Abstract
The interaction between the poly(A)-binding protein (PABP) and eukaryotic translational initiation factor 4G (eIF4G), which brings about circularization of the mRNA, stimulates translation. General RNA-binding proteins affect translation, but their role in mRNA circularization has not been studied before. Here, we demonstrate that the major mRNA ribonucleoprotein YB-1 has a pivotal function in the regulation of eIF4F activity by PABP. In cell extracts, the addition of YB-1 exacerbated the inhibition of 80S ribosome initiation complex formation by PABP depletion. Rabbit reticulocyte lysate in which PABP weakly stimulates translation is rendered PABP-dependent after the addition of YB-1. In this system, eIF4E binding to the cap structure is inhibited by YB-1 and stimulated by a nonspecific RNA. Significantly, adding PABP back to the depleted lysate stimulated eIF4E binding to the cap structure more potently if this binding had been downregulated by YB-1. Conversely, adding nonspecific RNA abrogated PABP stimulation of eIF4E binding. These data strongly suggest that competition between YB-1 and eIF4G for mRNA binding is required for efficient stimulation of eIF4F activity by PABP.
Keywords: eIF4F, mRNA circularization, PABP, translation initiation, YB-1
Introduction
Translational control of gene expression in most circumstances occurs at the level of initiation, in which the 80S ribosome is recruited to the mRNA and positioned at the initiation codon. This mechanism is ATP- and GTP-dependent and requires the participation of initiation factors (Merrick, 2004). The cap structure at the mRNA 5′ end and the poly(A) tail at the 3′ end are key structures responsible for ribosome recruitment to the mRNA, and their effect on translation is synergistic. This synergy is universal as it was demonstrated in yeast, plant, and mammalian systems (for reviews, see Jacobson, 1996; Sachs, 2000) and was also recapitulated in vitro (Gebauer et al, 1999; Bergamini et al, 2000; Michel et al, 2000; Svitkin et al, 2001a; Rifo et al, 2007).
The mechanism by which the mRNA 5′ cap and 3′ poly(A) tail synergize to stimulate translation has been studied extensively using biochemical and genetic approaches, yet is not fully understood. The m7G cap structure and the poly(A) tail are recognized by eIF4E and the poly(A)-binding protein (PABP), respectively. PABP is an evolutionarily conserved protein, which binds the poly(A) tract with a periodicity of ∼27 nucleotides (Baer and Kornberg, 1983). eIF4E is the cap-binding subunit of the eIF4F complex, which also includes eIF4A, an RNA-dependent ATPase/RNA helicase, and eukaryotic translational initiation factor 4G (eIF4G), a high-molecular-weight protein that functions as a scaffold for binding eIF4E and eIF4A (Gingras et al, 1999). Importantly, eIF4G also interacts with PABP (Tarun and Sachs, 1996; Imataka et al, 1998) and the 40S ribosome-binding initiation factor eIF3 (Hershey and Merrick, 2000; Morino et al, 2000). Circularization of an mRNA through the molecular bridge cap–eIF4E–eIF4G–PABP–poly(A) was observed with recombinant yeast proteins, eF4G, eIF4E, and PABP (Wells et al, 1998). Depletion of PABP from Krebs-2 mouse ascites cell extracts results in a reduction of 48S and 80S ribosome initiation complex formation, which is rescued by the addition of recombinant PABP (Kahvejian et al, 2005). Furthermore, the importance of the intact bridging complex eIF4E–eIF4G–PABP for efficient crosslinking of eIF4E to the m7G cap was demonstrated (Kahvejian et al, 2005). Finally, the translational repressor PABP-interacting protein 2 (Paip2) exerts its activity by displacing PABP from the poly(A) tail and eIF4G (Khaleghpour et al, 2001; Karim et al, 2006). These findings define PABP as a bona fide initiation factor (although it is apparently not released from the mRNA in contrast to other initiation factors), and emphasize the importance of mRNA circularization for translation initiation.
In addition to PABP, all cytoplasmic messenger ribonucleoproteins (mRNPs) contain an mRNA packaging protein, YB-1 (Blobel, 1972). YB-1 possesses high affinity for single-stranded RNA and DNA. At high concentrations, YB-1 functions as a general translation repressor that interferes with eIF4F–mRNA interaction (Minich et al, 1993; Davydova et al, 1997; Evdokimova et al, 2001; Nekrasov et al, 2003). In addition, YB-1 stabilizes mRNA (Evdokimova et al, 2001) and has several documented activities in transcription (Kohno et al, 2003).
In this study, we show that YB-1 and other RNA-binding protein availability controls the PABP-stimulatory activity of eIF4F and assembly of ribosome initiation complexes. We propose that YB-1 competes with eIF4G for binding to the mRNA. PABP relieves this competition by enhancing the binding of eIF4G to the mRNA and stabilizing the eIF4E–cap complex. These data and our earlier published study (Svitkin et al, 1996) implicate YB-1 as an essential factor for the stringent translational control of eukaryotic mRNAs by the 5′ cap and 3′ poly(A) tail.
Results
Stimulation of 48S initiation complex formation by PABP in a reconstituted system
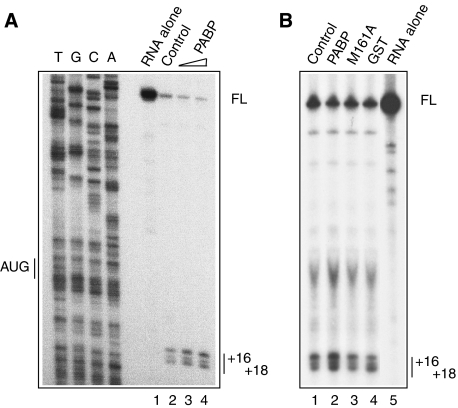
Experiments were first conducted to define the requirement for PABP activity in an in vitro reconstitution of 48S ribosomal complex formation. Upon incubation of β-globin mRNA, ATP, GTP, initiator Met-tRNAi, eIF1, eIF1A, eIF2, eIF3, eIF4A, eIF4B, eIF4F, and 40S ribosomal subunits, a 48S ribosomal complex can be formed on the initiation codon of the mRNA (Pisarev et al, 2007). The formation of the initiation complex can be measured by toe-printing, which is based on the inhibition of primer extension by reverse transcriptase on the template mRNA by the 48S ribosomal complex at positions +16 to +18 downstream of the initiation codon (Pestova et al, 1996). If PABP stimulates eIF4F activity, then it should stimulate the assembly of the 48S complex. Indeed, under standard conditions of reconstitution (Pisarev et al, 2007) a small but reproducible enhancement of 48S complex formation by PABP was evident (∼1.6-fold enhancement at 15 μg/ml PABP; Figure 1A, compare lanes 3 and 2). Increasing the concentration of PABP in the reaction mixture failed to further stimulate 48S complex formation (Figure 1A, lane 4). GST and a mutant PABP, M161A, which cannot interact with eIF4G (Kahvejian et al, 2005), showed no activity, thus providing negative controls for this experiment (Figure 1B). The effect of PABP on 48S complex formation in the reconstituted system is feeble as compared with the strong stimulation of ribosome binding by PABP in crude cell extracts (Kahvejian et al, 2005). This suggests that a factor(s) required for PABP function is lacking in the reconstituted system. We therefore used crude cell extracts to search for this factor(s).
Figure 1.
