Summary
The lantibiotic haloduracin consists of two post-translationally processed peptides, Halα and Halβ, that act in synergy to provide bactericidal activity. An in vitro haloduracin production system was utilized to examine the biological impact of disrupting individual thioether rings in each peptide. Surprisingly, the Halα B-ring, which contains a highly conserved CTLTXEC motif, was expendable. This motif has been proposed to interact with haloduracin’s predicted target, lipid II. Exchange of the glutamate residue in this motif for alanine or glutamine did completely abolish antibacterial activity. This study also established that Halα-Ser26 and Halβ-Ser22 escape dehydration, requiring revision of the Halβ structure previously proposed. Extracellular proteases secreted by the producer strain can remove the leader peptide, and the Halα cystine that is dispensable for bioactivity protects Halα from further proteolytic degradation.
Introduction
Lantibiotics are a class of cyclic peptides produced by Gram-positive bacteria. They are ribosomally synthesized as inactive precursor peptides that undergo post-translational modification to generate their mature, biologically active forms. More than 50 lantibiotics are known to date, most of which function as antimicrobial peptides (Chatterjee et al., 2005b; Cotter et al., 2005; Willey and van der Donk, 2007). Notably, the most extensively studied member, nisin, has been used worldwide for decades in the food industry without the development of widespread antibiotic resistance (Delves-Broughton et al., 1996). Moreover, other lantibiotics such as mersacidin, lacticin 3147, and microbisporicin are effective against methicillin-resistant Staphylococcus aureus and other multidrug resistant pathogens (Castiglione et al., 2008; Galvin et al., 1999; Kruszewska et al., 2004).
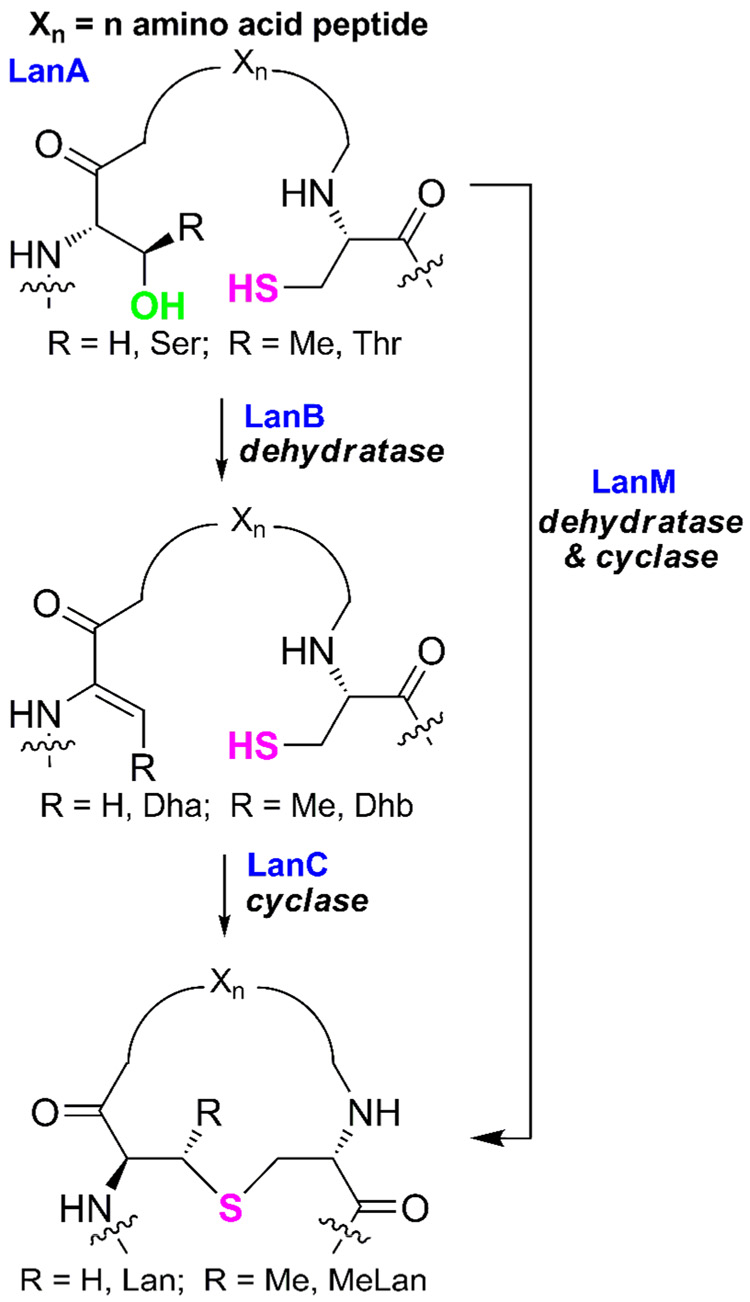
All lantibiotics contain the characteristic lanthionine (Lan) and/or methyllanthionine residues (MeLan) and also typically the unsaturated amino acids dehydroalanine (Dha) and dehydrobutyrine (Dhb) that are introduced by post-translational modification. Lantibiotic precursor peptides (LanA) consist of a structural region that will be transformed into the antimicrobial peptide and an N-terminal leader sequence that does not undergo modification. Specific Ser and Thr residues in the structural region are enzymatically dehydrated to yield Dha and Dhb, respectively (Figure 1). Through a Michael-type addition, Cys residues react with these dehydro amino acids to form the thioether rings, Lan from Ser and MeLan from Thr. The precise roles of the leader peptide are still under debate (Furgerson Ihnken et al., 2008; Levengood et al., 2007; Patton et al., 2008), but following modification it must be removed by proteolysis to generate the active lantibiotic (Chen et al., 2001; Corvey et al., 2003; Li et al., 2006; McClerren et al., 2006; van der Meer et al., 1994; Xie et al., 2004).
FIGURE 1. General overview of the common steps in lantibiotic biosynthesis.
LanA, lantibiotic precursor peptide.
A subclass of lantibiotics consists of the two-peptide systems. These compounds are formed from two precursor peptides that are each post-translationally modified to create two distinct products that act in synergy to provide bactericidal activity (Garneau et al., 2002). Currently, seven two-component lantibiotics are known, including the best-studied example, lacticin 3147 (Ryan et al., 1999), and the most recently identified member, haloduracin (Lawton et al., 2007; McClerren et al., 2006). For haloduracin, produced by the Gram-positive alkaliphilic bacterium Bacillus halodurans C-125, the unmodified peptides HalA1 and HalA2 are each dehydrated and cyclized by two dedicated bifunctional enzymes, HalM1 and HalM2, respectively. During this process, one of the four Ser/Thr residues in the structural region of HalA1 and one of the eight Ser/Thr residues in the structural peptide of HalA2 escape dehydration by their respective modification enzymes. Proteolytic removal of the leader results in the mature, active components, Halα and Halβ. The HalA1 peptide is predicted to have 32% and 44% amino acid sequence identity with the precursor peptides from mersacidin (P43683) and lacticin 3147 A1 (O87236), respectively. Both mersacidin and the mature A1 component of lacticin 3147 bind to lipid II, an essential precursor in peptidoglycan biosynthesis (Brötz et al., 1998; Wiedemann et al., 2006a). A sequential model has been proposed for the bactericidal action of two-peptide lantibiotics in which the α (A1) peptide first binds to lipid II and the α:lipid II complex serves as a docking site for the β (A2) peptide. Once recruited, the extended conformation of the β component is believed to facilitate pore formation in the bacterial membrane (Morgan et al., 2005; Wiedemann et al., 2006a).
Herein the structural features essential for biological activity of haloduracin were investigated using a recently developed in vitro biosynthetic system (McClerren et al., 2006). A number of haloduracin analogues were constructed and the impact of the alterations on antibiotic activity was assessed. In addition, this study suggests that some or all of the proteolytic process that removes the leader peptides may be performed by extracellular proteases secreted by the B. halodurans producing strain.
