Summary
We introduce a simple-to-use graphical tool that enables researchers to easily prepare time-of-flight mass spectrometry data for analysis. For ease of use, the graphical executable provides default parameter settings experimentally determined to work well in most situations. These values can be changed by the user if desired. PrepMS is a stand-alone application made freely available (open source), and is under the General Public License (GPL). Its graphical user interface, default parameter settings, and display plots allow PrepMS to be used effectively for data preprocessing, peak detection, and visual data quality assessment.
INTRODUCTION
Time-of-flight (TOF) mass spectrometry (MS) data is commonly used in efforts to discover disease-related biomarkers from subject samples (e.g. urine, saliva, or serum). In the biomarker discovery field, a common ionization platform is matrix-assisted laser desorption ionization (MALDI), and surface-enhanced laser desorption ionization (SELDI) is one popular variant (Adam, et al., 2002; Conrads, et al., 2004; Koomen, et al., 2005; Pawlik, et al., 2005; Schaub, et al., 2004).
Reproducibility of biomarker identification depends in part on careful data preprocessing (Baggerly, et al., 2004). Morris, et al. (2005) outline the following basic TOF MS data preprocessing steps: spectral calibration, including signal interpolation to impose a common time scale across spectra; spectral denoising, baseline correction and normalization; peak detection; and peak quantification. They provide a set of the Matlab scripts that implement some of the methods described in their paper (http://bioinformatics.mdanderson.org/cromwell.html). The user interested in implementing their methods will need good knowledge of Matlab, as well as access to commercially licensed software. In this note, we present a stand-alone compiled application, PrepMS, that combines into a single executable: wavelet denoising, baseline correction, and peak detection algorithms described in Morris et al. (2005); Matlab Bioinformatics Toolbox function msresample to perform interpolation; and graphical output for data quality assessment and results visualization. By providing the user with a simple graphical interface, PrepMS is accessible to both bioinformaticians and basic scientists for the purposes of TOF MS data preprocessing.
PROGRAM OVERVIEW
PrepMS is a fully automated stand-alone application. It is written in Matlab, but is provided as a stand-alone executable. PrepMS is platform independent: it can run on Linux and Windows alike. The program provides a graphical interface to the preprocessing algorithm. The user is required to provide some simple parameters, the first of which is the location of the tab-delimited 2 column data files: columns 1 and 2 contain mass to charge (m/z) ratios, and corresponding intensity values, respectively. The algorithm starts by removing the header lines (if present) from input files and reading the input files into memory.
Calibration is typically accomplished experimentally using a sample of known molecular weights, resulting in peaks that are reasonably well aligned across spectra. However, it is not uncommon to acquire different numbers of intensities within a common m/z window from one spectrum to the next as a consequence of changes in instrument calibration. Based on the quadratic relationship between mass and time, a second preprocessing step interpolates intensities to impose a common time scale across all spectra (Morris, et al., 2005). The user can shrink or lengthen all spectra to the shortest or longest spectrum, respectively, or specify the number of points to be interpolated. Additionally, TOF MS data can be susceptible to shifts in m/z over the course of acquiring multiple spectra (Yasui, et al., 2003). PrepMS can align the spectra by accepting any number of reference peak m/z values from the user or by using the top five peaks detected in the mean spectrum.
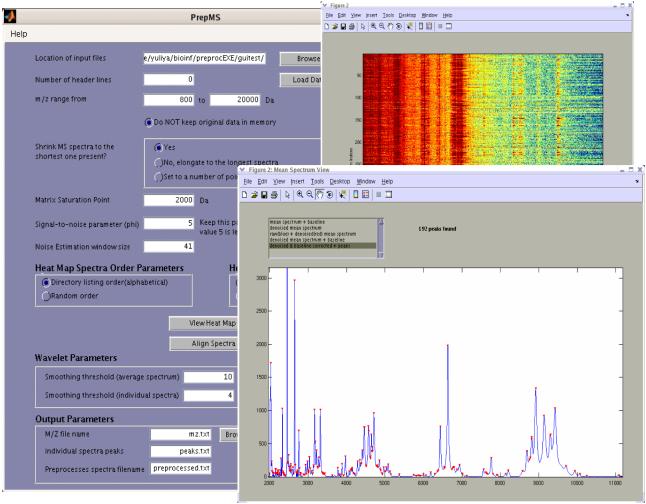
The algorithm then removes from every spectrum all intensities below a user-specified m/z threshold to eliminate matrix noise, a large-amplitude matrix signal that can swamp the biological signal at low m/z values. A simple click of a button, “View Heat Map”, displays the heat map of the ranked or log-transformed intensities to visually assess peak alignment (Figure 1). Spectra can be structured in random or directory listing (alphabetical) order. This step allows the user to identify possible machine- or day-specific effects that could shift peak locations along the m/z scale (Baggerly, et al., 2004).
Fig. 1.
Snapshot of the stand-alone PrepMS executable. Window with control parameters shown on the left. On the right are graphics of the mean spectrum with detected peaks shown in red, and a heat map of ranked intensities with spectra in random order.
Following Morris, et al. (2005), PrepMS conducts peak detection using the average spectrum rather than individual spectra. Using the mean spectrum increases peak detection reliability while simultaneously eliminating the need to match peaks across spectra. Furthermore, by borrowing strength across spectra, peak locations that would otherwise be undetected in individual spectra are identifiable from the mean.
Spectral denoising separates the electrical and chemical noise from signal, thereby enhancing subsequent feature detection and quantification. Coombes, et al. (2005) use the undecimated wavelet transform (UDWT) as implemented in the Rice Wavelet Toolbox (http://www.dsp.ece.rice.edu/software/rwt.shtml) to accomplish spectral denoising, and this approach is adopted by Morris, et al. (2005). Similarly, PrepMS denoises the mean spectrum using the UDWT based on a hard-thresholding algorithm that sets to zero all wavelet coefficients less than a specified threshold, leaving coefficients greater than that threshold unchanged (Coombes, et al., 2005).
