, Ub, Ubext or UbextI44A (I44A) and the stability of Ub-P-βgal was measured by βgal activity assay. The asterisk (*) indicates statistical significance between Ubext and UbextI44A (p = 0.0007).
Challenging the UPS Decreases Cellular Tolerance to Ubext
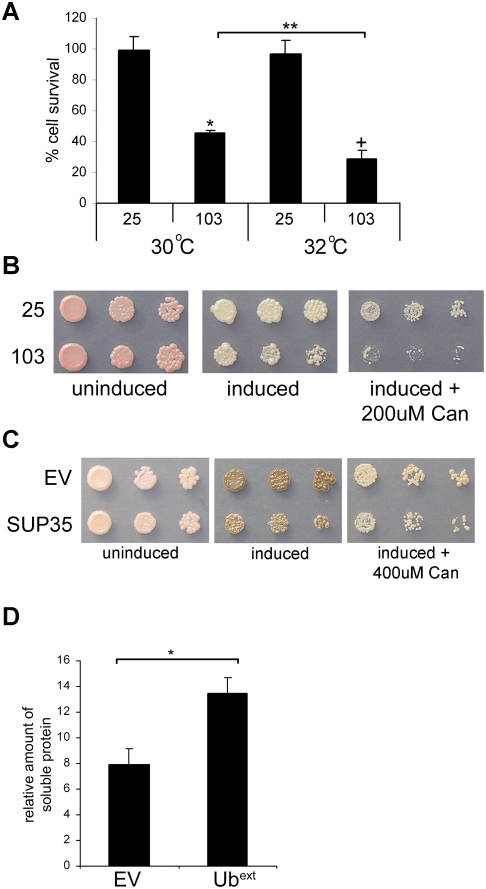
The UPS is required for the removal of misfolded proteins. Failure to remove misfolded proteins can lead to aggregation and have detrimental phenotypic consequences. Since the expression of Ubext exacerbates UPS defects, we next analyzed whether the tolerance to misfolded proteins was decreased in cells expressing Ubext. Canavanine is an arginine analog which becomes incorporated into newly synthesized proteins and causes misfolding [49]. Serial dilutions of cells expressing Ubext were spotted onto solid medium containing canavanine. Ubext-expressing cells showed impaired growth on canavanine containing medium (Figure 6). This suggests that Ubext interferes with the ability of the UPS to degrade natural substrates and challenges cell viability when presented with misfolded proteins.
Figure 6. Ubext expression increases cellular sensitivity to misfolded proteins.
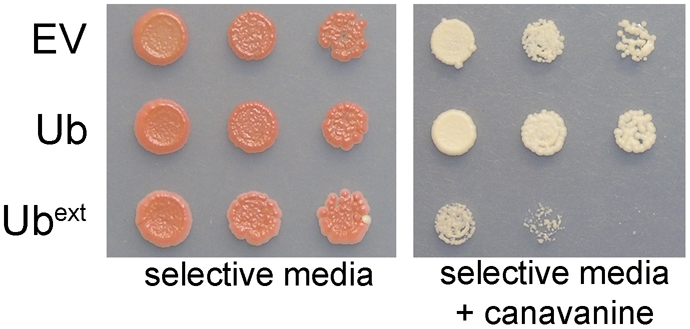
Ubext-expressing cells cannot tolerate excess misfolded proteins generated by the incorporation of canavanine. Serial dilutions of cells containing EV, Ub, or Ubext were spotted onto selective medium and selective medium containing 400 µM canavanine.
Expression of Ubext Affects the Cellular Tolerance to Toxic Aggregates but Does Not Affect Protein Aggregation
We next asked whether misfolded proteins that aggregate would present an additional challenge to cells expressing Ubext. Using tools and properties uniquely available in the yeast system, we sought to determine if Ubext affects protein aggregation by evaluating both toxic and non-toxic protein aggregates. Since cell death associated with toxic protein aggregates makes it difficult to evaluate the potential contribution of UPS dysfunction, the use of non-toxic aggregates in yeast could provide additional insight as to the direct effects of Ubext. UBB+1 enhanced the aggregation and toxicity of a polyglutamine-expanded protein in cultured mammalian cells [24]. To perform similar experiments in our yeast model, we used a galactose-inducible expanded Huntingtin (Htt) polyglutamine construct, TOXIC-Q103, which creates a toxic protein aggregate [50],[51]. Cells expressing Ubext could only tolerate a very low amount of TOXIC-Q103, and even with minimal induction, Ubext-expressing cells grew much worse in comparison to control cells (Figure 7A). Interestingly, the expression of UbextI44A did not result in the same enhanced protein aggregate toxicity (data not shown). Thus, partially alleviating the UPS impairment by altering Ubext protein interactions relieved the enhanced toxicity.
Figure 7. Ubext expression does enhance the toxicity of polyglutamine expanded protein but does not affect protein aggregation.
(A) Cells containing empty vector (EV), Ub, or Ubext were transformed with a galactose-inducible TOXIC-Q103 construct, which induces cell death in the presence of galactose. Serial dilutions of transformants were spotted onto selective medium (uninduced) and selective media containing either 0.1% or 0.3% galactose. (B) Pre-existing non-toxic HttQ103-GFP aggregates were not altered in the presence of Ubext. Cells transformed with non-toxic HttQ103-GFP and EV or Ubext were analyzed by fluorescence microscopy. The abundance and pattern of aggregates (dots) was evaluated as described in Materials and Methods using three independent cultures for each sample. Data are expressed as a percentage of the total cells containing aggregates. (C) TOXIC-Q103 protein is not stabilized in the presence of Ubext. Cells expressing TOXIC-Q103 in the absence (EV) or presence of Ubext were treated with cycloheximide and harvested at the indicated times post translational shut off (in minutes). Cell lysates were analyzed by western blot for the expression of TOXIC-Q103, (which is CFP tagged) using an anti-GFP antibody. Relative protein abundance was quantified as a ratio of the total (below). The membrane was reprobed with an anti-Pgk1 antibody to show protein loading. (D) TOXIC-Q103 protein aggregates do not cause proteasomal impairment. pGal-Ub-P-LacZ containing cells with and without Ubext were transformed with galactose-inducible Q25 or TOXIC-Q103 constructs. The transformants were grown in selective medium containing galactose for 24 hours and the stability of the Ub-P-βgal substrate was measured by βgal activity assay.
To determine whether Ubext expression might affect the aggregates themselves, we imaged a non-toxic version of a polyglutamine-expanded Htt protein fused to GFP (HttQ103-GFP) [52]. Evaluation of these protein aggregates eliminates the complication of cell death associated with toxic aggregates. Previous studies have demonstrated that genetic manipulations, such as altering chaperone levels, can change the abundance and pattern of polyglutamine-GFP aggregates in cells [53]. Thus, we tested whether UPS dysfunction caused by the expression of Ubext would change the aggregate distribution. Neither the abundance nor the pattern of HttQ103-GFP aggregates was altered in cells expressing Ubext (Figure 7B). Thus, although the expression of Ubext did enhance the cellular susceptibility to toxic aggregates, it did not grossly alter the formation or maintenance of non-toxic polyglutamine protein aggregates.
One mechanism by which Ubext could be enhancing the toxicity of TOXIC-Q103 could involve stabilization of the protein, as the level of expression directly correlates to the amount of toxicity. The stability of TOXIC-Q103 protein was evaluated from cells expressing Ubext after protein translation was inhibited by cycloheximide. No drastic stabilization of TOXIC-Q103 protein was apparent in cells expressing Ubext (Figure 7C).
We next asked whether the TOXIC-Q103 aggregates themselves caused UPS impairment. The stability of the UPS reporter protein, Ub-P-βgal, was monitored in cells containing TOXIC-Q103 aggregates in comparison to a non-pathological polyQ25 protein. No stabilization of the reporter was observed in cells harboring the toxic aggregates (Figure 7D). In addition, the UPS impairment caused by Ubext was not further increased by the presence of TOXIC-Q103 (Figure 7D). Thus, the enhanced toxicity of TOXIC-Q103 caused by Ubext is not due to additive effects on UPS impairment.