Results
Structures of Halα and Halβ
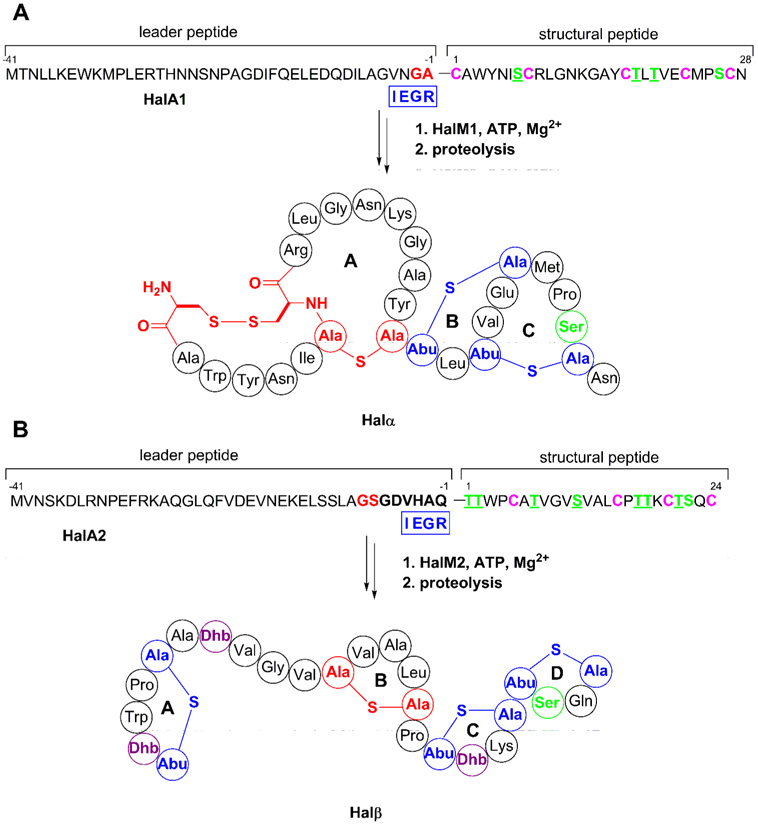
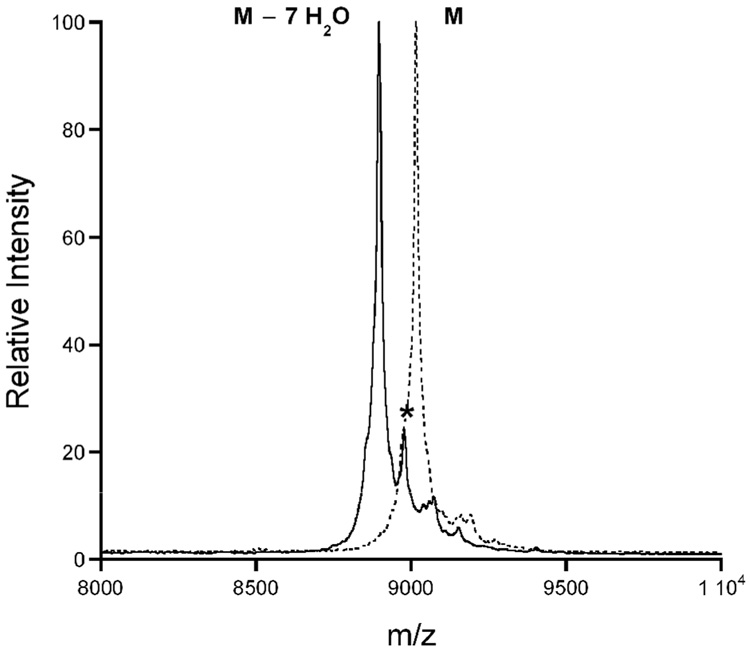
Based on the mass of haloduracin prepared enzymatically in vitro as well as the products isolated from B. halodurans C-125, one Ser/Thr residue escapes dehydration in both Halα and Halβ (McClerren et al., 2006). In order to identify the locations of these residues and to examine their biological significance, specific Ser/Thr residues were mutated in the substrate peptides to Ala. The mutant substrates were subjected to HalM1 or HalM2 and analyzed by MALDI-TOF MS. For HalA1, three out of four residues undergo dehydration. Comparison of its sequence to that of other lantibiotics suggested that Ser26 escaped dehydration (McClerren et al., 2006) (Figure 2A). To test this hypothesis the S26A mutant of HalA1-Xa was constructed by site-directed mutagenesis and processed in vitro with HalM1. As discussed previously (McClerren et al., 2006), this substrate contains a Factor Xa recognition site for leader peptide removal. Upon subjection to HalM1 under standard assay conditions (McClerren et al., 2006), three dehydrations were still observed by MS (Figure S1A, Supporting Information), thus indicating that Ser26 is indeed the residue that is not acted upon by HalM1. For Halβ, it was established that seven of eight Ser/Thr residues undergo dehydration in the structural region of HalA2 (McClerren et al., 2006). Based on homology to other lantibiotics it was proposed that Thr18 was not dehydrated. However, when the T18A mutant of HalA2-Xa was treated with HalM2 a product with 6-fold dehydration was observed (Figure S1B). This finding strongly suggests that Thr18 is normally dehydrated by HalM2. FTMS/MS data obtained previously narrowed down the other possible candidates for the residue that is not dehydrated to Thr17, Thr21, or Ser22 (McClerren et al., 2006). Thr17 and Thr21 are believed to be involved in methyllanthionine rings that are highly conserved in two-component lantibiotics (McClerren et al., 2006). Thus, HalA2-Xa S22A was constructed and subjected to HalM2 catalysis. MS analysis showed that this mutant was dehydrated 7-fold (Figure 3), demonstrating that Ser22 escapes dehydration by HalM2 in wild-type HalA2. The structure for Halβ has been revised to reflect these data (Figure 2B).
FIGURE 2. Biosynthesis of Halα (A) and Halβ (B).
In substrates designated HalA1-Xa or HalA2-Xa, the four amino acids at positions −1 to −4 were replaced by IEGR (boxed in blue). In (B), the revised structure of Halβ is shown based on the results in this study.
Figure 3. MALDI-TOF MS of HalA2-Xa S22A.
HalA2-Xa S22A before (dashed line) and after incubation with HalM2 (solid line). The asterisks (*) indicate a phosphorylated peptide (Chatterjee et al., 2005a).
The cyclization reaction is more difficult to examine because it does not involve a change in mass. In previous studies, techniques were developed that report on noncyclized cysteines through derivatization of their thiol groups (Li and van der Donk, 2007; Li et al., 2006; McClerren et al., 2006; Paul et al., 2007). Control reactions with unmodified HalA1 and HalA2 substrates indicated that iodoacetamide (IAA) was more effective for HalA1 whereas p–hydroxymercuribenzoic acid (PHMB) (Li and van der Donk, 2007; Paul et al., 2007; Pitts and Summers, 2002) demonstrated better derivatization efficiency for HalA2. Hence, throughout this study IAA was used to test cyclization of HalA1 mutants whereas PHMB was used for HalA2. The reasons behind the different derivatization efficiency of these peptides were not investigated but are likely related to their very different solubilities. The products of the enzyme assays with HalA1-Xa S26A, HalA2-Xa T18A, and HalA2-Xa S22A were treated with reductant followed by thiol derivatizing agent and monitored by MALDI-TOF MS, showing that none of the assay products displayed adducts (beyond the two expected for the cysteine residues of the disulfide in Halα) and demonstrating that these mutants were cyclized (Figure S1C–E). Removal of the leader peptide for evaluation of antimicrobial activity of the haloduracin analogues produced was accomplished using the previously described Factor Xa engineered system (McClerren et al., 2006). Briefly, in substrates denoted HalA1/2-Xa the four amino acids in the leader peptides immediately prior to the structural region of HalA1 (Val-Asn-Gly-Ala) and HalA2 (Val-His-Ala-Gln) were replaced with the Factor Xa recognition site (Ile-Glu-Gly-Arg, Figure 2). After treatment of the HalM1 and HalM2 enzymatic products with Factor Xa and confirmation of proteolytic processing by MALDI-TOF MS, agar diffusion assays against a haloduracin sensitive strain, Lactococcus lactis CNRZ 117, were performed. The data demonstrated that the two residues that escape dehydration (Halα Ser26 and Halβ Ser22) are not necessary for antibacterial activity as replacement with Ala still resulted in zones of growth inhibition. Furthermore, when an Ala residue was installed in place of Dhb18 of Halβ bioactivity was retained (Figure 4, regions 14–16).
