Abstract
The PTEN tumour suppressor gene is induced by the early growth response 1 (EGR1) transcription factor, which also transactivates p53, p73, and p300/CBP as well as other proapoptotic and anti-cancer genes. Here, we describe a novel Akt–EGR1–alternate reading frame (ARF)–PTEN axis, in which PTEN activation in vivo requires p14ARF-mediated sumoylation of EGR1. This modification is dependent on the phosphorylation of EGR1 at S350 and T309 by Akt, which promotes interaction of EGR1 with ARF at K272 in its repressor domain by the ARF/Ubc9/SUMO system. EGR1 sumoylation is decreased by ARF reduction, and no EGR1 sumoylation is detected in ARF−/− mice, which also exhibit reduced amounts of PTEN. Our model predicts that perturbation of any of the clinically important tumour suppressors, PTEN, EGR1, and ARF, will cause some degree of dysfunction of the others. These results also explain the known negative feedback regulation by PTEN on its own synthesis through PI3 kinase inhibition.
Keywords: EGR1, p14ARF/p19ARF, PTEN, sumoylation, tumour suppression
Introduction
PTEN (phosphatase and tensin homologue, deleted on chromosome 10) is one of the most frequently lost tumour suppressors in human cancer (Li et al, 1997; Teng et al, 1997). The PTEN gene can be damaged by mutation (Priulla et al, 2007) or silenced by epigenetic mechanisms (Mirmohammadsadegh et al, 2006), and PTEN protein stability or function can be reduced by other mechanisms (Priulla et al, 2007). However, in many cancers, the PTEN gene is intact, but appears to be transcriptionally silent. PTEN is a rapidly degraded protein with a half-life (T1/2) of only 2–4 h (depending on cell type), and it appears that many of the genetic alterations found in PTEN in cancer cells further accelerate this rapid degradation (Davies et al, 1999). Thus, PTEN function is critically dependent on de novo synthesis to replenish the pool. We discovered that the early growth response gene 1 (EGR1) transcription factor (Sukhatme et al, 1988) binds directly to a consensus EGR1-binding motif in the PTEN promoter, activates gene transcription, and is necessary for upregulation of PTEN mRNA in response to UV and γ-irradiation and other stress stimuli (Virolle et al, 2001). PTEN transcription can also be induced by p53 (Stambolic et al, 2001), which binds to a site in the promoter close to the EGR1-binding site. Taken together with reports that PTEN and p53 can form a complex (Mayo and Donner, 2002) and our finding that EGR1 transactivates p53 and p73 (Yu et al, 2007), which also transactivates p53, it seems that EGR1, PTEN, and p53 form an intimately connected regulatory network, the understanding of which could be vital to targeted cancer therapy. EGR1 is receiving much attention recently because of its wide range of activities as a transcription factor. Remarkably, EGR1 can exert an effect either as a growth promoter or as a tumour suppressor. EGR1 can also be induced by mutant p53 to contribute to gain of tumour-transforming function (Weisz et al, 2004). In contrast, EGR1 is also upregulated by growth factor addition to normal cells. This may result in cell proliferation or, as described here, suppress cell proliferation or induce apoptosis of transformed or cancer cells; and the alternate reading frame (ARF) protein expedites this latter effect.
The ARF protein (p19ARF in mouse and p14ARF in human) is a product of ARF of the CDKN2A locus. ARF functions mainly as a tumour suppressor essential for ARF–MDM2–p53 pathway (Zhang et al, 1998) and the Rb–E2F-1 pathway (Dimri et al, 2000). ARF has a p53-independent function, promoting the sumoylation of several ARF-interacting proteins, such as Topoisomerase I (Karayan et al, 2001), MDM2 (Xirodimas et al, 2002), Werners helicase (Woods et al, 2004) as well as p53 (Chen and Chen, 2003), and the proteasome-dependent degradation of CtBP (Paliwal et al, 2007).
Here, we show that IGF-1-activated AKT leads to phosphorylation of EGR1 followed by sumoylation of EGR1 by ARF, thereby producing a new modified molecule that directly transactivates the PTEN promoter. The resulting PTEN provides one of many pathways showing how EGR1 can stimulate growth or apoptosis, through specific pathways for each different outcome.
Results
PTEN is not induced in tissues of the EGR1−/− or ARF−/− mouse models
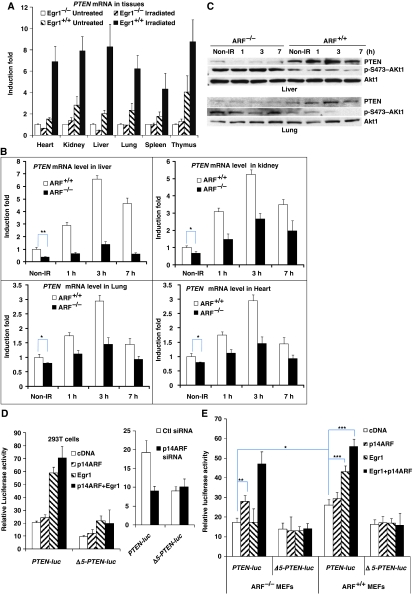
As shown in Figure 1A, PTEN was not induced in any of the studied tissues of EGR1−/− mice at 2.5 h after 5 Gy of γ-irradiation, a time when EGR1 induction is known to be high. In contrast, PTEN expression in wild-type mice was strongly increased two- to four-fold in those tissues. This result suggests that EGR1 is a major transcriptional inducer of the PTEN gene in vivo. We also found that mice lacking ARF show low basal levels and a reduced induction of PTEN mRNA after γ-irradiation compared with wild-type mice, in several tissues (including kidney, heart, liver, and lung) examined over a time course of 7 h (Figure 1B; Supplementary Figure S1). The liver of the ARF−/− mouse 3 h after irradiation was the most sensitive, where PTEN mRNA expression was reduced five-fold compared with normal, and three other tissues examined were also affected, showing reduced PTEN. These findings were verified at the protein levels. PTEN proteins were upregulated and phosphorylated-Akt proteins were subsequently decreased in ARF+/+ liver and lung. In contrast, PTEN proteins were slightly increased, concomitant with sustained Akt phosphorylation in ARF−/− tissues (Figure 1C). Thus, both EGR1 and ARF appear to be important for the full induction of PTEN transcription in living mouse tissues.
Figure 1.
PTEN is not induced in tissues of the EGR1−/− or ARF−/− mouse models. (A) Analysis of PTEN mRNA levels by qRT–PCR in six tissues from 8-week-old, healthy EGR1−/− and wild-type mice that were γ-irradiated with 5 Gy and killed 2.5 h later. (B) A similar qRT–PCR analysis for PTEN mRNA in four tissues: liver, kidney, lung, and heart (and also spleen, data not shown) from ARF−/− 129sv/BL6 mice and wild-type mice of the same strain that were γ-irradiated with5 Gy and killed 0, 1, 3, or 7 h later. (C) Western blot analysis for PTEN, Akt1, and phospho-Akt1 in two tissues liver and lung from the same mice in (B). Luciferase activity measured 24 h after transfection into 293T cells (D) ARF−/− and ARF+/+ MEFs (E) with the indicated DNA constructs and siRNAs. The PTEN-luciferase promoter construct was described earlier (Virolle et al, 2001) and the same reporter Δ5-PTEN-luc lacking most EGR1-binding sites is shown in Supplementary Figure S2. The data represent the mean and triplicate determinations±s.d. (*P<0.05, **P<0.01, ***P<0.001).
