Abstract
We used a transgene system to study spreading of RNA-directed DNA methylation (RdDM) during transcriptional gene silencing in Arabidopsis thaliana. Forward and reverse genetics approaches using this system delineated a stepwise pathway for the biogenesis of secondary siRNAs and unidirectional spreading of methylation from an upstream enhancer element into downstream sequences. Trans-acting, hairpin-derived primary siRNAs induce primary RdDM, independently of an enhancer-associated ‘nascent' RNA, at the target enhancer region. Primary RdDM is a key step in the pathway because it attracts the secondary siRNA-generating machinery, including RNA polymerase IV, RNA-dependent RNA polymerase2 and Dicer-like3 (DCL3). These factors act in a turnover pathway involving a nascent RNA, which normally accumulates stably in non-silenced plants, to produce cis-acting secondary siRNAs that induce methylation in the downstream region. The identification of DCL3 in a forward genetic screen for silencing-defective mutants demonstrated a strict requirement for 24-nt siRNAs to direct methylation. A similar stepwise process for spreading of DNA methylation may occur in mammalian genomes, which are extensively transcribed in upstream regulatory regions.
Keywords: Dicer-like3, methylation spreading, Pol IV, RNA-directed DNA methylation, secondary siRNAs
Introduction
Spreading of silent chromatin along a chromosome is a feature of many epigenetic processes in eukaryotes but the mechanisms, including the potential roles of non-coding RNAs, are still under investigation (Talbert and Henikoff, 2006; Clark, 2007; Pauler et al, 2007; Kwon and Workman, 2008). In fission yeast, an RNAi-mediated pathway fosters spreading of histone H3 lysine 9 methylation in heterochromatic DNA repeats (Locke and Martienssen, 2006; Iida et al, 2008; Zhang et al, 2008). The process of RNA-directed DNA methylation (RdDM) in Arabidopsis offers an opportunity to study spreading of cytosine methylation facilitated by the RNAi machinery and siRNAs in euchromatic portions of the genome.
RNA-directed DNA methylation is a small RNA-mediated epigenetic modification that is highly developed in flowering plants (Bei et al, 2007). The hallmarks of RdDM include methylation of cytosines in all sequence contexts (CG, CNG, CNN, where N is A, T or C) and restriction of methylation to the region of RNA–DNA sequence homology. The establishment and maintenance of RdDM require for the most part conventional DNA cytosine methyltransferases, histone-modifying enzymes and nuclear-localized RNAi proteins (Matzke et al, 2007; Chan, 2008). In addition, several plant-specific proteins are required, most notably subunits of two novel RNA polymerases termed Pol IV and Pol V (Pikaard et al, 2008; Wierzbicki et al, 2008). Pol IV and Pol V share the same second largest subunit, NRPD2/NRPE2, but are distinguished by their unique largest subunits, NRPD1 and NRPE1, respectively. Pol IV is needed to produce and/or amplify the small RNA trigger, whereas Pol V acts downstream of this step to facilitate de novo methylation at the small RNA-targeted site (Pikaard et al, 2008). RdDM can silence transposons, but the potential reversibility of this process suggests broader functions in stress responses and plant development (Penterman et al, 2007; Chan, 2008; Hollick, 2008).
To identify components of the RdDM machinery that are important for development, we established a two-component transgene silencing system based on an enhancer that is active in shoot and root meristem regions (Kanno et al, 2008). A forward genetic screen to recover mutants defective in RdDM of the target enhancer and silencing of a downstream GFP reporter gene has identified so far four dms (defective in meristem silencing) mutants. These include the previously identified SNF2-like factor DRD1 (DMS1) and the Pol V subunits, NRPE2a (DMS2) and NRPE1 (DMS5) (Kanno et al, 2004, 2005). Whether Pol V transcribes extensively in this transgene system or acts primarily to open chromatin at the siRNA-targeted site to expose DNA to cytosine methyltransferases is unknown. A recent study detected Pol V-dependent intergenic transcripts that may interact with siRNAs to induce methylation of homologous endogenous sequences (Wierzbicki et al, 2008). DMS3 is a novel structural-maintenance-of-chromosomes hinge domain-containing protein that has an unknown function in RdDM (Kanno et al, 2008).
A distinctive feature of the meristem silencing system is the production of 24-nt secondary siRNAs that are associated with spreading of methylation downstream of the enhancer region originally targeted by primary siRNAs, a phenomenon termed transitivity (Voinnet, 2008). Secondary siRNA biogenesis is correlated with the disappearance of a longer ‘nascent' transcript that overlaps with primary siRNAs. To explain these observations, we proposed a hypothetical pathway of secondary siRNA formation involving Pol IV as well as unidentified Argonaute (AGO), RNA-dependent RNA polymerase (RDR) and Dicer-like (DCL) activities (Kanno et al, 2008).
Formation of approximately 21–22-nt secondary siRNAs and spreading of methylation within transcribed regions have been observed during post-transcriptional gene silencing (PTGS) in plants and shown to require RDR6 and transcription of the target gene (Vaistij et al, 2002; Van Houdt et al, 2003; Eamens et al, 2008). By contrast, our meristem silencing system provides a well-defined system for analysing the genetic requirements for the biogenesis of 24-nt secondary siRNAs and spreading of methylation (secondary RdDM) within upstream regulatory regions during transcriptional gene silencing (TGS). Here, we report the findings of experiments designed to investigate possible functions for two RNA-dependent RNA polymerases, RDR2 and RDR6, as well as NRPD1 and AGO4 in 24-nt secondary siRNA biogenesis, secondary RdDM and silencing of the GFP reporter gene. We also report the identification of a dcl3 mutant in a forward genetic screen.