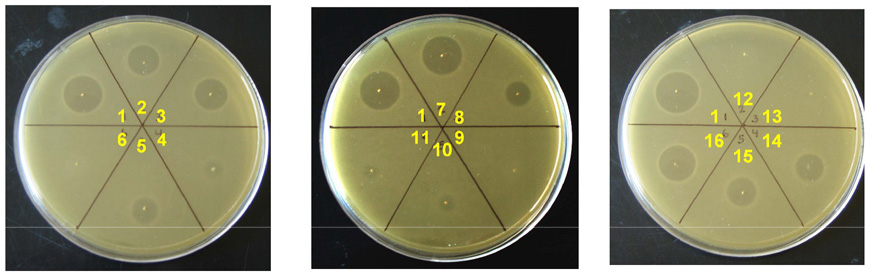
FIGURE 4. Antimicrobial activity assays for haloduracin mutants against the indicator strain L lactis CNRZ 117.
In vitro refers to compound produced with purified HalM enzymes and subsequent proteolysis with Factor Xa. In vivo refers to Halα or Halβ isolated and purified from B. halodurans C-125. Region 1, Halα + Halβ (both in vivo); Region 2, Halα (in vitro) and Halβ (in vivo); Region 3, Halα C8A (in vitro) + Halβ (in vivo); Region 4, Halα C17A (in vitro) + Halβ (in vivo); Region 5, Halα C23A (in vitro) + Halβ (in vivo); Region 6, Halα C27A (in vitro) + Halβ (in vivo); Region 7, Halα (in vivo) +Halβ (in vitro); Region 8, Halα (in vivo) + Halβ C5A (in vitro); Region 9, Halα (in vivo) + Halβ C15A (in vitro); Region 10, Halα (in vivo) + Halβ C20A (in vitro); Region 11, Halα (in vivo) + Halβ C24A (in vitro); Region 12, Halα E22A (in vitro) + Halβ (in vivo); Region 13, Halα E22Q (in vitro) + Halβ (in vivo); Region 14, Halα S26A (in vitro) + Halβ (in vivo); Region 15, Halα (in vivo) + Halβ Dhb18Ala (in vitro); Region 16, Halα (in vivo) + Halβ S22A (in vitro).
Thioether ring disruption in Halα and Halβ
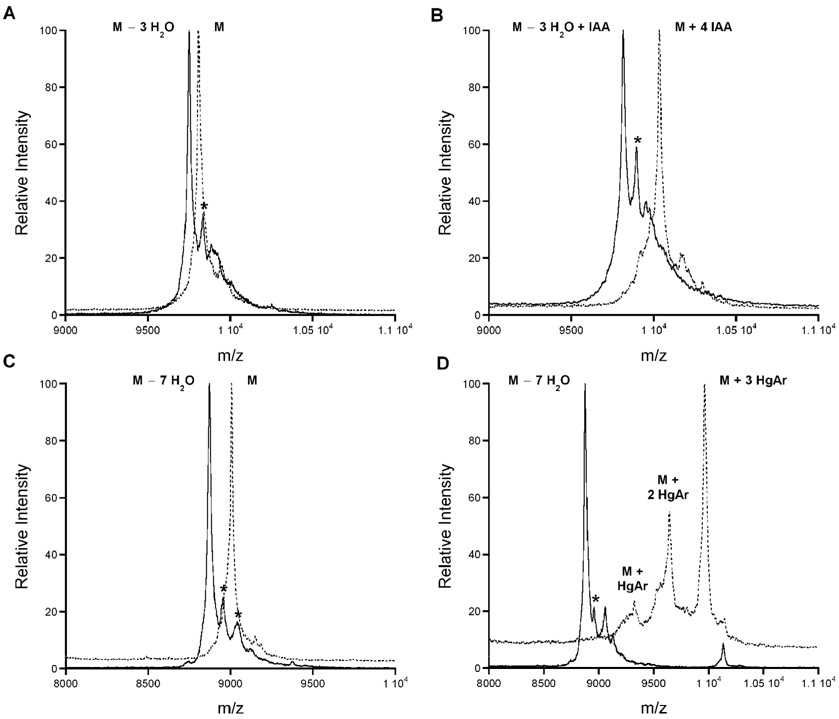
To evaluate the significance of each sulfide ring in Halα and Halβ, they were disrupted individually by converting Cys residues to Ala in the substrate peptides. Previously it was shown that the cystine linkage in Halα was not required for bacterial growth inhibition (McClerren et al., 2006). Even when the Cys thiols were alkylated by treatment with iodoacetamide (IAA), a zone of inhibition was observed in agar diffusion assays. The nonessential nature of the cystine was confirmed in this study by generation of the HalA1-Xa C8A peptide and incubation with HalM1 resulting in a 3-fold dehydrated product (Figure 5A). Subsequent analysis with IAA for cyclization as discussed above demonstrated that the mutant contained a single reactive Cys (Cys1) indicating that three Cys residues were unavailable for alkylation (Figure 5B), a result consistent with formation of three thioether rings. The HalM1 product was treated with Factor Xa protease to remove the leader sequence, and tested against the bacterial indicator strain resulting in a zone of inhibition (Figure 4, region 3) confirming that the disulfide is dispensable.
FIGURE 5. Representative MALDI-TOF MS data of the HalA1-Xa Cys-to-Ala mutants treated with HalM1.
The asterisks (*) indicate phosphorylated products (Chatterjee et al., 2005a). (A) HalA1-Xa C8A before (dashed line) and after incubation with HalM1 (solid line). (B) HalA1-Xa C8A treated with TCEP/IAA before (dashed line) and after HalM1 (solid line). (C) HalA2-Xa C5A before (dashed line) and after incubation with HalM2 (solid line). (D) HalA1-Xa C5A treated with TCEP/PHMB before (dashed line) and after HalM2 (solid line).
Using a similar strategy, the A-, B-, and C-rings were disrupted by mutating Cys17, Cys23, and Cys27 of HalA1-Xa to Ala, respectively. MALDI-TOF MS analysis of the assay products prepared by incubation of the HalA1-Xa Cys-to-Ala mutants with HalM1 showed that 3-fold dehydration occurred for all (Figure S2), and IAA analysis revealed that all of these HalM1-processed mutants contained two reactive Cys residues corresponding to Cys1 and Cys8. The IAA-derivatization studies suggest that all other Cys residues underwent the anticipated cyclization in these mutants. Subsequent removal of the leader peptide and qualitative antimicrobial assays demonstrated that the C-ring was essential for bioactivity because the C27A mutant did not produce a zone of inhibition (Figure 4, region 6). Surprisingly, the B-ring proved to be expendable as Halα-C23A gave a very clear inhibition zone when antibiotic activity was assessed in three independent experiments (Figure 4, region 5). A very small but reproducible zone was observed for the C17A mutant suggesting the A-ring is important but not absolutely essential (region 4).
Evaluation of the importance of the Lan and MeLan rings in Halβ was similarly completed. The A-, B-, C-, and D-rings were individually disrupted by conversion of Cys5, Cys15, Cys20, and Cys24 of HalA2 to Ala, respectively. Incubation with HalM2 and analysis by MALDI-TOF MS revealed 7 dehydrations for all four mutants as shown for the C5A mutant in Figure 5C (for all other mutants, see Figure S3). Subsequent analysis with PHMB for cyclization showed that the mutants C5A, C20A, and C24A did not contain any free thiols suggesting they underwent complete cyclization as shown for C5A in Figure 5D (for other mutants see Figure S3). However, C15A did display multiple adducts derived from PHMB (Figure S3C) indicating that disruption of the B-ring also affected ring formation of one or more of the other rings. The consequences of removal of one or more rings on antimicrobial activity were evaluated by agar diffusion assays after leader peptide removal. The mutant that lacked the A-ring (C5A) retained antibiotic activity (Figure 4, region 8). No zone of inhibition was observed for the C15A mutant of Halβ and only very weak zones of growth inhibition were observed for the C20A and C24A mutants (Figure 4, regions 9–11).