Enhanced Cellular Toxicity Is Observed with a Second Toxic Protein
To evaluate the generality of the effects of Ubext on the phenotypic response to toxic protein aggregates, we used a yeast prion protein. Prion proteins in yeast form ordered aggregates that are not harmful to the cells [54]–[56]. Sup35p, an essential translation termination factor, is the protein determinant of the yeast prion [PSI+] [55]. The aggregated prion state of Sup35p, [PSI+], causes read through of stop codons in translated mRNAs (nonsense suppression). The percentage of read through is low and generally has no deleterious effects to cells grown in rich medium [54]. The presence of the [PSI+] prion can be monitored in a strain carrying an ade1-14 mutant allele with a premature stop codon [57]. In [psi−] cells, Sup35p is soluble and functional, and translation is terminated at the premature stop codon in ade1-14. Thus, [psi−] ade1-14 cells cannot grow on medium lacking adenine and when grown on rich medium they appear red due to the accumulation of an intermediate in the adenine biosynthetic pathway. Conversely, aggregated Sup35p in [PSI+] cells limits the amount of functional Sup35p, thereby causing nonsense suppression of the ade1-14 premature stop codon and translation of full-length Ade1 protein. These cells are adenine prototrophs and appear white on rich medium. As such, one can evaluate the functional state of Sup35p as it relates to protein aggregation by monitoring the color of the yeast colony. Cells can be maintained stably as [psi−], but they can be induced to become [PSI+] by over expressing the Sup35 protein.
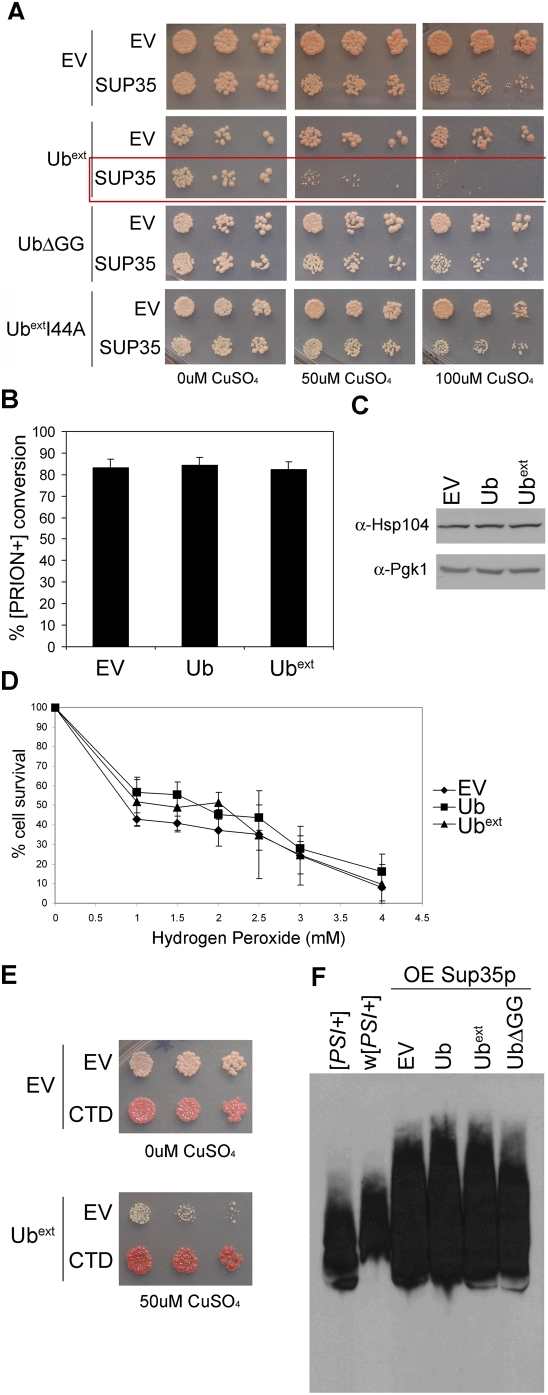
The [PSI+] prion state is not toxic, however, over expression of Sup35p in [PSI+] cells inhibits cell growth due to the lack of sufficient translation termination [58]–[60]. As one would expect, the over expression of Sup35p is not toxic to [psi−] cells. Thus, the toxicity results from too much aggregation of Sup35p in the prion state. These toxic aggregates provide a means to assess the effects of aggregation of a protein of known function in combination with UPS dysfunction. Since most toxic protein aggregates cause cell death by unknown mechanisms, analyzing the Sup35p aggregates in [PSI+] cells provides a unique opportunity to dissect the contributions of the toxic protein aggregates and UPS dysfunction. To evaluate the effects of UPS dysfunction on toxic protein aggregates, [PSI+] cells harboring a copper-inducible SUP35 were transformed with Ubext and assayed for cell viability (Figure 8A). Ubext-expressing [PSI+] cells were more susceptible to the over expression of Sup35p (Figure 8A, red box ). The expression of Ubext did not increase basal levels of Sup35p, as determined by SDS-PAGE and western blot analysis (data not shown). Intriguingly, the expression of a different mutant ubiquitin protein, which caused UPS impairment similar to Ubext (data not shown), UbΔGG, did not enhance the toxicity of Sup35p over expression to the same extent (Figure 8A, compare fourth row to sixth row). These results show that Ubext enhances the toxicity of protein aggregates by a mechanism that cannot be solely attributed to its effects on UPS impairment, since UbΔGG did not have the same effect. Furthermore, the hydrophobic domain mutant, UbextI44A, did not result in the same sensitivity to over expressed Sup35p in [PSI+] cells (Figure 8A). This suggests that the mechanism by which Ubext enhances the toxicity of protein aggregates requires interactions with other proteins via the hydrophobic domain.
Figure 8. Expression of Ubext enhances the susceptibility of cells to toxic Sup35p aggregates but does not affect Sup35p aggregation.
(A) [PSI+] cells expressing Ubext show reduced cell viability with lower induction of Sup35p. [PSI+] cells containing empty vector (EV), Ubext, UbΔGG, or UbextI44A were transformed with a copper-inducible SUP35 or EV and analyzed for growth by spotting serial dilutions onto selective media containing 0, 50, or 100 µM CuSO4. At 300 µM CuSO4, [PSI+] cells over expressing Sup35p alone are not viable (not shown). (B) Prion conversion or induction was not enhanced in cells expressing Ubext. [psi−] cells expressing pSup35 or the control (EV) were transformed with empty vector (EV), Ub or Ubext and were analyzed for [PSI+] prion formation by monitoring colony color (the appearance of pink colonies). The graph represents the average of three independent cultures in which approximately 2,000 colonies per culture were evaluated for conversion. (C) Hsp104 protein levels are not enhanced in Ubext-expressing cells. Protein lysate from cells containing EV, Ub, or Ubext were subject to SDS-PAGE and western blot using an anti-Hsp104 antibody. Pgk1p expression was analyzed as a loading control. (D) The expression of Ubext did not alter cell survival in the presence of oxidative stress. Cells containing EV, Ub, or Ubext were treated with increasing concentrations of hydrogen peroxide (H2O2) and the number of viable cells was graphed as a percentage of the untreated. (E) The C-terminal domain of Sup35p (CTD) rescued the enhance susceptibility caused by Ubext in [PSI+] cells over expressing Sup35p. Upper: [PSI+]-mediated nonsense suppression is alleviated by expression of the CTD. [PSI+] cells containing EV show more nonsense suppression (the colony color is light pink). However, [PSI+] cells expressing the CTD display efficient translation termination and the colonies are red. Lower: [PSI+] cells expressing Ubext in addition to excess Sup35p (induced by 50 µM copper) are rescued from death by the expression of the CTD. (F) Sup35 protein aggregates were not altered by the presence of Ubext. Sup35p aggregates in strong [PSI+] ([PSI+]) and a weak strain variant of [PSI+] (w[PSI+]) were analyzed by SDD-AGE. The difference in Sup35p aggregate size of these prion strain variants can be appreciated by this method (compare [PSI+] to w[PSI+]). Sup35p aggregates from cells expressing excess Sup35p (OE Sup35p) and expressing Ub, Ubext, UbΔGG or containing an EV control were analyzed by SDD-AGE and western blot with an anti-Sup35 antibody.