Results
Secondary siRNA biogenesis, DNA methylation and ‘nascent' RNA accumulation
We used a reverse genetics approach to test several factors proposed to be involved in secondary siRNA biogenesis in our transgene TGS system (Figure 1A–C). RDR2 is needed to produce 24-nt ‘heterochromatic' siRNAs (Xie et al, 2004); RDR6 is required to generate 21–22-nt secondary siRNAs in the PTGS pathway (Vaistij et al, 2002); AGO4 is needed for de novo DNA methylation (Chan et al, 2004) and for RNA slicing at some target loci (Qi et al, 2006); NRPD1, the largest subunit of Pol IV, is required for biogenesis of 24-nt heterochromatic siRNAs (Herr et al, 2005; Onodera et al, 2005). DCL3 is a dicer activity that produces 24-nt ‘heterochromatic' siRNAs (Xie et al, 2004) and is discussed later in the context of a forward genetic screen.
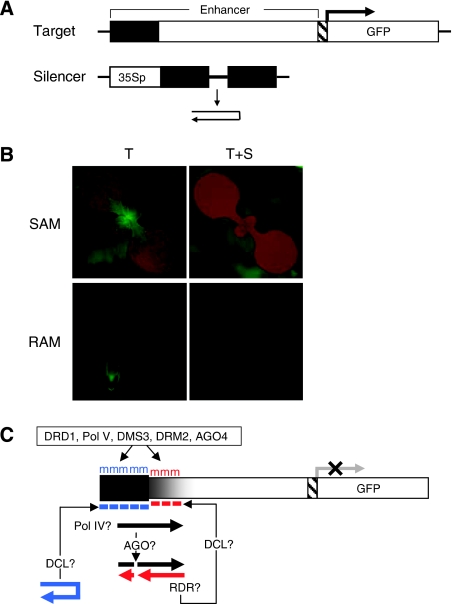
Figure 1.
Transgene silencing system and hypothetical model for spreading of DNA methylation through secondary siRNAs. (A) The target locus (T) contains a meristem-specific enhancer (region targeted for methylation shown in black) placed upstream of a minimal promoter (hatched) and GFP-coding region. The unlinked silencer locus (S) contains a transcribed inverted DNA repeat of distal enhancer sequences encoding a hairpin RNA. The hairpin RNA trigger is diced into short interfering (si) RNAs, which induce de novo methylation of the target enhancer in trans. There is a short tandem repeat in the target enhancer (Kanno et al, 2008), which has an unknown function in spreading of methylation and silencing. 35Sp: 35S promoter of cauliflower mosaic virus. (B) Transgenic seedlings containing only the target locus (T) display GFP fluorescence in root and shoot apical meristem regions (RAM and SAM, respectively) (left); in seedlings containing the target and silencer loci (T+S), GFP fluorescence is abolished (right). (C) In the hypothetical model tested in this study, primary siRNAs (blue dashes) originating from DCL processing of the hairpin RNA trigger induce primary RdDM (blue ‘m') of the targeted enhancer region (black box) and may guide AGO slicing of an overlapping nascent RNA (black arrow), which is proposed to be transcribed by Pol IV from the methylated DNA template. The Pol IV-generated nascent RNA, or cleavage fragments thereof, provide substrates for RDR; the resulting double-stranded RNA is diced to generate 24-nt secondary siRNAs (red dashes) that induce secondary RdDM (red ‘m') in the downstream region (black shade). GFP expression is silenced. Primary and secondary RdDM require DRD1, Pol V and DMS3 (Kanno et al, 2008) and are presumably catalysed by the de novo methyltransferase DRM2 (Cao et al, 2003) with the participation of AGO4 (Chan et al, 2004).
We introgressed the rdr2-1, rdr6-1, nrpd1-7 and ago4-1 mutations into the target (T)-silencer (S) line by crossing doubly homozygous plants (T/T;S/S) with the respective homozygous mutant (m/m). The resulting F1 progeny (genotype T/-; S/-; M/m) were self-fertilized to generate a segregating F2 population. To determine whether any of the mutants release GFP silencing, F2 seeds were sown on sterile medium and the number of GFP-positive seedlings was determined approximately 20 days after germination. If a mutation releases silencing, approximately one-third of the F2 progeny should be GFP positive; if a mutation does not release silencing, only around 18.5% of the F2 progeny should be GFP positive. With the exception of ago4-1, which appeared to partially release GFP silencing, none of the mutations alleviated silencing of the GFP reporter gene (Supplementary Table 1). We will first describe experiments with the three mutants that do not release GFP silencing: rdr2, rdr6 and nrpd1.
To obtain rdr2, rdr6 and nrpd1 mutant plants and their wild-type siblings for the analysis of secondary siRNAs, DNA methylation and the nascent RNA, we genotyped F3 progeny to obtain the following genotypes: T/T; S/(S);m/m and T/T; S/(S);M/M. We used only F3 progeny that were homozygous for the target locus to avoid dosage effects on nascent RNA synthesis that might alter the amount of secondary siRNAs independently of any effect of a specific mutation. The silencer locus could be either homozygous or hemizygous, as in both states sufficient primary siRNAs are produced to induce GFP silencing.
All wild-type and mutant plants harbouring the silencer locus contained primary siRNAs, which were present in three size classes (21-, 22- and 24-nt), that presumably result from multiple DCL enzymes acting on abundant hairpin RNAs in plants (Figure 2A) (Fusaro et al, 2006). By contrast, 24-nt secondary siRNAs were detected in wild-type plants and in the rdr6 mutant, but not in rdr2 and nrpd1 plants (Figure 2B). The genotypes of the rdr2, rdr6 and nrpd1 mutants were confirmed by testing for endogenous siRNA02, which is dependent on RDR2 and NRPD1 (Figure 2C), and tasiRNA255, which is dependent on RDR6 (Figure 2D).