A highly conserved glutamate residue in Halα is essential for bioactivity
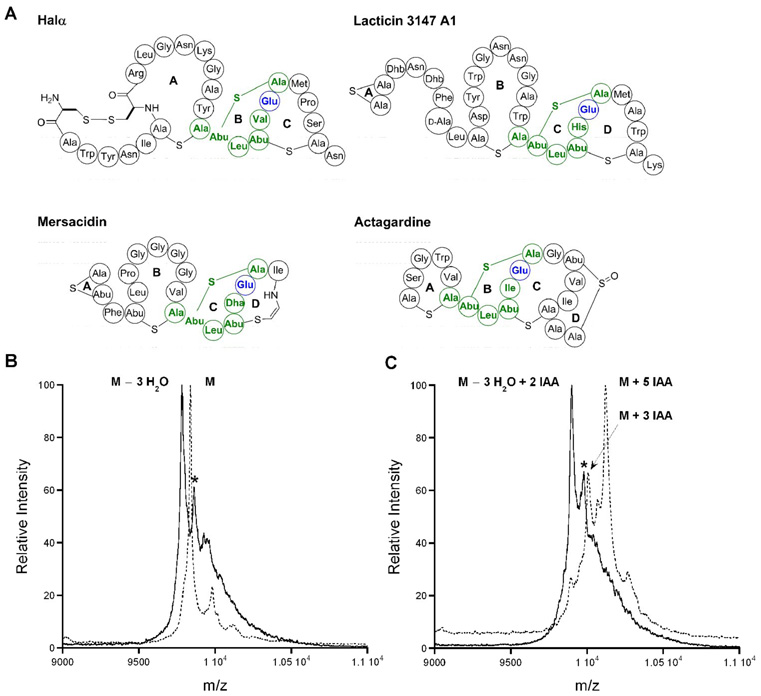
It was recently demonstrated that a highly conserved glutamate in mersacidin (Glu17) was essential for binding to its target lipid II (Brötz et al., 1998; Hsu et al., 2003; Szekat et al., 2003). Due to the high degree of homology between the C-ring of mersacidin and the B-ring of Halα (Figure 6A), the corresponding Glu22 could play a similar role in the mode of action of haloduracin. Therefore, Glu22 in HalA1-Xa was mutated to Ala and Gln. Upon treatment with HalM1, these mutants underwent three dehydrations as expected (Figure 6B). Cyclization was confirmed using IAA treatment under reducing conditions followed by MS analysis. The two IAA adducts observed were assigned to the two Cys residues that form the disulfide ring of HalA1-Xa (Figure 6C). Bioactivity analysis after removal of the leader peptide with Factor Xa showed that antibacterial activity was completely abolished under the conditions used (Figure 4, regions 12 and 13).
FIGURE 6. The importance of Glu22 for bioactivity.
(A) Structures of Halα, lacticin 3147 A1, mersacidin, and actagardine. Residues colored in green represent the proposed lipid II binding motif (CTLTXEC) with the Glu residue examined in this study colored in blue. (B) HalA1-Xa E22Q before (dashed line) and after incubation with HalM1 (solid line). The asterisks (*) indicate phosphorylated peptides (Chatterjee et al., 2005a). (C) HalA1-Xa E22Q treated with TCEP/IAA before (dashed line) and after HalM1 (solid line).
Proteolytic processing
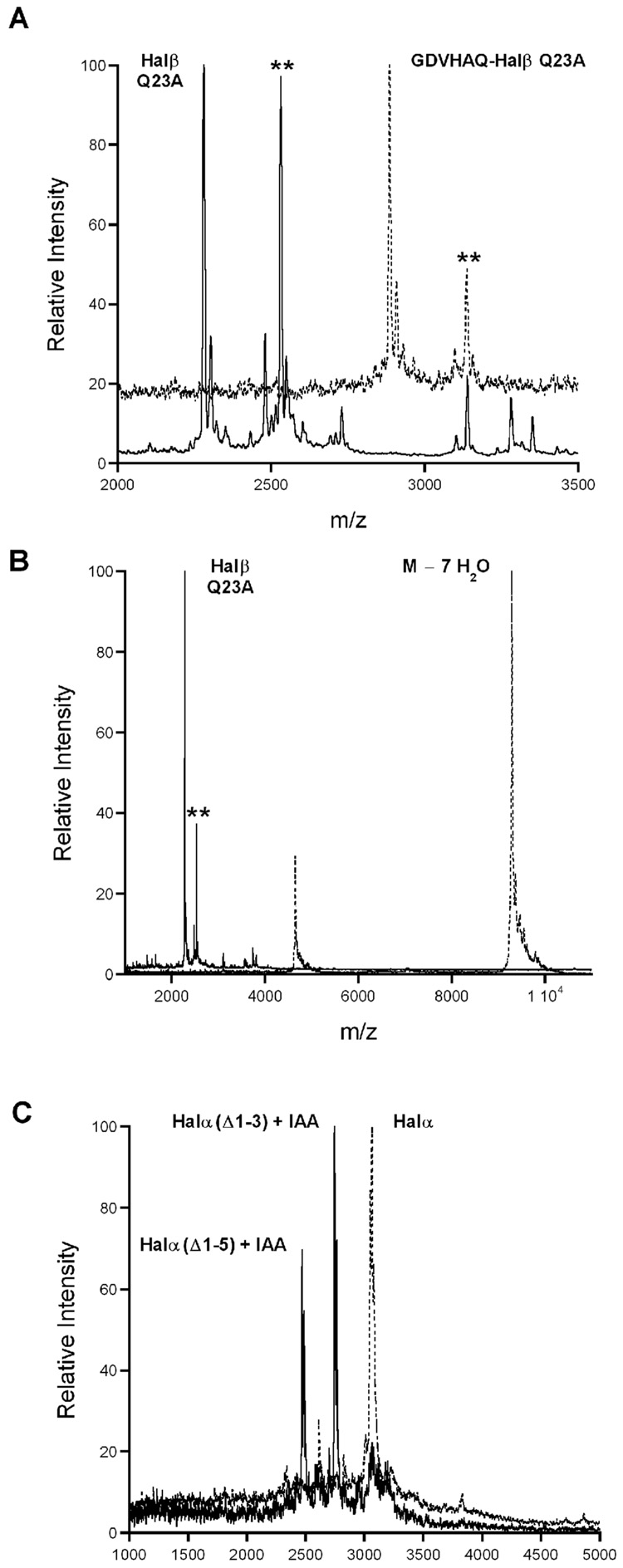
Leader peptide removal for both HalA1 and HalA2 is proposed to be achieved in vivo by a single enzyme termed HalT (Lawton et al., 2007; McClerren et al., 2006). The predicted cleavage site for LanT protease domains is after a double Gly recognition motif (GG, GA, or GS sequences) (Håvarstein et al., 1995; Nes and Tagg, 1996). For HalA1, proteolysis behind the predicted cleavage site Gly–2Ala–1 (Figure 2A) was confirmed by mass spectrometry on haloduracin α produced by B. halodurans C-125 (McClerren et al., 2006). On the other hand, for HalA2 the cleavage was predicted to take place after Gly–8Ser–7 (Figure 2B), yet the mass spectrometric data on Halβ produced by B. halodurans C-125 demonstrated that residues Gly–6 through Gln–1 were also removed (Figure 2B). Therefore, if HalT cleaves after the GlySer sequence, then an additional N-terminal proteolytic step must occur. The biological significance of the removal of the residues GDVHAQ was examined by the generation of a mutant of HalA2, designated HalA2-XaGS, with the Factor Xa cleavage signal immediately following the predicted HalT cleavage signal at Gly–8Ser–7 (Figure S4A). This construct was incubated with HalM2 to yield the expected 7-fold dehydrated product (Figure S4B) and was further processed by treatment with Factor Xa. When the resulting product designated GDVHAQ-Halβ was tested for antibiotic activity in the presence of Halα, inhibition zones comparable to in vivo and in vitro prepared haloduracin were observed (Figure S4D). This data indicated that the additional six amino acids on the N-terminus of this mutant product do not interfere with the bioactivity of haloduracin.