We evaluated whether the aggregation of Sup35 is altered by the expression of Ubext. A previous study demonstrated that altering ubiquitin levels by either increasing the expression of ubiquitin or preventing its recycling caused an increase in the formation of the [PSI+] prion [61]. Furthermore, deletion of a ubiquitin conjugating enzyme also enhanced [PSI+] induction [62]. Thus, there is genetic precedence for perturbations of the UPS affecting prion protein aggregation. We asked whether the presence of Ubext would alter the spontaneous formation of aggregated Sup35p and change cells from [psi−] to [PSI+]. We did not observe a change in the spontaneous conversion rate (data not shown), which we have measured to be ∼1 in 105 in our strain [63]. We next evaluated the induction of the [PSI+] prion state in the presence and absence of Ubext by over expressing Sup35p in [psi−] cells. Since Ubext perturbs the UPS, one might predict an effect on the induction of protein aggregation. To the contrary, the expression of Ubext did not enhance the induction of [PSI+] (Figure 8B).
Expression of Ubext Does Not Cause a Stress Response
The enhanced toxicity of protein aggregates caused by Ubext could be the result of a general stress response elicited in cells expressing Ubext. The expression of a heat shock element (HSE)-LacZ reporter fusion was evaluated in Ubext-expressing cells and no increase in transcription from the HSE promoter at 30°C or at a sub-lethal heat stress of 37°C was observed (data not shown). We next asked whether the presence of Ubext increased the translation of a stress-inducible heat shock protein. Protein lysate from Ubext-expressing cells and control cells showed similar levels of Hsp104p (Figure 8C), a stress-responsive chaperone. Finally, we tested the tolerance of the cells to oxidative stress. Cells challenged with hydrogen peroxide showed no change in survival in the presence of Ubext (Figure 8D). These results suggest that Ubext expression in yeast neither induces a general stress response nor preconditions the yeast to exogenous insult. Therefore, the enhanced susceptibility of Ubext-expressing cells to toxic aggregates is not likely the result of Ubext inducing a general stress.
Restoration of Translation Termination Rescues Enhanced Toxicity Caused by Ubext
Overcoming the enhanced protein aggregate toxicity induced by Ubext expression could shed light on the mechanism by which Ubext exerts its affects. In attempts to alleviate the Ubext-enhanced aggregate toxicity we conducted a genomic over expression screen using the toxicity caused by over expression of Sup35p in [PSI+] cells. We uncovered two rescuing factors, HSP104 and SUP45. Both of these proteins alleviate the toxicity by affecting Sup35p aggregation and the associated phenotypic readout. Over expression of Hsp104p affects the Sup35p aggregates [64] and Sup45p can sequester Sup35p away from the aggregates [65]. To verify that the enhanced protein aggregate toxicity in the presence of Ubext can be overcome by altering nonsense suppression, we over expressed the C-terminal domain (CTD) of Sup35p, which is sufficient for translation termination but cannot aggregate and form or join the prion state [58],[66]. We found that the expression of the CTD not only restored translation termination of [PSI+] cells (Figure 8E, upper), but also alleviated the enhanced toxicity caused by the expression of Ubext (Figure 8E, lower). These results demonstrate that alleviating the primary deficit in the cells (i.e. the effects of [PSI+]) is sufficient to overcome toxicity even in the presence of a modifier (Ubext).
Model to Explain Cellular Affects of Ubext on Aggregate Toxicity
We next asked whether Ubext affected the toxic Sup35p aggregates, since the enhanced cellular toxicity caused by Ubext and excess Sup35p is [PSI+]-dependent. We assayed Sup35p aggregates by semi-denaturing detergent agarose gel electrophoresis (SDD-AGE) [67]. This technique allows large protein aggregates to migrate into the gel and can resolve aggregates of different sizes, as demonstrated by a strain variant of [PSI+] (weak [PSI+]), which harbors larger Sup35p aggregates than our [PSI+] starting strain (Figure 8F). We observed no change in the size of Sup35p aggregates from cells over expressing Sup35p in combination with Ubext or UbΔGG. One possible explanation for the enhanced toxicity in the presence of Ubext could relate to a change in the degradation of misfolded Sup35p. As such, we asked whether Ubext was promoting the accumulation of ubiquitinated-Sup35p. We reprobed the SDD-AGE membrane with an anti-ubiquitin antibody but did not find any ubiquitinated Sup35p by this approach. In additional attempts to look for ubiquitination of Sup35p, we purified Sup35 aggregates [68] but again were unable to detect any ubiquitinated Sup35 protein (data not shown). Other researchers have also noted an inability to identify ubiquitinated-Sup35p [61],[62]. Thus, we conclude that although Ubext affects the ability of cells to tolerate toxic Sup35p over expression, it is unlikely a direct consequence of blocking the ubiquitination and degradation of Sup35p.
We also evaluated whether the polyglutamine-expanded Htt proteins are ubiquitinated. We were unable to detect ubiquitinated polyglutamine protein in yeast by immunoprecipitation, SDD-AGE or immunofluorescence (data not shown). The inability to find ubiquitinated polyglutamine protein has also been noted previously [52],[69],[70]. Therefore, as with toxic Sup35p, Ubext is affecting the tolerance to TOXIC-Q103 aggregates by an indirect means.
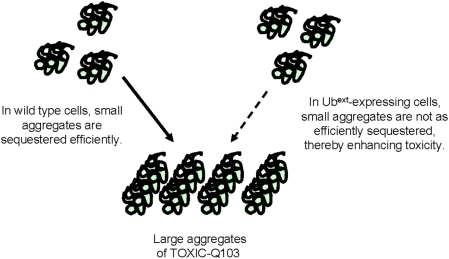
How could Ubext be affecting the toxicity of protein aggregates if those proteins are not subject to ubiquitination and degradation by the UPS? One possible explanation of the effects of Ubext on protein aggregate toxicity could be due to a change in the ability to efficiently sequester the toxic proteins into large aggregates (Figure 9). A toxic polyglutamine protein expressed in yeast was rendered non-toxic when sequestered into a single, large aggresome-like structure [70]. Furthermore, a non-toxic polyglutamine protein, which localizes to an aggresome-like structure, became dispersed in ubiquitination-deficient cells. We hypothesize that Ubext alters the localization of toxic proteins into the large aggregate structures due to its effects on UPS function. The enhanced toxicity could be the consequence of a reduced ability to sequester toxic soluble oligomer species (Figure 9).
Figure 9. Model for Ubext affects on toxic protein aggregates.
We propose that enhanced protein aggregate toxicity in Ubext-expressing cells is due to the inability of misfolded amyloidogenic proteins to be properly sequestered. The small soluble oligomers are more toxic than the large insoluble protein aggregates. UPS impairment caused by the expression of Ubext may hinder the rapid sequestration or retention of toxic oligomers into large protein aggregates.
Enhancing Protein Aggregate Toxicity by Increasing the Burden on the UPS
Based on our hypothesis, we predict that protein aggregate toxicity can be affected by perturbations in ubiquitination or by overwhelming the UPS in general. We took advantage of a temperature-sensitive ubiquitin activating enzyme (E1) mutant (uba1-204) [71] to evaluate the effect of an overall reduction in ubiquitination on the phenotypic response to TOXIC-Q103 aggregates. UBA1 is an essential gene responsible for the first step of the ubiquitination cascade. At the restrictive temperature, the uba1-204 mutant limits substrate ubiquitination. A recent study demonstrated that polyglutamine protein aggregate patterns were altered in cells expressing the uba1-204 mutant [70]. uba1-204 cells expressing TOXIC-Q103 or the control (Q25) were grown in inducing conditions at the permissive (30°C) and restrictive temperatures (32°C) and colony survival was measured (Figure 10A). Cells expressing TOXIC-Q103 showed approximately 50% survival in comparison to those expressing Q25, and this survival was further decreased in conditions of limiting ubiquitination (i.e. 32°C). To directly compare the affect of Ubext expression on the TOXIC-Q103 aggregates, we measured colony survival as performed above. Cells harboring TOXIC-Q103 aggregates in the presence of Ubext allowed for only a 7% survival in comparison to TOXIC-Q103 aggregates alone (56% survival). Thus, Ubext is a more potent modifier of toxic protein aggregates than perturbations in ubiquitination.
Figure 10. Protein aggregate toxicity is enhanced by perturbations of the UPS and protein aggregate solubility is enhanced by Ubext.
