I. Introduction
Bacterial transcription is carried out by a multisubunit RNA polymerase holoenzyme, Eσ, comprised of the core enzyme E (subunits α2, β, β′ and ω), and one of several σ specificity subunits (a major “housekeeping” sigma factor, such as E. coli σ70, or one of a variable number of alternative sigma factors present in different bacterial species) [1–4]. While the response of RNAP to specialized DNA-binding transcription factors plays a critical role in determining the cell’s transcription program under different growth conditions, the promoter sequences with which RNAP interacts are variable and are a major determinant of the wide range of strength and regulation of gene expression. RNAP-promoter complex formation and transcription initiation in the absence of transacting factors are referred to as basal promoter function. In this chapter we focus on methods for observing and studying basal promoter function as a key component of the overall mechanism of transcription regulation. The mechanism of action of regulatory factors (activators or repressors) must ultimately be understood in terms of how they alter the interactions of RNAP with a particular promoter.
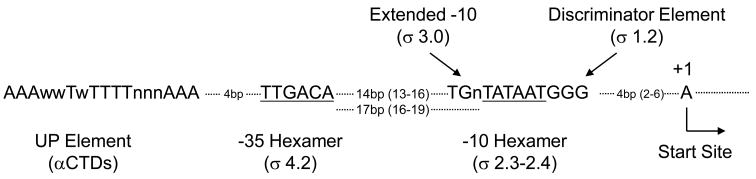
An RNAP-promoter complex capable of transcription initiation (an open complex; RPO) is formed through interactions that involve several regions of RNAP and that span 70–80bp of promoter DNA (~−60 to +20 with respect to the transcription start site, +1). Each RNAP subunit, with the exception of ω, participates in these interactions, although a majority of the sequence specific interactions occur with the σ subunit. In an E. coli Eσ70-promoter complex, sequence-specific interactions with σ70 occur at the −10 element (with σ70 regions 2.3–2.4), the extended −10 element (σ70 region 3.0), the −35 element (σ70 region 4.2), and the discriminator element immediately downstream of the −10 hexamer (σ70 region 1.2) (Fig. 1; [1,5]). In addition, the C-terminal domain of the α subunit can interact sequence-specifically with the UP element, located upstream of the −35 hexamer [6]. Sequence non-specific DNA contacts are mediated by several regions of the large β andβ′ subunits in the active site channel and downstream of the start site (as observed in elongation complexes; [7,8]) and in the spacer region between the −10 and −35 hexamers [9].
Fig. 1.
(A) Consensus sequences for Eσ70 recognition elements and RNAP regions that interact with these elements: UP Element with α subunit C-terminal domains (αCTDs, the α subunit DNA binding domains), −35 hexamer with σ70 4.2, extended −10 element with σ70 3.0, −10 hexamer with σ70 2.3–2.4, and discriminator region element withσ70 1.2. The transcription start site is indicated as (+1). The most common spacing between recognition elements, in bp, with (range) is indicated. For a recent review, see [5]. Both the spacing of the −35 element with respect to the extended −10 element (14bp), and with respect to the −10 element (17bp) are shown.
Promoters vary widely in overall strength (~ 4 orders of magnitude) and in the extent of similarity to the consensus sequences for the recognition elements [1,10,11]. The −10 element is the most highly conserved of the promoter elements, but similarities to consensus usually are found in one or more of the other recognition elements as well [6,12,13]. The contribution of each recognition element to promoter function depends upon the context in which it is found.
In this review we discuss several methods for investigating RNAP-promoter complex formation, including preliminary identification of the promoter itself using in vivo methods, followed by the characterization of properties of complex formation with RNAP in vitro. Formation of the RNAP-promoter complex can be strongly affected by several parameters in vitro, and effects of these conditions vary with different promoter sequences. These parameters will be discussed as well.
2. Promoter Identification and Activity Determination In Vivo
While bioinformatic analyses can predict some promoters correctly, definitive identification of promoters from sequence information alone remains difficult [14,15]. In addition, while in vitro methods that examine RNA polymerase-promoter complex formation can be very informative, they can identify interactions that do not correspond to promoters that function in vivo (e.g., “tight-binding sites”, or end-binding sites; [16,17]). Thus, correct identification of a promoter is established most convincingly through the use of multiple approaches, both in vivo and in vitro. These may include identification of the transcription start site using purified in vivo RNA, demonstration of promoter activity in vivo by using a promoter region fragment fused to a reporter gene, and in vitro transcription and RNAP-promoter binding studies.
More than one promoter may exist for expression of a particular gene in response to different growth conditions and may be recognized by RNAP holoenzyme forms containing different sigma subunits. Promoters may be located in an intergenic region immediately upstream of the gene of interest, or within or upstream of an adjacent open reading frame, and transcripts may be monocistronic or polycistronic. Information from annotated sequence databases can be consulted, where available, to identify previously characterized or predicted regulatory regions (e.g.: EcoCyc, [18]; Regulon Database: [19]). Data from experimental determinations of RNA polymerase binding sites across the genome using chromatin immunoprecipitation and microarray analysis (ChIP-chip) may in some cases suggest a location on which to focus for promoter analysis [20,21].
2.1. Identification of the In Vivo Transcription Start Site
Identification of the 5′ end of in vivo mRNA can provide precise information about the location of the promoter, if care is taken to avoid ends generated by RNA processing (or degradation) rather than the true 5′ end (e.g., see [22]). Combining the identification of a transcription start site from the mRNA 5′ end with other assays of promoter function in vivo and in vitro (see below) together can demonstrate the location of the promoter convincingly.
Several methods have been employed to identify the transcription start site, including S1 nuclease mapping, primer extension, and 5′ RACE (rapid amplification of cDNA ends) [23]. The strength of the promoter, and thus the abundance of the transcript to be analyzed, should influence the choice of method. Abundant transcripts can be detected readily by methods that do not require amplification of the signal (S1 nuclease or primer-extension), while low abundance transcripts may require signal amplification, either by cloning the promoter region on a multicopy plasmid, or by amplification of cDNA copies produced from the transcript (5′ RACE).
2.1.1. Extraction of Total Cellular RNA
Each of these methods requires extraction of total RNA from cell cultures, followed by different procedures for characterizing the 5′ end of the transcript of interest. Success depends heavily on the quality of the RNA extracted. Care must be taken to prevent degradation of short-lived RNAs, such as might occur if lysis of cells is not coupled with exposure to denaturants that will inactivate RNA processing/degradation enzymes. Collecting cells by centrifugation prior to lysis can also lead to changes in cell physiology and transcription activity and therefore should be avoided if possible.
The method for total RNA extraction from E. coli cultures used in our laboratory involves immediate transfer of a cell culture sample (7 ml of culture grown in LB or MOPS defined media) into boiling lysis buffer (1.5 ml of 1 M NaCl, 2.5% SDS, 0.05 M EDTA) in a 50 ml conical polypropylene tube [24]. Samples are incubated in the boiling water bath for 1.5 min, then extracted twice with 4 ml of a 1:1 phenol-chloroform mixture (we use water-saturated phenol, which is acidic, not buffer neutralized phenol). 5 ml of the aqueous phase is brought to 0.3 M sodium acetate and precipitated with 10 ml of prechilled 100% ethanol. After overnight precipitation at −20°C, RNA is pelleted by centrifugation at 3200 RPM for 30 min at 4°C in 50 ml conical tubes (in a swinging bucket rotor). The RNA pellet is dissolved in 0.4 ml of 10 mM Tris-Cl, pH 8.0, brought to 0.3 M sodium acetate, re-precipitated with ethanol (−80°C for 10 min) in a 1.5ml microfuge tube, and centrifuged at 4°C for 20min. The pellet is washed thoroughly with cold ethanol, then air dried, resuspended in ~25–100 μl of 10 mM TrisCl, pH 8.0, and stored at −20°C prior to analysis. RNA concentration is determined by UV absorbance. A modification of this procedure was described for total RNA extraction from B. subtilis cultures as well [25]. Commercially available RNA extraction kits are also commonly used (e.g., Qiagen RNeasy with additional Protect Bacteria Kit, which purifies only RNAs of 200nt or larger) and manufacturers’ suggestions for use with bacterial RNA extraction should be followed to ensure immediate exposure of cells to denaturant during the lysis step.
2.1.2. Primer Extension of Total RNA
For analysis of specific 5′ ends in the extracted total RNA by primer extension with a reverse transcriptase (e.g., M-MLV Reverse transcriptase; [24, 26]), the primer should anneal to the transcript ≈75–100nt from the predicted transcription start site, such that the size of the cDNA extension product can be determined at single nucleotide resolution on a denaturing sequencing gel by comparison with sequencing reactions prepared using the same primer. The primer is 32P-labeled at the 5′ end with γ-32P-ATP and T4 polynucleotide kinase, and purified using a Qiagen Qiaquick nucleotide removal kit, modifying the manufacturer’s procedure to include several washes of the columns with buffer PE prior to eluting the labeled primer. Alternative methods for use of 5′-fluorescent tagged rather than radiolabeled primers have also been described (e.g., [27]).
To anneal the labeled primer with the transcript, combine in a 25μl final volume: ~ 5μg of resuspended total RNA sample, 5 μl of 5X M-MLV RT buffer (Promega; or appropriate buffer for other reverse transcriptases), and ≥ 0.5 pmol 32P-labeled primer. (This amount of total RNA is sufficient to detect transcripts from moderately strong to strong promoters). Heat at 80°C for 10 min, and cool slowly to 30–40°C (over 20–30 minutes). Bring the sample to 48–49°C and add (as 5 μl of a combined master mix): 1 μl M-MLV RT 5X buffer, 0.5 μl M-MLV RT, 1 μl 0.1 M DTT and 2.5 μl of a stock of 5 mM each dNTP. Incubate for 30 min at 48–49°C, and terminate the reaction by adding 30 μl of formamide stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanole). It is important to carry out the extension reaction at a relatively high temperature (compatible with the reverse transcriptase being used) to reduce potential effects of RNA secondary structure and increase the yield of complete extension products. Modified versions of M-MLV reverse transcriptase lacking RNaseH activity (e.g., M-MLV Point, Promega; or Superscipt II, Invitrogen), can also be used and favor the synthesis of full length cDNA products by preventing cleavage of the RNA template. Products of the extension reaction are analyzed by denaturing acrylamide gel electrophoresis. Sequencing reactions, prepared using the same labeled primer, serve to identify the length of the primer extension product, and thus the 5′ end of the original mRNA (see Fig. 2; [26]). A primer extension reaction illustrating the transcription start site for the E. coli rRNA promoter rrnB P1 (detected in a transcript produced from an rrnB P1-lacZ fusion) is shown in Figure 2.