Effect of PABP on 48S ribosomal complex formation in a reconstituted system. (A) Ribosomal 40S subunits were incubated with rabbit globin mRNA under standard reaction conditions (see Materials and methods). Recombinant PABP was present in the reaction mixtures at 15 μg/ml (lane 3) and 30 μg/ml (lane 4) concentrations. In lane 2, no PABP was added. In lane 1, no reaction components were present except for the mRNA. The assembly of 48S ribosomal complexes with β-globin mRNA was analysed by toe-printing. Side-by-side sequence analysis of pBS− (β-globin) (Morino et al, 2000) with the use of the same primer is shown on the left. Formation of the 48S complex was quantified. With the value for lane 2 being set as 100, the values for lanes 3 and 4 were 158 and 163, respectively. (B) Negative controls. 48S ribosomal complex formation was analysed as above. PABP wild-type, PABP mutant (M161A) or GST was present in the reaction mixtures at 15 μg/ml concentrations where indicated. In lane 1, no proteins were added. In lane 5, no reaction components were present except for the mRNA. The relative amounts of 48S complexes, with the value for lane 1 being set as 100, were 150, 90, and 90, for lanes 2–4, respectively.
Reduction of eIF4F activity in 80S ribosome initiation complex formation in PABP-depleted extracts
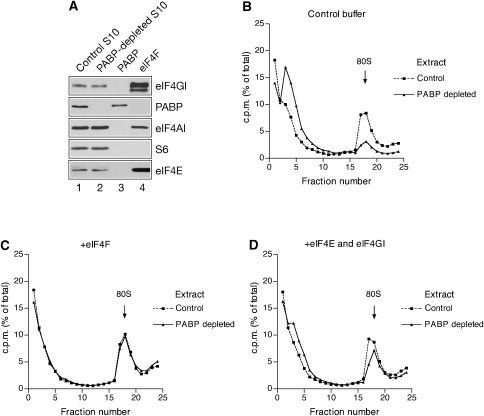
48S and 80S ribosome initiation complex formation is diminished in cell extracts lacking PABP (Kahvejian et al, 2005). We used this assay to discover factors that modulate the PABP dependence of translation initiation. A Krebs-2 ascites cell extract was depleted of PABP by using a GST–Paip2 affinity column (Svitkin and Sonenberg, 2004). Western blotting with an anti-PABP antibody (Afonina et al, 1997) did not reveal any residual PABP in the extract after depletion (Figure 2A). Importantly, neither eIF4F (eIF4GI, eIF4AI, and eIF4E) nor 40S ribosomal subunits (as exemplified by S6 ribosomal protein) were co-depleted with PABP (Figure 2A; Svitkin and Sonenberg, 2004; Thoma et al, 2004). It is noteworthy that a 5-μl aliquot of control S10 extract and 0.1 μg of recombinant PABP produced signals of similar intensity (Figure 2A; compare lanes 1 and 3). This suggests that the endogenous PABP concentration in the S10 extract is about 20 μg/ml (or 10 μg/ml in reaction mixtures, which contain 50% v/v of the S10 extract; see below). Control and PABP-depleted extracts were incubated in the presence of radiolabelled globin mRNA and cycloheximide, and analysed by velocity sedimentation in sucrose gradients. 80S ribosome recruitment decreased to ∼16% of control after PABP depletion leading to the enhanced association of mRNA with free mRNPs (fractions 3 and 4 of the gradient) (Figure 2B). Supplementing PABP-depleted extracts with recombinant PABP at close to physiological concentration (10 μg/ml) restored 80S ribosome initiation complex formation, further excluding the possibility of co-depletion of initiation factors (data not shown and Kahvejian et al, 2005). To ascertain that the lack of PABP diminishes eIF4F activity (Kahvejian et al, 2005), we investigated whether excess eIF4F would relieve the suppression of 80S initiation complex formation caused by the absence of PABP. The addition of eIF4F to the reaction mixtures completely rescued 80S initiation complex formation in the depleted extract (Figure 2C). This restoration was not caused by PABP that could contaminate our eIF4F preparation. Western blotting failed to reveal the presence of PABP in eIF4F (Figure 2A, lane 4). Moreover, the addition of recombinant eIF4E and eIF4GI (84–1599), which corresponds to the full-size eIF4GI except for the first 83 amino acids, to reconstitute eIF4F significantly increased ribosome binding in the PABP-depleted extract (from ∼16 to 66% of the control level; Figure 2D). eIF4E alone was less stimulatory than the eIF4E/4GI complex, whereas eIF2 was completely inactive (data not shown). Thus, PABP serves to enhance eIF4F activity in ribosome binding, although an unlikely possibility that PABP and eIF4F have redundant functions in translation initiation cannot be excluded by these data alone.
Figure 2.
Rescue of 80S ribosome binding to globin mRNA in PABP-depleted extracts. (A) Western blot analysis of control or PABP-depleted Krebs-2 S10 extract (5 μl) using antibodies against eIF4GI, PABP, eIF4AI, S6 ribosomal protein, and eIF4E as indicated. Lanes 3 and 4 show blots of recombinant PABP (0.1 μg) and eIF4F (1.5 μg), respectively. (B–D) 80S ribosome initiation complex profiles of control (squares, dashed line) and PABP-depleted (triangles, solid line) Krebs-2 cell extracts supplemented with control buffer (B), eIF4F (48 μg/ml) (C), or a combination of recombinant eIF4E (6 μg/ml) and eIF4GI (84–1599) (30 μg/ml) (D). 80S ribosome binding to 32P-labelled globin mRNA was analysed as described in Materials and methods. Arrows indicate peaks corresponding to mRNA in complex with 80S ribosomes. Relative efficiencies of ribosome binding in PABP-deleted versus control extract were 16, 98, and 66% for (B–D), respectively. The data are representative of four experiments.
General RNA-binding proteins enhance PABP dependency of translation initiation
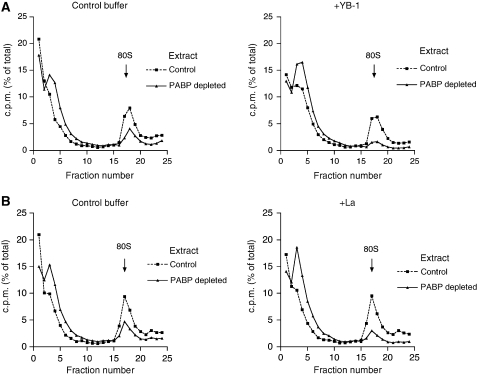
We showed earlier that general RNA-binding proteins enhance cap- and eIF4E-dependency of translation (Svitkin et al, 1996). However, the role of these proteins in PABP/poly(A)-mediated translation remained unknown. A role in the disruption of eIF4G–mRNA interaction was previously assigned to YB-1 (Evdokimova et al, 2001; Nekrasov et al, 2003). We therefore examined how YB-1 affects 80S initiation complex formation in the absence and presence of PABP. Strikingly, YB-1 preferentially inhibited ribosome binding in PABP-depleted as compared with control extract. In the presence of YB-1 (40 μg/ml), the reduction of 80S complex formation caused by PABP depletion was 6.7-fold as compared with 2.7-fold observed with control buffer (Figure 3A). The difference between the PABP-depleted and control extracts in supporting ribosome binding was even greater (10-fold) in the presence of 80 μg/ml YB-1 (data not shown). La autoantigen, which is a promiscuous RNA-binding protein (Wolin and Cedervall, 2002), also preferentially inhibited 80S ribosomal complex formation in PABP-depleted as compared with control extracts (Figure 3B). However, as compared with YB-1, La was required at ∼2.5-fold higher molar concentration to exert a similar effect. Notably, supplementing PABP-depleted extracts with YB-1 or La increased the abundance of free mRNPs (fractions 3 and 4). PTB also increased PABP dependency of 80S initiation complex formation but to an extent lower than La (data not shown). These observations indicate that several RNA-binding proteins can affect PABP responsiveness of translation initiation, although their individual contribution to this regulation is apparently less significant than that of YB-1.
Figure 3.