(A) Limiting ubiquitination also decreases cellular survival in the presence of TOXIC-Q103. Cellular viability of TOXIC-Q103 (103) or the Q25 control expressed in ts uba1-204 cells was measured at the permissive temperature (30°C) and a restrictive temperature (32°C). The graph represents the percentage of viable cells from the inducing plates compared to cells grown on non-inducing medium. The asterisk (*) indicates statistical significance between 25 and 103 at 30°C (p = 0.0007), the cross (+) indicates statistical significance between 25 and 103 at 32°C (p = 0.0003), and the double asterisk (**) indicates statistical significance between 103 at 30°C and 32°C (p = 0.0068). (B) Increasing misfolded proteins enhanced toxicity in the presence of TOXIC-Q103. Cell expressing TOXIC-PQ103 (103) and the Q25 control (25) were spotted onto inducing medium and inducing medium containing 200 µM canavanine (Can). (C) The cellular susceptibility of over expressed Sup35p in [PSI+] cells in the presence of canavanine is not as detrimental as the co-expression of Ubext. Cells expressing excess Sup35p (induced with 200 µM CuSO4) were spotted onto plates containing 400 µM canavanine (Can). Sup35 over expressing cells are slightly less viable in the presence of 400 µM canavanine. All cells died at higher concentrations of CuSO4 and canavanine. (D) Cells expressing Ubext contain more soluble TOXIC-Q103 protein. Cells expressing TOXIC-Q103 in the presence of Ubext or absence (EV) were lysed and the soluble protein was analyzed by western blot after high speed ultracentrifugation. Densitometry was performed to determine the amount of soluble TOXIC-Q103 protein normalized to the total protein for each sample and graphed in relative arbitrary units. Three independent cultures for each sample were analyzed. The asterisk (*) denotes statistical significance (p = 0.0052).
Since decreased ubiquitination had an affect on the protein aggregate toxicity, we asked if protein aggregate toxicity could also be enhanced by increasing the burden on the UPS. We measured the viability of cells expressing TOXIC-Q103 or over expressing Sup35p in the presence of canavanine. Serial dilutions of cells expressing Q25 and TOXIC-Q103 were spotted onto inducing media containing canavanine. The effects of the glutamine expansion on cell viability can be seen on inducing plates and in the presence of a UPS burden (canavanine) the toxicity is enhanced (Figure 10B). Over expressed Sup35p in [PSI+] cells also shows toxicity and in the presence of canavanine the toxicity is slightly enhanced (Figure 10C). However, canavanine is less potent at enhancing the toxicity of over expressed Sup35p in comparison to the effect of Ubext (Figure 8A). Nonetheless, perturbations to the UPS in general do appear to enhance protein aggregate toxicity. We propose that this is due to a change in efficient sequestration of toxic proteins into insoluble aggregates (Figure 9).
Since Ubext enhanced the toxicity of TOXIC-Q103, we tested whether Ubext-containing cells were compromised in their ability to sequester or retain TOXIC-Q103 in the insoluble aggregates. Protein lysates from Ubext and controls cells (EV) were subjected to high speed ultracentrifugation and analyzed to determine whether Ubext influences the amount of soluble TOXIC-Q103. Serial dilutions of the total and resulting soluble fraction were applied to PVDF and visualized by western blot. The amount of soluble protein as normalized to total protein was determined by densitometry (Figure 10D). The amount of soluble TOXIC-Q103 was higher in Ubext-expressing cells than wild type cells. Thus, the enhanced toxicity of TOXIC-Q103 in Ubext-expressing cells correlates to an increased pool of soluble protein and supports the model proposed in Figure 9.
Ubext Alters Ubiquitination Patterns
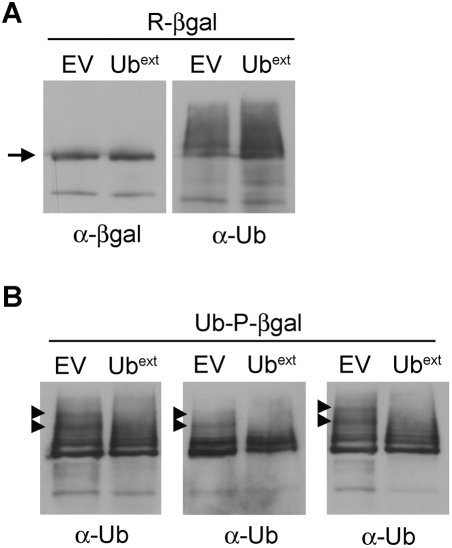
Since altered ubiquitination affected the distribution of expanded polyglutamine proteins [70] and enhanced the cellular susceptibility to toxic polyglutamine aggregates (Figure 10A), we asked whether Ubext has a direct effect on the ubiquitination of proteasome substrates. In light of the fact that the toxic protein aggregates are not ubiquitinated, we evaluated the ubiquitination pattern of the UPS reporters. To compare the ubiquitination of these constructs with and without the expression of Ubext, we utilized a temperature-sensitive proteasome mutant strain (pre1-1 pre2-2) [28]. This strain is defective in proteolysis and when grown at the restrictive temperature, R-βgal and Ub-P-βgal accumulate (Figure 11A). Striking substrate ubiquitination can be observed in pre1-1 pre2-2 cells expressing Ubext and control cells after IP. When we compared the R-βgal substrate ubiquitination in EV and Ubext-containing cells, we did not discern any difference in the ubiquitination pattern (Figure 11A). However, a subtle yet reproducible ubiquitination pattern difference was seen with the Ub-P-βgal substrate (Figure 11B). Three independent IP experiments are shown and two ubiquitinated-βgal bands appear in control cells (EV) which are absent or greatly reduced in Ubext-expressing cells. The altered ubiquitination pattern of some UPS substrates in the presence of Ubext could change the ability of these proteins to be processed by the proteasome. Furthermore, such changes could be an important modifier of the cellular effects of toxic protein aggregates.
Figure 11. Ubext alters the ubiquitination pattern of a UPS substrate.
(A) R-βgal ubiquitination pattern is not altered in cells expressing Ubext. pGalUb-R-LacZ was transformed into proteasome mutant cells (pre1-1 pre2-2) expressing Ubext or EV and R-βgal was analyzed by immunoprecipitation (IP). Membranes were probed with anti-βgal and anti-ubiquitin antibodies. Arrow indicates full length βgal protein. (B) Ub-P-βgal ubiquitination is affected in cells expressing Ubext. Ub-P-βgal IPs were performed as in A. A subtle but reproducible difference in ubiquitination pattern was observed. Three independent IPs are shown. Arrowheads highlight distinct bands present in the EV lanes that are absent in Ubext lanes.
Discussion
We created a novel model of UBB+1 by constitutively expressing an analogous mutant ubiquitin protein in yeast to investigate the causal relationship between this proteasomal inhibitor and protein aggregation. We demonstrated that the Ubext mutant was not functional as ubiquitin and was not deleterious to the cells. Importantly, the expression of Ubext in yeast caused impairment of the UPS. Since proteasome dysfunction can lead to protein aggregation, we were intrigued that the presence of Ubext served to neither induce nor alter non-toxic protein aggregates in yeast. However, the expression of Ubext rendered the cells more susceptible to toxic protein aggregates, and this could not be attributed to an increase in general stress elicited by Ubext. We propose that the reduced UPS functionality and altered ubiquitination of UPS substrates in Ubext-expressing cells creates an environment in which toxic amyloidogenic proteins either cannot join or are not maintained as large insoluble aggregates. As a result, protein aggregate toxicity is enhanced due to an increase in soluble or oligomeric toxic protein. Thus, this yeast model system revealed that Ubext is a phenotypic modifier of toxic protein aggregates. This genetically tractable model provides a platform to further dissect how UBB+1 affects the cellular tolerance to toxic protein aggregates.
The mechanism of UPS impairment caused by UBB+1 is not well understood. We asked whether Ubext causes a reduction in proteasome activity. Using an unstable ubiquitin-independent substrate (GFP-ODC) [37], we observed no significant change in the activity of the proteasome in Ubext-expressing cells. Based on this result, we suggest that Ubext is not clogging the core of the proteasome and propose that Ubext is interacting with other components of the ubiquitin processing cascade or with the regulatory cap of the proteasome. We hypothesized that disrupting the interaction of Ubext with component(s) of the ubiquitin processing pathway would alleviate the proteasomal impairment. Mutational analysis revealed that ubiquitin conjugation and the hydrophobic patch affect the extent to which Ubext causes UPS impairment. Interestingly, the effects were distinct with different substrates. This supports the idea that Ubext is interacting with multiple components of the UPS; reduction of its interaction via the hydrophobic patch or elimination of its ubiquitination weakened some of the observed effects but not others.