Figure 2.
RNA profiles in rdr2, rdr6, nrpd1 and ago4 mutants. Northern blots of small RNAs: (A) primary siRNAs derived from hairpin RNA trigger; (B) secondary siRNAs; (C) siRNA02; (D) tasiRNA255. The arrowheads at the right indicate 21- and 24-nt-size classes. Ethidium bromide staining of the major RNA on the gels is shown as a loading control. The same blot was reprobed in B–D. (E) RT–PCR detection of nascent RNA; (F) RT–PCR of an actin control; (G) GFP expression. The DNA lane is amplified genomic DNA. Abbreviations: T/T;−/−, non-silenced plants containing a homozygous target locus; T/T;S/(S), silenced plants containing a homozygous target locus and either a hemizygous or homozygous silencer locus; WT, wild-type transgenic plant; Col-0, wild-type non-transgenic plants. For each of the four mutants tested (small letters), results from wild-type siblings (capital letters) are shown for comparison. The quantitative differences in primary siRNAs (A) are most likely due to the fact that RNA was prepared from pools of F2 or F3 plants that could be either homozygous or hemizygous for the silencer locus.
The absence of the secondary siRNAs in the rdr2 and nrpd1 mutants was correlated with a strong reduction of secondary RdDM (Figure 3A and B). By contrast, secondary RdDM was present at wild-type levels in the rdr6 mutant (Figure 3C), which also contained a wild-type level of secondary siRNAs (Figure 2B). Primary RdDM remained at wild-type levels in the rdr2 and nrpd1 mutants (Figure 3A and B) and is apparently sufficient for silencing GFP expression, as indicated by the failure of rdr2 and nrpd1 mutations, which eliminate secondary siRNAs and secondary RdDM, to release GFP silencing (Supplementary Table 1).
Figure 3.
Analysis of DNA methylation at the targeted enhancer and downstream region by bisulphite sequencing in rdr2, rdr6, nrpd1 and ago4 mutants. The graphs show the percentage of methylation at individual cytosines in mutant plants (top) and their wild-type siblings (bottom). (A) rdr2/RDR2; (B) nrpd1/NRPD1; (C) rdr6/RDR6; (D) ago4/AGO4. The black bar represents the enhancer region targeted by primary siRNAs (primary RdDM); the shaded grey bar indicates the downstream region targeted by secondary siRNAs (secondary RdDM). Black lines: CG methylation; blue lines: CNG methylation; red lines: CNN methylation. Residual secondary RdDM in rdr2 may be due to low levels of 24-nt siRNAs produced by RDR6/DCL3 activities (Gasciolli et al, 2005; Eamens et al, 2008). The results are from at least 10 cloned sequences. Original data are shown in Supplementary Figure 4.
The nascent RNA accumulates in the wild-type target line lacking the silencer locus but is greatly reduced or undetectable in rdr2, rdr6 and nrpd1 mutants and their wild-type siblings that contain silencer-encoded primary siRNAs (Figure 2E). In plants containing primary siRNAs and secondary siRNAs (wild-type plants and the rdr6 mutant), the absence of the nascent RNA can be explained by invoking the proposed turnover pathway involving AGO, RDR and DCL proteins (Figure 1C). This explanation does not account for the absence of the nascent RNA in nrpd1 and rdr2 mutants, which also lack secondary siRNAs despite wild-type levels of primary siRNAs and primary RdDM. One possibility is that the nascent RNA is cleaved by an AGO protein in the region overlapping with primary siRNAs. However, a 3′-cleavage fragment was not detected with the primers used (Figure 2E).
An alternate explanation is that the nascent RNA is not transcribed in nrpd1 plants because of impaired Pol IVa function. Interestingly, the nascent RNA can be detected in the non-silenced target line in the nrpd1 background (Figure 2E), indicating that it is transcribed by an RNA polymerase other than Pol IV—presumably Pol II—in the absence of primary siRNAs and primary RdDM. A hypothetical explanation for the missing nascent RNA in the rdr2 mutant is that a Pol IV-generated transcript, or cleavage products thereof, is rapidly turned over by nuclear exonucleases, such as XRN2 and XRN3 (Gy et al, 2007), unless copied into double-stranded RNA by RDR2.
The effects of the ago4-1 mutation on GFP silencing are complex because reactivation of GFP expression was not uniformly observed in both the root and shoot meristem regions of all seedlings. However, when considering seedlings in which reactivation occurred in both meristematic regions, there was a good correspondence between homozygosity for the ago4-1 mutation and a GFP-positive phenotype (Supplementary Table 1). A contribution of AGO4 to GFP silencing is consistent with the substantial loss of primary RdDM in ago4-1 plants (Figure 3D). The ago4-1 mutant lacked detectable secondary siRNAs (Figure 2B) and consequently had negligible secondary RdDM (Figure 3D); as expected from the absence of secondary siRNAs, accumulation of the nascent RNA was observed in ago4-1 plants (Figure 2E).
Identification of a dcl3-null mutation in a forward genetic screen
In a forward genetic screen using this transgene TGS system to identify silencing-defective mutants (Kanno et al, 2008), we identified one allele of a new complementation group, dms6. The dms6-1 mutant displayed an unusual pattern of accumulation of siRNA1003 (originating from 5S rDNA repeats) resembling that observed in a dcl3-1 mutant, which contains a T-DNA insertion in the DCL3 gene (Xie et al, 2004). The most striking feature of this pattern is an irregular ladder of RNAs that migrate above the 24-nt siRNA1003 observed in wild-type plants (Figure 4A). Given the similar patterns of siRNA1003 accumulation observed in dms6-1 and the known dcl3-1 mutant, we sequenced the DCL3 gene in the dms6-1 mutant and indeed detected a mutation that introduces a premature stop codon at amino acid 130, which is near the N-terminal part of the DCL3 protein (Figure 4, bottom); consequently, dms6-1, which is most likely a null allele of DCL3, has been renamed dcl3-5.