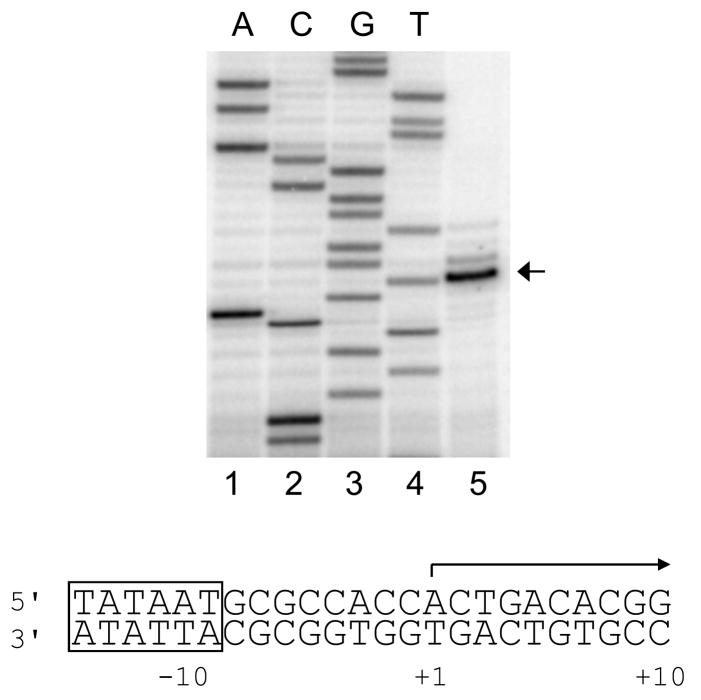
Fig. 2.
Transcription start site position(s) for the E. coli rrnB P1 promoter determined by primer extension using total cellular RNA prepared from a strain carrying an rrnB P1-lacZ fusion on a λ prophage. The primer annealed to the transcript downstream of the promoter endpoint in the fusion. Sequence marker lanes, prepared with the same labeled primer as used for primer extension are shown in lanes 1–4. Primer extension product is shown in lane 5. The sequence of the rrnB P1 promoter region is shown below the gel, for reference. Data are from P. Changdrangsu and R. Gourse. (see also [28]).
Primer extension can also be used for quantitative assessment of RNA transcript levels as a function of growth phase, growth condition, mutations affecting putative transcription factors, or mutations in the promoter itself [25,26,28]. If RNA products with short half-lives are produced (e.g., using promoter-lacZ fusions in a ΔlacZ host, where the transcript has a 1–3 minute half-life), this experiment can provide an approximation of relative RNA synthesis rates. A molar excess of labeled primer over transcript should be used (determined empirically by testing a series of primer concentrations). The use of a purified RNA recovery marker, added during RNA extraction from the culture (see above), can serve as a control to normalize for recovery during the extraction procedure, for primer extension, and for gel loading. The recovery marker RNA can be produced from a construct designed to contain the same primer binding site, but located at a different distance from the promoter, and can be extracted from a strain carrying this construct (see [28]). An aliquot of the extracted total RNA from this marker strain is added to test samples during extraction of the test RNA.
To amplify weak signals not detected readily by the methods described above, the promoter region (as well as mutant promoter derivatives for comparison) can be inserted into a multicopy plasmid (e.g., [28]) upstream of a unique sequence to be transcribed (e.g., lacZ, using a ΔlacZ host strain, or other sequence not present in the host chromosome). Total RNA is prepared from cultures carrying these plasmids, and used in the primer extension assays as described.
2.1.3. 5′ RACE
An alternative method (5′ RACE; 5′ amplification of cDNA ends) can also be used to determine the 5′ end of lower abundance transcripts produced from weaker promoters (e.g., [15,22,29,30]). The method involves primer extension (1st strand cDNA synthesis) from extracted total RNA, followed by PCR amplification of the cDNA products using procedural modifications that create a primer binding site flanking the 3′ end of the cDNA (or, alternatively, of the 5′ end of the RNA). Products of this amplification can be sequenced directly, or cloned and sequenced to identify the 5′ mRNA end. Several versions of 5′ RACE have been described, including classic RACE and new RACE. In classic RACE procedures, a polyA sequence is added to the 3′ end of the 1st strand cDNA product using terminal transferase. Forward primers for the second strand synthesis reaction are designed to include a unique sequence followed by a polyT tract (to anneal to the polyA tail). PCR is then carried out with a thermostable DNA polymerase, using the unique sequence forward primer, and a gene-specific reverse primer. In the new RACE procedure, the RNA sample is treated with tobacco acid pyrophosphatase (TAP) to remove the γ and β phosphates from the 5′ end, leaving a 5′ monophosphate. (A control RNA sample not treated with TAP can be used to distinguish mRNA processing products with 5′ monophosphate ends from true 5′ ends.) An RNA oligonucleotide (adaptor RNA) is then ligated to 5′ monophosphate ends using T4 RNA ligase, and the modified RNA is used as a template for second strand cDNA synthesis (primer extension) and subsequent amplification using a thermostable DNA polymerase, a forward primer to the RNA oligo (adaptor primer, containing the RNA oligo sequence) and a gene-specific reverse primer (GSP). The junction of the ligated RNA oligo with the transcribed RNA, determined from sequencing the amplified DNA, identifies the 5′ transcript end [15,29,30].
2.2 Detecting In Vivo Promoter Activity using Promoter-Reporter Fusions
In vivo support for the correct identification of a promoter from a 5′ mRNA end (see above) can be obtained from fusions of fragments containing the presumptive promoter to a reporter gene (e.g., β-galactosidase, galK, gfp, cat; [31]). For initial confirmation that the region of interest contains a functional promoter, the promoter fragment (amplified from the chromosome using region specific primers encoding restriction sites for cloning) should be large enough to include potential upstream regulatory sequences (i.e. at least 100 to 150 bp upstream of the presumptive transcription start site), and the downstream junction should not create an inadvertent in-frame fusion to the reporter gene. Construction of fusions with substitutions in the proposed promoter consensus sequence serves to confirm the position of the promoter within the inserted fragment. For further study of promoter regulation, fusions of a set promoter fragments with variable lengths of DNA upstream of the start site can be used to define the location of sequences that affect promoter function (e.g., [32,33]).
Expression from fusions can be monitored by assay of the protein product (e.g., enzymatic assay, fluorescence, or antibiotic resistance), or by quantitative primer extension of the RNA transcribed by the fusion [25,26]. The β-galactosidase (lacZ) gene is frequently used in these studies because its product is very stable and is readily detected in cultures or on indicator plates, and it can be quantified easily [31,33,34]. Activity from the presumptive promoter region should be compared to the background activity from the fusion vector lacking a promoter insert and from fusions made from mutant derivatives of the promoter region. Mutations at the most highly conserved promoter positions (e.g., in the TTG of the −35 hexamer or in the first, second, or sixth positions of the −10 hexamer, TA---T) should reduce or abolish transcription and confirm the position of the promoter.
Promoter-reporter fusions can be constructed using multicopy plasmid vectors or by insertion of the fusion into the host cell chromosome in single copy. Two examples of multicopy vectors that encode a promoterless lac operon include pRS415 [35] and the broad host range plasmid pRW50 [36]. Reporter gene activity determined in strains carrying these fusion plasmids can confirm the presence of an active promoter, but potential effects of variable growth conditions or regulatory gene mutations on plasmid copy number may limit their usefulness for the study of promoter regulation.
Integration of the promoter-reporter fusion into the chromosome provides a more suitable system for analysis of regulation and can be accomplished using one of several different methods. Bacteriophage λ vectors have been used frequently. One such system ([37]; referred to as “system I” in [33]), suitable for the analysis of very strong promoters such as stable RNA promoters, involves ligation of a promoter fragment to two large purified DNA fragments containing the left and right “arms” of bacteriophage λ. The ligated products are packaged in vitro into λ phage particles using commercial packing extracts (Promega), and strains lysogenic for λ integrated into the attB site of the host chromosome are obtained from strains infected with these phage. Since λ lysogens can potentially contain two or more tandemly integrated λ prophage, monolysogens must be distinguished from multiple copy lysogens. This is most often done in our laboratory by comparison of reporter activity from several independent isolates. Alternatively, multiple lysogens can be identified by PCR amplification of a unique fragment from the junction between tandem copies of lambda (an attP-containing fragment not present in single copy lysogens; [38]). However, since the attP junction fragment is also present in bacteriophage that result from low level spontaneous phage induction in cultures of lambda lysogens, care must be taken to pellet and wash the bacterial cells several times to remove such phage prior to conducting the PCR amplification [38].
An alternative bacteriophage λ fusion system (developed by Simons and colleagues, [35]; referred to as “system II” in [33]) utilizes a promoter-lacZ fusion constructed on a plasmid as an intermediate step. The fusion is then crossed onto a promoterless λ phage vector by in vivo homologous recombination between regions flanking the promoter insertion site on the phage and the plasmid. Phage that have acquired the promoter are detected as blue-plaque formers on X-gal indicator plates, and strains monolysogenic for these phage are purified from turbid plaque centers as blue colony formers. This system is useful for many promoters, but it is not suitable for very strong (e.g., full length rRNA) promoters since they appear to be unstable in the plasmid intermediate stage and accumulate suppressor mutations [33]. A useful feature of this fusion system is the presence of tandem transcription terminators located just upstream of the promoter cloning site. Read-through from transcripts originating upstream of the cloned promoter is blocked by these rrnT1 terminators, resulting in a very low background activity in this system (<1 Miller unit, compared with 20 Miller units or more for “system 1”; [33]), and therefore more accurate assay of activity from weak promoters.
Another useful method for construction of chromosomal fusions does not use a bacteriophage vector, but employs homologous recombination mediated by plasmid-encoded λ Red recombinase to direct the insertion of a linear fragment containing the fusion into a particular target site in the chromosome (e.g., the chromosomal lac operon; [39]). In this system, fusions are constructed on a plasmid intermediate (pPK7035) that encodes lacI, followed by an antibiotic resistance gene (kanR), a transcriptional terminator, the promoter cloning site, and a portion of the lacZ gene. A linear fragment containing the kanR gene, terminator, and the promoter-lacZ fusion junction is amplified from the plasmid using primers to lacI (upstream) and lacZ (downstream), and electroporated into a host strain containing the λ Red expression plasmid. Recombinants are selected for kanamycin resistance, screened for lacZ activity and for the correct structure of the insertion by PCR amplification from the chromosome, and confirmed by DNA sequencing. The resulting chromosomal fusions can be moved easily from one strain background to another by P1 transduction, without having to establish copy number status (as is required for λ lysogens). Since a plasmid intermediate is involved in their construction, this method may not be suitable for very strong promoters, as described above. This method is analogous to those used for creating specific gene inactivation/antibiotic cassette replacement mutations by transformation of a linear fragment into a strain expressing λ homologous recombination functions (λ Red) from a plasmid or from a defective λ prophage [40, 41].