Previous studies have investigated the connection between UPS dysfunction and protein aggregation, especially in the context of protein conformational disorders [72]. It remains difficult, however, to discern the precise nature of the causal relationship between protein aggregation and proteasomal impairment. Evidence that UBB+1 and other disease-associated mutations in the UPS can cause proteasomal impairment and increase protein aggregation supports the idea that proteasome dysfunction plays a stimulatory role in protein aggregation. However, in some cases, such as that with mutant Parkin in familial Parkinson's Disease, decreased UPS function is not associated with protein aggregation [8]. Using non-toxic protein aggregates in yeast, we have demonstrated that a UBB+1-like protein, Ubext, neither induced nor changed protein aggregates. Our results provide evidence that a compromised UPS does not necessarily affect protein aggregation per se but can cause phenotypic effects by decreasing cellular tolerance to deleterious protein aggregates.
We hypothesize that Ubext is altering the sequestration of aggregated proteins (Figure 9). Due to the altered substrate ubiquitination and the general UPS impairment caused by Ubext, misfolded proteins are not efficiently degraded and somehow perturb the sequestration of amyloidogenic proteins into the insoluble aggregates which may have a protective function. How the UPS functionality plays a role in the ability of the cell to efficiently sequester non-ubiquitinated proteins remains to be elucidated. One recent study suggests that different cellular compartments retain aggregates of ubiquitinated and non-ubiquitinated proteins and a reduction in UPS activity can cause a change in this localization [69]. If proper localization of aggregated proteins protects the cell from smaller toxic oligomeric species [73],[74], then the inability of toxic oligomers to be efficiently sequestered would be deleterious (Figure 9). Indeed, the expression of Ubext resulted in an increase in the relative amount of soluble TOXIC-Q103 protein (Figure 10D) and the combination of Ubext and TOXIC-Q103 was more deleterious to cell survival (Figure 7A). Further evidence to support the idea that the redistribution of aggregates can lead to cell death comes from a recent report investigating the nature of the aggregates formed in response to the expression of expanded polyglutamine protein in yeast [70]. A single large aggregate, an aggresome-like structure, was formed by polyglutamine proteins that were not toxic to the cells. When the large aggregate was unable to form, multiple small aggregates were observed and the appearance of these correlated with toxicity. Thus, the single large aggregate appears to be protective against polyglutamine protein aggregate toxicity. Among the cellular factors found that could disrupt the formation of the single aggregate when mutated were two ubiquitin-associated proteins. Furthermore, limiting general cellular ubiquitination by the uba1-204 mutant also disrupted the formation of the large aggregate [70]. We show that uba1-204 enhanced the cellular toxicity of the toxic polyglutamine aggregates used in our study (Figure 10A). Taken together, the data support the proposed model of the effect of Ubext on protein aggregate toxicity (Figure 9).
Since Ubext causes UPS impairment and a change in ubiquitination of substrates, this could cause the mis-handling or redistribution of some ubiquitin-conjugated proteins and hinder toxic protein aggregates from being rapidly sequestered, resulting in enhanced cell death (Figure 9). Thus, even though the toxic protein aggregates may not be substrates of the UPS, perturbations in the processing of normal UPS substrates may affect cellular tolerance to toxic aggregates. Our data suggest that all perturbations in the UPS are not equally potent at altering the cellular tolerance to toxic aggregates. Therefore, we conclude that the magnitude of the enhanced protein aggregate toxicity in the presence of the extended mutant ubiquitin is exceptional. This is likely due to its interactions with other proteins and supports further that UBB+1 may be a potent disease modifier.
Since protein conformational disorders result from a combination of cellular perturbations, often including the unknown affects of aging, then eliminating individual modifiers or enhancers may prove useful for disease therapy. Obviously, alleviating the primary causative agent, when known, could prove to be the most beneficial. For example, when we used the Sup35p toxic aggregate model we were able to rescue the Ubext-enhanced toxicity by restoring the loss of function caused by Sup35p sequestration into aggregates. However, in many protein conformational diseases, the function of the proteins found in the aggregates and cellular toxicity is not understood. Therefore, investigating ways to alleviate the effects of known modifiers represents an important therapeutic avenue for disease treatment and prevention. The insight gained by developing a yeast model of UBB+1 has provided a means to further investigate the role of protein aggregate compartmentalization in toxicity, which may underlie some of the effects observed in cells or tissues experiencing chronic UPS impairment. The identification of UBB+1-interacting proteins may allow for the elucidation of the mechanism whereby a natural modifier of UPS function affects cellular tolerance to toxic protein aggregates.
Materials and Methods
Strains
Yeast strains were grown and manipulated by standard techniques [75]. Unless otherwise indicated, all yeast strains used in this study were derivatives of 74-D694 (MATa or MATα ade1-14 trp1-289 his3Δ-200 ura3-52 leu2-3,112) [64]. The Δubi4 strain was created by PCR amplification of the antibiotic resistance marker KanMX4 with primers A and B and subsequent transformation of the resulting product into 74-D694. For all primer sequences, see Table 1. The Δubp14 strain was created by PCR amplification of BY4741 Δubp14 genomic DNA with primers C and D and subsequent transformation of the resulting product into 74-D694. The proteasome mutant strain, WCG4-11/22a (MATa his3-11,15 leu2-3,112 ura3 pre1-1 pre2-2) and control strain, WCG4a (MATa his3-11,15 leu2-3,112 ura3) were a kind gift of P. Coffino [37]. The 74-D694 [PSI+]-inducible prion strain [psi−] [RNQ+] and the weak [PSI+] strain variant were a kind gift from S. Liebman [76]. A 74-D694 [PSI+] [RNQ+] strain was used in the PQ toxicity study. The uba1-204 strain was a kind gift from R. Deshaies [71].
Table 1. Primer sequences.
| Code | Used to make (enzyme) | Sequence (5′ orientation) |
| A | ubi4 deletion | GTATTACCCGGCTTCGCGAAAATAGTGAACGTCATAGTATAAGACGATTCATCGATGAATTCGAGCTCG |
| B | ubi4 deletion | GGGGTATATATAGAGAGGCTCCGGGTTTTGCCACCTTTGAATTCGCCTGCCGTACGCTGCAGGTCGAC |
| C | ubp14 deletion | CACTTGATGAAATCACAGTGAAAAGCG |
| D | ubp14 deletion | CGATAGATTTGATCATACACATATAATGC |
| E | 5′ Ub (XbaI) | GCT CTA GAA TGC AGA TTT TCG TCA AGA C |
| F | 3′ Ubext (BamHI) | CGGGATCCTTAACAAAGATCTGCATACCACCTTAGCCTTAGCACAAGATGTAAGG |
| G | 5′ Ub (BamHI) | CGGGATCCATGCAGATTTTCGTCAAGAC |
| H | 3′ Ub (SalI) | GCGTCGACTCAACCACCTCTTAGCCTTAG |
| I | 5′ Ubext K11R (BamHI) | CCGGATCCATGCAGATTTTCGTCAAGACTTTGACCGGTAGAACCATAACATTGG |
| J | 3′Ubext (SalI) | CGGTCGACTTAACAAAGATCTGCATACCACCTTAGCCTTAGCACAAGATGTAAGG |
| K | 5′ Ubext K29R* | GAAGGTATCCCTCCAGATCAAC |
| L | 3′ Ubext K29R* | CTTGTCTTGAATTcTCGACTTAACGTTGTCGATG |
| M | 5′ Ubext K48R* | ACGGTAGAACGCTGTCTG |
| N | 3′ Ubext K48R * | CTTCTAGCTGtcTACCGGCAAAG |
| O | 3′ Ubext K63R∧ | GATGTAAGGTGGACTCCCTCTGAATGTTGTAATC |
| P | 3′ Ubext+GG (SalI) | GGCGGTCGACTTAACCACCACAAAGATCTGCATACCAC |
| Q | 3′ UbΔGG (SalI) | GGCGGTCGACTTATCTTAGCCTTAGCACAAG |
| R | 3′ UbextI44A∧ | CTTACCGGCAAAGGCCAATCTTTGTTG |
| S | 5′ UbextI44A∧ | CAACAAAGATTGGCCTTTGCCGGTAAG |
*: Used in three-way ligation with cut p423TEF vector.