Figure 4.
RNA profile in the dcl3-5 mutant. Northern blots of small RNAs (A–C; E–J) and RT–PCR detection of the nascent RNA (D). (A) siRNA1003; (B) primary siRNAs; (C) secondary siRNAs; (D) nascent RNA (top) and actin control (bottom); (E) 45S rDNA siRNAs; (F) siRNA02; (G) solo LTR siRNAs; (H) Tag2 siRNAs; (I) Sat5 siRNAs. The arrowheads at the left indicate 21- and 24-nt size classes. In panels G–I, the lanes were pieced together from the same gel, as indicated by the dividing white lines. At the bottom of each blot, ethidium bromide staining of the major RNA on the gels is shown as a loading control. Bottom: domain structure of the DCL3 protein (1531 amino acids; Schauer et al, 2002) and position of the dcl3-5 mutation. It is unclear why a dcl3 mutation was recovered in this mutant screen and not a previous one based on silencing a seed-specific promoter (Kanno et al, 2004, 2005), but it may reflect the enhanced sensitivity of the present screen, which uses detection of enhanced GFP in colourless root tips.
In the dcl3-5 mutant, 24-nt primary siRNAs derived from the hairpin RNA trigger are eliminated, whereas 21- and 22-nt primary siRNAs are present at slightly elevated levels (Figure 4B), probably through the compensatory action of DCL4 and DCL2, respectively (Fusaro et al, 2006). However, despite their increased abundance, the 21- and 22-nt siRNAs are unable to efficiently induce RdDM of the target enhancer region (Figure 5A). The dcl3-5 mutation also reduces methylation of endogenous sequences including a Tag2 transposon-related sequence (Figure 5B) and 5S rDNA repeats (Supplementary Figure 2).
Figure 5.
Analysis of DNA methylation at the targeted enhancer and an endogenous Tag2 element by bisulphite sequencing in the dcl3-5 mutant. The graphs show the percentage of methylation at individual cytosines in the dcl3-5 mutant (top) and wild-type plants (bottom) in the following sequences. (A) Targeted enhancer region (black bar) and downstream region (shaded grey bar); (B) Tag2 element. Black lines: CG methylation; blue lines: CNG methylation; red lines: CNN methylation. The results are from eight cloned sequences. Original data are shown in Supplementary Figure 4. The nearly complete loss of asymmetric CNN methylation in both sequences indicates a defect in de novo methylation; the residual CG and CNG methylation is probably due to siRNA-independent maintenance of symmetrical methylation (Aufsatz et al, 2002).
Secondary siRNAs are undetectable in the dcl3-5 mutant (Figure 4C); however, it is unlikely that the absence of secondary siRNAs is a direct result of DCL3 deficiency, as initiation of secondary siRNA biogenesis depends on pre-existing primary RdDM, which is greatly reduced in the dcl3-5 mutant (Figure 5A). Nevertheless, the fact that the secondary siRNAs are 24-nt in length suggests that they are indeed products of DCL3 and able to induce methylation. The absence of secondary siRNAs is consistent with the presence of the nascent RNA in dcl3-5 plants (Figure 4D).
We tested the accumulation of additional endogenous 24-nt siRNAs in the dcl3-5 mutant. A pattern of larger (>24-nt) siRNAs, similar to those observed for siRNA1003, as well as faint smaller siRNAs were observed from 45S rDNA repeats (Figure 4E). Other endogenous 24-nt siRNAs that are derived from sequences located in euchromatic chromosome arms, including siRNA02, a retrotransposon solo LTR and the Tag2 element analysed for methylation are mainly present as 21- and 22-nt size classes in dcl3-5 plants (Figure 4F–H). Interestingly, siRNAs derived from SAT5, a repetitive sequence family comprising several dispersed single copies in euchromatin and a tandem repeat block in the pericentromeric heterochromatin on chromosome 5 (L Daxinger and M Matzke, unpublished data) appear as prominent 21–22-nt siRNAs as well as a ladder of larger RNAs resembling those seen for siRNAs originating from 5S and 45S rDNA (Figure 4I).
Discussion
We have defined a stepwise pathway for biogenesis of 24-nt secondary siRNAs and unidirectional spreading of DNA methylation in Arabidopsis. Hairpin-derived primary siRNAs induce primary RdDM at the target enhancer region. This step initiates the Pol IV-RDR2-dependent turnover of a nascent RNA to produce secondary siRNAs, which trigger secondary RdDM in the downstream region (Figure 6). The stepwise scenario is supported by the observations that primary and secondary siRNA formation, as well as the corresponding RdDM steps, occur sequentially and can be uncoupled genetically.
Figure 6.