General RNA-binding proteins increase PABP dependence of 80S ribosome binding to globin mRNA in cell extracts. 80S ribosome initiation complex profiles of control (squares, dashed line) and PABP-depleted (triangles, solid line) Krebs-2 cell extracts. (A) Reaction mixtures were supplemented with control buffer (left panel) or YB-1 (40 μg/ml; right panel). Relative efficiencies of ribosome binding in PABP-deleted versus control extract were 37 and 15% for the left and right panels, respectively. (B) Reaction mixtures were supplemented with control buffer (left panel) or La (110 μg/ml; right panel). Relative efficiencies of ribosome binding in PABP-deleted versus control extract were 33 and 14% for the left and right panels, respectively. The data are representative of three experiments.
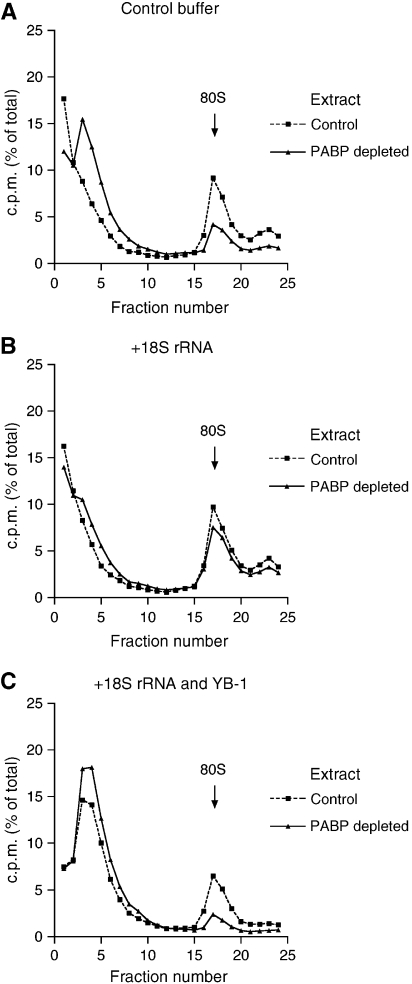
On the basis of the results above, one would expect that sequestering RNA-binding proteins with a nonspecific RNA should relax the control of 80S initiation complex formation by PABP. We therefore compared the effect of PABP depletion on 80S complex assembly in the absence and presence of a nonspecific RNA. We chose 18S ribosomal RNA (rRNA) because of its ability to stimulate ribosome binding in crude cell extracts (Weber et al, 1979). Strikingly, 18S rRNA preferentially stimulated ribosome binding in the PABP-depleted extract to restore it to control levels (compare Figure 4B with 4A). The addition of YB-1 together with 18S rRNA restored the inhibition of ribosome binding in the PABP-depleted extract (Figure 4C). This result is consistent with the ability of YB-1 to interact with 18S rRNA.
Figure 4.
18S rRNA relieves the inhibition of 80S initiation complex formation in PABP-depleted Krebs-2 cell extracts. 80S ribosome initiation complex profiles of control (squares, dashed line) and PABP-depleted (triangles, solid line) Krebs-2 extracts supplemented with control buffer (A), 18S rRNA (25 μg/ml) (B) or a combination of 18S rRNA (25 μg/ml) and YB-1 (150 μg/ml) (C) are shown. Relative efficiencies of ribosome binding in PABP-deleted versus control extract were 32, 78, and 16% for (A–C), respectively. The data are representative of three experiments.
To examine the correlation between the efficiency of 80S ribosome initiation complex formation and 40S ribosomal subunit recruitment, 40S ribosome binding studies were carried out with extracts supplemented with GMPPNP (Kahvejian et al, 2005). The amount of 40S ribosomal subunits assembled on the mRNA was ∼2-fold lower in PABP-depleted than in control extract (Supplementary Figure S1A). YB-1 inhibited, whereas 18S rRNA stimulated, 48S ribosome initiation complex formation and, importantly, their effects were much greater in the PABP-depleted than in control extract (Supplementary Figure S1B and C). Consequently, the reduction in 48S ribosome initiation complex formation due to PABP depletion was higher (∼5-fold) in the presence of YB-1 as compared with control (∼2-fold) and essentially abrogated in the presence of 18S rRNA. Interestingly, in this analysis, we noticed PABP-dependent formation of complexes migrating faster than the 40S marker. The origin of these complexes is not known; they may arise from binding of two or more 40S ribosomal subunits to the mRNA.
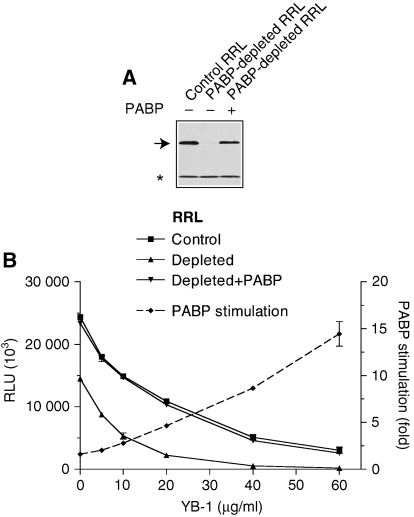
YB-1 renders translation in a rabbit reticulocyte lysate PABP dependent
Translation in RRL has been amply documented to be less dependent on the cap and poly(A) tail as compared with Krebs-2 ascites cell extract (Munroe and Jacobson, 1990; Svitkin et al, 1996; Imataka et al, 1998). We previously attributed the cap independence of RRL to the paucity of general RNA-binding proteins relative to eIF4F (Svitkin et al, 1996). We reasoned that in a similar manner translation in RRL is relatively PABP independent because of a high eIF4F/general RNA-binding protein ratio. It is conceivable that in the absence of competition the recruitment of eIF4F by the mRNA is highly efficient and thus not upregulated by the PABP/poly(A) complex. To assess this hypothesis, we studied whether excess of YB-1 would render translation in RRL PABP-dependent. Endogenous PABP was completely removed from RRL by retention on a GST–Paip2–Sepharose resin (Figure 5A). Aliquots of control and PABP-depleted RRL were supplemented with increasing amounts of YB-1 and then programmed for translation with a capped and polyadenylated luciferase (Luc) mRNA. In the absence of YB-1, the translation ratio for the non-depleted versus depleted RRL was 1.7 (Figure 5B), consistent with the minor contribution of the PABP/poly(A) complex to translation efficiency in RRL. YB-1 inhibited translation in both systems in a dose-dependent manner. However, the magnitude of the inhibition was significantly greater in the PABP-depleted than in control RRL (e.g. 46-fold versus 4.8-fold at 40 μg/ml of YB-1). When the depleted RRL was reconstituted with recombinant PABP (10 μg/ml), the resistance of translation to YB-1 inhibition was restored to control levels (Figure 5B). It is noteworthy that because the reconstituted and control RRL produced similar signals when analysed for PABP by western blotting, the concentration of added PABP was close to physiological levels. The translation stimulation by PABP increased from 1.6- to 14-fold in the presence of increasing concentrations of YB-1. Thus, YB-1 renders translation in RRL PABP-dependent.
Figure 5.
YB-1 augments PABP dependence of translation in RRL. (A) Western blot analysis of PABP in control and PABP-depleted RRL reaction mixtures (5 μl), either not supplemented (−) or supplemented (+) with recombinant PABP (10 μg/ml), as indicated. An arrow and an asterisk indicate the positions of PABP and a cross-reactive protein (which served as an internal loading control), respectively. (B) Increasing concentrations of YB-1 were added to control or PABP-depleted RRL programmed with capped Luc(A+) mRNA. Where indicated, the translation in PABP-depleted RRL was carried out in the presence of recombinant PABP (10 μg/ml). After incubation at 30°C for 60 min, luciferase levels were measured. The relative luciferase units (RLUs) reported are for 1-μl aliquots of translational samples and are averages of three assays. Standard deviation from the mean (when larger than the individual data symbol) is shown. The dotted line represents stimulation of luciferase synthesis in PABP-depleted RRL by recombinant PABP.