∧: Used in bridge-PCR and cloned into cut p423TEF vector.
Plasmids
All plasmids were created using standard molecular biology protocols [77] and verified by DNA sequencing. For primer sequences, refer to Table 1. Where appropriate, the enzyme used is listed parenthetically. To create p413TEFUbext, ubiquitin was PCR amplified from 74-D694 genomic DNA using primers E and F and cloned into p413TEF [78] at XbaI and BamHI. To create p413TEFUb, ubiquitin was PCR amplified from 74-D694 genomic DNA using primers G and H and cloned into p413TEF at BamHI and SalI. Ubext was subcloned from p413TEFUbext to p423TEF and p426TEF at SpeI and BamHI. Ubiquitin was subcloned from p413TEFUb to p423TEF and p426TEF at SalI and BamHI. All Ubext amino acid substitutions (p423TEFUbextK11R, UbextK29R, UbextK48R, UbextK63R, UbextK29/48R, UbextI44A) were created using either three-way ligation or bridge PCR into p423TEF using p423TEFUbext as a template (except for the p423TEFUbextK29/48R mutant which utilized p423TEFUbextK29R) and following standard molecular biology techniques [77]. p423TEFUbext+GG was created by PCR amplification of ubiquitin DNA with primers G and P and cloned into p423TEF at BamHI and SalI. p423TEFUbΔGG was created by PCR amplification of ubiquitin DNA with primers G and Q and cloned into p423TEF at BamHI and SalI. The 4xHSE-LacZ plasmid was a kind gift of S. Lindquist. In vivo UPS functionality was measured using Ub-X-LacZ reporters: pGal-Ub-M-LacZ, pGal-Ub-R-LacZ, and pGal-Ub-P-LacZ [30]. The ubiquitin-independent proteasome substrates, p416ADH1GFP-mODC and p416ADH1GFP-mODCC441A were a kind gift from P. Coffino [37]. The UBI4promoter-LacZ reporter was a kind gift from M. Altmann [79]. [PSI+] induction assays used the inducer plasmid pEMBL Sup2 (referred to as pSup35 in this manuscript) [58]. Non-toxic polyglutamine aggregation assays used p416GPD polyQ103-GFP [52], referred to as HttQ103-GFP in this manuscript. Toxic polyglutamine aggregation assays employed p416Gal FLAG103Q-CFP (referred to as TOXIC-Q103) and p416Gal FLAG25Q-CFP (referred to as Q25) (kind gift M. Duennwald) [50],[51]. For the toxicity assay in [PSI+] cells, Sup35p was over expressed from a copper inducible promoter. pRS315Cup-SUP35 was generated by cloning Cup1-SUP35 between XhoI and SacI. pRS316-TEF-CtermSup35 contains only the C-terminal domain (amino acids 254–685) of Sup35 and was created by subcloning TEF-CtermSup35 from pRS306TEF-CtermSup35 [80] at HindIII and SacI.
Protein Analyses
Protein lysates were analyzed by standard SDS-PAGE. Protein lysis followed the β-galactosidase assay (see below). The following antibodies were used: Ubiquitin (PD41) (Santa Cruz sc-8017), Hsp104 (kind gift of S. Lindquist), GFP (kind gift of M. Linder), β-galactosidase (Promega Z378A), Pgk1 (Molecular probes A6457), and Sup35 (kind gift of S. Lindquist) [81]. Large Sup35 protein aggregates were separated by SDD-AGE as previously described [82] with modifications previously described [63]. Sup35p over expression was achieved by growing the cultures in 50 µM copper sulfate overnight. Immunoprecipitations were carried out as previously described [83] using 5 µl of mouse anti-β-galactosidase. TOXIC-Q103 protein stabilization was measured after a six hour induction (2% galactose and 1% raffinose containing media) in the presence of 0.5 mg/ml cycloheximide in cultures with equal numbers of cells.
The relative amount of TOXIC-Q103 soluble protein was determined by slot blot. Cells containing TOXIC-Q103 and either EV or Ubext were grown overnight in selective medium, washed in inducing medium containing 2% galactose/1% raffinose and induced for 14–16 hours. Cells were harvested and lysed with glass beads in PEB (250 mM Tris HCl pH 7.5, 50 mM KCl, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 10 mM PMSF, 5 µg/ml Aprotinin, Roche Protease cocktail inhibitor (Roche)). Equal protein (100 µg) from EV and Ubext-containing cells was subjected to ultracentrifugation (80,000 rpm for 30 minutes at 4°C). Serial dilutions of the supernatant and total fractions (diluted 1/10) were applied to activated PVDF and probed with an anti-GFP antibody. The supernatant fraction and corresponding total fractions were quantified using Image J software and graphed as normalized arbitrary units.
β-Galactosidase Assays
UPS functionality was determined by the degradation of Ub-LacZ fusions [30] using Galacto-light™ (Applied Biosystems). Cells containing pGal-Ub-M-LacZ, pGal-Ub-R-LacZ and pGal-Ub-P-LacZ were grown in selective medium for 24 hours. The cultures were washed three times in selective medium containing 2% galactose / 1% raffinose and grown overnight in the 2% galactose / 1% raffinose. The cultures were harvested and lysed in Galacto-light Lysis Solution using glass beads. Cell lysate was pre-cleared for 30 seconds at 6,000 rpm at 4°C. In a flat bottom, black-sided 96-well dish, 70 µl of Galacto Reaction Buffer was added to 10 µl of protein lysate and incubated for 60 minutes at room temperature. Luminescence was read immediately after the addition of 100 µl of Light Emission Accelerator. Luminescence values were normalized to protein concentration as determined by Bradford reagent (BioRad). Error bars in all βgal activity assays represent the standard deviation from three independent cultures for each sample. The TOXIC-Q103 protein βgal activity assay was conducted as described above using a TRP1 version of pGal-Ub-P-LacZ (subcloned into p424Gal vector) with a 24 hour induction. All statistical analyses were conducted using Student's T-Test.
Microscopy
Polyglutamine aggregation was monitored by GFP fluorescence in a 74-D694 [PSI+] [RNQ+] strain background. Three independent samples of mid-log phase cells containing p416GPD polyQ103-GFP [52] and p423TEF EV or p423TEF Ubext were visualized. Individual fluorescent cells were evaluated for a single aggregate, few aggregates (2–3 per cell) or multiple aggregates (greater than 3 aggregates per cell) as previously described [53]. Approximately 200 cells were analyzed for each sample in triplicate. Error bars represent the standard deviation.
Phenotypic Analysis
Hydrogen peroxide resistance
An equal number of mid-log phase cells containing p423TEF EV, p423TEF Ub or p423TEF Ubext were treated with various concentrations (1 mM, 1.5 mM, 2 mM, 2.5 mM, 3 mM, and 4 mM) of hydrogen peroxide for 30 minutes at 30°C in liquid selective medium. Cells were diluted (1∶5000) and plated on selective medium. Viable cells were counted and normalized to the non-treated sample. Error bars represent the standard deviation of three independent cultures for each construct in each condition.
Proteasome mutant strain synthetic lethality
The proteasome mutant strain (WCG11-22a) and control strain (WCGa) containing p423TEF EV, p423TEF Ub or p423TEF Ubext were grown overnight in selective medium at 30°C and five-fold serial dilutions of the cultures were spotted on selective medium and grown at 30°C and 37°C.
Canavanine treatment
Cells containing p423TEF EV, p423TEF Ub or p423TEF Ubext were grown overnight in selective medium. Five fold serial dilutions of cultures were spotted onto selective medium containing 200 or 400 µM canavanine. Cells containing TOXIC-Q103, Q25, pRS315EV or pRS315Cup-Sup35 were grown overnight in selective medium. Five fold serial dilutions of cultures were spotted onto selective media containing the indicated amount of copper sulfate and canavanine or galactose and canavanine.