We investigated the possibility that N-terminal trimming of the GDVHAQ-Halβ product occurred on the agar plate by extracellular proteases from the reporter strain during the bioassay and that the trimmed Halβ product was actually responsible for the observed zone of inhibition. Furthermore, we investigated whether the producer strain B. halodurans C-125 secretes a protease that could perform N-terminal trimming of longer Halβ products. For this purpose, the HalA2-XaGS Q23A mutant was generated. The Q23A mutation was introduced so that Halβ generated by processing of GDVHAQ-Halβ could be distinguished from Halβ produced endogenously by B. halodurans C-125. This mutation was chosen since an Ala is present at this position in lacticin 3147 A2. The peptide was expressed in E. coli, purified, and subjected to HalM2 modification resulting in seven dehydrations and complete cyclization (Figures S5, A and B). Subsequent Factor Xa cleavage yielded GDVHAQ-Halβ Q23A as observed by MALDI-TOF MS. Incubation with the L. lactis CNRZ 117 indicator strain did not result in processing of GDVHAQ-Halβ Q23A (Figure 7A). However, the culture supernatant from the B. halodurans C-125 producer strain did contain one or more proteases that cleanly removed the GDVHAQ sequence resulting in Halβ Q23A (Figure 7A). In a second experiment, full length HalA2-XaGS Q23A processed by HalM2 was incubated with the culture supernatant. Once more, proteolytic processing occurred removing the entire leader sequence and producing Halβ Q23A (Figure 7B). To examine the fate of Halα under these conditions, purified Halα was incubated with the culture supernatant of B. halodurans C-125 with and without pre-treatment with the reductant tris(2-carboxyethyl)phosphine (TCEP) in order to reduce the N-terminal disulfide and subsequent alkylation by IAA. Interestingly, the Halα that was not treated with TCEP/IAA did not undergo any proteolysis as judged by MALDI-TOF MS (Figure S5D), but the sample pre-incubated with TCEP/IAA was proteolytically processed to peptides corresponding to loss of the N-terminal three and five amino acids (Figure 7C).
Figure 7. Proteolytic processing of haloduracin.
(A) HalA2-XaGS Q23A treated with HalM2 and Factor Xa resulted in GDVHAQ-Halβ Q23A. This peptide was used to monitor proteolytic processing. GDVHAQ-Halβ treated with supernatant of L. lactis CNRZ 117 (dashed line) or B. halodurans C-125 (solid line). GDVHAQ-Halβ Q23A calcd 2,883 Da; obsd 2,886 Da. A TCEP adduct was observed at 3,136 Da (Δm = 250 Da, calcd 3,133 Da) and is labeled with a double asterisk (**). Halβ Q23A calcd 2,276 Da; obsd 2,281 Da. A TCEP adduct was observed at 2,532 Da (Δm = 250 Da, calcd 2,526 Da) and is labeled with a double asterisk (**). (B) HalA2-XaGS Q23A treated with HalM2 and then supernatant of L. lactis CNRZ 117 (dashed line). calcd 9,290 Da (M + H − 7 H2O); obsd 9,290 Da. HalA2-XaGS Q23A treated with HalM2 and then supernatant of B. halodurans C-125 (solid line). calcd 2,276 Da (Halβ Q23A); obsd 2,279 Da. (C) Halα isolated from B. halodurans C-125 was treated with TCEP/IAA. The resulting bisalkylated peptide was then incubated with supernatant of L. lactis CNRZ 117 (dashed line) or B. halodurans C-125 (solid line). Halα + 2 IAA calcd 3,163 Da; obsd 3,165 Da. Halα (Δ1–3) + IAA calcd 2,745 Da; obsd 2748. Halα (Δ1–5) + IAA calcd 2,468 Da; obsd 2469.
Discussion
The ribosomal origin of the lantibiotics has provided a convenient means to conduct structure-activity relationship studies via the generation of analogues by mutagenesis of the prepeptides. Such mutagenesis studies have been carried out both in vivo in the producer strains or a suitable heterologous host, and in reconstituted in vitro systems (Chatterjee et al., 2005b; Cotter et al., 2005; Lubelski et al., 2007). In this study, haloduracin mutants were produced in vitro and analyzed for antibacterial activity with a focus on the post-translational modifications. The outcome with the S26A mutant of HalA1 confirmed the previous hypothesis based on sequence homology with other lantibiotics that this residue is not dehydrated in Halα. On the other hand, the results with both HalA2 T18A and S22A mutants are inconsistent with the previously proposed structure of Halβ based on sequence homology (McClerren et al., 2006). Both mutants strongly suggest that Ser22 in Halβ is not dehydrated, a result that was confirmed for wild-type HalA2 by postsource decay MS in a study on the directionality and processivity of HalM2 (unpublished results). The observation that the residues that escape dehydration in both Halα and Halβ are serines is in line with a previous study that compared all known lantibiotics and concluded that Ser residues escape dehydration with a much higher frequency than Thr residues (Rink et al., 2005). The underlying reason for this trend is not known. In the case of haloduracin, the flanking residues of Ser26 in Halα (Pro and Cys) and Ser22 in Halβ (Thr/Dhb and Gln) are not obvious deactivating residues. For instance, a Pro flanking Thr17 and Thr/Dhb flanking Thr2 and Thr17 in HalA2 did not negatively affect dehydration by HalM2. Although the molecular logic of why lantibiotic synthetases sometimes skip a Ser/Thr is not known, a potential evolutionary reason could be that the unmodified Ser/Thr in question is important for the biological activity of the final compound. It appears, however, that this is not the case for Ser26 in Halα and Ser22 in Halβ as shown by antimicrobial assays of analogues in which these residues were mutated. Similarly, the Thr17, Thr19, and Thr23 residues that remain unmodified in lacticin 3147 A2 (but have no homologous residues in haloduracin) were not critical for its biological activity (Cotter et al., 2006).
The corollary of Ser22 escaping dehydration in Halβ is that Thr18 must be dehydrated, in contrast to the homologous residue in lacticin 3146 A2 (Thr23), which is not dehydrated according to NMR studies (Martin et al., 2004). The differences between these two lantibiotics illustrate that the use of sequence homology to predict the structure of lantibiotics is perilous. This finding is relevant for the currently structurally uncharacterized two-component lantibiotics Smb and BHT for which the genes were recently reported. These predict 7 (BHT) or 8 (Smb) Ser/Thr in the A-peptide and 7 Ser/Thr in the B-peptide (Hyink et al., 2005; Yonezawa and Kuramitsu, 2005) (Figure S6), but the masses observed for the mature peptides suggest only 5 of these residues are dehydrated in each peptide for Smb (Petersen et al., 2006). The BHT/Smb peptides have sequence homology with both haloduracin and lacticin 3147 (Figure S6), but based on the structures of these two lantibiotics it cannot be concluded which residues escape dehydration in Smb/BHT.
The thioether rings in lantibiotics are believed to be the basis for their biological activity and in general disruption of a ring results in abolishment of antimicrobial activity (Bierbaum et al., 1996; Chatterjee et al., 2006; Chatterjee et al., 2005b; Chen et al., 1998; Kuipers et al., 1996; Ottenwälder et al., 1995; van Kraaij et al., 2000). At present only limited information is available regarding the importance of the Lan and MeLan rings in each peptide of two-component lantibiotics. Ala-scanning and random mutagenesis have been applied to the genes encoding the precursor peptides of lacticin 3147, resulting in a large amount of valuable data (Cotter et al., 2006; Field et al., 2007). However, the absence of bioactivity of several of the mutants involving the thioether rings resulted from abolished production and hence provided no direct information regarding their importance for activity. In the current study, a comprehensive examination of the importance of each (methyl)lanthionine in haloduracin for biological activity was performed, taking advantage of its in vitro reconstituted biosynthesis. These experiments show that mutation of Cys17, Cys23, and Cys27 resulted in Halα analogues in which the A-, B-, and C-rings were disrupted, respectively, without any evidence that the remaining rings were not formed. The qualitative antimicrobial activity of these mutants shows that the C-ring of Halα is essential for activity. Its A-ring is important but not absolutely essential with a weak zone of inhibition observed. On the other hand, the B-ring of Halα is not required. The retention of bioactivity upon disrupting the B-ring of Halα was surprising given its high level of conservation in comparison with mersacidin (C-ring) (Chatterjee et al., 1992), actagardine (B-ring) (Zimmermann et al., 1995), and lacticin 3147 A1 (C-ring) (Ryan et al., 1999) (Figure 6). These three lantibiotics bind to the peptidoglycan precursor lipid II (Brötz et al., 1998; Wiedemann et al., 2006b; Zimmermann and Jung, 1997) and the CTLTXEC motif encompassing the conserved rings is believed to be important for this activity. For instance, alanine scanning mutagenesis carried out for lacticin 3147 A1 in vivo (Cotter et al., 2006) showed this region was highly sensitive to alteration with respect to bioactivity. In addition, when the C-ring in lacticin 3147 A1 was opened, bioactivity was abolished (Cotter et al., 2006; Field et al., 2007). At present it is not known why disruption of the B-ring of Halα is not detrimental to bioactivity, especially since the conserved Glu residue in the CTLTXEC motif is critical. The abolishment of antimicrobial activity for Halα E22A and E22Q is in agreement with previous studies on mersacidin in which the Glu was substituted with Ala and antibiotic activity was lost (Szekat et al., 2003).