3. RNAP-Promoter Complex Formation In Vitro
The interaction of purified RNA polymerase with a promoter to form a complex in vitro can be detected and studied by several methods, including in vitro transcription, gel mobility shift (EMSA), footprinting, or filter binding assays. The process of RNAP-promoter complex formation is affected greatly by several parameters, including salt concentration, temperature, template topology, and RNA polymerase concentration. The optimum conditions for RNAP complex formation with a particular promoter must be determined empirically. Considerations for these parameters are discussed below. Defining the requirements for RNAP interaction with a particular promoter in the absence of added factors forms a basis for understanding steps in complex formation and transcription initiation that may be affected by trans-acting activators or inhibitors of the process.
E. coli RNAP is often used to examine promoter function in vitro, not only for E. coli promoters, but also for those from other species, and can be purified by standard procedures [42]. E. coli RNAP is also commercially available from Epicentre as holoenzyme, or core enzyme to which a purified sigma factor can be added. This approach may suffice for some purposes for work with promoters from other species, but it may not reflect recognition and regulation of promoters accurately in the relevant host. An alternative option is the use of RNAP purified from the actual host organism. This approach may be facilitated by the use of either a histidine tagged subunit (e.g. H6-tagged chromosomally-encoded B. subtilis β′; [25]), or immunoaffinity chromatography using a resin linked to a polyol-responsive antibody to an RNAP subunit. Antibodies that cross react with the β or β′ subunits from many bacterial species have been described, and RNAP can be eluted from these commercially available resins under mild conditions ([43,44]; Neoclone, Madison WI). In cases where promoters recognized by alternative sigma factors are required, core RNAP and the sigma subunit can be purified separately and reconstituted into holoenzyme in vitro.
3.1. In Vitro Transcription
3.1.1 Solution Conditions for Formation of the Complex
Several solution variables must be optimized for detection and study of RNAP-promoter complexes at a particular promoter, since optimal conditions will vary with the promoter sequence. These variables will be considered in the in vitro transcription section, but they are also applicable to the other in vitro assays described below.
To monitor the formation of an RNAP-promoter complex by in vitro transcription, the promoter DNA template can be provided as either a supercoiled plasmid or a DNA fragment. A supercoiled DNA template offers advantages for detection of the transcript. Promoters are often affected strongly by supercoiling, either positively or negatively, but in a majority of cases are more active on negatively supercoiled DNA [45,46]. Transcripts from some promoters may be difficult to detect using linear fragment templates. Plasmid vectors that are convenient for use in in vitro transcription contain a polylinker cloning site (for directional insertion of a promoter fragment) and a transcription terminator (located a known and relatively short distance downstream of the insertion site). Examples of such vectors include pRLG770 [47) and pSR [48]. The inserted promoter produces a transcript of defined length that can be identified by comparison with transcription reactions carried out with the vector lacking the promoter insertion. These vectors, if constructed with plasmids containing a ColEI origin of replication, also produce a readily detectable transcript from the plasmid replication region, the 110 nt RNA I transcript [49] that can serve as a size marker and in some cases as an internal control in the reaction (see Fig. 3). The length of the sequence downstream of the transcription start site in the inserted fragment containing the promoter should be chosen to produce a transcript that differs from the RNAI transcript and other transcripts generated by the plasmid. Under most conditions (and especially at higher RNAP concentrations and at low salt concentrations), some relatively large transcripts are produced from these vectors (see Fig. 3) and may complicate detection of the transcript of interest if their sizes overlap.
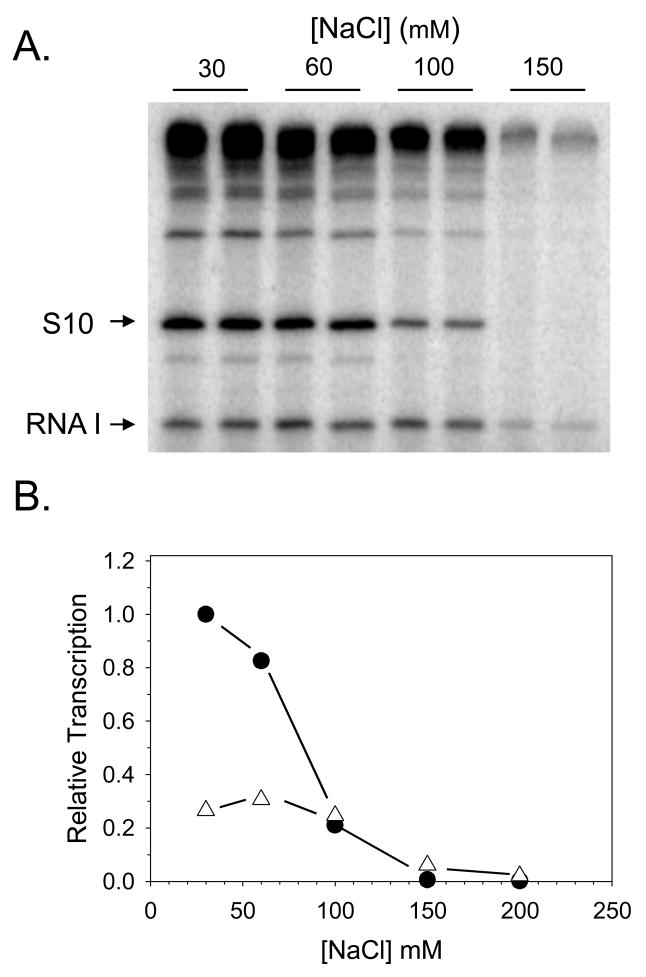
Fig. 3.
In vitro transcription from the multicopy plasmid vector pRLG770 [32] containing the E. coli S10 promoter (−100 to +50). (A) Single round transcription reactions were carried out in buffer containing different final concentrations of NaCl. Complexes were preformed with 10 nM RNAP and 1 nM plasmid DNA at 30°C, followed by addition of the competitor heparin (to 10 μg/ml) and NTP substrates. Duplicate samples were carried out at each of the indicated NaCl concentrations, and products were separated on a 5% acrylamide-7M urea, 1X TBE gel. Transcription products from the S10 promoter (~~190 nt), and from the vector-encoded RNA I promoter (110nt) are indicated. (B) Data from (A) were quantified using ImageQuant software. Average transcription levels from duplicate lanes are plotted relative to that from the S10 promoter at the highest NaCl concentration (30 mM). Filled circles: S10 promoter transcripts; open triangles: RNA I promoter transcripts. Data are from J. Lemke and R. Gourse, unpublished.
Whereas linear promoter fragment templates may be easier to prepare (as restriction fragments or PCR products), transcription from these templates often is much weaker than from supercoiled plasmid templates, leading to additional difficulties with interpretation. Problems can include production of additional transcripts that do not correspond to promoter-initiated events, since RNAP binds to the ends of DNA fragments (particularly when they do not contain blunt ends; [16,17]) and initiates transcription from these ends, producing end-to-end or end-to-terminator transcripts. End-bound RNAP complexes are less stable than most promoter-specific complexes and can be eliminated by adding a competitor. Competitors like the polyanion heparin can be added after formation of the complex and prior to addition of the substrate NTPs, if the promoter complex is stable to heparin. Correct identification of the transcript of interest from linear templates also can be complicated by the occurrence of transcripts that are longer than full fragment length, produced as a result of “wrap around” transcription or “template hopping” (see [50]). These RNA species can arise from transcripts initiated at promoters, as well as from end-bound complexes.
The choice of temperature can also affect the formation of an RNAP-promoter complex very significantly, and thus the production of promoter-specific transcripts. The rate of formation of a competitor-resistant (open) complex shows a large dependence on temperature [51,52]. Although early intermediates (RPC) in the formation of transcriptionally-active promoter complexes can occur at low temperatures where strand separation typically does not occur (e.g., 0°C–12°; [53]), temperatures above ~18–20 degrees are generally required for efficient formation of open complexes (RPO) [54]. However, at certain promoters, open complexes can form at much lower temperatures (e.g., [55]).
The rate of formation of an RNAP-promoter complex shows an even greater dependence on both the concentration and the type of salt present in the reaction (greater even than for most other protein-DNA interactions; [1,56,57]. For example, in studies with the λPR promoter, increasing the NaCl concentration from 0.19 M to 0.27 M decreased the overall association rate constant for complex formation by two orders of magnitude [56]. The use of glutamate rather than chloride as the anion (e.g. KGlutamate vs KCl) significantly increased the rate of binding of RNAP to the λPR promoter at a given salt concentration, but for both salts, increasing the concentration resulted in reduced rates of binding [57]. (KGlutamate effects were examined since glutamate is the most abundant anion in vivo, and therefore considered more physiologically relevant [57]). Promoters vary widely in their salt-sensitivity. Some display an extreme salt-sensitivity (e.g., E. coli rRNA promoters; [58]), whereas others require relatively high salt concentrations for optimal function in vitro (e.g, [59]). The well-characterized promoters lacUV5 and λPR typically have been studied in vitro at KCl concentrations of ~100 mM [56,60,61]. The in vitro transcription experiment shown in Fig. 3 illustrates the responses of two promoters to changes in salt concentration. Transcription from both the ribosomal protein S10 (rpsJ) promoter and the RNA1 promoter was optimal at lower salt concentrations, but transcription from the S10 promoter decreased much more than from the RNA I promoter as the salt concentration increased. As a result, although the S10 transcript was much more abundant than the RNA I transcript at 30 mM NaCl, their relative amounts became similar at 100 mM NaCl, and they were reversed at 150 mM.
The RNAP concentration required to detect transcripts depends upon the strength of the particular promoter being tested as well as on solution conditions. Very strong promoters may require low nM final RNAP concentrations, while other promoters may require much higher concentrations [62].
3.1.2. In Vitro Transcription Reactions
In vitro transcription reactions are carried out under either single-round or multiple-round conditions. In single-round experiments, RNAP is often preincubated with the promoter in the absence of substrates to form a stable (open) complex, after which a competitor like heparin is added to sequester unbound or unstably bound RNAP. A single round of transcription is then initiated from the prebound open complexes by addition of NTP substrates (see below for additional detail). Under these conditions, the level of transcription reflects the equilibrium occupancy of the promoter by RNAP at the time of competitor and substrate addition. It should be noted that not all promoters form stable, competitor resistant complexes. Heparin is usually used at a final concentration of ~10–100 μg/ml for most promoters, but some promoter complexes decay very rapidly, and/or are actively disrupted by heparin. A complex in which heparin does not disrupt RNAP-DNA interactions should display the same decay rate at a range of heparin concentrations (e.g., 10–200 μg/ml; see decay rate measurements below). Stabilization of complexes can in some cases be achieved by formation of a short transcript (e.g., the 5-mer transcript formed at rrnB P1 in the presence of the first two NTPs, ATP and CTP; [58,63]).