To verify that ARF directly affects the EGR1-driven PTEN transcription, we tested if EGR1 can stimulate PTEN transcription in ARF−/− mouse embryo fibroblasts (MEFs) by using a luciferase reporter containing the promoter region of the PTEN gene (Virolle et al, 2001). Although expression of EGR1 or EGR1+p14ARF stimulated PTEN-luc in 293T cells (Figure 1D) and ARF+/+ MEFs (Figure 1E), there was no response to EGR1 alone in ARF−/− MEFs (Figure 1E). Re-expression of p14ARF restored the low basal activity of PTEN-luc in ARF−/− MEFs to the same level of the activity in ARF+/+ MEFs (Figure 1E). Interestingly, there was no major increase in luciferase activity upon addition of exogenous p14ARF either in ARF+/+ MEFs or in 293T cells. When comparing EGR1 alone with EGR1+p14ARF, there were notable increases in the induction of PTEN promoter activity in ARF−/− MEFs, whereas there was no statistical significance in 293T cells, which contain abundant levels of endogenous p14ARF (Karayan et al, 2001). Conversely, the activity of PTEN-luc in 293T cells was decreased when p14ARF was knocked down by p14ARF-siRNA (Figure 1D). As a control, a similar luciferase reporter gene lacking the most efficient EGR1-binding sites (Δ5-PTEN-luc) in the PTEN promoter (Supplementary Figure S2) was much less or not induced by exogenous EGR1 or p14ARF or EGR1+p14ARF or p14ARF-siRNA. Taken together, these data demonstrate that the ARF tumour suppressor is required for EGR1-driven PTEN transcription.
EGR1 interacts with ARF
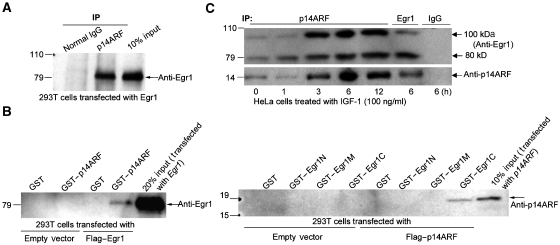
We asked if EGR1 interacts with p14ARF. First, 293T cells were transfected with EGR1 expression plasmid and lysed in M-RIPA buffer (containing 1% of NP40). Lysate was immunoprecipitated with anti-p14ARF and western blotted with anti-EGR1, indicating that EGR1 binds to p14ARF (Figure 2A). Next, equal amounts of bacterially expressed GST or GST–p14ARF fusion proteins and Glutathione-Sepharose beads were added to each lysate of 293T cells transfected with empty vector or with Flag-EGR1. The proteins attached on the GST–p14ARF beads were recovered and washed to remove non-linked proteins. Immunoblots with anti-EGR1 to identify the proteins on the beads also indicated that EGR1 binds to p14ARF (Figure 2B, left panel). Furthermore, we determined what portion of EGR1 binds to p14ARF by making GST–EGR1 fragment constructs that contained N-terminal fragment (1–278), or middle fragment (274–421), or C-terminal fragment (416–543). Anti-p14ARF immunoblot of material bound to the indicated GST or GST–EGR1 fusion proteins in lysates from 293T cells transfected with empty vector or Flag–p14ARF revealed that only fragment EGR1-C binds to p14ARF (Figure 2B, right panel), thus identifying this portion as the binding region for p14ARF. ARF has been shown to target interacting proteins, such as HDM2 for sumoylation (Xirodimas et al, 2002), suggesting a similar post-translational modification to EGR1.
Figure 2.
EGR1 interacts with ARF. (A) Anti-EGR1 immunoblot of normal IgG (control) or anti-p14ARF immunoprecipitates from 293T cells transfected with EGR1. Lane 3 contains input cell lysate (in M-RIPA buffer). (B) Left panel: equal amounts of bacterially expressed GST or GST–p14ARF fusion proteins and Glutathione-Sepharose beads were incubated with lysates of 293T cells transfected with empty vector or vector-EGR1. The proteins associated with GST–p14ARF, bound on the Glutathione-Sepharose beads, were washed five times with the same M-RIPA buffer before western blotting. The input is the cell lysate of cells transfected with EGR1. Right panel: anti-p14ARF immunoblot of material bound to the indicated GST or GST–EGR1 fragments N(1–278), M(274–421), or C(416–543) fusion proteins in lysates (in M-RIPA buffer) from 293T cells transfected with empty vector or Flag–p14ARF. (C) Anti-EGR1 and p14ARF immunoblot of anti-p14ARF or EGR1 or IgG immunoprecipitates from HeLa cells treated with IGF-1 for the indicated times 0–12 h (with NEM-RIPA buffer).
To examine endogenous EGR1 interacting with any bound proteins, we used HeLa cells treated with IGF-1 after starvation in low serum medium. Cells were collected with N-ethylmaleimide (NEM)-PBS buffer and the cell pellets were directly lysed in NEM-RIPA buffer. Immunoprecipitation of the lysates with anti-p14ARF was performed at the indicated times (0–12 h) after IGF-1 addition. HeLa cell immunoprecipitates were immunoblotted to detect EGR1. The interaction between endogenous EGR1 and ARF over 3–12 h was increased and gave the expected normal size of 80 kDa EGR1 band after stimulation of HeLa cells with IGF-1. Interestingly, an additional 100 kDa band was also seen (Figure 2C). The size increase indicated that the larger EGR1 protein most likely consisted of sumoylated EGR1, and that the EGR1–ARF complex might also interact with a sumoylating agent, such as Ubc9 (Rizos et al, 2005; Tago et al, 2005; Jakobs et al, 2007b).
ARF promotes the sumoylation of EGR1
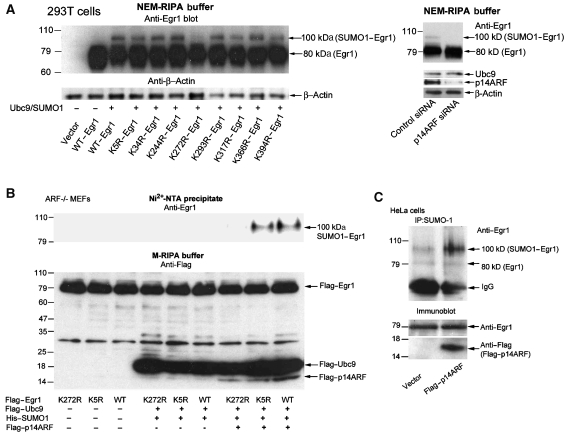
As ARF was shown to associate with Ubc9, which is a E2-conjugating enzyme essential for sumoylation ligase, and to direct the sumoylation of binding partners (Jakobs et al, 2007a), we asked if ARF/Ubc9 can sumoylate EGR1. We then performed in vivo sumoylation assays, which also allowed for the determination of the precise location of the sumoylated site in EGR1 by mutating the most likely lysine residues (Supplementary Figure S3) for analysis of a reactive lysine. In the presence of Ubc9 and SUMO1, the relative molecular weight of EGR1 shifted from 80 to 100 kDa (Figure 3A, left panel; Supplementary Figure S4). This shift of EGR1 was most clearly absent in the K272R (lane 7). Similarly, the 293T cells were cotransfected with Ubc9/SUMO1/EGR1 and control siRNA or p14ARF siRNA. Forty eight hours after transfection, the cells were collected, directly lysed in NEM-RIPA buffer, and used for western blot analysis. As shown in Figure 3A (right panel), the 100 kDa sumoylation of EGR1 was significantly reduced by p14ARF siRNA compared with control siRNA. These data suggest that ARF targets EGR1 for sumoylation at K272, but other lysine residues, such as K5, could also be involved.
Figure 3.