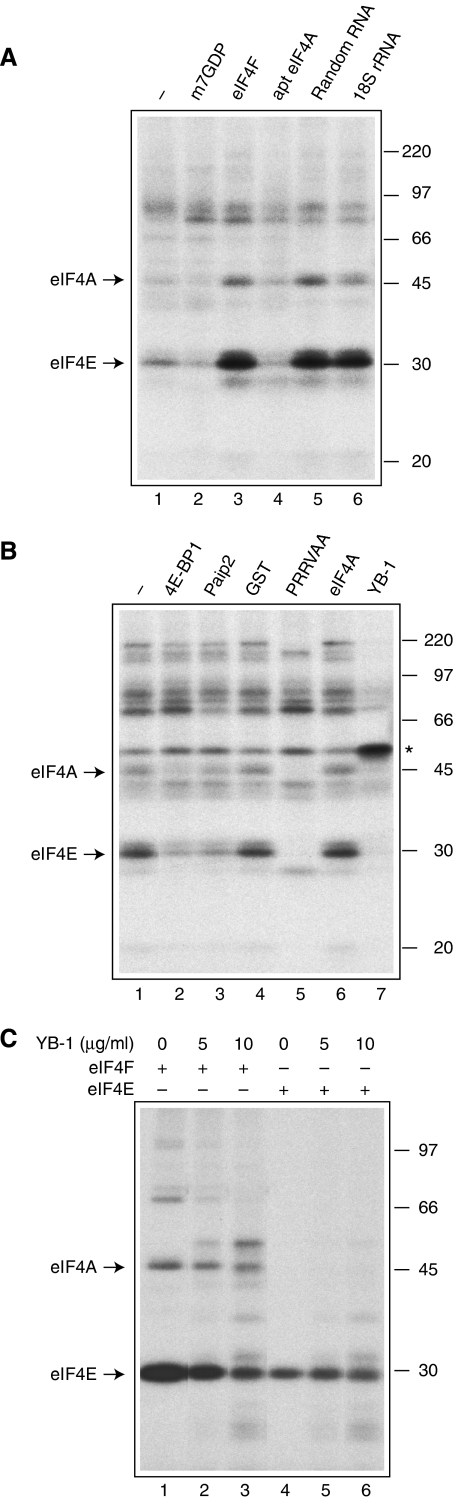
PABP confers competitive advantage to eIF4G over YB-1 in RNA binding
The interaction of eIF4E with the m7G cap structure can be analysed by crosslinking (Sonenberg and Shatkin, 1977; Sonenberg et al, 1978). We applied a chemical crosslinking technique (Sonenberg and Shatkin, 1977) to investigate the competition between initiation factors and YB-1 for mRNA binding and the effect of PABP on this competition. The experiments were carried out in control and PABP-depleted RRL with Luc(A+) mRNA, which was 32P-labelled at the 5′ m7G cap. In control RRL, eIF4E and eIF4A crosslinked specifically to the m7G cap, as the crosslinking was inhibited by the cap analogue, m7GDP (Figure 6A, compare lanes 2 and 1). The addition of eIF4F resulted in increased crosslinking of eIF4E and eIF4A, further establishing their correct identification (Figure 6A, lane 3). To corroborate the eIF4E crosslinking assay as a read-out of eIF4F activity, the activity of eIF4F was inhibited by the addition of either an aptamer to eIF4A (Oguro et al, 2003) or the PRRVAA eIF4A dominant-negative mutant (Svitkin et al, 2001b). Both eIF4A inhibitors impaired eIF4E crosslinking to the cap (Figure 6A, lane 4 and 6B, lane 5). Inhibition of eIF4E crosslinking was also caused by the addition of 4E-BP1 (which disrupts the eIF4E–eIF4G interaction) and Paip2 (which disrupts eIF4G–PABP and PABP–poly(A) interactions), but not by GST (Figure 6B, lanes 2–4), in agreement with the requirement of an eIF4E–eIF4G–PABP–poly(A) bridging complex for increased interaction of eIF4E with the m7G cap (Haghighat and Sonenberg, 1997; Kahvejian et al, 2005). Strikingly, the random oligonucleotide RNA pool (Oguro et al, 2003), which was used as a negative control for the eIF4A aptamer-mediated inhibition, and 18S rRNA strongly stimulated eIF4E crosslinking (Figure 6A, lanes 5 and 6). Thus, outcompeting nonspecific protein–mRNA interactions with the aid of exogenous RNA yields more eIF4E–cap complexes. In contrast, excess YB-1 (30 μg/ml) abrogated eIF4E crosslinking to the cap structure (Figure 6B, lane 7). YB-1 (migrating as a ∼50-kDa polypeptide) crosslinked to the cap at the expense of eIF4E. Measurable crosslinking of YB-1 to the cap is most likely a consequence of a high mass ratio of YB-1 to mRNA (9:1), which engenders saturated complexes (Skabkin et al, 2004). As we have shown earlier, YB-1 binding is not inhibited by cap analogues, attesting to a cap nonspecific nature of this interaction (Evdokimova et al, 2001). We also assessed the effect of YB-1 on eIF4E crosslinking in a system containing pure factors: eIF4F or eIF4E. In this system, crosslinking of eIF4E was more efficient as it was a component of the eIF4F complex (Figure 6C, compare lanes 1 and 4) (see also Lee et al, 1985; Haghighat and Sonenberg, 1997). Importantly, the inhibition of eIF4E crosslinking by YB-1 could be recapitulated in a reaction mixture containing pure eIF4F, but not eIF4E (Figure 6C). This suggests that YB-1 inhibits eIF4E binding indirectly by inhibiting eIF4G–mRNA complex formation.
Figure 6.
Initiation factor crosslinking to the m7G cap structure as affected by eIF4F inhibitors. (A) RRL chemical crosslinking pattern. RRL was preincubated at 30°C for 2 min with control buffer (−), 0.6 mM m7GDP (to offset magnesium chelating by m7GDP, an extra 0.6 mM MgCl2 was included in the reaction mixture), 100 μg/ml eIF4F, 25 μg/ml aptamer to eIF4A, 25 μg/ml random RNA (random nucleotide pool RNA; see Oguro et al, 2003) or 50 μg/ml 18S rRNA, as indicated. After the addition of oxidized 32P cap-labelled Luc(A+) mRNA, the reaction mixtures were incubated at 30°C for 10 min. (B) RRL was preincubated with control buffer (−), GST-4E-BP1 (40 μg/ml), GST–Paip2 (40 μg/ml), GST (40 μg/ml), eIF4A PRRVAA dominant-negative mutant (30 μg/ml), eIF4A wild-type (30 μg/ml) or YB-1 (30 μg/ml), as indicated. Incubation with mRNA was as above. (C) Pure eIF4F or eIF4E (3 pmol) was incubated with mRNA in the presence of the indicated concentrations of YB-1. Labelled proteins were analysed by SDS–PAGE and autoradiography. Arrows indicate the positions of eIF4E and eIF4A. On (B), an asterisk indicates the position of YB-1. The positions of the 14C-methylated molecular weight markers (GE Healthcare) are shown at the right.