In multiple round in vitro transcription experiments, reactions are initiated by the addition of RNAP. Theoretically, many rounds of initiation might be expected, but in practice only a few rounds are usually observed. No competitor is added to the reaction. Multiple-round experiments are usually sufficient to detect effects of promoter mutations, RNAP substitutions, solution variables, NTP concentration (see below), or trans-acting factors. Effects on the rate of complex formation are sometimes masked in single-round experiments where protocols often call for preincubation of RNAP with the promoter.
A typical multiple round in vitro transcription reaction (25 μl volume) might include the following: a supercoiled plasmid template containing the promoter of interest at ~1 nM concentration (~20–50 ng of a 5kb plasmid), in a reaction buffer containing a final concentration of 10 mM Tris-Cl, pH 7.9, 100 mM KCl, 10 mM MgCl2, 1 mM DTT, 100 μg/ml of BSA, 200μM ATP, 200μM GTP, 200μM CTP, 10μM UTP and 2–4 μCi of α32P-UTP. The reaction is initiated by the addition of RNAP to a final concentration of 1–50 nM (determined empirically), incubated at 25°C, 30°C or 37°C for 15 minutes, and terminated by the addition of an equal volume of a combination of stop reaction/gel loading solution containing 95% formamide, 20 mM EDTA, pH 8.0, and 0.05% Bromophenol blue. The sample is transferred immediately to ice (to reduce potential degradation by RNAse), or for longer term storage is kept at −20°C, prior to gel electrophoresis. A denaturing gel containing 7 M Urea and 5.5% acrylamide (19:1 acrylamide:bis acrylamide) and 1X TBE (90 mM Tris, 90 mM Boric Acid, 2.5 mM EDTA) is generally suitable for resolving transcripts. Following electrophoresis, the gel can be dried and exposed to a phosphorimager screen (or to X-ray film), and the bands can be quantified.
Transcription initiation from some promoters is affected by the concentration of the first nucleotide in the transcript [25,26,64]. E. coli rRNA promoters form very unstable open complexes with RNAP (see also below), and are regulated in vitro and in vivo by concentration of the initiating NTP.
To minimize transcript degradation by RNAse contaminants, it is generally satisfactory to use double-distilled water and clean plasticware (tubes and tips), to wear gloves, and to avoid touching tube lids or pipet tip ends. If necessary, solutions can be treated with DEPC to inactivate RNAses. Any glassware used to prepare or store buffers should be baked at high temperatures, and touching glass gel plates with ungloved hands should be avoided.
3.2. Gel Mobility Shift Assay (EMSA)
RNAP-promoter complexes formed with a labeled promoter fragment can be detected as a species with much slower electrophoretic mobility than the free DNA fragment on a native (nondenaturing) acrylamide gel. In addition to binding to promoter sequences, RNAP also binds to DNA in a sequence-independent manner (like other DNA binding proteins; see [65]). RNAP also binds to the ends of DNA fragments [16,17]. Nonspecifically bound RNAPs are generally in rapid equilibrium with free DNA, and therefore are displaced by competitors like heparin or (unlabeled) duplex DNAs. Without competitors, aggregates of DNA-bound RNAPs that typically will not enter the gel during electrophoresis can form at higher RNAP concentrations [66]. However, even complexes containing only a single RNAP can be bound sequence-independently. Therefore, appropriate controls (e.g. promoter mutants) should be used to verify the specificity of detected complexes. This is especially critical for promoter complexes that are unstable to heparin challenge (see above).
EMSA assays can be useful for detection of the contributions of activator or repressor proteins, RNAP concentration, and other components to the process of RNAP-promoter complex formation, and have been used to define equilibrium or rate constants for RNAP-promoter binding (e.g., [67,68]). For quantitative analysis, it should be determined whether the complex formed is stable over the time frame of the gel run (the gel matrix is often assumed to provide a “caging” or stabilizing effect on DNA-protein complexes).
RNAP-promoter complexes for EMSA assays can be assembled as described above for in vitro transcription reactions, but with a labeled promoter fragment and without substrates. 32P-end-labeling of fragments is done either by filling in a recessed 3′ end with DNA polymerase Klenow fragment or by labeling a 5′ end of the fragment (or primer(s) used for PCR amplification) with T4 polynucleotide kinase. Standard procedures can be employed for these labeling reactions (see also footprinting section below). Alternatively, a non-radiolabeled fragment can be prepared by PCR with a fluorescently tagged primer (e.g., FAM; [27]), permitting the DNA to be used and detected at a low (nM) concentration, as with a radiolabeled fragment.
The mobility of the RNAP-promoter complex observed in EMSA may depend upon the temperature of the gel during electrophoresis. As described above, complex formation is temperature-dependent, and different intermediates may exist in a population at equilibrium at some temperatures and for some promoters. The behavior of the lacUV5 promoter-RNAP complex in gel mobility shift experiments was analzyed in detail by Straney and Crothers [66,69]). Complexes exhibiting different mobilities were observed, presumably reflecting different conformations of the complex, and the relative abundance of these complexes varied as a function of the temperature of the gel. Two species of relatively slow mobility (upper and lower) were present at higher temperatures (30°C and 37°C), and a faster migrating complex was observed at low temperature. These complexes were also characterized by footprinting, and their capacity for abortive product synthesis [69]. For other promoters, the complex may migrate on the gel as a single species under the conditions examined (e.g., [68]).
Gel shift experiments involving ternary complexes (RNAP, promoter, and a transcription factor or short transcript) have also been described. The mobility of the presumed ternary complex may differ from that of the binary complex, but the protein composition of the complex should be verified to determine whether the altered mobility reflects the presence of the additional protein rather than a conformational variant of the binary complex. The presence of an additional protein factor can sometimes be confirmed by carrying out the EMSA assay with radiolabeled protein factor, rather than labeled promoter fragment (e.g., [70]).
3.3. Footprinting of the RNAP-Promoter Complex
In this section we focus on useful features of different footprinting reagents as probes of RNAP-promoter complexes, and we provide detailed procedures for DNAseI and hydroxyl radical footprinting of RNAP-promoter complexes. Footprinting an RNAP-promoter complex can address several important questions about the nature of the complex. It can confirm the location of the complex (by defining upstream and downstream boundaries of protection against enzymatic or chemical probes), identify particular promoter positions that are critical to complex formation or that are in very close proximity to RNAP (chemical protection or interference footprints), and determine whether the complex is open (RPO), and what region of the DNA is single-stranded. Footprints also can be useful for characterizing properties of intermediate complexes if the intermediates can be enriched in a population formed under specific solution conditions or with mutant promoters or RNAPs (e.g., closed complexes can be enriched at low temperature or with certain mutant RNAPs, and later intermediates can be enriched by other methods; [53, 71–73]).
Most footprinting reactions are carried out under equilibrium conditions, and use digestion or chemical modification times of ~30 seconds or longer. The complex formed at equilibrium and under specific solution conditions for a particular promoter may be an open complex, an earlier intermediate complex, or the population may contain a mixture of different complexes. Recent work has described conditions for real-time footprints carried out on a millisecond time frame using X-ray generated hydroxyl radicals [71]. These experiments examine transient intermediates that occur during the process of complex formation.
Several footprinting reagents have been used to characterize RNAP-promoter complexes. DNAse I is frequently used to identify the upstream and downstream boundaries of protection by RNAP (see Fig. 4A). It can be used over a range of temperatures and solution conditions, and its application is relatively straightforward. DNAseI recognizes DNA through the minor groove [74], and although cleavage is not sequence-specific, cleavage efficiency shows considerable variation at different sites and is particularly poor in A-or T-tracts. A typical open complex footprint encompasses 60–80 bp of DNA (−60 or −40 to +20 with respect to the transcription start site), but the precise boundaries vary with the promoter and the solution conditions [75]. Occupancy of intermediate complexes can be observed with DNAseI at some promoters in the absence of a competitor (Fig. 4; e.g., [72,76]. Characteristic sites of enhanced DNAseI cleavage are often observed within the DNAseI footprint [1,75], and are interpreted to reflect kinking or distortion of the DNA in the complex.
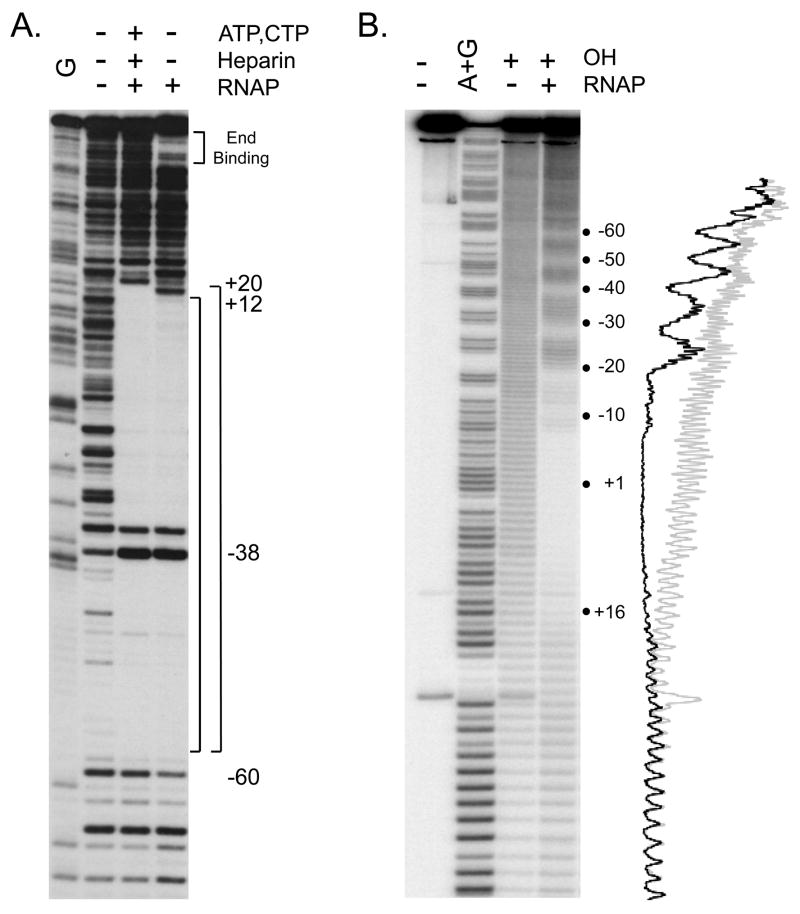
Fig. 4.