ARF promotes the sumoylation of EGR1. (A) Left panel: 293T cells cotransfected with wild-type or mutated EGR1 constructs with or without Ubc9/SUMO1 were directly lysed in NEM-RIPA buffer 48 h after transfection. Right panel: 293T cells cotransfected with Ubc9/SUMO1/EGR1 and control siRNA or p14ARF siRNA were lysed in NEM-RIPA buffer 48 h after transfection. Western blot was analysed with indicated antibodies. (B) Anti-EGR1 blot of Ni2+-NTA precipitates from ARF−/− MEFs (passage 10) transfected with Flag–EGR1 (K272R, K5R, and wild type) alone and with Flag–UBC9/His6–SUMO1 and with or without Flag–p14ARF, as indicated (upper panel); anti-Flag blot of total lysates in NEM-RIPA buffer (lower panel). (C) Anti-EGR1 blot of anti-SUMO1 immunoprecipitates from HeLa cells transfected with empty vector or Flag–p14ARF (upper panel). Lower panel: anti-EGR1 or anti-Flag immunoblot of total lysates.
Next, we asked if mutants K272R–EGR1 and K5R–EGR1 compared with WT–EGR1 could become sumoylated in ARF−/− MEFs. We used an assay in which His6-tagged SUMO1 is expressed in cells, followed by cell lysis in a denaturing 6 M guanidinium-HCl buffer and precipitation of the tagged SUMO1 with Ni2+-NTA beads. Any SUMO1-conjugated proteins (e.g., EGR1) can then be detected in this precipitate by western blotting. Indeed, transfection of cells with EGR1 and His6–SUMO1/Ubc9/p14ARF resulted in a strong reactivity with anti-EGR1 antibodies in the Ni2+-NTA precipitate (Figure 3B). The EGR1 reactivity resided in one band of Mr ∼100 kDa, consistent with modified EGR1 that was covalently conjugated with one molecule of SUMO1 (SUMO1–EGR1). Sumoylation occurred only when EGR1 was wild type or contained one mutation at K5R, but no sumoylation was detectable when a K272R mutation was expressed, indicating that K272 of EGR1 is required for EGR1 sumoylation by the Ubc9/SUMO1/ARF system. Moreover, in the absence of p14ARF, EGR1 (WT or K5R) was not sumoylated even in conditions where Ubc9/SUMO1 was overexpressed, and as expected, this event was rescued by re-introducing p14ARF into ARF−/− MEFs. These data strongly suggest that ARF is required for EGR1 sumoylation.
Confirmation of the requirements for the sumoylation of EGR1 was also made in HeLa cells that were transfected with Flag–p14ARF or empty vector. The lysates were immunoprecipitated with anti-SUMO1 and western blotted to show that the 100-kDa-sumoylated EGR1 was present at high levels only in the presence of high levels of p14ARF in the p14ARF-transfected cells (Figure 3C).
Sumoylation of EGR1 is required for PTEN induction
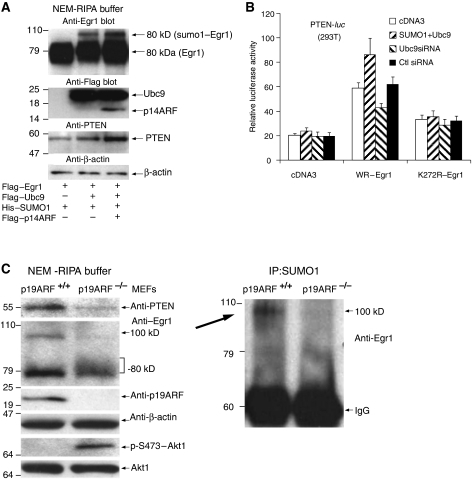
As shown in Figure 4A (Supplementary Figure S5), an additional 100 kDa SUMO1–EGR1 was seen, and PTEN protein levels were also increased in 293T cells transfected with Ubc9, SUMO1, EGR1, and p14ARF (lane 2 and 3), compared with transfection with EGR1 alone (lane 1). Notably, these conditions were also associated with a clearly increased amount of PTEN protein (lane 3). Thus, for conditions where either ARF was depleted (Figure 3A, right panel) or ARF was overexpressed (Figures 3C and 4A), the results showed reciprocal effects on EGR1 sumoylation, which appeared to be important for PTEN transactivation.
Figure 4.
Sumoylation of EGR1 is required for PTEN induction. (A) Anti-EGR1 blot of 293T cells transfected with Flag–EGR1 alone or with Ubc9/SUMO1 and p14ARF as indicated (upper panel). Anti-Flag (second panel), anti-PTEN (third panel), and anti-β-actin (bottom panel) blots of the same experiment. (B) Activation of the PTEN-luc reporter by WT–EGR1 and by K272R–EGR1 in the presence of Ubc9/SUMO1 or Ubc9 siRNA. The 293T cells were transfected with PTEN-luc and SUMO1/Ubc9 or control siRNA or Ubc9 siRNA, and empty vector cDNA3, or WT-EGR1, or K272R-EGR1, as indicated. Luciferase activity was measured 24 h later. (C) Left panel: immunoblots for PTEN, EGR1, p19ARF, β-actin, Akt1, and phospho-Akt1 from ARF−/− (passage 12) and ARF+/+ (passage 5) MEFs. Right panel: anti-EGR1 blot of anti-SUMO1 immunoprecipitates from the same lysates.
We used the K272R mutant to test if EGR1 that cannot be sumoylated at this site can still stimulate PTEN transcription. The 293T cells were transfected with PTEN-luc and SUMO1/Ubc9 or Ubc9 siRNA or control siRNA, and empty vector cDNA3, or WT–EGR1, or K272R–EGR1, as indicated in Figure 4B. In the presence of WT–EGR1, compared with empty vector and control siRNA, the activity of PTEN-luc with SUMO1/Ubc9 was significantly increased, whereas the activity of PTEN-luc with Ubc9 siRNA was much reduced, indicating that sumoylation of EGR1 is controlling EGR1 activity on PTEN transcription. In contrast, in the presence of K272R–EGR1, the activities of PTEN-luc were much reduced. Compared with expression of WT–EGR1 with empty vector, expression of the mutant EGR1 suppressed transactivation of PTEN-luc to similar levels under four conditions of cotransfection with empty vector cDNA3, SUMO1/Ubc9, control siRNA, or Ubc9 siRNA. This result suggests that sumoylation at the K272 residue contributes to EGR1-mediated PTEN transcription.
We examined the function of EGR1 sumoylation on PTEN expression in ARF+/+ and ARF−/− MEFs. Only the ARF+/+ cells formed the 100-kDa-sumoylated EGR1 that was accompanied by 80 kDa EGR1 and ARF as well as PTEN but less phospho-Akt (Figure 4C; Supplementary Figure S6). In contrast, in ARF−/− embryonic fibroblasts, the 100 kDa band of EGR1 was absent and PTEN levels remained low, coinciding with sustained phospho-Akt. Thus, we concluded that EGR1 sumoylation in cells depends on the presence of ARF, and furthermore that induction of PTEN depends on the modification of EGR1 at K272.