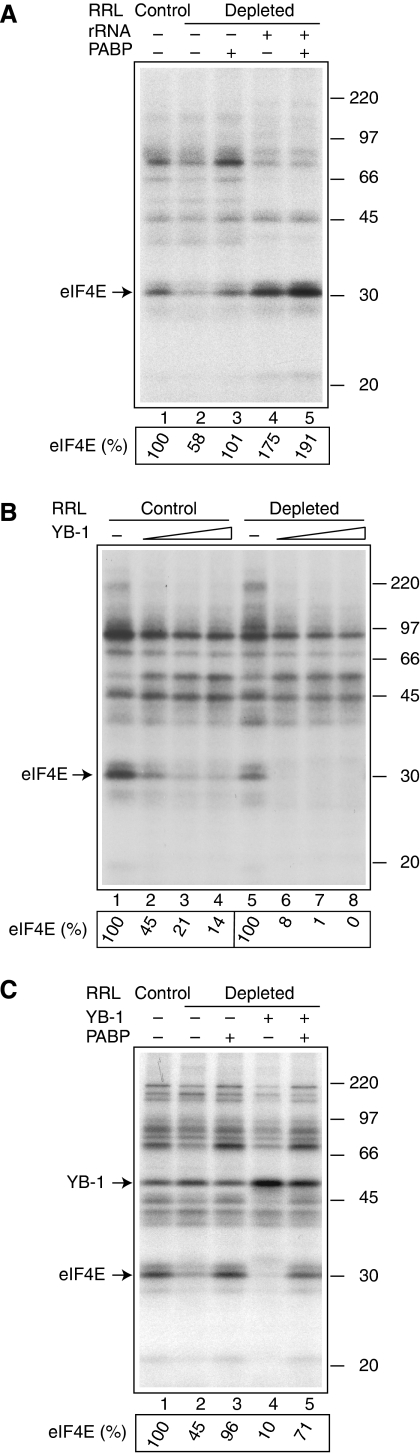
Binding of PABP to eIF4G increases the binding of eIF4E to the cap (Kahvejian et al, 2005). This stimulation requires tethering of PABP to the poly(A) tail, as it is significantly higher with poly(A+) then poly(A−) mRNA (see Figure 5 in Kahvejian et al, 2005). To investigate the role of general RNA-binding proteins in this interaction, we investigated whether their sequestration by 18S rRNA would weaken the PABP stimulation of eIF4E–cap interaction. PABP depletion reduced the association of eIF4E with the m7G cap (1.7-fold) (Figure 7A, compare lanes 2 and 1) and adding recombinant PABP to the depleted RRL reversed this inhibition (compare lanes 3 and 2). Adding 18S rRNA to the depleted extract caused an increase (three-fold) in the association of eIF4E with the m7G cap (Figure 7A, compare lanes 4 and 2). Significantly, this increase was refractory to further stimulation by added PABP (Figure 7A, compare lanes 5 and 4). By comparison, PABP stimulated the eIF4E–cap interaction 1.7-fold in the absence of added 18S rRNA (Figure 7A, compare lanes 3 and 2). Thus, limiting the availability of RNA-binding proteins renders eIF4E–cap interaction resistant to stimulation by PABP. An alternative possibility is that crosslinking of eIF4E is already maximal in the presence of 18S rRNA (Figure 7A, lane 4), so that addition of PABP in lane 5 cannot further stimulate it.
Figure 7.
Effect of YB-1 availability on PABP dependence of eIF4E crosslinking to the m7G cap structure in RRL. (A) 18S rRNA enhances eIF4E crosslinking and makes it refractory to stimulation by PABP. Control or PABP-depleted RRL was preincubated with PABP (10 μg/ml) and 18S rRNA (50 μg/ml) where indicated. Incubation with oxidized 32P cap-labelled Luc(A+) mRNA was as described for Figure 6A. Relative efficiencies of eIF4E crosslinking are indicated at the bottom. The value obtained for control RRL with no added proteins was set as 100%. (B) YB-1 dose–response of eIF4E crosslinking as affected by PABP depletion. Crosslinking was carried out with control or PABP-depleted RRL preincubated with the increasing concentrations of YB-1 (15 μg/ml, lanes 2 and 6; 20 μg/ml, lanes 3 and 7; and 25 μg/ml, lanes 4 and 8). In lanes 1 and 5, the reaction mixtures were supplemented with buffer alone. Relative amounts of radioactivity in eIF4E are given below. Values obtained in the absence of added YB-1 were set as 100%. (C) YB-1 enhances the stimulation of eIF4E–cap interaction by PABP. Crosslinking was carried out with control and PABP-depleted RRL preincubated with PABP (10 μg/ml) or YB-1 (10 μg/ml) where indicated. Quantification of eIF4E bands was as for (A). Arrows indicate the positions of eIF4E and YB-1. The positions of the 14C-methylated protein molecular weight markers (GE Healthcare) are shown at the right.
As we have shown above that YB-1 exerted the strongest effect among several RNA-binding proteins on the stimulation of translation initiation by PABP, we wished to examine the effect of YB-1 on the PABP/eIF4F synergism in stimulating eIF4E–cap interaction. The addition of YB-1 to control and PABP-depleted RRL decreased the binding of eIF4E to the cap in a dose-dependent manner (Figure 7B). However, consistent with the potentiation of translational inhibition by YB-1 after PABP depletion (Figure 5B), YB-1 reduced binding of eIF4E to the cap more potently in the absence than in the presence of PABP. For instance, 15 μg/ml YB-1 caused a 12-fold inhibition of eIF4E crosslinking in PABP-depleted RRL versus only 2.2-fold inhibition in control RRL. Thus, PABP enhances the resistance of eIF4E binding to inhibition by YB-1. Accordingly, in PABP-depleted RRL, the stimulation of eIF4E crosslinking by recombinant PABP was greater in the presence than in the absence of added YB-1 (i.e. 7.1-fold versus 2.1-fold; Figure 7C, compare lanes 5 and 4, and compare lanes 3 and 2, respectively). Thus, YB-1, by decreasing eIF4F–mRNA binding, enhances the dependence of eIF4F on PABP for eIF4E–cap interaction. The endogenous concentration of YB-1 in RRL reaction mixtures is about 30 μg/ml as estimated by western blotting (data not shown). Thus, YB-1 exerts a differential effect on translation and eIF4E crosslinking in control and PABP-depleted RRL when added at close to physiological concentrations. A band corresponding to YB-1 was prominent in the samples supplemented with YB-1 (Figure 7C, lanes 4 and 5). However, YB-1 better associated with the m7G cap structure when eIF4E binding was reduced by PABP depletion (Figure 7C, compare lanes 4 and 5). Thus, the lack of PABP reduces the ability of eIF4F to compete with YB-1 for mRNA binding.
Discussion
Efficient translation in eukaryotic cells occurs when the mRNA is in a closed-loop conformation (Jacobson, 1996; Kahvejian et al, 2001). The precise mechanism by which the 3′ poly(A) tail functions synergistically with the 5′ cap to stimulate translation is not fully understood. Cooperative binding of the eIF4G/4E complex and PABP to the mRNA engenders a more stable eIF4E association with the cap structure than eIF4G/4E binding alone (Kahvejian et al, 2005). Besides serving as an additional anchorage site for eIF4F on the mRNA, the PABP/poly(A) complex serves (especially in yeast) to increase eIF4F binding through allosteric changes in eIF4G/4E (Gross et al, 2003). In addition to the promotion of eIF4F binding and 40S ribosome subunit recruitment, PABP was suggested to stimulate 60S ribosomal subunit joining and ribosome recycling (Jacobson, 1996; Searfoss et al, 2001). The latter activities of PABP are not addressed in the present study.