[PSI+] induction
Three independent cultures of 74-D694 [psi−] [RNQ+] cells containing pEMBL Sup2 [58] in addition to p423TEF EV, p423TEF Ub or p423TEF Ubext were grown overnight in selective medium to an OD600∼1.5. The cultures were diluted and plated on YPD, where ∼2,000 colonies were scored for prion induction. Error bars represent the standard deviation.
Toxic polyglutamine aggregation
[PSI+] [RNQ+] cells containing p416Gal FLAG103Q CFP or p416Gal FLAG25Q CFP [50],[51] and p423TEF EV, p423TEF Ub, or p423TEF Ubext were grown overnight in selective medium. The cultures were diluted five-fold and spotted on selective media containing 0.1% or 0.3% galactose with 1% raffinose.
Toxic Sup35p over expression
74-D694 [PSI+] [RNQ+] cells containing pRS315Cup-EV or pRS315Cup-Sup35 and p423TEF EV, p423TEF Ubext, p423TEF UbΔGG or p423TEF UbextI44A were grown overnight in selective medium. Cultures were diluted five-fold and spotted on selective medium containing 50 µM or 100 µM copper sulfate. pRS315TEF-CtermSup35 or control pRS315 EV were transformed into Ubext-expressing cells containing pRS315Cup-Sup35 and plated on selective medium containing 50 µM copper sulfate.
Acknowledgments
We thank the members of the True lab, C. Weihl, D. Harris, J. Cooper, M. Linder, and J. Weber for helpful discussions and critical reading of the manuscript. We are grateful for the technical assistance provided by J. Purdy and R. Bouttenot. We thank many people for reagents, equipment and advice: P. Coffino, S. Liebman, R. Deshaies, S. Lindquist, M. Duennwald, A. Varshavsky, M. Linder, J. Glover, K. Weilbacher, and M. Hochstrasser.
Footnotes
The authors have declared that no competing interests exist.
This research was supported by National Institutes of Health grant F31 NS054513 (EMHT), the Alzheimer's Association (HLT) and the Alzheimer's Disease Research Center (HLT).
References
- 1.Goedert M, Spillantini MG. A century of Alzheimer's disease. Science. 2006;314:777–781. doi: 10.1126/science.1132814. [DOI] [PubMed] [Google Scholar]
- 2.Jentsch S. The ubiquitin-conjugation system. Annu Rev Genet. 1992;26:179–207. doi: 10.1146/annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- 3.Johnson ES, Bartel B, Seufert W, Varshavsky A. Ubiquitin as a degradation signal. Embo J. 1992;11:497–505. doi: 10.1002/j.1460-2075.1992.tb05080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz AL, Ciechanover A. The ubiquitin-proteasome pathway and pathogenesis of human diseases. Annu Rev Med. 1999;50:57–74. doi: 10.1146/annurev.med.50.1.57. [DOI] [PubMed] [Google Scholar]
- 6.Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 7.Alves-Rodrigues A, Gregori L, Figueiredo-Pereira ME. Ubiquitin, cellular inclusions and their role in neurodegeneration. Trends Neurosci. 1998;21:516–520. doi: 10.1016/s0166-2236(98)01276-4. [DOI] [PubMed] [Google Scholar]
- 8.Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 9.Maraganore DM, Lesnick TG, Elbaz A, Chartier-Harlin MC, Gasser T, et al. UCHL1 is a Parkinson's disease susceptibility gene. Ann Neurol. 2004;55:512–521. doi: 10.1002/ana.20017. [DOI] [PubMed] [Google Scholar]
- 10.Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 11.van Leeuwen FW, Hol EM, Fischer DF. Frameshift proteins in Alzheimer's disease and in other conformational disorders: time for the ubiquitin-proteasome system. J Alzheimers Dis. 2006;9:319–325. doi: 10.3233/jad-2006-9s336. [DOI] [PubMed] [Google Scholar]
- 12.van Leeuwen FW, de Kleijn DP, van den Hurk HH, Neubauer A, Sonnemans MA, et al. Frameshift mutants of beta amyloid precursor protein and ubiquitin-B in Alzheimer's and Down patients. Science. 1998;279:242–247. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- 13.van Leeuwen FW, Burbach JP, Hol EM. Mutations in RNA: a first example of molecular misreading in Alzheimer's disease. Trends Neurosci. 1998;21:331–335. doi: 10.1016/s0166-2236(98)01280-6. [DOI] [PubMed] [Google Scholar]
- 14.van Leeuwen FW, Fischer DF, Benne R, Hol EM. Molecular misreading. A new type of transcript mutation in gerontology. Ann N Y Acad Sci. 2000;908:267–281. doi: 10.1111/j.1749-6632.2000.tb06654.x. [DOI] [PubMed] [Google Scholar]
- 15.van Den Hurk WH, Willems HJ, Bloemen M, Martens GJ. Novel frameshift mutations near short simple repeats. J Biol Chem. 2001;276:11496–11498. doi: 10.1074/jbc.M011040200. [DOI] [PubMed] [Google Scholar]
- 16.Hol EM, van Leeuwen FW, Fischer DF. The proteasome in Alzheimer's disease and Parkinson's disease: lessons from ubiquitin B+1. Trends Mol Med. 2005;11:488–495. doi: 10.1016/j.molmed.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Fischer DF, De Vos RA, Van Dijk R, De Vrij FM, Proper EA, et al. Disease-specific accumulation of mutant ubiquitin as a marker for proteasomal dysfunction in the brain. Faseb J. 2003;17:2014–2024. doi: 10.1096/fj.03-0205com. [DOI] [PubMed] [Google Scholar]
- 18.De Vrij FM, Sluijs JA, Gregori L, Fischer DF, Hermens WT, et al. Mutant ubiquitin expressed in Alzheimer's disease causes neuronal death. Faseb J. 2001;15:2680–2688. doi: 10.1096/fj.01-0438com. [DOI] [PubMed] [Google Scholar]
- 19.Van Leeuwen FW, Hol EM, Hermanussen RW, Sonnemans MA, Moraal E, et al. Molecular misreading in non-neuronal cells. Faseb J. 2000;14:1595–1602. doi: 10.1096/fj.14.11.1595. [DOI] [PubMed] [Google Scholar]
- 20.Lam YA, Pickart CM, Alban A, Landon M, Jamieson C, et al. Inhibition of the ubiquitin-proteasome system in Alzheimer's disease. Proc Natl Acad Sci U S A. 2000;97:9902–9906. doi: 10.1073/pnas.170173897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Tijn P, de Vrij FM, Schuurman KG, Dantuma NP, Fischer DF, et al. Dose-dependent inhibition of proteasome activity by a mutant ubiquitin associated with neurodegenerative disease. J Cell Sci. 2007;120:1615–1623. doi: 10.1242/jcs.03438. [DOI] [PubMed] [Google Scholar]
- 22.Lindsten K, de Vrij FM, Verhoef LG, Fischer DF, van Leeuwen FW, et al. Mutant ubiquitin found in neurodegenerative disorders is a ubiquitin fusion degradation substrate that blocks proteasomal degradation. J Cell Biol. 2002;157:417–427. doi: 10.1083/jcb.200111034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer DF, van Dijk R, van Tijn P, Hobo B, Verhage MC, et al. Long-term proteasome dysfunction in the mouse brain by expression of aberrant ubiquitin. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.06.009. In press. [DOI] [PubMed] [Google Scholar]
- 24.de Pril R, Fischer DF, Maat-Schieman ML, Hobo B, de Vos RA, et al. Accumulation of aberrant ubiquitin induces aggregate formation and cell death in polyglutamine diseases. Hum Mol Genet. 2004;13:1803–1813. doi: 10.1093/hmg/ddh188. [DOI] [PubMed] [Google Scholar]
- 25.Ozkaynak E, Finley D, Solomon MJ, Varshavsky A. The yeast ubiquitin genes: a family of natural gene fusions. Embo J. 1987;6:1429–1439. doi: 10.1002/j.1460-2075.1987.tb02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Treger JM, Heichman KA, McEntee K. Expression of the yeast UB14 gene increases in response to DNA-damaging agents and in meiosis. Mol Cell Biol. 1988;8:1132–1136. doi: 10.1128/mcb.8.3.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- 28.Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf DH. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J Biol Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- 29.Amerik A, Swaminathan S, Krantz BA, Wilkinson KD, Hochstrasser M. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. Embo J. 1997;16:4826–4838. doi: 10.1093/emboj/16.16.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 31.Dohmen RJ, Madura K, Bartel B, Varshavsky A. The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc Natl Acad Sci U S A. 1991;88:7351–7355. doi: 10.1073/pnas.88.16.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seufert W, Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. Embo J. 1990;9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varshavsky A. The N-end rule pathway of protein degradation. Genes Cells. 1997;2:13–28. doi: 10.1046/j.1365-2443.1997.1020301.x. [DOI] [PubMed] [Google Scholar]