PTEN transactivation by IGF-1-induced pathway of Akt–EGR1–ARF–PTEN
To test the function of ARF-dependent modification of EGR1 in the regulation of PTEN, we tested serum-starved HeLa cells with IGF-1. We measured the mRNA and protein levels of the involved proteins, Akt phosphorylation, EGR1, and EGR1 sumoylation in a time course study from 0 to 24 h after IGF-1 addition (Figure 5A). Addition of IGF-1 rapidly activated the Akt kinase, accompanied by EGR1 mRNA and protein at 1 h. Whereas the EGR1 mRNA levels quickly subsided, the level of EGR1 protein was more sustained and there was a second, smaller, induction of EGR1 mRNA and a clear increase in EGR1 protein at 12–24 h. ARF mRNA was induced with slow kinetics, and peaked at 3 h after IGF-1 addition. The increase in ARF protein was slower. Interestingly, SUMO1–EGR1 appeared with kinetics that paralleled the induction of ARF, whereas the induction of PTEN mRNA and protein followed closely behind (Figure 5A). Unexpectedly, addition of the PI3K (and hence Akt) inhibitor, LY294002, reduced the expression of all genes except EGR1, which was boosted, because EGR1 transcription is also induced by alternate pathways (Shin et al, 2006). In contrast, the levels of β-actin were unchanged during the time course. A similar time-course analysis of ARF+/+ and ARF−/− embryonic fibroblasts (Figure 5B) also showed that ARF was necessary for sumoylation of EGR1 and induction of PTEN.
Figure 5.
PTEN transactivation by Akt–EGR1–ARF–PTEN axis. (A) Left panels: EGR1, ARF, and PTEN mRNA levels assessed by qRT–PCR from HeLa cells treated for the indicated times with 100 ng/ml of IGF-1 (black symbols) or LY294002 plus IGF-1 (open symbols). Right panels: immunoblots for phospho-Akt, EGR1, EGR1 in anti-SUMO immunoprecipitates, ARF, PTEN, and β-actin from the same IGF-1-treated cells. The second panel in each group is from cells treated with 20 μM of the PI3K inhibitor LY294002. SUMO1 immunoprecipitations were performed in the presence of N-ethylmaleimide to block desumoylation. (B) Immunoblots of lysates from ARF+/+ and ARF−/− embryonic fibroblasts treated as in (A). (C) EGR1 and p14ARF location as a function of time after addition of IGF-1. Confocal microscopy of EGR1 (red), p14ARF (green), and DNA (blue) in cells treated with IGF-1 for the indicated times. Note that some EGR1s and ARFs colocalize at peak between 3 and 6 h. These confocal images are not taken with equal exposure times and therefore protein levels cannot be compared between panels, for example, it does not show here that EGR1 protein levels are much lower at t=0 than at t=1 h. The purpose of this experiment was to visualize the location of EGR1 and ARF, rather than quantitate. The lower row of images are parallel samples treated with the PI3K inhibitor LY294002 where p14ARF is much reduced. All individual and merged stains are shown in Supplementary Figure S7.
Parallel HeLa cell samples were processed for immunofluorescence staining and confocal microscopy (Figure 5C; Supplementary Figure S7) to visualize the localization of EGR1 and ARF. EGR1 is largely cytoplasmic at t=0, but almost exclusively nuclear at 3 and 6 h. Furthermore, yellow (green+red) and white (green+red+blue) pixels occur at these same time points in and around the predominantly green areas, which most likely represent ARF-containing nucleoli. This revealed that IGF-1 induced accumulation of EGR1 in the nucleus (from a partly cytosolic location), where some EGR1 colocalized with ARF (which is also induced by IGF-1, Figure 5A and Supplementary Figure S8) in nucleoli and perhaps elsewhere in the nucleus, consistent with the time course of protein expression and co-immunoprecipitation (Figure 2C). The response was much reduced in LY294002-treated cells. These results support the conclusion that ARF-dependent sumoylation occurs in the nucleoli and is associated with increased expression of PTEN protein.
Akt has an important function in the regulation of PTEN transcription
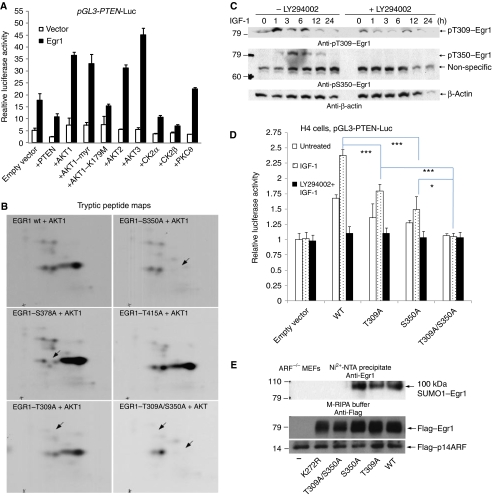
It is known that PTEN negatively regulates its own transcription (Birle et al, 2002), whereas Akt (all three isoforms) augments EGR1-dependent PTEN transcription (Figure 6A). Co-expression of a kinase-inactive mutant of Akt1 (K179M) had little effect. In contrast, casein kinase 2 (CK2) α or β subunits caused a decrease in EGR1 activity, whereas PKCθ had a small effect. These results support the notion that Akt has an important function in the regulation of PTEN transcription, and that PTEN may provide negative feedback on its own transcription (presumably through Akt).
Figure 6.
Akt-phosphorylated EGR1 is necessary for EGR1 sumoylation and PTEN induction. (A) Transactivation of the PTEN-luc reporter by EGR1 in the presence of PTEN, Akt, or other protein kinases. The pGL3–PTEN–luc construct was transfected into 293T cells with empty pcDNA3 vector or EGR1 in the same vector together with expression plasmids for PTEN or the indicated kinases. Luciferase activity was measured 24 h later. Note that PTEN decreases PTEN transcription, whereas all three forms of Akt increase PTEN transcription. Akt1–K179M is a kinase-inactive mutant of Akt1, whereas Akt1myr is constitutively active. All kinases were well expressed (not shown). (B) Tryptic peptide maps of the indicated mutant EGR1-M proteins phosphorylated by Akt1. Identification of S350 and T309 as phosphorylation sites in the Zn-finger region of EGR1. (C) Phosphorylations of EGR1 at S350 and T309 in cells. Lysates from HeLa cell samples in parallel as Figure 5A were blotted with anti-pS350 or anti-pT309 or anti-β-actin. (D) Transactivation of the PTEN-luc reporter by point-mutated EGR1. The PTEN-luc construct was transfected into H4 cells together with empty vector or with the wild type or the indicated point-mutants of EGR1. Twenty-four hours after transfection, serum-free medium was used for 18 h, and to one group LY294002 (20 μM) was added for 1 h pretreatment. Luciferase activity was measured 12 h later after addition of IGF-1 (100 ng/ml). (E) EGR1 phosphorylation at two sites T309 and S350 by Akt is necessary for EGR1 sumoylation. Anti-EGR1 blot of Ni2+-NTA precipitates from ARF−/− MEFs (passage 10) transfected with UBC9/His6–SUMO1 and Flag–p14ARF together with (or without) indicated mutants or wild-type Flag–EGR1. Anti-Flag blot of total lysates (lower two panels) in M-RIPA buffer. This experiment is similar to Figure 3B.