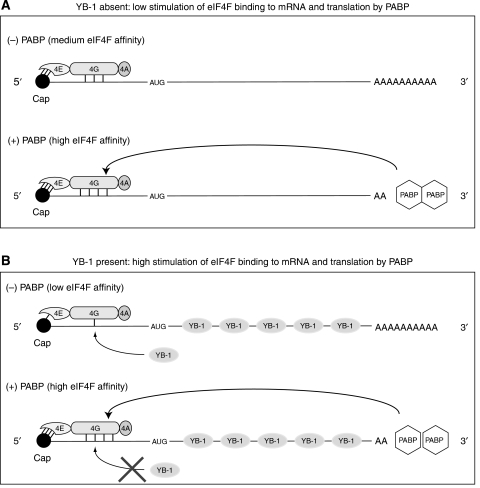
Previous models for translational control by PABP have not taken into consideration general RNA-binding proteins, of which YB-1 is a prominent member, in the regulation of eIF4F access to mRNA. Here, we showed that to respond to PABP, the interaction of eIF4F with the mRNA needs to be downregulated by RNA-binding proteins. In a non-competitive system, containing purified translation components and globin mRNA, the efficiency of reconstitution of 48S translation initiation complexes is high, most likely reaching a plateau (Pisarev et al, 2007), and thus, is only mildly responsive to PABP (Figure 1). In contrast, in Krebs-2 cell extracts where general RNA-binding proteins are abundant, the assembly of ribosomal initiation complexes is largely PABP dependent (Figure 2; Supplementary Figure S1; Kahvejian et al, 2005). Adding YB-1 or La to a Krebs-2 cell extract enhances PABP dependence of ribosome binding even further (Figure 3). A model based on these results and according to which the PABP/poly(A) complex counteracts YB-1-mediated inhibition of eIF4F–mRNA interaction is presented in Figure 8. When YB-1 is limiting, the recruitment of eIF4F by mRNA occurs efficiently and could be only slightly potentiated by PABP (Figure 8A). When abundant, YB-1 diminishes the eIF4F–mRNA interaction thereby creating favourable conditions for the modulation of this interaction by PABP (Figure 8B). It is possible that this model does not apply to all capped mRNAs. An exception could take the form of mRNAs that bear an extensive 5′-proximal secondary structure. Because YB-1 has single-stranded RNA-binding specificity (Minich et al, 1993), it may not be bound efficiently to such 5′ untranslated regions (UTRs). In this case, the 5′ UTR secondary structure could downregulate eIF4F binding to enable PABP-dependent translation. Experiments reported here used an mRNA with a moderately structured 5′ UTR (that of β-globin mRNA) (Auron et al, 1982). It remains to be seen whether an extensive 5′-proximal secondary structure can circumvent the requirement for YB-1 for the PABP/poly(A) responsiveness of translation.
Figure 8.
Model explaining how YB-1 increases mRNA affinity response of eIF4F to PABP. It is postulated that the default binding of eIF4F to the capped 5′ end of an mRNA is relatively efficient (A; medium affinity) and, thus, the ability of PABP to stimulate this binding is limited (A; medium to high affinity transition). When YB-1 competes with eIF4G for mRNA binding, the basal affinity of eIF4F for the mRNA is decreased allowing eIF4F binding to vary over a wider range in response to PABP (B; low to high eIF4F affinity transition). The high-affinity binding of eIF4F to the mRNA conferred by PABP precludes or greatly reduces competition from YB-1 (crossed lines). The number of connecting lines between eIF4F subunits and mRNA denotes binding affinity.
Translation in the most popular in vitro translation system, micrococcal nuclease-treated RRL, poorly exhibits cap- and poly(A) tail-mediated synergistic stimulation (Munroe and Jacobson, 1990; Wakiyama et al, 1997). In contrast, RRL that was partially depleted of ribosomes by ultracentrifugation (Michel et al, 2000) or not nuclease treated (Rifo et al, 2007) shows the cap/poly(A) synergism. However, ribosome depletion of RRL dramatically diminishes its translational efficiency and the reason for the improved characteristics of the latter system is not clear. The results presented here and elsewhere (Svitkin et al, 1996) strongly indicate that cap and poly(A) independence of translation in RRL is a consequence of limiting general RNA-binding proteins, and relatively high concentration of eIF4F. The addition of YB-1 to RRL dramatically increases cap and poly(A) dependencies of translation (Figure 5; Svitkin et al, 1996). Thus, nonspecific RNA–protein interactions could be considered not just as a means to inhibit global protein synthesis but also one to impose a stringent translational control by the mRNA 5′ cap and 3′ poly(A). An additional contribution of mRNA coating by RNA-binding proteins to high fidelity of translation is that it disables spurious internal initiation sites to prevent the so-called ‘trash internal initiation' (Merrick, 2004) and to ensure a precise delivery of eIF4F to the 5′ end of the mRNA (Svitkin et al, 1996; Pisarev et al, 2002).
Assigning to the conserved RNA-binding protein YB-1 a role as a critical regulator of eIF4F recruitment by an mRNA and, consequently, cap- and poly(A)-dependent translation is based on the following findings: (1) YB-1 is very abundant in the cytoplasm. By contrast, most other general RNA-binding proteins, such as La and PTB, are predominantly nuclear. In addition, in vitro, YB-1 exaggerated the PABP depletion-mediated inhibition of 80S initiation complex formation at lower concentrations than La and PTB; (2) YB-1 is the major core protein of mRNPs, the association of which with the mRNA includes binding near the 5′ cap (Figure 6B; Evdokimova et al, 2006a); (3) YB-1 competes with eIF4G for binding to the mRNA, resulting in a decreased eIF4E–5′ cap association and inhibition of translation (Figure 6C; Davydova et al, 1997; Evdokimova et al, 2001; Nekrasov et al, 2003). Consistent with competition between YB-1 and eIF4G for binding to the mRNA, YB-1, but no other proteins, was shown to UV crosslink to the mRNA cap structure in a RRL where the function of eIF4F was inhibited (Evdokimova et al, 2001). Thus, although several RNA-binding proteins share the potential to enhance the dependency of translation on eIF4E/cap and PABP/poly(A) complexes, the regulatory mechanism operative with YB-1 is prevailing under physiological conditions. To demonstrate the contribution of YB-1 to PABP-dependent translation more directly, we attempted to immunodeplete YB-1 from Krebs-2 extract. However, YB-1 depletion with available antibodies was only partial.
The abundance and activity of YB-1 are under control. First, YB-1 is downregulated at the mRNA level by the phosphoinositide 3-kinase (PI3K) pathway (Bader et al, 2003). Second, an acidic protein, YBAP1, can displace YB-1 from the mRNA to stimulate translation (Matsumoto et al, 2005). Third, YB-1 is targeted for phosphorylation at Ser-102 by the serine/threonine protein kinase Akt and the phosphorylated YB-1 exhibits reduced affinity for mRNA (Evdokimova et al, 2006b). Reduced YB-1 function in some cancer cells along with the complementary eIF4E/4G upregulation is believed to enhance otherwise inefficient translation of mRNAs, the activity of which is highly eIF4E dependent (Evdokimova et al, 2006a). In addition, a prediction from this work is that the impaired YB-1 function in some cancer cells may relax the translational control imposed by the m7G cap and poly(A) tail structures.
Materials and methods
Proteins and antibodies
Recombinant PABP, wild-type or mutant (M161A), YB-1, and La proteins were expressed and purified essentially as described (Svitkin et al, 1994; Evdokimova et al, 2001; Kahvejian et al, 2005). For expression and purification of recombinant eIF4GI (84–1599) and eIF4E, see Yanagiya et al, submitted. Native eIF4F and eIF2 were purified from RRL (Pisarev et al, 2007). Primary antibodies against PABP, eIF4GI, eIF4AI, and eIF4E were described (Khaleghpour et al, 2001; Svitkin et al, 2005). Anti-S6 ribosomal protein monoclonal antibody (5G10) was from Cell Signaling. Secondary anti-rabbit and anti-mouse HRP-conjugated antibodies were from GE Healthcare.