The baseline correction step estimates and removes the baseline artifact, a smooth additive component of the signal that is attributable in part to charge accumulation (Malyarenko, et al., 2005). The baseline is viewed graphically as an elevation of the horizontal axis that decays with increasing time of flight.
Peak locations are identified by the m/z positions of local maxima with corresponding intensities exceeding a pre-specified signal to noise (S/N) threshold, φ. Following recommendations by Morris, et al. (2005), the default setting for φ is 5/√n, where n is the number of spectra used to construct the average spectrum. Local noise, N, is estimated using MAD computed from the wavelet-based noise estimates in a window comprised of 41 m/z locations by default.
At each peak location found in the mean spectrum, PrepMS quantifies intensities for individual spectra. Specifically, individual spectra are denoised using the UDWT, and a monotone minimum baseline is estimated and removed. Here, the wavelet smoothing parameter η is set to a smaller default value of 4 as compared to 10 for the mean spectrum. In general, η should be set to a value lower than that for the mean spectra. Spectra are then normalized to total ion current by dividing peak intensities by the sum of all intensities for a given spectrum. At each peak location, quantification is based on the maximum observed normalized intensity in the window bounded to the left and right by local minima used in identifying feature m/z locations.
The resulting peaks, the corresponding m/z values, and the preprocessed individual spectra are stored in the tab-delimited files peaks.txt, mz.txt, and preprocessed.txt respectively. Alternative file names can be specified by the user. In addition, the mean, denoised and baseline corrected spectrum is displayed, and detected peaks are identified with red triangles positioned at the peak intensities in the mean spectrum plot (Figure 1). Other plots are available, for example, the baseline can be plotted with the mean spectrum or mean denoised spectrum as a red line by using a selection list box at the top of the figure window. Individual spectra can be viewed in the second display window with the baseline and/or peaks identified. Basic graph manipulation tools allow zooming in and out of particular regions of the spectrum, as well as saving figures as various image file types.
In conclusion, PrepMS is a graphical user-friendly TOF MS data preprocessing tool that implements a robust peak identification algorithm based on the mean spectrum reported by Morris, et al. (2005). Sensible default parameters eliminate the need to understand peak detection algorithms and the details of wavelet denoising. In summary, PrepMS provides a straight-forward fully automated graphical user interface for TOF MS data preprocessing.
ACKNOWLEDGEMENTS
YVK is supported by NLM training grant 1-T15-LM07438. EGH is partially supported by NIH/NIDCR grant K25 DE016863. JSA is supported by NHLBI Proteomics Initiative N01-HV-28181 (http://proteomics.musc.edu). JSM and KAB are supported by R01 grant CA-107304 from the NIH/NCI. AJS is supported by NIH/NIDDK R01 DK064371. The authors thank Dr. Daniel Knapp for comments that substantially improved the manuscript.
Footnotes
Availability: Stand-alone executable files and Matlab toolbox are available for download at: http://sourceforge.net/projects/prepms
REFERENCES
- Adam BL, et al. Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res. 2002;62:3609–3614. [PubMed] [Google Scholar]
- Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDITOF protein patterns in serum: comparing datasets from different experiments. Bioinformatics. 2004;20:777–785. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]
- Conrads TP, et al. High-resolution serum proteomic features for ovarian cancer detection. Endocr Relat Cancer. 2004;11:163–178. doi: 10.1677/erc.0.0110163. [DOI] [PubMed] [Google Scholar]
- Coombes KR, et al. Improved peak detection and quantification of mass spectrometry data acquired from surface-enhanced laser desorption and ionization by denoising spectra with the undecimated discrete wavelet transform. Proteomics. 2005;5:4107–4117. doi: 10.1002/pmic.200401261. [DOI] [PubMed] [Google Scholar]
- Koomen JM, et al. Plasma protein profiling for diagnosis of pancreatic cancer reveals the presence of host response proteins. Clin Cancer Res. 2005;11:1110–1118. [PubMed] [Google Scholar]
- Malyarenko DI, et al. Enhancement of sensitivity and resolution of surface-enhanced laser desorption/ionization time-of-flight mass spectrometric records for serum peptides using time-series analysis techniques. Clin Chem. 2005;51:65–74. doi: 10.1373/clinchem.2004.037283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JS, Coombes KR, Koomen J, Baggerly KA, Kobayashi R. Feature extraction and quantification for mass spectrometry in biomedical applications using the mean spectrum. Bioinformatics. 2005;21:1764–1775. doi: 10.1093/bioinformatics/bti254. [DOI] [PubMed] [Google Scholar]
- Pawlik TM, et al. Significant differences in nipple aspirate fluid protein expression between healthy women and those with breast cancer demonstrated by time-of-flight mass spectrometry. Breast Cancer Res Treat. 2005;89:149–157. doi: 10.1007/s10549-004-1710-4. [DOI] [PubMed] [Google Scholar]
- Schaub S, et al. Urine protein profiling with surface-enhanced laser-desorption/ionization time-of-flight mass spectrometry. Kidney Int. 2004;65:323–332. doi: 10.1111/j.1523-1755.2004.00352.x. [DOI] [PubMed] [Google Scholar]
- Yasui Y, et al. A data-analytic strategy for protein biomarker discovery: profiling of high-dimensional proteomic data for cancer detection. Biostatistics. 2003;4:449–463. doi: 10.1093/biostatistics/4.3.449. [DOI] [PubMed] [Google Scholar]