(A) DNAse I footprints of RNAP complexes formed with the E. coli rrnB P1 promoter. The promoter fragment (−88 to +50) was 32P-end labeled at the 3′ end of the template strand. Complexes were formed with 10 nM RNAP at 37°C in buffer containing 30 mM KCl, 10 mM Tris-Cl pH7.9, 10 mM MgCl2, 1 mM DTT and 100 μg/ml BSA, and were digested with DNase I (0.6 ug/ml) for 30 sec. The sample in lane 3 also contained the two initiating nucleotides ATP (500 μM) and CTP (50μM), resulting in formation of a heparin stable complex, RPAC [32, 58], and heparin was added to this sample prior to DNAse I digestion. The upstream boundary of protection against DNaseI digestion (~−60) is the same in lanes 3,4 and the downstream boundary differs (+20 for RPAC, lane 3; ~+12 for the complex formed in the absence of ATP and CTP, lane 4). Position of DNaseI enhancement is noted at −38. Binding of RNAP to the end of the promoter fragment is indicated in lane 4, but is not seen in the presence of heparin (lane 3). The sequence marker (G ladder; lane 1) was prepared by dimethyl sulfate modification, followed by strand cleavage with piperidine [80]. (B) Hydroxyl radical footprint of an RNAP-lacUV5 promoter complex. The lacUV5 fragment (−130 to +40) was 32P- labeled at the 3′ end of the non-template strand, and was incubated with 15 nM RNAP at 37°C in a buffer containing 110 mM KCl, 10 mM Tris-Cl pH 8.0, 10 mM MgCl2, 1 mM DTT, followed by hydroxyl radical cleavage as described in text. The RNAP-promoter complex was then separated from free DNA by retention on a nitrocellulose filter and eluted from the filter (see text). Lane 1: promoter fragment, no digestion; lane 2: A+G marker prepared as described in text; lane 3: promoter fragment digested with hydroxyl radicals in absence of RNAP; lane 4: RNAP-promoter complex digested with hydroxyl radicals. Traces of gel lanes 3 (light gray) and 4 (black) were generated as described in text.
A higher degree of resolution (single bp) is obtained with hydroxyl radical protection footprints, in which RNAP protects the DNA backbone against cleavage in several distinct regions (see Fig. 5B; [32,53]. In an open complex, five clusters of protected positions (centered at ~ 10bp intervals at −50, −40, −30, −20 and −10) correlate with the alignment of promoter DNA along the surface of the enzyme in this region [9]. Promoter DNA from ~−10 to ~+16, which includes the region of DNA strand separation, is completely protected against cleavage, corresponding to its enclosure within the main channel of the enzyme. Advantages of this method include its lack of pronounced sequence specificity, single bp resolution, and the wealth of information that can be obtained about specific protected positions. However, the efficiency of cleavage is low, and is reduced further if the buffer contains components that quench hydroxyl radicals (particularly glycerol, which inhibits cleavage at very low concentrations of >0.5%; [77]). Hydroxyl radical footprints can be difficult to interpret from simple visual inspection, and scans of gel lanes are crucial for obtaining the most information from these footprints (see below).
Fig. 5.
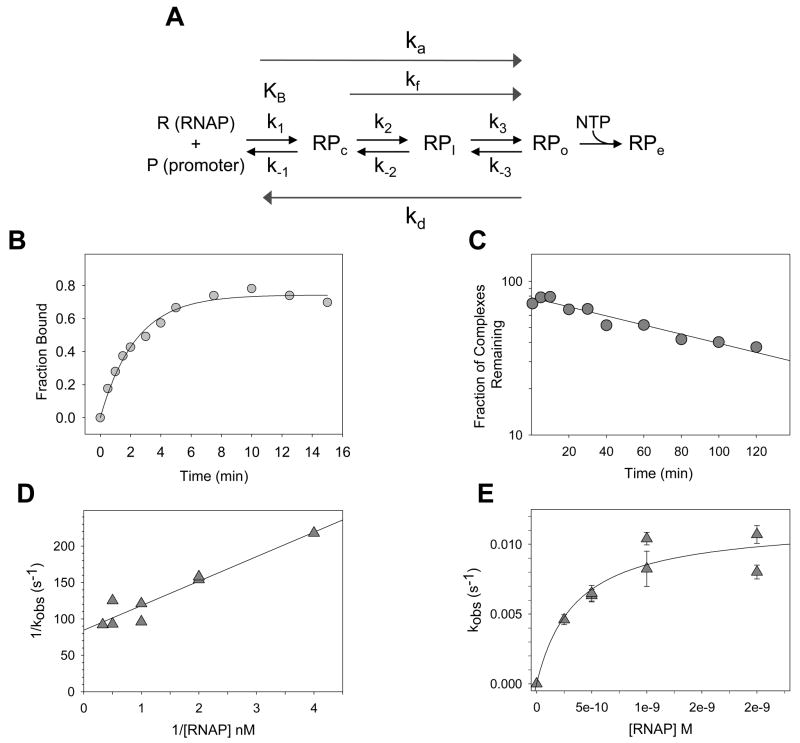
Analysis of kinetic parameters of RNAP-promoter complex formation. A) Multiple steps in the process of forming an RNAP-promoter open complex. R: (RNAP); P: promoter DNA; RPc: closed complex, competitor-sensitive; RPI: intermediate complex, DNA strands closed; RPo: open complex, DNA strands open. RPe: elongation complex. NTP: nucleotide substrates. RPo is a competitor (heparin) resistant complex at many, but not all, promoters. KB (or K1): equilibrium constant for formation of RPc; ka: composite second order association rate constant; kf (or ki): RNAP concentration-independent isomerization rate constant (may reflect rates for multiple intermediate steps at some promoters); kd: composite dissociation rate constant. (see [1,2,52]). (B) Time course for formation of a heparin resistant RNAP complex at the lacUV5 promoter. The 32P end-labeled fragment containing the E. coli lacUV5 promoter (−130 to +40) was incubated with 5 nM RNAP at 30°C in buffer containing 110 mM KCl, 10 mM Tris-Cl pH 8.0, 10 mM MgCl2, 1 mM DTT, 100 μg/ml bovine serum albumin (BSA). Aliquots were removed at time intervals, heparin was added (to 10 μg/ml final), and samples were filtered through nitrocellulose filters [86]. The rate constant, kobs, for formation of the complex at this RNAP concentration (7 × 10−3 s−1) was determined from Eq. 1, as described in text. (C) Determination of the dissociation rate of an RNAP-lacUV5 promoter complex. Complexes were formed with a 32P end-labeled lacUV5 promoter fragment (−130 to +40) and 10 nM RNAP at 30°C in the buffer described for panel (B). Competitor (heparin, final concentration 10 μg/ml) was added at t=0, and aliquots were filtered at time intervals. A semi-log plot of complexes remaining as a function of time (in minutes) is shown. The dissociation rate (kd) determined from a single exponential fit of the data was 2.3 × 10−4 (s−1). (D) Tau plot (1/kobs vs 1/[RNAP]) of observed rate constants, kobs, for formation of a heparin resistant RNAP-promoter complex (RPAC) with a 32P end-labeled fragment containing the E. coli rrnB P1 promoter (−88 to +50), determined at a series of RNAP concentrations. Heparin (10 μg/ml) stable complexes at rrnB P1 are formed in the presence of the first two NTP substrates (ATP, CTP), which produces a 5-mer transcript [58,63]. Complexes were formed at 25°C in buffer containing 45 mM NaCl, 10 mM Tris-Cl pH 8.0, 10 mM MgCl2, 1 mM DTT, 100 μg/ml bovine serum albumin (BSA), 500 μM ATP and 50μM CTP. Values for kobs were determined as shown in panel B, using Eq. (1). The overall association rate constant, ka, determined from a fit of the data to Eq. (2), was ~3.0 × 107 M−1 s−1, and kf was 1.2 × 10−2 s−1. (E) Nonlinear plot (kobs vs. [RNAP]) of the same data as in (D). ka, from fit of data to Eq. (3) was 3.3 × 107 M−1 s−1, and kf was 1.2 × 10−2 s−1.
Potassium permanganate (KMnO4) can be used to identify single-stranded regions within the RNAP-promoter complex and can be performed either in vitro or in vivo. KMnO4 reacts with unpaired (or distorted) thymine residues (and in rare cases other bases), and the modifications can be detected either by primer extension or by strand cleavage at high temperature and alkaline pH ([32,55,78].
Information about base-specific interactions between RNAP and promoter DNA can be obtained from chemical modification studies carried out as protection experiments (the preformed complex is modified), or as interference experiments (premodified DNA is used to form complexes, and modified positions that are critical for complex formation are identified as those that block formation of the complex; [79]). Dimethyl sulfate (DMS) has been used for both protection and interference studies with RNAP. Purine bases are methylated by DMS (at position N7 of guanine in the DNA major groove, or N3 of adenine in the minor groove), and can be detected by strand cleavage at the modified positions or by primer extension (e.g., [13,32,79,80]). Specific purine bases that are critical contacts to RNAP in the recognition elements described above have been detected by DMS footprinting ([13,32,79,81]. In addition, methylation of single-stranded cytosines has been used as an alternative method for detection of strand separation in the complex (32,82).
Interference studies have also been carried out with RNAP using promoter fragments with randomly-positioned bases that contain modifications (e.g., dUTP substitutions; [81]; see also [83] for review of template-directed interference footprinting procedures) or fragments with randomly-positioned missing bases or missing nucleoside (e.g., [84]). These experiments can be used to scan the promoter for positions that are critical for complex formation.
3.3.1. DNA Fragment Preparation for Footprinting
The quality of footprinting data is greatly improved by DNA fragment preparation methods that ensure a homogeneous, single-labeled end and the absence of nicks or other chemical damage. To reduce DNA damage, use freshly purified plasmid DNA, and minimize the time of exposure to phenol during preparation. (Prepare a Tris-Cl pH 8.0-buffered phenol stock just before use, since neutralized phenol oxidizes more rapidly than water-saturated phenol). Store DNA samples at −20°C in buffered solution. DNA fragments can be prepared by restriction digestion of plasmid DNA, or by PCR amplification from a plasmid template. Results are often better with plasmid-derived fragments, although we have had success with PCR fragments prepared by a small number of cycles of amplification (15 cycles) using a relatively large amount (~400 ng) of plasmid as the template for PCR [81]. To avoid heterogeneity at the ends of PCR fragments, use primers that encode restriction sites such that fragment ends can be generated by restriction digestion. (Include an additional 4–5nt of flanking random sequence 5′ to the restriction site in the primer to facilitate digestion.) Purify the PCR fragment further by phenol extraction and ethanol precipitation or by using a Qiagen kit prior to digestion and labeling to eliminate reagents (dNTPs, DNA polymerase) that may interfere with reactions or alter the products.