- 34.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, et al. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 35.Pegg AE. Recent advances in the biochemistry of polyamines in eukaryotes. Biochem J. 1986;234:249–262. doi: 10.1042/bj2340249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghoda L, van Daalen Wetters T, Macrae M, Ascherman D, Coffino P. Prevention of rapid intracellular degradation of ODC by a carboxyl-terminal truncation. Science. 1989;243:1493–1495. doi: 10.1126/science.2928784. [DOI] [PubMed] [Google Scholar]
- 37.Hoyt MA, Zhang M, Coffino P. Ubiquitin-independent mechanisms of mouse ornithine decarboxylase degradation are conserved between mammalian and fungal cells. J Biol Chem. 2003;278:12135–12143. doi: 10.1074/jbc.M211802200. [DOI] [PubMed] [Google Scholar]
- 38.Zhang M, Pickart CM, Coffino P. Determinants of proteasome recognition of ornithine decarboxylase, a ubiquitin-independent substrate. Embo J. 2003;22:1488–1496. doi: 10.1093/emboj/cdg158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyazaki Y, Matsufuji S, Murakami Y, Hayashi S. Single amino-acid replacement is responsible for the stabilization of ornithine decarboxylase in HMOA cells. Eur J Biochem. 1993;214:837–844. doi: 10.1111/j.1432-1033.1993.tb17987.x. [DOI] [PubMed] [Google Scholar]
- 40.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilkinson KD, Ventii KH, Friedrich KL, Mullally JE. The ubiquitin signal: assembly, recognition and termination. Symposium on ubiquitin and signaling. EMBO Rep. 2005;6:815–820. doi: 10.1038/sj.embor.7400506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 44.Lee DH, Sherman MY, Goldberg AL. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4773–4781. doi: 10.1128/mcb.16.9.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Nocker S, Sadis S, Rubin DM, Glickman M, Fu H, et al. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 47.Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C. Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc Natl Acad Sci U S A. 1996;93:861–866. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sloper-Mould KE, Jemc JC, Pickart CM, Hicke L. Distinct functional surface regions on ubiquitin. J Biol Chem. 2001;276:30483–30489. doi: 10.1074/jbc.M103248200. [DOI] [PubMed] [Google Scholar]
- 49.Fowden L, Lewis D, Tristram H. Toxic amino acids: their action as antimetabolites. Adv Enzymol Relat Areas Mol Biol. 1967;29:89–163. doi: 10.1002/9780470122747.ch3. [DOI] [PubMed] [Google Scholar]
- 50.Duennwald ML, Jagadish S, Giorgini F, Muchowski PJ, Lindquist S. A network of protein interactions determines polyglutamine toxicity. Proc Natl Acad Sci U S A. 2006;103:11051–11056. doi: 10.1073/pnas.0604548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci U S A. 2006;103:11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci U S A. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gokhale KC, Newnam GP, Sherman MY, Chernoff YO. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J Biol Chem. 2005;280:22809–22818. doi: 10.1074/jbc.M500390200. [DOI] [PubMed] [Google Scholar]
- 54.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 55.Tuite MF, Cox BS. Propagation of yeast prions. Nat Rev Mol Cell Biol. 2003;4:878–890. doi: 10.1038/nrm1247. [DOI] [PubMed] [Google Scholar]
- 56.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 57.Chernoff YO, Uptain SM, Lindquist SL. Analysis of prion factors in yeast. Methods Enzymol. 2002;351:499–538. doi: 10.1016/s0076-6879(02)51867-x. [DOI] [PubMed] [Google Scholar]
- 58.Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, et al. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 59.Dagkesamanskaya AR, Ter-Avanesyan MD. Interaction of the yeast omnipotent suppressors SUP1(SUP45) and SUP2(SUP35) with non-mendelian factors. Genetics. 1991;128:513–520. doi: 10.1093/genetics/128.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chernoff YO, Inge-Vechtomov SG, Derkach IL, Ptyushkina MV, Tarunina OV, et al. Dosage-dependent translational suppression in yeast Saccharomyces cerevisiae. Yeast. 1992;8:489–499. doi: 10.1002/yea.320080702. [DOI] [PubMed] [Google Scholar]
- 61.Chernova TA, Allen KD, Wesoloski LM, Shanks JR, Chernoff YO, et al. Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due to depletion of the free ubiquitin pool. J Biol Chem. 2003;278:52102–52115. doi: 10.1074/jbc.M310283200. [DOI] [PubMed] [Google Scholar]
- 62.Allen KD, Chernova TA, Tennant EP, Wilkinson KD, Chernoff YO. Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J Biol Chem. 2007;282:3004–3013. doi: 10.1074/jbc.M609597200. [DOI] [PubMed] [Google Scholar]
- 63.Tank EM, Harris DA, Desai AA, True HL. Prion protein repeat expansion results in increased aggregation and reveals phenotypic variability. Mol Cell Biol. 2007;27:5445–5455. doi: 10.1128/MCB.02127-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 65.Derkatch IL, Bradley ME, Liebman SW. Overexpression of the SUP45 gene encoding a Sup35p-binding protein inhibits the induction of the de novo appearance of the [PSI+] prion. Proc Natl Acad Sci U S A. 1998;95:2400–2405. doi: 10.1073/pnas.95.5.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 68.Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol Biol Cell. 2008;19:2433–2443. doi: 10.1091/mbc.E08-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y, Meriin AB, Zaarur N, Romanova NV, Chernoff YO, et al. Abnormal proteins can form aggresome in yeast: aggresome-targeting signals and components of the machinery. Faseb J. 2008 doi: 10.1096/fj.08-117614. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ghaboosi N, Deshaies RJ. A conditional yeast E1 mutant blocks the ubiquitin-proteasome pathway and reveals a role for ubiquitin conjugates in targeting Rad23 to the proteasome. Mol Biol Cell. 2007;18:1953–1963. doi: 10.1091/mbc.E06-10-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lindsten K, Dantuma NP. Monitoring the ubiquitin/proteasome system in conformational diseases. Ageing Res Rev. 2003;2:433–449. doi: 10.1016/s1568-1637(03)00031-x. [DOI] [PubMed] [Google Scholar]
- 73.Ferreira ST, Vieira MN, De Felice FG. Soluble protein oligomers as emerging toxins in Alzheimer's and other amyloid diseases. IUBMB Life. 2007;59:332–345. doi: 10.1080/15216540701283882. [DOI] [PubMed] [Google Scholar]
- 74.Walsh DM, Selkoe DJ. A beta oligomers - a decade of discovery. J Neurochem. 2007;101:1172–1184. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- 75.Guthrie C, Fink G. Guide to Yeast Genetics and Molecular and Cell Biology. San Diego: Elsevier Academic Press; 2004. [Google Scholar]
- 76.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sambrook J, Russell DW. Molecular cloning : a laboratory manual. Cold Spring Harbor, , N.Y.: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 78.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 79.Danaie P, Altmann M, Hall MN, Trachsel H, Helliwell SB. CLN3 expression is sufficient to restore G1-to-S-phase progression in Saccharomyces cerevisiae mutants defective in translation initiation factor eIF4E. Biochem J. 1999;340 (Pt 1):135–141. [PMC free article] [PubMed] [Google Scholar]
- 80.True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 81.Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 82.Bagriantsev S, Liebman SW. Specificity of prion assembly in vivo. J Biol Chem. 2004;279:51042–51048. doi: 10.1074/jbc.M410611200. [DOI] [PubMed] [Google Scholar]
- 83.Ho AK, Shen TX, Ryan KJ, Kiseleva E, Levy MA, et al. Assembly and preferential localization of Nup116p on the cytoplasmic face of the nuclear pore complex by interaction with Nup82p. Mol Cell Biol. 2000;20:5736–5748. doi: 10.1128/mcb.20.15.5736-5748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]