Stepwise pathway for secondary siRNA biogenesis and spreading of methylation. Proteins whose functions have been demonstrated in this study are shown in white letters on dark boxes. Top: in non-silenced plants containing the target locus (T) only, the targeted enhancer region (black box) is unmethylated and the nascent RNA, which normally does not interfere with GFP expression, is presumably transcribed by Pol II because it is still detectable in an nrpd1 mutant. Bottom: in the presence of the silencer locus (S), GFP expression is silenced by primary RdDM (blue ‘m'), which is induced by hairpin-derived, DCL3-dependent 24-nt primary siRNAs (blue dashes). Primary RdDM attracts the secondary siRNA-generating machinery, which includes Pol IV, RDR2 and DCL3. In this model, Pol IV transcribes the methylated template to produce the nascent RNA, which is copied and diced by RDR2 and DCL3, respectively, to produce 24-nt secondary siRNAs (red dashes) that guide secondary RdDM (red ‘m') in the downstream region (black shade). Pol IV may also transcribe the double-stranded RNA to sustain secondary siRNA production once the cycle is initiated (Supplementary Figure 2). In both cases, Pol IV transcribes a precursor of secondary siRNAs. Whether AGO cleavage of the nascent RNA is required to provide substrates for RDR2 is still not known. siRNA-guided de novo methylation requires DRD1, DMS3 and Pol V (Kanno et al, 2008) and is presumably catalysed by DRM2 (Cao et al, 2003).
Our study supports a previous conjecture that primary RdDM is required to initiate 24-nt secondary siRNA formation. This requirement was initially suggested by the absence of secondary siRNAs in nrpe1 and drd1 mutants, which still produce primary siRNAs and the overlapping nascent RNA but lack primary RdDM (Kanno et al, 2008). The nascent RNA stably accumulates in the absence of primary RdDM in non-silenced plants but, in that instance, is presumably a Pol II transcript (Figure 6) because it is still made in an nrpd1 mutant. The key function of primary RdDM appears to be in attracting the secondary siRNA-generating machinery, which includes Pol IV and RDR2. The contribution may be direct if primary RdDM provides a template that is preferentially recognized by Pol IV, which has been proposed to transcribe methylated DNA (Herr et al, 2005; Onodera et al, 2005). In this model, Pol IV would transcribe the nascent RNA from the methylated template (Figure 6). An indirect contribution of primary RdDM is also possible if the nascent RNA needs to be sliced by AGO4 to provide substrates for RDR2. AGO4, which associates with WG/GW repeats in the C-terminal domain of NRPE1 (El-Shami et al, 2007), would be brought in together with Pol V during the primary RdDM step. Owing to the partial effect of the ago4-1 mutation on primary RdDM and the probable redundancy of AGO4 and AGO6 (Zheng et al, 2007), we could not discern whether AGO slicing of the nascent RNA is necessary for the biogenesis of secondary siRNAs. This issue can be tested in the future by using ago4 (and ago6) mutants deficient in catalytic PIWI domain but not the siRNA-binding PAZ domain (Qi et al, 2006).
Whereas both of the RdDM steps require the same factors (Pol V-DRD1-DMS3-AGO4-DRM2) (Figure 6), the two DCL3-dependent 24-nt siRNA populations differ in their genetic requirements. Cis-acting secondary siRNAs, but not trans-acting hairpin-derived primary siRNAs, require Pol IV and RDR2 for their formation. Owing to the strong correlation between the appearance of secondary siRNAs and the disappearance of the nascent RNA, we infer that the nascent RNA is directly or indirectly transcribed by Pol IV and/or RDR2 during secondary siRNA formation. Secondary RdDM occurs only in the presence of secondary siRNAs, which are generated after primary RdDM is induced. Primary RdDM can be established and maintained independently of secondary RdDM, as observed in rdr2 and nrpd1 mutants. The dispensability of RDR2 (and RDR6) for primary RdDM differs from the situation in fission yeast, where heterochromatin formation induced in trans by hairpin-derived siRNAs nevertheless requires RDR-dependent synthesis of double-stranded RNA in cis at the target locus (Iida et al, 2008). The basis of this difference is unknown but, as discussed below, it may reflect whether siRNAs interact with target DNA or RNA sequences.
Although the nascent RNA is clearly involved in the formation of secondary siRNAs that induce secondary RdDM, our data suggest that it does not contribute to primary RdDM or GFP silencing. The evidence for this is that the nascent RNA is greatly reduced or undetectable in rdr2 and nrpd1 mutants that lack secondary siRNAs, yet primary RdDM is present at wild-type levels and the GFP reporter gene is silenced in these mutants. Thus the nascent RNA acts only in the non-essential pathway of secondary siRNA biogenesis. This finding, together with the sharp boundary between primary and secondary RdDM, can be used to argue that primary siRNAs induce primary RdDM by interacting directly with the DNA target sequence and not with a nascent ‘enhancer-associated' RNA (Han et al, 2007). The dispensability of secondary siRNAs and secondary RdDM for GFP silencing in our system is consistent with the fact that we have not recovered nrpd1 or rdr2 mutants in our forward genetic screen (this study; Kanno et al, 2008). Interestingly, both nrpd1 and nrpe1 mutants were recovered in an independent forward screen using a hairpin RNA to transcriptionally silence a 35S promoter-driven GFP transgene (Eamens et al, 2008). The unusual requirement for Pol IV in hairpin RNA-triggered silencing may reflect the need to amplify low levels of 35S promoter siRNAs.
RDRs and direction of methylation spreading
During PTGS, spreading of RdDM into transcribed regions is associated with the production of approximately 21–22-nt secondary siRNAs that require RDR6 and transcription of target locus for their biogenesis (Vaistij et al, 2002; Eamens et al, 2008). The observation that formation of 24-nt secondary siRNAs in our TGS system is unaffected in an rdr6 mutant but is abolished in an rdr2 mutant clearly distinguishes the two pathways of secondary siRNA biogenesis. A further difference is that NRPD1 and NRPE1 are dispensable for initiating secondary siRNA formation in a PTGS system that is triggered by a virus (Eamens et al, 2008), whereas both of these factors are required for secondary siRNAs and secondary RdDM in our TGS system. Although it is relatively easy to distinguish primary and secondary siRNAs that are derived from the transgene sequences, it is less clear how to distinguish endogenous primary and secondary siRNAs. On the basis of our results with the transgene system, we suggest that endogenous secondary siRNAs are those that require DRD1/Pol V-mediated DNA methylation, and hence will decrease in abundance in drd1 and nrpe1 mutants (Supplementary Figure 3).