Prepare fragments that are 150–200 bp in length, such that the predicted RNAP binding site is flanked on either side by unbound sequence (≈40–50 bp). (RNAP typically protects 60–80bp in footprints.) A longer distance between the labeled end and the protein binding site will reduce the resolution in the region of interest on sequencing gels.
Label approximately 5–10 pmol of fragment at either the 5′ or the 3′ end of one strand. For 3′ end labeling, “fill in” only the first position at a 5′ overhanging restriction site end with a DNA polymerase [Sequenase (USB) or DNA polymerase Klenow fragment] and one radiolabeled dNTP (the first templated position). Avoid filling in all templated positions, since incomplete reactions will produce a heterogenous population of labeled DNA molecules. For example, to label a HindIII end, use only α-32P-dATP in the reaction, and no other dNTPs. Avoid using sites such as EcoRI where the first two templated dNTPs are the same, since this can result in fragment length heterogeneity. Use only a brief labeling time. For example, for a Sequenase fill-in reaction, incubate for only 5 min. at only 30°C in a 25 μl final volume: 5–10 pmol of DNA fragment, 7 μl of α 32P-dNTP (~3000Ci/mmol, 10 μCi/μl; e.g., α 32P-dATP for a HindIII site), 1.8 μl 0.1 M DTT, 5 ul of 5X Sequenase reaction buffer and 5 units of Sequenase (USB). After labeling, add 20 μl of 7.5 M ammonium acetate, and precipitate with two volumes of 100% ethanol, using 20 μg of molecular biology grade glycogen (Roche) as carrier. To avoid precipitation of salts, do not prechill the sample at −20°C or −80°C prior to centrifugation. Centrifuge immediately for 5–10 min. in a microcentrifuge at 4°C, and rinse the pellet well with 70% ethanol to remove traces of salt that might interfere with subsequent steps. If needed, digest at a second restriction site near the opposite end of the fragment to eliminate any unwanted labeling in the opposite strand.
Alternatively, the fragment may be labeled at one dephosphorylated 5′ end using T4 polynucleotide kinase and γ-32P ATP (or the 5′ end of one primer can be labeled prior to a PCR reaction to generate the fragment). The following procedures are used in our lab for 5′ end labeling of fragments: dephosphorylate the 5′ end using Calf Intestinal Alkaline Phosphatase (CIAP) in a 70 μl reaction volume containing the DNA fragment (~5–10 pmol), 1X manufacturer’s reaction buffer, and 0.2 units CIAP. Incubate for 30 min at 37°C, add an additional 0.2 units of CIAP, and continue incubation for 30 min. Add sodium acetate to 0.3 M final concentration, phenol extract the sample, ethanol precipitate the aqueous phase (with addition of 20 μg of glycogen as carrier), and rinse the pellet well with 70% ethanol to remove all traces of phenol and salt. (Alternatively, use a phosphatase that can be completely heat inactivated, and eliminate the phenol extraction steps.) To label the 5′ end, combine the resuspended dephosphorylated DNA fragment, 5 μl of 10X T4-polynucleotide kinase reaction buffer, 6 μl γ-32P ATP, 20 units (2 μl) T4 polynucleotide kinase, and 2 μl 0.1 M DTT in a final volume of 50 μl, and incubate for 30 min at 37°C. Add sodium acetate to 0.3 M, 20 μg of glycogen as carrier for precipitation, and 125 μl of 100% ethanol. Centrifuge and rinse the pellet well with 70% ethanol as described above. Digest with a second restriction enzyme to produce a fragment with a single labeled end.
Gel purify the labeled fragment to eliminate other unwanted labeled fragments. Use a non-denaturing acrylamide gel (5%, 19:1 acrylamide:bis). The position of the labeled band can be determined from a phosphorimager scan using a print made at 100% of the actual size of the gel as a template. Elute DNA from the minced gel slice by diffusion (overnight, 4°C) into buffer appropriate for the next steps (e.g., 2–5 ml of 0.2 M NaCl, 20 mM Tris-Cl pH 7.4, 1 mM EDTA, the loading buffer for ElutipD purification mini columns). We have had good success with Elutip D columns (Whatman) for rapid concentration and purification of DNA fragments for footprints. The gel and subsequent Elutip D purification steps eliminate enzymes, unincorporated nucleotides, incomplete length PCR fragments, as well as soluble gel materials.
3.3.2. General Footprinting Reaction Conditions
Solution conditions for forming the RNAP-promoter complex should take into consideration requirements for the cleavage reagent (e.g., MgCl2 for DNAseI; little or no glycerol for hydroxyl radical; see below), and should be compatible with high occupancy of the complex (see temperature and salt conditions described above for in vitro transcription reactions). The end-labeled promoter fragment is usually used at a concentration of ~0.5–1.0 nM and should provide an amount of radioactivity (~20,000 DPM) that allows detection of the gel image in a reasonable amount of time (e.g., overnight). The exact concentration of DNA (in the low nM range) is not critical, assuming that RNAP is present in excess molar concentration. The optimum RNAP concentration is dictated by its binding constant for the particular promoter under the solution conditions used, and this is generally in a range higher than the DNA concentration needed for detection of radioactivity (≥5 nM). A series of RNAP concentrations should be tested.
A clear footprint signal requires conditions in which close to 90–100% occupancy of the RNAP-promoter complex has been achieved. If this cannot be attained by increasing the RNAP concentration or altering the solution conditions, it may be necessary to isolate the complex away from free DNA on a gel (EMSA), or by nitrocellulose filtration of the complex after digestion with the footprinting reagent (e.g. DNAseI or hydroxyl radical) (see Fig. 4B). The DNA in the complex is then eluted from the gel, concentrated and purified using an ElutipD column, as described above, before running on a sequencing gel. If the complex is isolated on a filter, it can be eluted either in Qiagen PB buffer and concentrated by a PCR Purification column, or eluted in 0.5 M NH4OAc, 0.01 M MgOAc, 0.1% SDS, 0.1 mM EDTA followed by ethanol precipitation. If isolation of the complex is necessary, scale up the reaction with ~10-fold more labeled fragment, since not all will be recovered.
3.3.3. DNAse I Footprinting
3.3.3.1.Titration of DNAseI
The final concentration of DNAseI typically used in our footprinting reactions is ≈0.5–2 μg/ml for a 30 second digestion. However, since the source and batch of DNAseI, the temperature and salt conditions, and the DNA fragment length are all variables that affect the extent of digestion, it is important to calibrate the concentration needed to give the appropriate level of partial digestion under the specific conditions used. Calibrating the amount of DNAseI to use, in the absence of RNAP, should prevent the very common problem of “overdigestion” of the control samples lacking RNAP. Calibrate the DNAseI stock by digesting free (unbound) DNA using a two-fold dilution series of DNAseI concentrations under the specific conditions to be used for footprinting. Based on results of the titration experiment, choose a final DNAseI concentration that will produce approximately one-third full length DNA and two-thirds digested DNA (reflecting an average of one cleavage per DNA molecule). Prepare fresh dilutions of bovine pancreatic DNAseI from the concentrated stock solution for each experiment, using the same dilution procedures each time to get reproducible extents of digestion. CaCl2 is often recommended for use with DNAseI, but it is not essential for activity. Small changes in DNA concentration in the ≈nM range do not affect the extent of DNAseI digestion. However, if high concentrations of competitor DNA are used, the titration should be carried out in the presence of the competitor.
3.3.3.2. DNAseI Footprinting Reactions
Use 25 μl reactions containing buffer and labeled DNA fragment (prepare a “master mix”, and aliquot it to different reaction tubes). Pre-equilibrate to the temperature to be used. Include the following control samples in the experiment: (1) no RNAP, no DNaseI (only RNAP dilution buffer, as a control for background nicks in the fragment); (2) RNAP at the highest concentration used, no DNAseI (as a control for nuclease activity in the RNAP preparation); (3) no RNAP but plus DNAseI (for comparison with the positions cleaved in the presence of RNAP).
Add RNAP (in a small volume, and at staggered 2 min intervals) to the prealiquoted DNA and buffer samples, and incubate at the desired temperature for ~10–15 min to allow binding to reach equilibrium. Add DNAseI (in a small volume) to each sample (at 2 min intervals, as for protein addition above), mix quickly and incubate for 30 sec. Stop reactions after 30 sec. by immediate addition of 75 μl of a stop solution calculated to bring the sampes to final concentrations of 10 mM EDTA and 0.3 M sodium acetate. Vortex and place samples on ice. When the set of reactions is completed, add glycogen (20 μg; Roche) to each sample as carrier for subsequent ethanol precipitation. Samples may be phenol extracted to remove RNAP, but this is not required. [For extraction, add 100 μl of freshly buffered phenol (an equal volume), vortex briefly, and centrifuge briefly (15 sec) to separate phases. Remove the aqueous phase to a clean tube.] Whether the samples are phenol extracted or not, they should be ethanol precipitated to concentrate the DNA and remove salts prior to gel loading. Add 0.3 ml 100% ethanol, and centrifuge at 4°C for ≈10min. Chilling at low temperatures prior to centrifugation is not necessary and is not recommended, since it may result in precipitation of salts that will interfere with gel electrophoresis. Rinse the DNA/glycogen pellets twice with 70% ethanol, and air or vacuum dry. [Note that the ethanol precipitation step will result in loss of the smallest DNA products (~15nt in length or shorter)]. Resuspend in 4–5 μl of gel loading solution (8 M Urea, 0.5X TBE, 0.05% BPB, 0.05% Xylene Cyanole).
3.3.4. Hydroxyl Radical Footprinting
Many of the considerations described above for DNAseI footprinting are applicable to hydroxyl radical footprinting. The following additions or modifications should also be made. (1) Use more DPM of radiolabeled fragment, since the efficiency of DNA cleavage by hydroxyl radicals is relatively low (use ≈5X more than for DNaseI reactions). (2) Pre-existing nicks in the fragment can present a greater problem due to inefficient DNA cleavage by hydroxyl radicals, so fragment integrity should be carefully checked by loading an equal amount of untreated labeled DNA on the sequencing gel. (3) Resolution of bands is more difficult, since cleavage occurs at every position, producing more bands than obtained with DNAseI. To improve resolution, be sure that the labeled end is relatively close to the binding site (~40 bp). Resolution can also be improved by running the gel longer, and by fixing the gel in 10% acetic acid, 10% methanol before drying. (4) Glycerol is a scavenger of hydroxyl radicals, so its final concentration in reaction buffers must be very low (0.5% or less; [77]), and control samples should contain the same concentration of glycerol as test samples.