S350 and T309 as phosphorylation sites of EGR1 by Akt1 in vitro
The stimulatory effect of Akt on PTEN transcription through EGR1 introduces the possibility that Akt may directly phosphorylate and stimulate EGR1. It is known that Akt regulates other transcription factors, such as the Forkhead family transcription factors. Indeed, Akt1 readily phosphorylated the central M portion (aa 274–421) of EGR1 (Supplementary Figure S9). The tryptic peptide maps of this phosphoprotein showed that Akt1 phosphorylated one major site, which migrated relatively far to the right on the maps (indicating that the peptide has a stronger positive charge than most peptides), and a few minor sites (Figure 6B). To identify this major phosphorylation site, we mutated a number of possible Akt phosphorylation sites and repeated the kinase reaction and tryptic peptide mapping experiment, which showed that the mutation of S350 to alanine eliminated the main phosphorylation site (upper panel). In contrast, the 4–5 additional weak spots were unchanged, showing that these represent other (minor) sites of in vitro phosphorylation and that the protein was still well folded. We also mutated the predicted Akt target sites S378, T415 (middle panel), T363, and T391 (data not shown), which in similar kinase reactions and tryptic peptide mapping experiments had no impact on phosphorylation by Akt. To help identify the minor site(s), we also analysed the Akt-phosphorylated EGR1M protein by LC-MS/MS and found that T309 was phosphorylated (Supplementary Figure S10). We mutated this site to determine if it corresponds to any (or all) of the minor peptides on the tryptic peptide maps. The mutation of T309 to alanine eliminated the minor phosphorylation sites, and the double mutation of T309A/S350A eliminated most of the spots (lower panel). To follow the phosphorylation of EGR1 in cells, we used phosphospecific antibodies. Both sites are relevant, as the phosphorylations of EGR1 at phospho-S350 and phospho-T309 were detected by specific antibodies (Supplementary Figure S11). Thus, S350 is the major site and T309 is the minor site in EGR1 for Akt1.
S350 and T309 as phosphorylation sites for Akt1 of EGR1 in vivo
Phospho-specific antibodies were used to study the dynamics of phosphorylation of EGR1 in cells. To test the function of the Akt-dependent phosphorylation of EGR1, HeLa cell samples treated with IGF-1 to activate Akt as in Figure 5A were used for western blot with anti-pS350 or anti-pT309. As shown in Figure 6C (middle panel), S350-phosphorylation of EGR1 occurred between 1 and 12 h, with a peak at 3–6 h after IGF-1 addition, and this phosphorylation was completely abolished in the presence of PI3K inhibitor, LY294002, which efficiently reduced Akt activation (see Figure 5A). This result suggests that S350 phosphorylation of EGR1 is controlled only by Akt. In contrast, T309 phosphorylation of EGR1 had a basal level and became much stronger with a peak at 1–6 h upon addition of IGF-1 and was still at the basal level even in the presence of the inhibitor LY294002 (Figure 6C, upper panel), indicating that T309 phosphorylation is controlled not only by Akt but also by other unknown protein kinases, probably PKC family, as this site contains the motif KXXXT. Although the phosphorylation kinetics of the two sites was somewhat different, both phosphorylations occurred in parallel as Akt phosphorylation peaked at 1 h and lasted for 24 h (Figure 5A). The sum of results indicates that Akt is responsible for phosphorylation of EGR1 at S350 and T309 following stimulation of IGF-1.
EGR1 phosphorylation by Akt is necessary for EGR1 sumoylation and PTEN induction
The positive effect of Akt on the EGR1-induced transactivation of PTEN-luc (Figure 6A) indicated that EGR1 phosphorylation by Akt might have a direct stimulatory effect on EGR1. Conversely, the negative effect of expressing active PTEN on the activity of PTEN-luc may simply be due to decreased Akt activity by de novo PTEN. To test this notion, we created the S350A, T309A, and T309A/S350A mutants of EGR1. Luciferase activity assays in 293T cells (Supplementary Figure S12) showed that S350A or T309A had reduced ability to transactivate PTEN-luc. T309A/S350A had much reduced ability, although it still had some ability to induce PTEN transcription, suggesting that additional endogenous EGR1 effects are involved.
To directly address the function of phosphorylation of EGR1 by Akt, we reconstituted the EGR1-negative H4 cells (derived from HT1080 fibrosarcoma cells) with wild-type EGR1 and its S350A, T309A, and S350A/T309A mutants, and measured the ability of IGF-1 to induce PTEN-luc activity. This study showed that the activity of WT–EGR1 in cells treated with IGF-1 was significantly increased compared with untreated cells, whereas cells treated with LY294002+IGF-1 had no activity (Figure 6D). Although two single mutants treated with IGF-1 still showed some ability, the S350A/T309A mutant, similar to empty vector, had no activity. Thus, phosphorylation of EGR1 at S350 and T309 by Akt are necessary for PTEN induction.
On the basis of the result of Figure 5C, it appears that inhibition of Akt does not prevent nuclear translocation of EGR1 and that there is a considerable lag between Akt activation and PTEN mRNA upregulation (Figure 5A), suggesting that Akt may induce one or several intermediate events involving EGR1 that occur in a sequence to induce PTEN transcription. We consider the induction of ARF and ARF-mediated sumoylation of EGR1 to be such intermediate events. Indeed, inhibition of Akt prevented sumoylation after IGF-1 stimulation (Figure 5A). Simultaneously, Akt is still active long after its peak activation (Figure 5A) and EGR1 is phosphorylated by Akt in a prolonged manner (Figure 6C). To test this hypothesis, we assessed the ability of mutants S350A, T309A, and S350A/T309A to become sumoylated in intact cells ARF−/− MEFs (Figure 6E). Both of the single mutants S350A and T309A were still able to be sumoylated, but the double mutant T309A/S350A was not sumoylated, suggesting that phosphorylation of EGR1 at two sites, S350 and T309, is required for sumoylation of EGR1 at K272 in cells.
No SUMO1–EGR1 and low expression of PTEN in ARF−/− mouse tissues
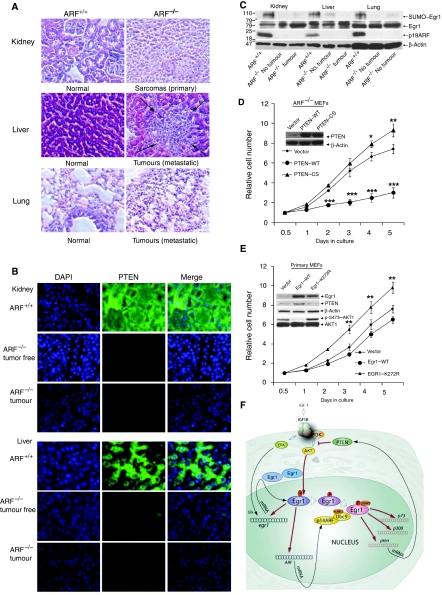
An important consequence of the lack of ARF in mice is the development of tumours in many tissues of ARF−/− mice starting at 2 months of age, whereas ARF+/+ mice remain healthy. To determine the relationship between tumourigenesis and PTEN induction by EGR1 sumoylation under the control of ARF, we analysed tumours that developed in several ARF−/− mice. The most common spontaneous tumours in ARF−/− mice are sarcomas and lymphomas (Kamijo et al, 1998). Primary sarcomas (Figure 7A), lymphomas (Supplementary Figure S13), and muscle tumours (Supplementary Figure S14) were observed in ARF−/− mice but not in ARF+/+ mice. Kidneys (Figure 7A) and inguinal mammary glands (Supplementary Figure S13) of ARF−/− mice were prone to tumour formation; liver and lung soon followed suit. Liver and lung metastases from sarcomas (Figure 7A), lymphomas (Supplementary Figure S13) as well as rhabdomyosarcoma with lung metastases (Supplementary Figure S14) were seen. In fact, mice lacking ARF develop various cancers with 100% penetrance (Kamijo et al, 1998). We performed immunofluorescence analysis of PTEN expression in mouse tissues and tumours (Figure 7B and Supplementary Figure S15), which revealed very low expression of PTEN in ARF−/− mouse tissues and tumours compared with ARF+/+ normal tissues. The 100-kDa-sumoylated EGR1 appeared only in ARF+/+ mouse tissues and was absent from ARF−/− mouse tissues and tumours (Figure 7C). Thus, it appears that induction of PTEN controlled by the ARF–EGR1 sumoylation pathway is highly relevant to tumourigenesis.
Figure 7.