Bidirectional spreading has been observed in a PTGS system (Vaistij et al, 2002), whereas spreading of methylation in our transgene system occurs in the 3′-direction only (Supplementary Figure 4). Spreading is probably unidirectional because the nascent RNA, which provides a template for RDR2, does not extend upstream of the targeted enhancer region; consistent with this, no secondary siRNAs originating from the upstream region are detectable (T Kanno, L Daxinger, M Matzke, unpublished data). For reasons that are not yet clear, spreading of methylation is not always observed in transgenic plants (Aufsatz et al, 2002; Kanno et al, 2004, 2005; Vogt et al, 2004). Methylation spreading that triggers silencing of endogenous genes in Arabidopsis has been reported recently, but the genetic requirements differ somewhat from those needed in our transgene system. Bidirectional spreading of DNA methylation and siRNAs from tandem repeats occurred at the SDC locus, but the two phenomena occurred independently and required the CG methyltransferase MET1 and Pol IV-RDR2-DCL3, respectively (Henderson and Jacobsen, 2008). Unidirectional spreading of methylation and 24-nt siRNAs from a LINE retrotransposon into an adjacent APC gene was observed in a ddm1 mutant (Saze and Kakutani, 2007). The number of plant genes that might be regulated by methylation spreading under normal or adverse conditions is not yet known (Woo and Richards, 2008).
Identification of a dcl3 mutant
Our study is the first to report the identification of a dcl3 mutation in a forward screen for silencing-defective mutants. The only other EMS-induced point mutation of DCL3 (dcl3-3) was isolated as an enhancer of silencing in a transgene PTGS system (Smith et al, 2007). In addition to DCL3, which generates 24-nt heterochromatic siRNAs (Xie et al, 2004), there are three other DCL activities in Arabidopsis: DCL1 produces miRNAs; DCL2 generates 22-nt virus-derived siRNAs and 24-nt stress-related nat-siRNAs; DCL4 generates 21-nt tasiRNAs and siRNAs involved in RNAi (Vazquez, 2006; Voinnet, 2008).
The dcl3-5 mutation identified in our study demonstrated that hairpin-derived 24-nt siRNAs are essential to induce primary RdDM of the transgene target sequence; 21- and 22-nt siRNAs, even when present at elevated levels in the dcl3-5 mutant, were unable to compensate for the loss of the 24-nt-size class, perhaps because they are too small or not loaded onto the right effector complex. The strict requirement for hairpin-derived 24-nt siRNAs to induce methylation in our system and in others (Wang et al, 2008) contrasts with a previous report that DCL2, DCL3 and DCL4 are partially redundant in de novo methylation (Henderson et al, 2006). The discrepancy is unlikely to reflect differences in DCL processing of transgenic hairpin RNAs versus RDR-derived double-stranded RNAs because the endogenous Tag2 element that we studied sustained a heavy loss of methylation in the dcl3-5 mutant despite the presence of 21–22-nt siRNAs. Perhaps other locus-specific effects that remain to be identified can account for the differing results.
We observed larger (>24-nt) small RNAs derived from 5S and 45S ribosomal DNA sequences and the SAT5 repetitive sequence in the dcl3-5 mutant. Indeed this feature allowed us to select DCL3 as a candidate gene for the dms6-1 mutant. By contrast, other endogenous 24-nt siRNAs are present mainly as 21- and 22-nt-size classes in dcl3 plants, presumably owing to the compensating activities of DCL4 and DCL2, respectively (Xie et al, 2004; Gasciolli et al, 2005; Fusaro et al, 2006; Henderson et al, 2006; Dunoyer et al, 2007). SAT5 repeats provide an interesting example in which prominent 21- and 22-nt siRNAs are observed in addition to larger (>24-nt) small RNAs. The former may originate from dispersed single copies of SAT5 that are present in euchromatic regions, whereas the latter might originate from a block of SAT5 repeats in the pericentromeric region on chromosome 5. The variable accumulation of different size classes of endogenous siRNAs observed in dcl3 mutants may thus reflect differential accessibility of DCL activities to double-stranded RNAs derived from dispersed versus tandem repeats.
RdDM in mammalian cells?
Mammals methylate their genomes but lack Pol IV subunits, RDRs and 24-nt siRNAs. However, there is evidence that Pol lI can transcribe RNA and hence act as an RDR (Lehmann et al, 2007; Chang et al, 2008). A DMS3-related protein, SmcHD1, is important for murine X chromosome inactivation, which involves non-coding RNAs, DNA methylation and spreading of silencing along the inactive X chromosome (Blewitt et al, 2008). Piwi-interacting RNAs (piRNAs) are a larger size class of small RNA (25–30 nt) that guide de novo methylation of retrotransposons in the male germ line in mice (Kuramochi-Miyagawa et al, 2008). It is thus possible to envision a similar pathway of RdDM and methylation spreading through regions of mammalian genomes that encode overlapping short and long non-coding RNAs (Kapronov et al, 2007).
Materials and methods
Nomenclature
In recently updated nomenclature (Wierzbicki et al, 2008), Pol IVa and Pol IVb are renamed Pol IV and Pol V, respectively. The largest subunits of Pol IV and Pol V, previously termed NRPD1a and NRPD1b, have been renamed NRPD1 and NRPE1, respectively. The shared second largest subunit, formerly NRPD2a, has the synonymous name NRPD2a/NRPE2a.