The qualitative SAR studies on Halβ resulted in removal of its A–D-rings. For the mutants disrupting the A-, C-, and D-rings, the rings not targeted by mutagenesis were still formed by HalM2 according to the PHMB data, but mutation of Cys15 resulted in disruption of cyclization for other rings in addition to the B-ring. These findings illustrate the danger of reaching conclusions regarding the importance of certain residues for bioactivity on the basis of mutagenesis studies without confirming that the mutation did not interfere with ring formation. Based on the experimental data, it can be concluded that the A-ring in Halβ is dispensable and that the C- and D-rings are important but not essential. No conclusions can be made regarding the importance of the B-ring since the observed lack of bioactivity of the Halβ C15A mutant could be due to disruption of multiple rings.
This study also provides some additional insights into proteolytic processing of haloduracin. Similarly to the two-component lantibiotics cytolysin and plantaricin W (Cox et al., 2005; Holo et al., 2001), the mass of the mature Halβ peptide is smaller than that predicted by the cleavage site for the N-terminal protease domain of HalT. Additional proteolysis for cytolysin is carried out by a designated protease, termed CylA, which was shown to be necessary for antibiotic activity (Cox et al., 2005). No CylA homologs have been identified in the B. halodurans C-125 genome, yet six amino acids must be removed from the N-terminus of the product that is expected to be secreted by HalT. The observed removal of these amino acids upon exposure to the supernatant of B. halodurans cell culture strongly suggests that one or more secreted proteases are responsible for this process. Interestingly, when Halα was first reduced, opening the disulfide ring at its N-terminus, followed by alkylation of Cys1 and Cys5 with IAA, it too became susceptible to additional N-terminal proteolysis. Since the disulfide is shown not to be important for antimicrobial activity in haloduracin or in plantaricin W (Holo et al., 2001; McClerren et al., 2006), it may be conserved to protect the peptide from such degradation in the extracellular environment. Similarly, the N-terminal lanthionine in lacticin 3147 A1, which is also not required for antimicrobial activity (Cotter et al., 2006), may protect this compound from proteolyis. In fact, the great majority of the lantibiotics in the two-component class have cyclic structures at or near their N-termini (Figure S6).
Significance
This study is the first systematic analysis of the SAR of the individual thioether rings in a two-component lantibiotic. Using an in vitro biosynthetic system the C-ring and Glu22 of Halα of haloduracin are shown to be essential for antimicrobial activity with wild type Halβ. The A-ring of Halα is important but not essential and, surprisingly given its high conservation in lantibiotics, the B-ring is not required. The A-, C-, and, D-rings of Halβ were important but not absolutely required for its synergistic activity with wild type Halα. The disulfide ring present at the N-terminus of Halα is not important for its antimicrobial activity but protects the compound from proteolytic degradation by proteases secreted by the producer strain. Collectively, these results provide new insights into the roles and importance of the crosslinks found in two-component lantibiotics. This study also highlights the value of using an in vitro biosynthetic system for SAR studies.
Experimental Procedures
Construction of all overexpression plasmids and procedures for peptide purification, are described in the Supporting Information along with tables of all masses of the ions observed in the mass spectra shown in this work.
Enzymatic Assays for Dehydration Activity
Purified HalA peptides were each dissolved in HalM assay buffer (50 mM Tris, pH 8.3 for HalA1 peptides and 50 mM MOPS, pH 7.2 for HalA2 peptides) to a final concentration of 0.3 mg/mL and MgCl2, ATP, and TCEP were added to final concentrations of 10 mM, 2.5 mM and 1 mM, respectively (final volume 20 µL). Wild-type and mutant HalA1-Xa were incubated in separate reactions with HalM1 whereas wild-type and mutant HalA2-Xa were incubated with HalM2 with a final enzyme concentration of ~ 0.2 mg/mL enzyme. Activity assays were incubated at 25 °C for 3 h and then quenched by addition of 5% TFA to a final concentration of 0.5% TFA and analyzed by MALDI-TOF MS. Samples were desalted prior to MS using 10 µL C-18 zip tips (Millipore) and eluted with 4 µL of α–hydroxyl cinnamic acid. From this solution, 3 µL was applied to the MALDI-TOF MS target and analyzed.
Evaluation of Cyclization Activity
For HalM1 assays, 1 µL of 100 mM iodoacetamide (IAA) was added directly to 20 µL of HalM1 assay (~ 5 mM final concentration) and incubated at 25 °C for 90 min in the dark. For HalA1 starting material samples, 0.3 mg/mL peptide was incubated in 50 mM Tris, pH 8.3 supplemented with 1 mM TCEP for 20 min prior to addition of IAA and the reaction was carried out as described above. Samples were desalted by C-18 zip tip and analyzed by MALDI-TOF MS. For HalM2 assays, 20 µL aliquots were dried via centrivap and resuspended in 6 µL of 10 mM TCEP and 4 M guanidine hydrochloride, pH 8. Samples were incubated at 25 °C for 20 min and then 3 µL of saturated p–hydroxymercuribenzoic acid (PHMB) solution was added and the reaction mixture was incubated at 25 °C for 2 h in the dark. For HalA2 starting material samples, 0.3 mg/mL peptide was dried via centrivap and PHMB assays were carried out as described above. Samples were checked for thiol modification by zip tip desalting and eluting in 4 µL of α–hydroxyl cinnamic acid to run MALDI-TOF MS.
Antibacterial Activity Assays
Larger scale enzymatic assays (100 µL) were carried out and then dried via centrivap. The products were resuspended in 100 µL of Factor Xa cleavage buffer (20 mM HEPES, pH 8.0, 100 mM NaCl, 2 mM CaCl2) and 3 µL of 1 mg/mL Factor Xa (New England Biolabs) was added. Cleavage reactions were incubated at 25 °C for 4 h and analyzed by MALDI-TOF MS. Samples were dried by centrivap, and resuspended in 5 µL of sterile water. Any insoluble material was removed by centrifugation at 14 krpm for 5 min in a benchtop microcentrifuge. Solid agar diffusion assays were used to assess bactericidal activity. An overnight culture of a haloduracin sensitive strain, Lactococcus lactis CNRZ 117 (Centre National de Recherches Zootechniques, Jouy-enJosas, France), was grown in M17 medium (Difco) supplemented with 0.5% glucose at 30 °C under static conditions for 15–20 h. Plates were prepared by addition of 500 µL of 20% glucose (0.5% final concentration) and 150 µL of dense culture to 20 mL of molten M17 agar (50 °C) that was then allowed to solidify in a Petri dish at 25 °C. Authentic haloduracin was isolated from Bacillus halodurans C-125 (American Type Culture Collection) as described previously (McClerren et al., 2006) and then further purified by RP-HPLC to separate Halα and Halβ (T. Oman, W. van der Donk, unpublished results). For positive control samples, purified Halα and Halβ peptides were combined in a 1:1 ratio (10 µL each of 10 µM solutions). For evaluation of the consequences of mutations, purified Halα or Halβ peptides (5 µL of 10 µM solution) were combined with in vitro prepared mutants (5 µL concentrated HalM products from 100 µL-scale assays). Samples were applied to small wells that were manually formed in the agar plate. The seeded strain was allowed to grow at 25 °C for 12 h and antibacterial activity was qualitatively determined by the presence or absence of a growth inhibition zone.