No SUMO1–EGR1 and low expression of PTEN in ARF−/− mouse tissues. (A) Upper panel: sections from an ARF−/− mouse (8 months old) with poorly differentiated sarcoma (primary tumours were in kidney) revealing fascicles of spindle cells, compared with sections from an ARF+/+ mouse normal kidney tissues. Middle panel: sections from the same ARF−/− mouse reveal liver metastasis (arrows) from sarcoma, compared with sections from ARF+/+ mouse liver tissues. Lower panel: sections from the same ARF−/− mouse reveal lung metastasis from the sarcoma, compared with sections from lung tissues of the ARF+/+ mouse. (B) Sections (normal and tumorous) from the same ARF−/−, ARF+/+ tissues as in Figure 7A were immunostained with antibody to PTEN (green) and DAPI (blue). (C) Immunoblot of anti-EGR1 in SUMO1 immunoprecipitates (upper panel), and immunoblot of anti-EGR1 (second panel), anti-p19ARF (third panel) and anti-β-actin (bottom panel) in the same tissue lysates. Note that sumoylated EGR1 is only highly expressed in ARF+/+ mouse tissues. New pathway suppresses cell proliferation by control of PTEN expression. ARF−/− MEFs infected with pBabe–PuroL–PTEN (WT), pBabe–PuroL–PTEN (C124S), pBabe–PuroL vector (D) and wild-type primary MEFs infected with pBabe–PuroL–EGR1 (WT), pBabe–PuroL–EGR1 (K272R), pBabe–PuroL vector (E) were seeded on plates at 4–5 × 103 cells in 96-well plates and cell numbers were determined by CyQUANT fluorescence assay at 0.5, 1, 2, 3, 4, 5 days after seeding. The cell numbers were normalized against the value at 0.5 day (Mean values±s.e.m., from three independent experiments, *P<0.05, **P<0.01, ***P<0.001). The related protein levels were shown by western blot (inset). (F) Schematic model of the PTEN transactivation pathway through a novel Akt–EGR1–ARF–PTEN axis.
New pathway suppresses cell proliferation by control of PTEN expression
To prove a tumour suppression function for ARF/EGR1/PTEN, we restored PTEN expression in ARF-null MEFs, in which PTEN expression is low (Figures 4C and 5B), using retroviruses expressing PTEN–WT, or PTEN–CS (C124S at the phosphatase catalytic centre) or retroviruses carrying the empty vector. As shown in Figure 7D, re-expression of PTEN is associated with both decreased cell proliferation and saturation density when compared with PTEN–CS and empty vector, and PTEN–CS expression increased to a higher saturation density than the empty vector, suggesting that re-expression of PTEN conferred a growth suppression effect in ARF−/− MEFs.
To test whether the sumoylation site K272 of EGR1 is required for PTEN induction and inhibition of cell proliferation, we infected primary wild-type MEFs with retrovirus expressing EGR1–WT or EGR1–K272R or retrovirus carrying the empty vector. As shown in Figure 7E, mutant EGR1–K272R expression significantly increased cell proliferation rate compared with the empty vector, whereas EGR1–WT expression decreased cell proliferation rate. Moreover, PTEN protein levels followed the order of EGR1–WT>empty vector>EGR1–K272R in infected primary MEFs, whereas phospho-AKT protein levels were in the reverse order. In contrast, expression of either EGR1–WT or EGR1–K272R in ARF−/− MEFs did not affect either cell proliferation or saturation density, suggesting that p19ARF is necessary for the effect of EGR1 on PTEN induction (Supplementary Figure S16). The combined results on the function of phosphorylation and sumoylation strongly support the conclusion that sequential modification of EGR1 by the ARF/EGR1/PTEN pathway suppresses cell growth/proliferation by control of PTEN expression.
In summary, in Figure 7F, IGF-1-induced PTEN mRNA upregulation is due to an unexpected pathway that starts with the Akt kinase and involves EGR1 phosphorylation and sumoylation by an ARF-dependent mechanism. Therefore EGR1 is unable to activate PTEN in ARF−/− cells and mice.
Discussion
Both EGR1 and p53 transcription factors have major functions in controlling cell proliferation, and together they can additively suppress transformed growth (de Belle et al, 1999). Whereas p53 expression is associated with tumour suppression and apoptosis, EGR1 has dual functions in cell proliferation and in the promotion of apoptosis (Huang et al, 1998; Krones-Herzig et al, 2005). There are also several instances of interaction between EGR1 and p53, and the most important is the ability of each of the two transcription factors to induce the transcription of the other (Yu et al, 2007). Although p53 is more frequently inactivated in cancer cells compared with EGR1, the latter can also induce the p53-related p73 (Yu et al, 2007) to cause tumour suppression. In some conditions, p53 also binds to EGR1 to interfere or modify transcriptional activities (Liu et al, 2001).
Although EGR1 was first shown to be activated by growth factors and to stimulate proliferation of cells, we here show that the activation of Akt by IGF-1 leads to the phosphorylation of EGR1 at T309 and S350. The activation of EGR1 continues by its migration to the nucleolus (Figure 5C), where ARF-mediated sumoylation of EGR1 occurs, and where ARF and nucleophosmin have accumulated (den Besten et al, 2006). Sumoylation of other transcription factors has been described before, for example, for p53 (Chen and Chen, 2003). The process requires interaction of ARF with Ubc9/SUMO1, to cause the 80 kDa EGR1 protein to become the reactive 100 kDa molecule, SUMO1-EGR1. This modified form in the nucleolus appears to have an increased ability to induce PTEN transcription, thus providing the tumour suppressor activity through PTEN. This is a notable pathway owing to the fact that IGF-1 can cause the switch of functions of its transcription factor target, EGR1, to exert an effect as an inhibitor of growth factor-stimulated cancer cells.
Our data demonstrate that EGR1 is important for PTEN transcription in response to irradiation or IGF-1 and that this activity of EGR1 is controlled by ARF and EGR1 sumoylation. This implies that loss of EGR1, or loss of ARF, in cancer cells would be expected to lead to reduced PTEN expression as well. Indeed, EGR1 is often downregulated or lost in human cancer and in immortalized cell lines, such as HT1080 human fibrosarcoma (Liu et al, 1999) and many glial and breast tumour cell lines (Huang et al, 1997). In EGR1-deficient cell lines, re-introduction of EGR1 suppresses transformation and tumorigenicity (Huang and Adamson, 1995; Liu et al, 1999). Allelic deletions of EGR1 have been associated with the premalignant condition, myelodysplastic syndrome (Le Beau et al, 1993), and with the development of acute myeloid leukaemia (Horrigan et al, 1996) and small cell and non-small cell lung carcinoma (Levin et al, 1995).
In both EGR1−/− and ARF−/− mice, p53 is poorly induced upon γ-irradiation. This is at least partly because of the function of EGR1 in the transcription of p53 (Krones-Herzig et al, 2005) and PTEN. Also at the protein level, p53 associates with nuclear PTEN protein (Mayo and Donner, 2002; Mao et al, 2003), as well as with EGR1 and ARF. In addition, the promoter region of the ARF gene contains several perfect-match EGR1-binding sites, and ARF transcription is readily induced by EGR1 (Supplementary Figure S8). Thus, these connections constitute an elaborate tumour suppressor network (Figure 7F), the perturbation of which appears to be common in human cancer. Further interactions, such as the ability of EGR1 to induce p53, together with a reverse positive feedback (Yu et al, 2007), also modify the resulting expression levels of the interacting genes in the network described here, thus affecting the time of onset of tumour formation in each tissue of ARF−/− mice.