Mutants
We used the following Arabidopsis thaliana mutants in this study: rdr2-1 (SAIL_1277H08; Xie et al, 2004), rdr6-1 (Elmayan et al, 1998), nrpd1-7 (formerly nrpd1a-7) (Smith et al, 2007), ago4-1 (Zilberman et al, 2003) and dcl3-1 (SALK_005512; Xie et al, 2004). All mutants are in the Columbia (Col-0) ecotype, with the exception of ago4-1, which is in the Landsberg erecta line. The mutants were genotyped using primers listed in Supplementary Table 2. Genotypes of rdr2 and rdr6 plants were confirmed by testing for the presence of siRNA02 and tasiRNA255, respectively (Figure 2). In addition, the nrpd1-7 and ago4-1 mutations were confirmed by sequencing the respective gene in DNA isolated from the genotyped plants.
Small RNA isolation and northern blot analysis
Small RNAs were isolated from pooled 21-day-old seedlings or inflorescences using the mirVana miRNA isolation kit (Ambion) and analysed by northern blot hybridization according to published procedures (Kanno et al, 2005; Huettel et al, 2006). To obtain enough plants of the proper genotype for rdr2, rdr6 and nrpd1 mutants and their wild-type siblings, we used plants from the F3 generation. For the ago4-1 mutant, we used plants of the F2 generation. We used end-labelled oligonucleotides to detect siRNA1003 (from 5S rDNA repeats), siRNAs from 45S rDNA repeats, tasiRNA 255 and siRNA02. We used end-labelled LNA (locked nucleic acid) oligonucleotides to detect siRNAs from Tag2 and SAT5. A riboprobe was used to detect siRNAs from the solo LTR (Huettel et al, 2006). Primer information for specific probes is shown in Supplementary Table 2.
Nascent RNA analysis
To detect nascent RNAs, total RNA was isolated from mixed floral inflorescences using Trizol (Invitrogen). A cDNA was synthesized according to the manufacturer's instructions using a First Strand cDNA Synthesis kit (Fermentas) using an oligonucleotide d(T) primer and 1 μg of total RNA. The primers are shown in Supplementary Table 2. These primers detect a portion of the nascent transcript starting directly downstream of the targeted enhancer region and extending to the end of the GFP coding region (see the ‘green primer' set in Supplementary Figure 1 of Kanno et al, 2008).
Bisulphite sequencing
We isolated genomic DNA from rosette leaves using a DNeasy Plant Maxi kit (Qiagen). Bisulphite treatment of DNA was conducted using a EpiTect Bisulphite kit (Qiagen) according to the manufacturer's instructions with several modifications as follows. Genomic DNA was predigested with HindIII and 500 ng of the DNA was treated in the reaction solution containing 35 μl of the DNA protection buffer. Following incubation for 2 min at 95°C, eight cycles of these thermocycler conditions were performed: for 1 min at 95°C and for 2 h at 75°C. Primers are shown in Supplementary Table 2. As a control for complete bisulphite conversion, exon 15 from the PHAVOLUTA locus that lacks methylated cytosines in the wild type was used (Bao et al, 2004; Reinders et al, 2008). At least five clones were sequenced from each mutant and wild-type control. Bisulphite conversion of the PHAVOLUTA sequence ranged in all cases between 99 and 100%. Original data from the target enhancer in the various mutants and the Tag2 sequence in dcl3-5 are shown in Supplementary Figure 4.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Table 1
Supplementary Table 2
Acknowledgments
We are grateful to David Baulcombe for providing the nrpd1-7 mutant, Jim Carrington for rdr2-1 and dcl3-1 and Hervé Vaucheret for rdr6-1. We thank Stephan Kirchmaier for assistance with bisulphite sequencing. This work was supported by the European Science Foundation (ESF) under the EUROCORES Programme EuroDyna, through contract no. ERAS-CT-2003-980409 of the European Commission, DG Research, FP6 and the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (FWF) (grant nos. I26-B03 and P20707-B03) and the European Union (contract HPRN-CT-2002-00257).
References
- Aufsatz W, Mette MF, van der Winden J, Matzke AJM, Matzke M (2002) RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA 99: 16499–16506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao N, Lye KW, Barton MK (2004) MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell 7: 653–662 [DOI] [PubMed] [Google Scholar]
- Bei Y, Pressman S, Carthew R (2007) SnapShot: small RNA-mediated epigenetic modifications. Cell 130: 756. [DOI] [PubMed] [Google Scholar]
- Blewitt M, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, Apedaile A, Hilton DJ, Dunwoddie SL, Brockdorff N, Kay GF, Whitelaw E (2008) SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet 40: 663–669 [DOI] [PubMed] [Google Scholar]
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE (2003) Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13: 2212–2217 [DOI] [PubMed] [Google Scholar]
- Chan SWL (2008) Inputs and outputs for chromatin-targeted RNAi. Trends Plant Sci 13: 383–389 [DOI] [PubMed] [Google Scholar]
- Chan SWL, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE (2004) RNA silencing genes control de novo DNA methylation. Science 303: 1336. [DOI] [PubMed] [Google Scholar]
- Chang J, Nie X, Chang HE, Han Z, Taylor J (2008) Transcription of hepatitis delta virus RNA by RNA polymerase II. J Virol 82: 1118–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ (2007) Action at a distance: epigenetic silencing of large chromosomal regions in carcinogenesis. Hum Mol Genet 16: R88–R95 [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A, Voinnet O (2007) Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat Genet 39: 848–856 [DOI] [PubMed] [Google Scholar]
- Eamens A, Vaistij FE, Jones L (2008) NRPD1a and NRPD1b are required to maintain post-transcriptional RNA silencing and RNA-directed DNA methylation in Arabidopsis. Plant J 55: 596–606 [DOI] [PubMed] [Google Scholar]
- Elmayan T, Balzergue S, Béon F, Bourdon V, Daubremet J, Guénet Y, Mourrain P, Palauqui JC, Vernhettes S, Vialle T, Wostrikoff K, Vaucheret H (1998) Arabidopsis mutants impaired in cosuppression. Plant Cell 10: 1747–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi MA, Jacobsen SE, Coke R, Lagrange T (2007) Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev 21: 2539–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro AF, Matthew L, Smith NA, Curtin SJ, Dedic-Hagan J, Ellacott GA, Watson JM, Wang MB, Brosnan C, Carroll BJ, Waterhouse PM (2006) RNA interference-inducing hairpin RNAs in plants act through the viral defense pathway. EMBO Rep 7: 1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H (2005) Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 15: 1494–1500 [DOI] [PubMed] [Google Scholar]