PABP depletion of Krebs-2 extracts and RRL
The preparation of extracts from Krebs-2 cells and their treatment with micrococcal nuclease were as described earlier (Svitkin and Sonenberg, 2007). Micrococcal nuclease-treated RRL was purchased from Promega. For the removal of PABP, extracts were incubated with the GST–Paip2 protein that was immobilized onto glutathione–Sepharose beads (Svitkin and Sonenberg, 2004). Mock-depleted extracts were treated with GST alone. For RRL, two cycles of treatment with GST–Paip2–Sepharose were necessary to obtain complete depletion of PABP. Western blot analyses of PABP depletion and eIF4GI, eIF4AI, eIF4E, and S6 ribosomal protein co-depletion were performed using Western Lightning chemiluminescence kit (Perkin-Elmer Life Sciences).
In vitro translation
Translation in RRL was carried out as recommended by the manufacturer (Promega). KCl (40 mM) was added to the RRL to enhance cap dependency of translation (Chu and Rhoads, 1978). The reaction mixtures (10 μl) included GST or GST–Paip2-treated RRL (70% v/v), amino acids, capped Luc(A+) mRNA (2 μg/ml) (Svitkin and Sonenberg, 2004) and other components as specified in the figure legends. Incubation was at 30°C for 1 h. Luciferase levels were determined in 3-μl aliquots of 100-fold diluted samples by enzymatic assay (Promega). A Lumat LB 9507 bioluminometer (EG&G Bertold) was used for the measurements.
Ribosome-binding assays
80S ribosome-binding studies were carried out using Krebs-2 cell extracts and 3′-end labelled globin mRNA (Kahvejian et al, 2005). The extracts (15 μl) were supplemented with cycloheximide (0.6 mM) and other components except for the mRNA and preincubated at 30°C for 2 min. After the addition of the mRNA (∼106 c.p.m., 60 ng), the reaction mixtures (30 μl) were incubated at 30°C for 15 min. Reactions were stopped by four-fold dilution with ice-cold buffer (HSB; 0.5 M NaCl, 0.03 M Mg(CH3COO)2, and 0.02 M HEPES-KOH, pH 7.5) (Lodish and Rose, 1977). 80S ribosomal complexes were resolved by centrifugation in 5-ml 15–30% sucrose gradients (prepared with HSB) (Kahvejian et al, 2005). For 40S ribosome-binding studies, GMPPNP (2 mM) was substituted for GTP and an extra MgCl2 (2 mM) was included in the reaction mixture. 48S initiation complexes were resolved on 10–30% sucrose gradients prepared with a low salt buffer (Kahvejian et al, 2005). Centrifugation was in an SW55 rotor at 54 000 r.p.m. at 4°C for 1 h 45 min. Fractions (0.2 ml) were collected from the top of the tubes and the radioactivity was counted. The area under the 80S or 48S peak (less background) was used to quantify ribosome binding (Kahvejian et al, 2005).
Chemical crosslinking assay
Uncapped Luc mRNA (Promega) was 3′ poly(A) extended by ∼200 nt using a poly(A) tailing kit (Ambion). Luc(A+) mRNA (4 μg) was radioactively labelled at the m7G cap using [α-32P]GTP, S-adenosyl methionine, and vaccinia virus guanylyltransferase (Ambion) according to the manufacturer's instructions. After purification and oxidation with NaIO4, the 32P cap-labelled RNA was used for crosslinking studies in RRL as described earlier (Sonenberg, 1981; Lee et al, 1983; Merrick and Sonenberg, 1997; Kahvejian et al, 2005; Supplementary data). Crosslinking of pure initiation factors (eIF4F or eIF4E) was performed in a buffer containing 12.5 mM HEPES-KOH, pH 7.3, 25 mM KCl, 50 mM KCH3COO, 1 mM MgCl2, 0.125 mM spermidine, 1 mM DTT, and 1 mM ATP (15 μl total reaction volume). Other conditions were as described above and in the legend to Figure 6C.
Assembly and toe-printing of 48S ribosomal complexes
Ribosomal 40S subunits, native and recombinant initiation factors (eIF1, eIF1A, eIF2, eIF3, eIF4A, eIF4B, and eIF4F), aminoacylated calf liver initiator tRNA (Met-tRNAiMet) were prepared as described (Pisarev et al, 2007). The reconstitution of 48S ribosomal complexes from purified components and native β-globin mRNA was performed as described earlier (Pisarev et al, 2007). Primer extension inhibition (toe-printing) analysis of 48S initiation ribosomal complex formation was performed with the use of AMV reverse transcriptase and the primer 5′-GCATTTGCAGAGGACAGG-3′ (Morino et al, 2000). cDNA was analysed by denaturing 6% PAGE. Toe prints were quantified by BAS-2000 phosphorimager (Fuji).
Supplementary Material
Supplementary Figure S1
Supplementary Information
Acknowledgments
We thank Sandra Perreault and Colin Lister for excellent technical assistance. This study was supported by a grant from the National Institutes of Health (NIH; GM66157) to NS, who is a Howard Hughes Medical Institute International Scholar. WCM was supported by an NIH grant GM26796.
References
- Afonina E, Neumann M, Pavlakis GN (1997) Preferential binding of poly(A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J Biol Chem 272: 2307–2311 [DOI] [PubMed] [Google Scholar]
- Auron PE, Rindone WP, Vary CP, Celentano JJ, Vournakis JN (1982) Computer-aided prediction of RNA secondary structures. Nucleic Acids Res 10: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG, Felts KA, Jiang N, Chang HW, Vogt PK (2003) Y box-binding protein 1 induces resistance to oncogenic transformation by the phosphatidylinositol 3-kinase pathway. Proc Natl Acad Sci USA 100: 12384–12389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer BW, Kornberg RD (1983) The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J Cell Biol 96: 717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamini G, Preiss T, Hentze MW (2000) Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6: 1781–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G (1972) Protein tightly bound to globin mRNA. Biochem Biophys Res Commun 47: 88–95 [DOI] [PubMed] [Google Scholar]
- Chu LY, Rhoads RE (1978) Translational recognition of the 5′-terminal 7-methylguanosine of globin messenger RNA as a function of ionic strength. Biochemistry 17: 2450–2455 [DOI] [PubMed] [Google Scholar]
- Davydova EK, Evdokimova VM, Ovchinnikov LP, Hershey JW (1997) Overexpression in COS cells of p50, the major core protein associated with mRNA, results in translation inhibition. Nucleic Acids Res 25: 2911–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V, Ovchinnikov LP, Sorensen PH (2006a) Y-box binding protein 1: providing a new angle on translational regulation. Cell Cycle 5: 1143–1147 [DOI] [PubMed] [Google Scholar]
- Evdokimova V, Ruzanov P, Anglesio MS, Sorokin AV, Ovchinnikov LP, Buckley J, Triche TJ, Sonenberg N, Sorensen PH (2006b) Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Mol Cell Biol 26: 277–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V, Ruzanov P, Imataka H, Raught B, Svitkin Y, Ovchinnikov LP, Sonenberg N (2001) The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J 20: 5491–5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Corona DF, Preiss T, Becker PB, Hentze MW (1999) Translational control of dosage compensation in Drosophila by sex-lethal: cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J 18: 6146–6154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963 [DOI] [PubMed] [Google Scholar]
- Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G (2003) Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115: 739–750 [DOI] [PubMed] [Google Scholar]
- Haghighat A, Sonenberg N (1997) eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J Biol Chem 272: 21677–21680 [DOI] [PubMed] [Google Scholar]