In addition to using ARF−/− MEFs to demonstrate the actions of ARF, we also used HeLa and 293T as readily transfectable cells that lack active p53. HeLa cells possess wild-type p53, but due to the expression of HPV-encoded E6 and E7 oncoproteins, both p53 and retinoblastoma functions are inactivated. p53, for example, undergoes accelerated degradation and is undetectable in these cells (Wesierska-Gadek et al, 2002). Similarly p53 in 293T cells is disabled by the E1B and SV40 T-antigen used to immortalize the cells, and is therefore non-functional (Tago et al, 2005; Sherr, 2006). We also tested the effect of the transfection of siRNA to p53 into HeLa cells to measure the effect on PTEN levels in growing HeLa cells. However, p53 protein was undetectable even in normal-growing HeLa cells. Therefore, in these cells, ARF is elevated independently of p53 and can be tested for the requirement of EGR1 for PTEN transcription in the absence of p53, which is also a known inducer of PTEN (Stambolic et al, 2001) and can also bind to EGR1(Liu et al, 2001). Using these cells, we examined why ARF is needed for EGR1 function as a transcription factor.
PTEN transcription is controlled by several pathways that involve many transcription factors. In this study, we describe a novel multiple-stage pathway of PTEN transcription through a novel AKT–EGR1–ARF–PTEN axis and demonstrate its importance in cells and mice. ARF−/− tissues expressed much less PTEN when compared with ARF+/+ tissues; however, PTEN in the former was still detectable at the protein level (Figure 1C) and also inducible at the mRNA level (Figure 1B) after γ-irradiation, which involves other pathways, for example, PTEN transcription can also be induced by p53. Interestingly, studies show that the tumour suppressor effects of ARF and PTEN cooperate to reduce sarcomas (Carrasco et al, 2006). PTEN+/−ARF−/− has effects beyond ARF−/− alone on histiocytic sarcoma (Carrasco et al, 2006), probably due to PTEN expression that was reduced rather than absent in ARF−/− tissues. Also, there are other complex interactions with these two proteins that may occur. Therefore, it could not be simply concluded that ARF loss is a phenotype copy of PTEN loss.
Finally, as shown in the summary in Figure 7F, the target gene product, PTEN, also has a negative activity on its own production at the stage of interfering with the phosphorylation of Akt as the first step in the promotion of EGR1 as a tumour suppressor, following the steps outlined above. Thus, some feedback inhibition of the tumour suppressor activity of PTEN follows this additional effect.
Materials and methods
Cell cultures and mice
Human embryonic kidney 293T cells, H4 cells, HeLa cells, EGR1−/− and EGR1+/+ MEFs, and ARF−/− and ARF+/+ MEFs were cultured in DMEM containing 10% FBS, penicillin, and streptomycin at 37°C and 5% CO2. The ARF−/− and ARF+/+ MEFs, prepared from ARF-null and wild-type 129 embryos, were gifts from Dr C Sherr. ARF−/− 129sv/BL6 mice were kindly provided by the NCI Mouse Repository (Frederick, MD, USA) with the permission of Dr C Sherr.
Generation of PTEN or EGR1 retrovirus and infection of cells
To generate a high titre of viral stocks, pBabe–PuroL, pBabe–PuroL–PTEN (WT), pBabe–PuroL–PTEN (C124S), pBabe–PuroL–EGR1 (WT), or pBabe–PuroL–EGR1(K272R) were transfected into the BOSC-23 packaging line using Lipofectamine 2000. Wild-type primary and ARF−/− MEFs were infected and transduced cells (colonies) were selected by puromycin (for ARF−/− MEFs, 2 μg/ml for 2 weeks; for primary MEFs, 2 μg/ml for 3–4 days). Cell proliferation ratios were determined by CyQUANT Cell Proliferation Assay Kit (Invitrogen/Molecular Probes).
Three methods for analysis of SUMO-modified proteins
Analysis of exogenous SUMO1–EGR1 by directly lysing in NEM-RIPA buffer.
Analysis of endogenous SUMO1–EGR1 by immunoprecipitation.
Analysis of His-tagged SUMO1 conjugates by using Ni2+-NTA beads.
Luciferase assays
H4 and 293T cells were inoculated 1 day before, and transfection was performed with the Lipofectamine 2000 (Invitrogen). For transfection into ARF−/− and ARF+/+ (passage 4–5) MEFs, were electroporated with MEF Nucleofector Kit 1 (from Amaxa Biosystems) according to the manufacture's protocol. Luciferase assay were performed as described previously (Yu et al, 2007).
Supplementary Material
Supplementary Data
Acknowledgments
We thank Dr CJ Sherr for the kind gift of ARF+/+ and ARF−/− mouse embryo fibroblasts. This study was supported by grants 5R01CA096949 (TM), R01CA78606 (GSF) and 5UO1CA114810 (SPECS) (DM) from NIH.
Conflict of interest The authors declare that they have no competing financial interests.
References
- Birle D, Bottini N, Williams S, Huynh H, deBelle I, Adamson E, Mustelin T (2002) Negative feedback regulation of the tumor suppressor PTEN by phosphoinositide-induced serine phosphorylation. J Immunol 169: 286–291 [DOI] [PubMed] [Google Scholar]
- Carrasco DR, Fenton T, Sukhdeo K, Protopopova M, Enos M, You MJ, Divicio D, Nogueira C, Stommel J, Pinkus GS, Fletcher C, Hornick JL, Cavenee WK, Furnari FB, DePinho RA (2006) The PTEN and INK4A/ARF tumor suppressors maintain myelolymphoid homeostasis and cooperate to constrain histiocytic sarcoma development in humans. Cancer Cell 9: 379–390 [DOI] [PubMed] [Google Scholar]
- Chen L, Chen J (2003) MDM2–ARF complex regulates p53 sumoylation. Oncogene 22: 5348–5357 [DOI] [PubMed] [Google Scholar]
- Davies MA, Koul D, Dhesi H, Berman R, McDonnell TJ, McConkey D, Yung WKA, Steck PA (1999) Regulation of Akt/PKB activity, cellular growth, and apoptosis in prostate carcinoma cells by MMAC/PTEN. Cancer Res 59: 2551–2556 [PubMed] [Google Scholar]
- de Belle I, Huang RP, Fan Y, Liu C, Mercola D, Adamson ED (1999) p53 and Egr-1 additively suppress transformed growth in HT1080 cells but Egr-1 counteracts p53-dependent apoptosis. Oncogene 18: 3633–3642 [DOI] [PubMed] [Google Scholar]
- den Besten W, Kuo ML, Tago K, Williams RT, Sherr CJ (2006) Ubiquitination of, and sumoylation by, the Arf tumor suppressor. Isr Med Assoc J 8: 249–251 [PubMed] [Google Scholar]
- Dimri GP, Itahana K, Acosta M, Campisi J (2000) Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14ARF tumor suppressor. Mol Cell Biol 20: 273–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrigan SK, Westbrook CA, Kim AH, Banerjee M, Stock W, Larson RA (1996) Polymerase chain reaction-based diagnosis of del (5q) in acute myeloid leukemia and myelodysplastic syndrome identifies a minimal deletion interval. Blood 88: 2665–2670 [PubMed] [Google Scholar]
- Huang RP, Adamson ED (1995) A biological role for Egr-1 in cell survival following ultra-violet irradiation. Oncogene 10: 467–475 [PubMed] [Google Scholar]