- Gy I, Gasciolli V, Lauressergues D, Morel J-B, Gombert J, Proux F, Proux C, Vaucheret H, Mallory AC (2007) Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19: 3451–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Kim D, Morris KV (2007) Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci USA 104: 12422–12427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE (2008) Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG methylation and initiate siRNA spreading. Genes Dev 22: 1597–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE (2006) Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 38: 721–725 [DOI] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120 [DOI] [PubMed] [Google Scholar]
- Hollick JB (2008) Sensing the epigenome. Trends Plant Sci 13: 398–404 [DOI] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJ, Matzke M (2006) Targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J 25: 2828–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida T, Nakayama J, Moazed D (2008) siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol Cell 31: 178–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Bucher E, Daxinger L, Huettel B, Böhmdorfer G, Gregor W, Kreil DP, Matzke M, Matzke AJM (2008) A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet 40: 670–675 [DOI] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJM (2005) Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37: 761–765 [DOI] [PubMed] [Google Scholar]
- Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJM (2004) Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol 14: 801–805 [DOI] [PubMed] [Google Scholar]
- Kapronov P, Willingham AT, Gingeras TR (2007) Genome-wide transcription and the implications for genomic organization. Nat Rev Genet 8: 413–423 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T. (2008) DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SH, Workman JL (2008) The heterochromatin protein 1 (HP1) family: put away a bias toward HP1. Mol Cells 26: 217–227 [PubMed] [Google Scholar]
- Lehmann E, Brueckner F, Cramer P (2007) Molecular basis of RNA-dependent RNA polymerase II activity. Nature 450: 445–449 [DOI] [PubMed] [Google Scholar]
- Locke SM, Martienssen RA (2006) Slicing and spreading of heterochromatic silencing by RNA interference. Cold Spring Harbor Symp Quant Biol 71: 497–503 [DOI] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJM (2007) Targets of RNA-directed DNA methylation in Arabidopsis. Curr Opin Plant Biol 10: 512–519 [DOI] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS (2005) Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622 [DOI] [PubMed] [Google Scholar]
- Pauler FM, Koerner MV, Barlow DP (2007) Silencing by imprinted noncoding RNAs: is transcription the answer? Trends Genet 23: 284–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penterman J, Uzawa R, Fischer RL (2007) Genetic interactions between DNA demethylation and methylation in Arabidopsis. Plant Physiol 145: 1549–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard CS, Haag JR, Ream T, Wierzbicki A (2008) Roles of RNA polymerase IV in gene silencing. Trends Plant Sci 13: 390–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ (2006) Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443: 1008–1012 [DOI] [PubMed] [Google Scholar]
- Reinders J, Delucinge Vivier C, Theiler G, Chollet D, Descombes P, Paszkowski J (2008) Genome-wide, high-resolution DNA methylation profiling using bisulfite-mediated cytosine conversion. Genome Res 18: 469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Kakutani T (2007) Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin remodelling factor DDM1. EMBO J 26: 3641–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A (2002) DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci 7: 487–491 [DOI] [PubMed] [Google Scholar]
- Smith L, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, Pikaard CS, Baulcombe DC (2007) An SNF2 protein associated with nuclear RNA silencing and the spread of silencing signal between cells in Arabidopsis. Plant Cell 19: 1507–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S (2006) Spreading of silent chromatin: inaction at a distance. Nat Rev Genet 7: 793–803 [DOI] [PubMed] [Google Scholar]
- Vaistij FE, Jones L, Baulcombe DC (2002) Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14: 857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt H, Bleys A, Depicker A (2003) RNA target sequences promote spreading of RNA silencing. Plant Physiol 131: 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F (2006) Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci 11: 460–468 [DOI] [PubMed] [Google Scholar]
- Vogt U, Pélissier T, Pütz A, Razvi F, Fischer R, Wassenegger M (2004) Viroid-induced RNA silencing of GFP-viroid fusion transgenes does not induce extensive spreading of methylation or transitive silencing. Plant J 38: 107–118 [DOI] [PubMed] [Google Scholar]
- Voinnet O (2008) Use, tolerance and avoidance of amplified silencing by plants. Trends Plant Sci 13: 317–328 [DOI] [PubMed] [Google Scholar]
- Wang MB, Helliwell CA, Wu LM, Waterhouse PM, Peacock WJ, Dennis ES (2008) Hairpin RNAs derived from RNA polymerase II and polymerase III promoter-directed transgenes are processed differently in plants. RNA 14: 903–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Richards EJ (2008) Signalling silence—breaking ground and spreading out. Genes Dev 22: 1719–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobson SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biology 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI (2008) Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15: 381–388 [DOI] [PubMed] [Google Scholar]
- Zheng X, Zhu J, Kapoor A, Zhu J-K (2007) Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J 26: 1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Table 1
Supplementary Table 2