Immediately before beginning the RNAP binding reactions, prepare a 1:1 mixture of freshly prepared 100 mM ferrous ammonium sulfate and 200 mM EDTA. After RNAP promoter complex formation (in a 50 μl reaction volume), add competitor such as heparin, if needed, and add quickly along the wall of the tube: 0.6 μl 100 mM sodium ascorbate (store in aliquots at −20°C ); 4.0 μl 0.15% hydrogen peroxide (freshly diluted from 30% stock); 6.0 μl 1:1 iron:EDTA mix (final 5 mM iron, 10 mM EDTA). Mix these reactants into samples and incubate for 2 min. Stop reactions by addition of 80 μl 20mM Thiourea, and 60 μl 0.3 M NaCl, 1 mM EDTA. Phenol extract (if needed), and ethanol precipitate the samples, adding glycogen (20 ug) as carrier. Wash pellets with 70% ethanol to remove salts, dry, and resuspend in gel loading solution as described above.
3.3.5. Sequence Markers and Gel Electrophoresis
An A+G sequence ladder can be generated with the labeled fragment using the formic acid depurination reaction [80]. Incubate 12μl of 32P-labeled DNA fragment (in distilled water, not buffer) with 50 μl formic acid at room temperature for ~7 min (calibrate the time required to give a ladder of products). Add sodium acetate (to 0.3 M), glycogen carrier, and 2–3 volumes of 100% ethanol. Centrifuge the DNA, wash the pellet with 70% ethanol to remove salts, and air dry. To cleave the DNA at depurinated positions, resuspend the pellet in 100 μl of 1 M piperidine, transfer to a small (200 μl) PCR tube, and incubate in a thermocycler with a heated lid at 90 degrees for 30 min. Transfer the sample back to an Eppendorf tube, and ethanol precipitate the DNA. Rinse the DNA pellet well (2–3X) with 70%, since residual piperidine will cause aberrant mobility of the fragments on the sequencing gel. Air dry the samples, and resuspend in gel loading solution (8 M Urea, 0.5X TBE, 0.05% BPB, 0.05% Xylene Cyanole).
8–10% acrylamide gels (19:1 acrylamide:bis; 0.4mm thickness) with 7M Urea and 0.5X TBE are recommended to produce sharp bands needed for resolving digestion products from the footprinting reactions. Samples should be heated to ~90 degrees for only ~1min immediately prior to loading. (Stagger the heating of samples, such that one sample is heating while another is being loaded). Pipet the 4–5 μl samples directly into the bottom of the well so that they layer onto the gel as a sharp band. Run the gel at ~2000V for a time determined by the length of fragment and the position of the RNAP binding site within the fragment. Gels can be fixed in 10% methanol, 10% acetic acid to improve band sharpness (fix for approximately 10–15 min. for a gel of 0.4mm thickness). This step is not essential but is particularly useful for hydroxyl radical footprinting experiments where band sharpness is critical for resolution. Gels should be dried before exposure to a phosphorimager screen, as this will also improve band sharpness. To reduce the occurrence of aberrant bands (such as reannealed duplex DNA, which migrates faster than the single stranded fragment), the use of the urea loading solution is recommended (rather than a formamide-based loading solution), and gels should be run at warmer temperatures, with prewarmed upper chamber buffer, if possible.
3.3.6. Analysis of Footprinting Profiles
Analysis of long, complex RNAP footprint profiles can be facilitated by scanning lanes from the gel image and creating a graph of each lane (in a program such as ImageQuant), and transferring the graph data points to another program (e.g., Excel) for normalization of signals to regions of the profile that are not affected by RNAP binding. Normalized data points can then be displayed using a graphing program (e.g., SigmaPlot) (see Fig. 4B). Overlaying of profiles permits direct comparison of signal strength of individual bands in different lanes (e.g., [81]). (To obtain accurate quantification of gel lanes, it is useful to eliminate gel problems that distort band width, such as tapering of lanes caused by salt contamination of samples. See above).
3.4 Determining Rates of Kinetic Parameters of RNAP-Promoter Complex Formation and Decay
Characterization of an RNAP-promoter complex formed under favorable in vitro solution conditions at equilibrium can provide valuable structural information about the complex. However, different promoters display very different rates of complex formation and decay. Kinetic analysis may identify steps in complex formation that are potential targets for positive or negative regulation by transcription factors.
The formation of an RNAP-promoter complex capable of initiating transcription is a multistep process, involving conformational changes in both promoter DNA (P) and RNAP (R) (Fig. 5A). An initial closed complex, RPc, isomerizes through intermediate steps (RPI), and at least two such kinetically significant intermediates on the pathway to open complex formation occur at the well characterized λPR and lacUV5 promoters; 25 [1,52,60,61]. The steps in complex formation are characterized by equilibrium and rate constants, and these can differ significantly from one promoter to another [1,2,4]. The equilibrium constant KB (or K1) describes occupancy of the initial closed complex, and the overall composite association rate ka is the product KBkf, where kf is the composite rate constant for the isomerization steps that occur after initial complex formation. The dissociation of the complex is described by the rate constant kd.
The rate constants for association of RNAP with a promoter to form a competitor-resistant complex can be determined by monitoring complexes present as a function of time after RNAP addition to the reaction (Fig. 5B). Aliquots of the reaction are removed at time intervals and are added to tubes containing either a competitor and NTP substrates (for detecting complexes by transcriptional readout; [85]), or just a competitor (for detecting complexes by retention on nitrocellulose filters; [52,60]. (Effects of additional DNA-binding protein factors on complex formation or decay cannot be determined by the filter binding assay, since the factor will also cause the labeled DNA to be retained on the filter irrespective of the status of the RNAP-promoter complex.) RNAP must be present in at least a 3-fold molar excess over the concentration of promoter DNA in these experiments, such that the reactions are pseudo first-order [60]. For each RNAP concentration, a time course of complex formation is carried out, and an observed rate constant (kobs ) is determined from Eq. (1):
| Eq. (1) |
where cpmplateau corresponds to the maximal level of complex formation observed. In instances where the maximum level of complex formation (plateau in the time course) does not reach the same level at both low and high RNAP concentrations, a correction for the reversibility of the reaction is made, such that kobs values are corrected for maximum complex formation observed in the set of determinations (see [52]).
The observed rate constants (kobs) for a series of RNAP concentrations are used to determine the overall composite association rate constant (ka), and the composite isomerization rate contant, kf. In a double reciprocal linear plot of 1/kobs vs 1/[RNAP], referred to as a Tau plot [60,61] (see example in Fig. 5D), the reciprocal of the slope defines ka, and the reciprocal of the y-intercept defines kf [Eq. (2)]. (KB may then be determined from the equation ka=KBkf).
| Eq. (2) |
Alternatively, ka and kf can be determined from a weighted nonlinear plot of kobs vs [RNAP] [52,73] [Eq. (3)] (see example in Fig. 5E). More accurate determination of kf can generally be obtained from this method [52].
| Eq. (3) |
The rate of dissociation (decay) of a preformed RNAP-promoter complex can also be determined (Fig. 5C), yielding a composite decay rate constant, kd [50,60–62,85] and the complex half-life. In this type of experiment, complexes are preformed, competitor (e.g., heparin) is added at t=0, and aliquots of the sample are removed at time intervals for analysis of complexes remaining by either a transcription readout or by filter binding.