- Huang RP, Fan Y, de Belle I, Niemeyer C, Gottardis MM, Mercola D, Adamson ED (1997) Decreased Egr-1 expression in human, mouse and rat mammary cells and tissues correlates with tumor formation. Int J Cancer 72: 102–109 [DOI] [PubMed] [Google Scholar]
- Huang RP, Fan Y, Peng A, Zeng ZL, Reed JC, Adamson ED, Boynton AL (1998) Suppression of human fibrosarcoma cell growth by transcription factor, Egr-1, involves down-regulation of Bcl-2. Int J Cancer 77: 880–886 [DOI] [PubMed] [Google Scholar]
- Jakobs A, Himstedt F, Funk M, Korn B, Gaestel M, Niedenthal R (2007a) Ubc9 fusion-directed SUMOylation identifies constitutive and inducible SUMOylation. Nucl Acids Res 35: e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs A, Koehnke J, Himstedt F, Funk M, Korn B, Gaestel M, Niedenthal R (2007b) Ubc9 fusion-directed SUMOylation (UFDS): a method to analyze function of protein SUMOylation. Nat Meth 4: 245–250 [DOI] [PubMed] [Google Scholar]
- Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ (1998) Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA 95: 8292–8297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayan L, Riou JF, Séité P, Migeon J, Cantereau A, Larsen CJ (2001) Human ARF protein interacts with topoisomerase I and stimulates its activity. Oncogene 20: 836–848 [DOI] [PubMed] [Google Scholar]
- Krones-Herzig A, Mittal S, Yule K, Liang H, English C, Urcis R, Soni T, Adamson ED, Mercola D (2005) Early growth response 1 acts as a tumor suppressor in vivo and in vitro via regulation of p53. Cancer Res 65: 5133–5143 [DOI] [PubMed] [Google Scholar]
- Le Beau MM, Espinosa R, Neuman WL, Stock W, Roulston D, Larson RA, Keinanen M, Westbrook CA (1993) Cytogenetic and molecular delineation of the smallest commonly deleted region of chromosome 5 in malignant myeloid diseases. Proc Natl Acad Sci USA 90: 5484–5488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin WJ, Press MF, Gaynor RB, Sukhatme VP, Boone TC, Reissmann PT, Figlin RA, Holmes EC, Souza LM, Slamon DJ (1995) Expression patterns of immediate early transcription factors in human non-small cell lung cancer. The Lung Cancer Study Group. Oncogene 11: 1261–1269 [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R (1997) PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275: 1943–1947 [DOI] [PubMed] [Google Scholar]
- Liu C, Yao J, de Belle I, Huang R-P, Adamson ED, Mercola D (1999) The transcription factor EGR-1 suppresses transformation of human fibrosarcoma HT1080 cells by coordinated induction of transforming growth factor-beta 1, fibronectin, and plasminogen activator inhibitor-1. J Biol Chem 274: 4400–4411 [DOI] [PubMed] [Google Scholar]
- Liu J, Grogan L, Nau MM, Allegra CJ, Chu E, Wright JJ (2001) Physical interaction between p53 and primary response gene Egr-1. Int J Oncol 18: 863–870 [DOI] [PubMed] [Google Scholar]
- Mao JH, Wu D, Perez-Losada J, Nagase H, DelRosario R, Balmain A (2003) Genetic interactions between Pten and p53 in radiation-induced lymphoma development. Oncogene 22: 8379–8385 [DOI] [PubMed] [Google Scholar]
- Mayo LD, Donner DB (2002) The PTEN, Mdm2, p53 tumor suppressor-oncoprotein network. Trends Biochem Sci 27: 462–467 [DOI] [PubMed] [Google Scholar]
- Mirmohammadsadegh A, Marini A, Nambiar S, Hassan M, Tannapfel A, Ruzicka T, Hengge UR (2006) Epigenetic silencing of the PTEN gene in melanoma. Cancer Res 66: 6546–6552 [DOI] [PubMed] [Google Scholar]
- Paliwal S, Kovi RC, Nath B, Chen Y-W, Lewis BC, Grossman SR (2007) The alternative reading frame tumor suppressor antagonizes hypoxia-induced cancer cell migration via interaction with the COOH-terminal binding protein corepressor. Cancer Res 67: 9322–9329 [DOI] [PubMed] [Google Scholar]
- Priulla M, Calastretti A, Bruno P, Azzariti A, Paradiso A, Canti G, Nicolin A (2007) Preferential chemosensitization of PTEN-mutated prostate cells by silencing the Akt kinase. Prostate 67: 782–789 [DOI] [PubMed] [Google Scholar]
- Rizos H, Woodruff S, Kefford RF (2005) p14ARF interacts with the SUMO-conjugating enzyme Ubc9 and promotes the sumoylation of its binding partners. Cell Cycle 4: 597–603 [DOI] [PubMed] [Google Scholar]
- Sherr CJ (2006) Divorcing ARF and p53: an unsettled case. Nat Rev Cancer 6: 663–673 [DOI] [PubMed] [Google Scholar]
- Shin SY, Bahk YY, Ko J, Chung IY, Lee YS, Downward J, Eibel H, Sharma PM, Olefsky JM, Kim YH, Lee B, Lee YH (2006) Suppression of Egr-1 transcription through targeting of the serum response factor by oncogenic H-Ras. EMBO J 25: 1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak TW (2001) Regulation of PTEN transcription by p53. Molecular Cell 8: 317–325 [DOI] [PubMed] [Google Scholar]
- Sukhatme VP, Cao X, Chang LC, Tsai-Morris C-H, Stamenkovich D, Ferreira PCP, Cohen DR, Edwards SA, Shows TB, Curran T, Le Beau MM, Adamson ED (1988) A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell 53: 37–43 [DOI] [PubMed] [Google Scholar]
- Tago K, Chiocca S, Sherr CJ (2005) Sumoylation induced by the Arf tumor suppressor: A p53-independent function. Proc Natl Acad Sci 102: 7689–7694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng DHF, Hu R, Lin H, Davis T, Iliev D, Frye C, Swedlund B, Hansen KL, Vinson VL, Gumpper KL, Ellis L, El-Naggar A, Frazier M, Jasser S, Langford LA, Lee J, Mills GB, Pershouse MA, Pollack RE, Tornos C et al. (1997) MMAC1/PTEN mutations in primary tumor specimens and tumor cell lines. Cancer Res 57: 5221–5225 [PubMed] [Google Scholar]
- Virolle T, Adamson ED, Baron V, Birle D, Mercola D, Mustelin T, de Belle I (2001) The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nat Cell Biol 3: 1124–1128 [DOI] [PubMed] [Google Scholar]
- Weisz L, Zalcenstein A, Stambolsky P, Cohen Y, Goldfinger N, Oren M, Rotter V (2004) Transactivation of the EGR1 gene contributes to mutant p53 gain of function. Cancer Res 64: 8318–8327 [DOI] [PubMed] [Google Scholar]
- Wesierska-Gadek J, Schloffer D, Kotala V, Horky M (2002) Escape of p53 protein from E6-mediated degradation in HeLa cells after cisplatin therapy. Int J Cancer 101: 128–136 [DOI] [PubMed] [Google Scholar]
- Woods YL, Xirodimas DP, Prescott AR, Sparks A, Lane DP, Saville MK (2004) p14 Arf promotes small ubiquitin-like modifier conjugation of Werners Helicase. J Biol Chem 279: 50157–50166 [DOI] [PubMed] [Google Scholar]
- Xirodimas DP, Chisholm J, Desterro JMS, Lane DP, Hay RT (2002) P14ARF promotes accumulation of SUMO-1 conjugated (H)Mdm2. FEBS Letts 528: 207–211 [DOI] [PubMed] [Google Scholar]
- Yu J, Baron V, Mercola D, Mustelin T, Adamson ED (2007) A network of p73, p53 and Egr1 is required for efficient apoptosis in tumor cells. Cell Death Differ 14: 436–446 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG (1998) ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92: 725–734 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data