- Hershey JWB, Merrick WC (2000) Pathway and mechanism of initiation of protein synthesis. In Translational Control of Gene Expression, Sonenberg N, Hershey JWB, Mathews MB (eds), pp 33–88. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Imataka H, Gradi A, Sonenberg N (1998) A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J 17: 7480–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A (1996) Poly(A) methabolism and translation: the closed-loop model. In Translational Control, Hershey JWB, Mathews MB, Sonenberg N (eds), pp 451–480. Plainview, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Kahvejian A, Roy G, Sonenberg N (2001) The mRNA closed-loop model: the function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb Symp Quant Biol 66: 293–300 [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N (2005) Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 19: 104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim MM, Svitkin YV, Kahvejian A, De Crescenzo G, Costa-Mattioli M, Sonenberg N (2006) A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc Natl Acad Sci USA 103: 9494–9499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N (2001) Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell 7: 205–216 [DOI] [PubMed] [Google Scholar]
- Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M (2003) The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays 25: 691–698 [DOI] [PubMed] [Google Scholar]
- Lee KA, Edery I, Sonenberg N (1985) Isolation and structural characterization of cap-binding proteins from poliovirus-infected HeLa cells. J Virol 54: 515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KA, Guertin D, Sonenberg N (1983) mRNA secondary structure as a determinant in cap recognition and initiation complex formation. ATP-Mg2+ independent cross-linking of cap binding proteins to m7I-capped inosine-substituted reovirus mRNA. J Biol Chem 258: 707–710 [PubMed] [Google Scholar]
- Lodish HF, Rose JK (1977) Relative importance of 7-methylguanosine in ribosome binding and translation of vesicular stomatitis virus mRNA in wheat germ and reticulocyte cell-free systems. J Biol Chem 252: 1181–1188 [PubMed] [Google Scholar]
- Matsumoto K, Tanaka KJ, Tsujimoto M (2005) An acidic protein, YBAP1, mediates the release of YB-1 from mRNA and relieves the translational repression activity of YB-1. Mol Cell Biol 25: 1779–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC (2004) Cap-dependent and cap-independent translation in eukaryotic systems. Gene 332: 1–11 [DOI] [PubMed] [Google Scholar]
- Merrick WC, Sonenberg N (1997) Assays for eukaryotic translation factors that bind mRNA. Methods 11: 333–342 [DOI] [PubMed] [Google Scholar]
- Michel YM, Poncet D, Piron M, Kean KM, Borman AM (2000) Cap-Poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J Biol Chem 275: 32268–32276 [DOI] [PubMed] [Google Scholar]
- Minich WB, Maidebura IP, Ovchinnikov LP (1993) Purification and characterization of the major 50-kDa repressor protein from cytoplasmic mRNP of rabbit reticulocytes. Eur J Biochem 212: 633–638 [DOI] [PubMed] [Google Scholar]
- Morino S, Imataka H, Svitkin YV, Pestova TV, Sonenberg N (2000) Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol 20: 468–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe D, Jacobson A (1990) mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol Cell Biol 10: 3441–3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov MP, Ivshina MP, Chernov KG, Kovrigina EA, Evdokimova VM, Thomas AA, Hershey JW, Ovchinnikov LP (2003) The mRNA-binding protein YB-1 (p50) prevents association of the eukaryotic initiation factor eIF4G with mRNA and inhibits protein synthesis at the initiation stage. J Biol Chem 278: 13936–13943 [DOI] [PubMed] [Google Scholar]
- Oguro A, Ohtsu T, Svitkin YV, Sonenberg N, Nakamura Y (2003) RNA aptamers to initiation factor 4A helicase hinder cap-dependent translation by blocking ATP hydrolysis. RNA 9: 394–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU, Shatsky IN (1996) Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol 16: 6859–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Thomas AA, Merrick WC, Ovchinnikov LP, Shatsky IN (2002) Positive and negative effects of the major mammalian messenger ribonucleoprotein p50 on binding of 40 S ribosomal subunits to the initiation codon of beta-globin mRNA. J Biol Chem 277: 15445–15451 [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Unbehaun A, Hellen CU, Pestova TV (2007) Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol 430: 147–177 [DOI] [PubMed] [Google Scholar]
- Rifo RS, Ricci EP, Decimo D, Moncorge O, Ohlmann T (2007) Back to basics: the untreated rabbit reticulocyte lysate as a competitive system to recapitulate cap/poly(A) synergy and the selective advantage of IRES-driven translation. Nucleic Acids Res 35: e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A (2000) Physical and functional interactions between the mRNA cap structure and the poly(A) tail. In Translational Control of Gene Expression, Sonenberg N, Hershey JWB, Mathews MB (eds), pp 447–465. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Searfoss A, Dever TE, Wickner R (2001) Linking the 3′ poly(A) tail to the subunit joining step of translation initiation: relations of Pab1p, eukaryotic translation initiation factor 5B (Fun12p), and Ski2p–Slh1p. Mol Cell Biol 21: 4900–4908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Kiselyova OI, Chernov KG, Sorokin AV, Dubrovin EV, Yaminsky IV, Vasiliev VD, Ovchinnikov LP (2004) Structural organization of mRNA complexes with major core mRNP protein YB-1. Nucleic Acids Res 32: 5621–5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N (1981) ATP/Mg++-dependent cross-linking of cap binding proteins to the 5′ end of eukaryotic mRNA. Nucleic Acids Res 9: 1643–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Morgan MA, Merrick WC, Shatkin AJ (1978) A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc Natl Acad Sci USA 75: 4843–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Shatkin AJ (1977) Reovirus mRNA can be covalently crosslinked via the 5′ cap to proteins in initiation complexes. Proc Natl Acad Sci USA 74: 4288–4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Herdy B, Costa-Mattioli M, Gingras AC, Raught B, Sonenberg N (2005) Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol Cell Biol 25: 10556–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Imataka H, Khaleghpour K, Kahvejian A, Liebig HD, Sonenberg N (2001a) Poly(A)-binding protein interaction with eIF4G stimulates picornavirus IRES-dependent translation. RNA 7: 1743–1752 [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Meerovitch K, Lee HS, Dholakia JN, Kenan DJ, Agol VI, Sonenberg N (1994) Internal translation initiation on poliovirus RNA: further characterization of La function in poliovirus translation in vitro. J Virol 68: 1544–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Ovchinnikov LP, Dreyfuss G, Sonenberg N (1996) General RNA binding proteins render translation cap dependent. EMBO J 15: 7147–7155 [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N (2001b) The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7: 382–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Sonenberg N (2004) An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol Biol 257: 155–170 [DOI] [PubMed] [Google Scholar]
- Svitkin YV, Sonenberg N (2007) A highly efficient and robust in vitro translation system for expression of picornavirus and hepatitis C virus RNA genomes. Methods Enzymol 429: 53–82 [DOI] [PubMed] [Google Scholar]
- Tarun SZ, Sachs AB (1996) Association of the yeast poly(A) tail binding protein with translation initiation factor eIF4G. EMBO J 15: 7168–7177 [PMC free article] [PubMed] [Google Scholar]
- Thoma C, Bergamini G, Galy B, Hundsdoerfer P, Hentze MW (2004) Enhancement of IRES-mediated translation of the c-myc and BiP mRNAs by the poly(A) tail is independent of intact eIF4G and PABP. Mol Cell 15: 925–935 [DOI] [PubMed] [Google Scholar]
- Wakiyama M, Futami T, Miura K (1997) Poly(A) dependent translation in rabbit reticulocyte lysate. Biochimie 79: 781–785 [DOI] [PubMed] [Google Scholar]
- Weber LA, Simili M, Baglioni C (1979) Binding of viral and cellular messenger RNAs to ribosomes in eukaryotic cell extracts. Methods Enzymol 60: 351–360 [DOI] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB (1998) Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 2: 135–140 [DOI] [PubMed] [Google Scholar]
- Wolin SL, Cedervall T (2002) The La protein. Annu Rev Biochem 71: 375–403 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Information