Proteolytic processing
HalA2-XaGS and HalA2-XaGS Q23A were incubated with HalM2 and treated with Factor Xa (50 µL scale). The resulting products, GDVHAQ-Halβ and GDVHAQ-Halβ Q23A, were analyzed for antimicrobial activity using solid agar diffusion assays as described above. In order to test if the six N-terminal amino acids are removed by an external protease, these products were incubated with the cell-free supernatants from B. halodurans C-125 and L. lactis CNRZ 117 cultures. B. halodurans C-125 was grown in brain heart infusion (BHI) medium (Bacto or BBL) for 96 h at 30 °C with vigorous agitation. Cultures were centrifuged (5000 × g, 10 min) and cell-free supernatant was used for assays with GDVHAQ-Halβ Q23A. In addition, the cells were washed with 1 M NaCl. After centrifugation, the cell-free 1 M NaCl wash was also used for assay with GDVHAQ-Halβ Q23A. Production of haloduracin from GDVHAQ-Halβ Q23A was analyzed by MALDI-TOF MS. Both cell-free supernatant and the 1 M NaCl wash contained proteolytic activity as discussed in the results section. L. lactis CNRZ 117 was grown as described above and its culture supernatant was used for analogous experiments as described for B. halodurans. Identical procedures were also utilized to monitor proteolytic processing of Halα, and HalM2-processed HalA2.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health, Grant GM58822. L.E.C. was supported by the National Institutes of Health under Ruth L. Kirschstein National Research Service Award 5 T32 GM070421 from the National Institute of General Medical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bierbaum G, Szekat C, Josten M, Heidrich C, Kempter C, Jung G, Sahl HG. Engineering of a novel thioether bridge and role of modified residues in the lantibiotic Pep5. Appl. Environ. Microbiol. 1996;62:385–392. doi: 10.1128/aem.62.2.385-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brötz H, Bierbaum G, Leopold K, Reynolds PE, Sahl HG. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob. Agents Chemother. 1998;42:154–160. doi: 10.1128/aac.42.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglione F, Lazzarini A, Carrano L, Corti E, Ciciliato I, Gastaldo L, Candiani P, Losi D, Marinelli F, Selva E, et al. Determining the structure and mode of action of microbisporicin, a potent lantibiotic active against multiresistant pathogens. Chem. Biol. 2008;15:22–31. doi: 10.1016/j.chembiol.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Chatterjee C, Miller LM, Leung YL, Xie L, Yi M, Kelleher NL, van der Donk WA. Lacticin 481 Synthetase Phosphorylates its Substrate during Lantibiotic Production. J. Am. Chem. Soc. 2005a;127:15332–15333. doi: 10.1021/ja0543043. [DOI] [PubMed] [Google Scholar]
- Chatterjee C, Patton GC, Cooper L, Paul M, van der Donk WA. Engineering Dehydro Amino Acids and Thioethers into Peptides using Lacticin 481 Synthetase. Chem. Biol. 2006;13:1109–1117. doi: 10.1016/j.chembiol.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Chatterjee C, Paul M, Xie L, van der Donk WA. Biosynthesis and Mode of Action of Lantibiotics. Chem. Rev. 2005b;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Chatterjee S, Lad SJ, Phansalkar MS, Rupp RH, Ganguli BN, Fehlhaber HW, Kogler H. Mersacidin, a new antibiotic from Bacillus. Fermentation, isolation, purification and chemical characterization. J. Antibiot. 1992;45:832–838. doi: 10.7164/antibiotics.45.832. [DOI] [PubMed] [Google Scholar]
- Chen P, Novak J, Kirk M, Barnes S, Qi F, Caufield PW. Structure-activity study of the lantibiotic mutacin II from Streptococcus mutans T8 by a gene replacement strategy. Appl. Environ. Microbiol. 1998;64:2335–2340. doi: 10.1128/aem.64.7.2335-2340.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Qi FX, Novak J, Krull RE, Caufield PW. Effect of amino acid substitutions in conserved residues in the leader peptide on biosynthesis of the lantibiotic mutacin II. FEMS Microbiol. Lett. 2001;195:139–144. doi: 10.1111/j.1574-6968.2001.tb10511.x. [DOI] [PubMed] [Google Scholar]
- Corvey C, Stein T, Dusterhus S, Karas M, Entian KD. Activation of subtilin precursors by Bacillus subtilis extracellular serine proteases subtilisin (AprE), WprA, and Vpr. Biochem. Biophys. Res. Commun. 2003;304:48–54. doi: 10.1016/s0006-291x(03)00529-1. [DOI] [PubMed] [Google Scholar]
- Cotter PD, Deegan LH, Lawton EM, Draper LA, O'Connor PM, Hill C, Ross RP. Complete alanine scanning of the two-component lantibiotic lacticin 3147: generating a blueprint for rational drug design. Mol. Microbiol. 2006;62:735–747. doi: 10.1111/j.1365-2958.2006.05398.x. [DOI] [PubMed] [Google Scholar]
- Cotter PD, Hill C, Ross RP. Bacterial lantibiotics: strategies to improve therapeutic potential. Curr. Protein Pept. Sci. 2005;6:61–75. doi: 10.2174/1389203053027584. [DOI] [PubMed] [Google Scholar]
- Cox CR, Coburn PS, Gilmore MS. Enterococcal cytolysin: a novel two component peptide system that serves as a bacterial defense against eukaryotic and prokaryotic cells. Curr. Protein Pept. Sci. 2005;6:77–84. doi: 10.2174/1389203053027557. [DOI] [PubMed] [Google Scholar]
- Delves-Broughton J, Blackburn P, Evans RJ, Hugenholtz J. Applications of the bacteriocin, nisin. Antonie van Leeuwenhoek. 1996;69:193–202. doi: 10.1007/BF00399424. [DOI] [PubMed] [Google Scholar]
- Field D, Collins B, Cotter PD, Hill C, Ross RP. A system for the random mutagenesis of the two-peptide lantibiotic lacticin 3147: analysis of mutants producing reduced antibacterial activities. J. Mol. Microbiol. Biotechnol. 2007;13:226–234. doi: 10.1159/000104747. [DOI] [PubMed] [Google Scholar]
- Furgerson Ihnken LA, Chatterjee C, van der Donk WA. In vitro Reconstitution and Substrate Specificity of a Lantibiotic Protease. Biochemistry. 2008;47:7352–7363. doi: 10.1021/bi800278n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvin M, Hill C, Ross RP. Lacticin 3147 displays activity in buffer against gram-positive bacterial pathogens which appear insensitive in standard plate assays. Lett. Appl. Microbiol. 1999;28:355–358. doi: 10.1046/j.1365-2672.1999.00550.x. [DOI] [PubMed] [Google Scholar]
- Garneau S, Martin NI, Vederas JC. Two-peptide bacteriocins produced by lactic acid bacteria. Biochimie. 2002;84:577–592. doi: 10.1016/s0300-9084(02)01414-1. [DOI] [PubMed] [Google Scholar]
- Håvarstein LS, Diep DB, Nes IF. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- Holo H, Jeknic Z, Daeschel M, Stevanovic S, Nes IF. Plantaricin W from Lactobacillus plantarum belongs to a new family of two-peptide lantibiotics. Microbiology. 2001;147:643–651. doi: 10.1099/00221287-147-3-643. [DOI] [PubMed] [Google Scholar]
- Hsu ST, Breukink E, Bierbaum G, Sahl HG, de Kruijff B, Kaptein R, van Nuland NA, Bonvin AM. NMR study of mersacidin and lipid II interaction in dodecylphosphocholine micelles. Conformational changes are a key to antimicrobial activity. J. Biol. Chem. 2003;278:13110–13117. doi: 10.1074/jbc.M211144200. [DOI] [PubMed] [Google Scholar]