Acknowledgments
We thank Pete Chandrangsu and Justin Lemke for Figs. 2 and 3, and members of our laboratory for helpful discussions. This work was supported by National Institutes of Health Grant R37 GM37048 to R.L.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Record MT, Jr, Reznikoff WS, Craig ML, McQuade KL, Schlax PJ. In: Escherichia coli and Salmonella Cellular and Molecular Biology. Neidhardt RC, et al., editors. Vol. 1. ASM Press; Washington D.C.: 1996. pp. 792–821. [Google Scholar]
- 2.deHaseth PL, Zupancic ML, Record MT., Jr J Bacteriol. 1998;180:3019–3025. doi: 10.1128/jb.180.12.3019-3025.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helmann JD, deHaseth PL. Biochemistry. 1999;38:5959–5967. doi: 10.1021/bi990206g. [DOI] [PubMed] [Google Scholar]
- 4.Gruber TM, Gross CA. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 5.Haugen SP, Ross W, Gourse R. Nature Rev Microbiol. 2008;6:507–516. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gourse RL, Ross W, Gaal T. Molecular Microbiology. 2000;37:786–695. doi: 10.1046/j.1365-2958.2000.01972.x. [DOI] [PubMed] [Google Scholar]
- 7.Korsheva N, Mustaev A. Current Opinion in Microbiol. 2001;4:119–125. doi: 10.1016/s1369-5274(00)00176-4. [DOI] [PubMed] [Google Scholar]
- 8.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Nature. 2007;447:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 9.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 10.Lisser S, Margalit H. Nucl Acids Res. 1993;21:1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. Nucleic Acids Res. 2007;35:771–788. doi: 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell JE, Zheng D, Busby SJW, Minchin SD. Nucleic Acids Res. 2003;31:4689–4695. doi: 10.1093/nar/gkg694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 14.Huerta AM, Collado-Vides J. J Mol Biol. 2003;333:261–278. doi: 10.1016/j.jmb.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 15.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. PLoS Biol. 2006;4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melancon P, Burgess RR, Record MT., Jr Biochemistry. 1982;21:4318–4331. doi: 10.1021/bi00261a022. [DOI] [PubMed] [Google Scholar]
- 17.Melancon P, Burgess RR, Record MT., Jr Biochemistry. 1983;22:5169–5176. doi: 10.1021/bi00291a017. [DOI] [PubMed] [Google Scholar]
- 18.Karp PD, Keseler IM, Shearer A, Latendresse M, Krummenacker M, Paley SM, Paulsen I, Collado-Vides J, Gama-Castro S, Peralta-Gil M, Santos-Zavaleta A, Penaloza-Spinola MI, Bonavides-Martinez C, Ingraham J. Nucleic Acids Res. 2007;35:7577–7590. doi: 10.1093/nar/gkm740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gama-Castro SV, Jimenez-Jacinto, Peralta-Gil M, Santos-Zavaleta A, Penaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muniz-Rascado L, Martinez-Flores I, Salgado H, Bonavides-Marinez C, Abreu-Goodger C, Rodriguez-Penagos C, Miranda-Rios J, Morett E, Merino E, Huerta AM, Revino-Quintanilla L, Collado-Vides J. Nucl Acids Res. 2008;36(Database Issue):D120–124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grainger D, Hurd D, Harrison M, Holdstock J, Busby SJW. Proc Natl Acad Sci. 2005;102:17693–17698. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reppas NB, Wade JT, Church GM, Struhl K. Mol Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 22.Bensing BA, Meyer BJ, Dunny GM. Proc Natl Acad Sci. 1996;93:7794–7799. doi: 10.1073/pnas.93.15.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Russell DW. Molecular Cloning, A Laboratory Manual. Ch 7. Vol. 1. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2001. [Google Scholar]
- 24.Schneider DA, Murray HD, Gourse RL. Methods in Enzymol. 2003;370:606–617. doi: 10.1016/S0076-6879(03)70051-2. [DOI] [PubMed] [Google Scholar]
- 25.Krasny L, Gourse RL. EMBO J. 2004;23:4473–4483. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray HD, Gourse RL. Mol Microbiol. 2004;52:1375–1387. doi: 10.1111/j.1365-2958.2004.04060.x. [DOI] [PubMed] [Google Scholar]
- 27.Lloyd AL, Marshall BJ, Mee BJ. J Microbiol Methods. 2005;60:291–298. doi: 10.1016/j.mimet.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Josaitis CA, Gaal T, Gourse RL. Proc Natl Acad Sci. 1995;92:1117–1121. doi: 10.1073/pnas.92.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scotto-Lavino E, Du G, Frohman MA. Nat Protoc. 2006;1:2555–2562. doi: 10.1038/nprot.2006.480. [DOI] [PubMed] [Google Scholar]
- 30.Scotto-Lavino E, Du G, Frohman MA. Nat Protoc. 2006;1:3056–3061. doi: 10.1038/nprot.2006.479. [DOI] [PubMed] [Google Scholar]
- 31.Silhavy TJ, Berman ML, Enquist LE. Experiments with GeneFusions. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y.: 1984. [Google Scholar]
- 32.Newlands JT, Ross W, Gosink KK, Gourse RL. J Mol Biol. 1991;220:569–583. doi: 10.1016/0022-2836(91)90101-b. [DOI] [PubMed] [Google Scholar]
- 33.Rao L, Ross W, Appleman JA, Gaal T, Leirmo S, Schlax PJ, Record MT, Jr, Gourse RL. J Mol Biol. 1994;235:1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- 34.Miller JH. A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y.: 1992. [Google Scholar]
- 35.Simons RW, Houman F, Kleckner N. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 36.Lodge J, Fear J, Busby S, Gunasekaran P, Kamini NR. FEMS Microbiol Lett. 1992;95:271–276. doi: 10.1016/0378-1097(92)90441-p. [DOI] [PubMed] [Google Scholar]
- 37.Gourse RL, deBoer HA, Nomura M. Cell. 1986;44:197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- 38.Powell BS, Court DL, Nakamura Y, Rivas MP, Turnbough CL., Jr Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang Y, Weber KD, Qui Y, Kiley PJ, Blattner FR. J Bacteriol. 2005;187:1135–1160. doi: 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Datsenko KA, Wanner BL. Proc Natl Acad Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. Proc Natl Acad Sci. 2000;97:5975–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burgess RR, Jendrisak JJ. Biochemistry. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- 43.Thompson NE, Hager DA, Burgess RR. Biochemistry. 1992;31:7003–7008. doi: 10.1021/bi00145a019. [DOI] [PubMed] [Google Scholar]
- 44.Bergendahl V, Thompson NE, Foley KM, Olson BM, Burgess RR. Protein Expression and Purification. 2003;31:155–160. doi: 10.1016/s1046-5928(03)00145-1. [DOI] [PubMed] [Google Scholar]
- 45.Amoyal M, Buc H. J Mol Biol. 1987;195:795–808. doi: 10.1016/0022-2836(87)90485-2. [DOI] [PubMed] [Google Scholar]
- 46.Su TT, McClure WR. J Biol Chem. 1994;269:13511–13521. [PubMed] [Google Scholar]
- 47.Ross W, Thompson JF, Newlands JT, Gourse RL. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolb A, Kotlarz D, Kusan S, Ishihama A. Nucl Acids Res. 1995;23:819–826. doi: 10.1093/nar/23.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davison J. Gene. 1984;28:1–15. doi: 10.1016/0378-1119(84)90082-9. [DOI] [PubMed] [Google Scholar]
- 50.Paul BJ, Berkmen MB, Gourse RL. Proc Natl Acad Sci. 2005;102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roe JH, Record MT., Jr J Mol Biol. 1985;184:441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]
- 52.Saecker RM, Tsodikov OV, McQuade KL, Schlex PE, Jr, Capp MW, Record MT., Jr J Mol Biol. 2002;319:649–671. doi: 10.1016/S0022-2836(02)00293-0. [DOI] [PubMed] [Google Scholar]
- 53.Schickor P, Metzger W, Werel W, Lederer H, Heumann J. EMBO J. 1990;9:2215–2220. doi: 10.1002/j.1460-2075.1990.tb07391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spassky A, Kirkegaard K, Buc H. Biochemistry. 1985;24:2723–2731. doi: 10.1021/bi00332a019. [DOI] [PubMed] [Google Scholar]
- 55.Burns HD, Belyaeva TA, Busby SJ, Minchin SD. Biochem J. 1996;317:305–311. doi: 10.1042/bj3170305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roe JH, Record MT., Jr Biochemistry. 1985;24:4721–4726. doi: 10.1021/bi00339a002. [DOI] [PubMed] [Google Scholar]
- 57.Leirmo S, Harrison C, Cayley DS, Burgess RR, Record MT., Jr Biochemistry. 1987;26:2095–2101. doi: 10.1021/bi00382a006. [DOI] [PubMed] [Google Scholar]
- 58.Gourse RL. Nucl Acids Res. 19898;16:9789–9809. doi: 10.1093/nar/16.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosenthal AZ, Kim Y, Gralla JD. Molecular Microbiol. 2008;68:907–917. doi: 10.1111/j.1365-2958.2008.06186.x. [DOI] [PubMed] [Google Scholar]
- 60.Roe JH, Burgess RR, Record MT., Jr J Mol Biol. 1984;176:495–521. doi: 10.1016/0022-2836(84)90174-8. [DOI] [PubMed] [Google Scholar]
- 61.Buc H, McClure WR. Biochemistry. 1985;24:2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- 62.Barker MM, Gaal T, Gourse RL. J Mol Biol. 2001;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- 63.Borukhov S, Sagitov V, Josaitis CA, Gourse RL, Goldfarb A. J Biol Chem. 1993;268:23477–23482. [PubMed] [Google Scholar]
- 64.Murray HD, Schneider DA, Gourse RL. Mol Cell. 2003;12:125–134. doi: 10.1016/s1097-2765(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 65.von Hippel PH. Cell. 2006;125:1027–1028. doi: 10.1016/j.cell.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 66.Straney DC, Crothers DM. Cell. 1985;43:449–459. doi: 10.1016/0092-8674(85)90175-8. [DOI] [PubMed] [Google Scholar]
- 67.Hershberger P, deHaseth PL. J Mol Biol. 1991;222:479–94. doi: 10.1016/0022-2836(91)90491-n. [DOI] [PubMed] [Google Scholar]
- 68.Zhang X, Reeder T, Schleif R. J Mol Biol. 1996;258:14–24. doi: 10.1006/jmbi.1996.0230. [DOI] [PubMed] [Google Scholar]
- 69.Straney DC, Crothers DM. J Mol Biol. 1987;193:279–292. doi: 10.1016/0022-2836(87)90219-1. [DOI] [PubMed] [Google Scholar]
- 70.Severinova E, Severinov K, Darst SA. J Mol Biol. 1998;279:9–18. doi: 10.1006/jmbi.1998.1742. [DOI] [PubMed] [Google Scholar]
- 71.Sclavi B, Zaychikov E, Rogozina A, Walther F, Buckle M, Heumann H. Proc Natl Acad Sci. 2005;102:4706–4711. doi: 10.1073/pnas.0408218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cook V, deHaseth PL. J Biol Chem. 2007;282:21319–21326. doi: 10.1074/jbc.M702232200. [DOI] [PubMed] [Google Scholar]
- 73.Davis CA, Capp MW, Record MT, Jr, Saecker RM. Proc Natl Acad Sci. 2005;102:285–290. doi: 10.1073/pnas.0405779102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suck D. Biopolymers. 1998;44:405–421. doi: 10.1002/(SICI)1097-0282(1997)44:4<405::AID-BIP5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 75.Ozoline ON, Tsyganov MA. Nucl Acids Res. 1995;23:4533–4541. doi: 10.1093/nar/23.22.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bartlett MS, Gaal T, Ross W, Gourse RL. J Mol Biol. 1998;279:331–345. doi: 10.1006/jmbi.1998.1779. [DOI] [PubMed] [Google Scholar]
- 77.Dixon WJ, Hayes JJ, Levin JR, Weidner MF, Dombroski BA, Tullius TD. Methods in Enzymology. 1991;208:380–413. doi: 10.1016/0076-6879(91)08021-9. [DOI] [PubMed] [Google Scholar]
- 78.Sasse-Dwight S, Gralla JD. (1991) Methods in Enzymology. 1991;208:146–168. doi: 10.1016/0076-6879(91)08012-7. [DOI] [PubMed] [Google Scholar]
- 79.Siebenlist U, Simpson RB, Gilbert W. Cell. 1980;20:269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- 80.Maxam A, Gilbert W. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 81.Ross W, Ernst A, Gourse RL. Genes & Devel. 2001;15:491–506. doi: 10.1101/gad.870001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kirkegaard K, Buc H, Spassky A, Wang JC. Proc Natl Acad Sci. 1983;80:2544–2548. doi: 10.1073/pnas.80.9.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Storek MJ, Ernst A, Verdine GL. Nature Biotechol. 2002;20:183–186. doi: 10.1038/nbt0202-183. [DOI] [PubMed] [Google Scholar]
- 84.Li X-Y, McClure WR. J Biol Chem. 1998;273:23558–66. doi: 10.1074/jbc.273.36.23558. [DOI] [PubMed] [Google Scholar]
- 85.Barker MM, Gaal T, Josaitis CA, Gourse RL. J Mol Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 86.Ross W, Gourse RL. Proc Natl Acad Sci. 2005;102:291–296. doi: 10.1073/pnas.0405814102. [DOI] [PMC free article] [PubMed] [Google Scholar]