- Hyink O, Balakrishnan M, Tagg JR. Streptococcus rattus strain BHT produces both a class I two-component lantibiotic and a class II bacteriocin. FEMS Microbiol. Lett. 2005;252:235–241. doi: 10.1016/j.femsle.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Kruszewska D, Sahl HG, Bierbaum G, Pag U, Hynes SO, Ljungh A. Mersacidin eradicates methicillin-resistant Staphylococcus aureus (MRSA) in a mouse rhinitis model. J. Antimicrob. Chemother. 2004;54:648–653. doi: 10.1093/jac/dkh387. [DOI] [PubMed] [Google Scholar]
- Kuipers OP, Bierbaum G, Ottenwälder B, Dodd HM, Horn N, Metzger J, Kupke T, Gnau V, Bongers R, van den Bogaard P, et al. Protein engineering of lantibiotics. Antonie van Leeuwenhoek. 1996;69:161–169. doi: 10.1007/BF00399421. [DOI] [PubMed] [Google Scholar]
- Lawton EM, Cotter PD, Hill C, Ross RP. Identification of a novel two-peptide lantibiotic, Haloduracin, produced by the alkaliphile Bacillus halodurans C-125. FEMS Microbiol Lett. 2007;267:64–71. doi: 10.1111/j.1574-6968.2006.00539.x. [DOI] [PubMed] [Google Scholar]
- Levengood MR, Patton GC, van der Donk WA. The Leader Peptide is not Required for Post-translational Modification by Lacticin 481 Synthetase. J. Am. Chem. Soc. 2007;129:10314–10315. doi: 10.1021/ja072967+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, van der Donk WA. Identification of essential catalytic residues of the cyclase NisC involved in the biosynthesis of nisin. J. Biol. Chem. 2007;282:21169–21175. doi: 10.1074/jbc.M701802200. [DOI] [PubMed] [Google Scholar]
- Li B, Yu J-PJ, Brunzelle JS, Moll GN, van der Donk WA, Nair SK. Structure and Mechanism of the Lantibiotic Cyclase Involved in Nisin Biosynthesis. Science. 2006;311:1464–1467. doi: 10.1126/science.1121422. [DOI] [PubMed] [Google Scholar]
- Lubelski J, Rink R, Khusainov R, Moll GN, Kuipers OP. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell. Mol. Life Sci. 2007 doi: 10.1007/s00018-007-7171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin NI, Sprules T, Carpenter MR, Cotter PD, Hill C, Ross RP, Vederas JC. Structural Characterization of Lacticin 3147, a Two-Peptide Lantibiotic with Synergistic Activity. Biochemistry. 2004;43:3049–3056. doi: 10.1021/bi0362065. [DOI] [PubMed] [Google Scholar]
- McClerren AL, Cooper LE, Quan C, Thomas PM, Kelleher NL, van der Donk WA. Discovery and in vitro biosynthesis of haloduracin, a new two-component lantibiotic. Proc. Natl. Acad. Sci. USA. 2006;103:17243–17248. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan SM, O'Connor PM, Cotter PD, Ross RP, Hill C. Sequential actions of the two component peptides of the lantibiotic lacticin 3147 explain its antimicrobial activity at nanomolar concentrations. Antimicrob Agents Chemother. 2005;49:2606–2611. doi: 10.1128/AAC.49.7.2606-2611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nes IF, Tagg JR. Novel lantibiotics and their pre-peptides. Antonie van Leeuwenhoek. 1996;69:89–97. doi: 10.1007/BF00399414. [DOI] [PubMed] [Google Scholar]
- Ottenwälder B, Kupke T, Brecht S, Gnau V, Metzger J, Jung G, Götz F. Isolation and characterization of genetically engineered gallidermin and epidermin analogs. Appl. Environ. Microbiol. 1995;61:3894–3903. doi: 10.1128/aem.61.11.3894-3903.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton GC, Paul M, Cooper LE, Chatterjee C, van der Donk WA. The Importance of the Leader Sequence for Directing Lanthionine Formation in Lacticin 481. Biochemistry. 2008;47:7342–7351. doi: 10.1021/bi800277d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M, Patton GC, van der Donk WA. Mutants of the Zinc Ligands of Lacticin 481 Synthetase Retain Dehydration Activity but Have Impaired Cyclization Activity. Biochemistry. 2007;46:6268–6276. doi: 10.1021/bi7000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen FC, Fimland G, Scheie AA. Purification and functional studies of a potent modified quorum-sensing peptide and a two-peptide bacteriocin in Streptococcus mutans. Mol. Microbiol. 2006;61:1322–1334. doi: 10.1111/j.1365-2958.2006.05312.x. [DOI] [PubMed] [Google Scholar]
- Pitts KE, Summers AO. The roles of thiols in the bacterial organomercurial lyase (MerB) Biochemistry. 2002;41:10287–10296. doi: 10.1021/bi0259148. [DOI] [PubMed] [Google Scholar]
- Rink R, Kuipers A, de Boef E, Leenhouts KJ, Driessen AJ, Moll GN, Kuipers OP. Lantibiotic structures as guidelines for the design of peptides that can be modified by lantibiotic enzymes. Biochemistry. 2005;44:8873–8882. doi: 10.1021/bi050081h. [DOI] [PubMed] [Google Scholar]
- Ryan MP, Jack RW, Josten M, Sahl HG, Jung G, Ross RP, Hill C. Extensive post-translational modification, including serine to D-alanine conversion, in the two-component lantibiotic, lacticin 3147. J. Biol. Chem. 1999;274:37544–37550. doi: 10.1074/jbc.274.53.37544. [DOI] [PubMed] [Google Scholar]
- Szekat C, Jack RW, Skutlarek D, Farber H, Bierbaum G. Construction of an expression system for site-directed mutagenesis of the lantibiotic mersacidin. Appl. Environ. Microbiol. 2003;69:3777–3783. doi: 10.1128/AEM.69.7.3777-3783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer JR, Rollema HS, Siezen RJ, Beerthuyzen MM, Kuipers OP, de Vos WM. Influence of amino acid substitutions in the nisin leader peptide on biosynthesis and secretion of nisin by Lactococcus lactis. J. Biol. Chem. 1994;269:3555–3562. [PubMed] [Google Scholar]
- van Kraaij C, Breukink E, Rollema HS, Bongers RS, Kosters HA, de Kruijff B, Kuipers OP. Engineering a disulfide bond and free thiols in the lantibiotic nisin Z. Eur. J. Biochem. 2000;267:901–909. doi: 10.1046/j.1432-1327.2000.01075.x. [DOI] [PubMed] [Google Scholar]
- Wiedemann I, Bottiger T, Bonelli RR, Wiese A, Hagge SO, Gutsmann T, Seydel U, Deegan L, Hill C, Ross P, et al. The mode of action of the lantibiotic lacticin 3147--a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol. Microbiol. 2006a;61:285–296. doi: 10.1111/j.1365-2958.2006.05223.x. [DOI] [PubMed] [Google Scholar]
- Wiedemann I, Bottiger T, Bonelli RR, Wiese A, Hagge SO, Gutsmann T, Seydel U, Deegan L, Hill C, Ross P, et al. The mode of action of the lantibiotic lacticin 3147--a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol Microbiol. 2006b;61:285–296. doi: 10.1111/j.1365-2958.2006.05223.x. [DOI] [PubMed] [Google Scholar]
- Willey JM, van der Donk WA. Lantibiotics: Peptides of Diverse Structure and Function. Annu. Rev. Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- Xie L, Miller LM, Chatterjee C, Averin O, Kelleher NL, van der Donk WA. Lacticin 481: in vitro reconstitution of lantibiotic synthetase activity. Science. 2004;303:679–681. doi: 10.1126/science.1092600. [DOI] [PubMed] [Google Scholar]
- Yonezawa H, Kuramitsu HK. Genetic analysis of a unique bacteriocin, Smb, produced by Streptococcus mutans GS5. Antimicrob. Agents Chemother. 2005;49:541–548. doi: 10.1128/AAC.49.2.541-548.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann N, Jung G. The three-dimensional solution structure of the lantibiotic murein-biosynthesis-inhibitor actagardine determined by NMR. Eur. J. Biochem. 1997;246:809–819. doi: 10.1111/j.1432-1033.1997.00809.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann N, Metzger JW, Jung G. The tetracyclic lantibiotic actagardine. 1H-NMR and 13C-NMR assignments and revised primary structure. Eur. J. Biochem. 1995;228:786–797. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.