Abstract
Human conducting airways contain two anatomically distinct epithelial cell compartments: surface epithelium and submucosal glands (SMG). Surface epithelial cells interface directly with the environment and function in pathogen detection, fluid and electrolyte transport, and mucus elevation. SMG secrete antimicrobial molecules and most of the airway surface fluid. Despite the unique functional roles of surface epithelia and SMG, little is known about the differences in gene expression and cellular metabolism that orchestrate the specialized functions of these epithelial compartments. To approach this problem, we performed large-scale transcript profiling using epithelial cell samples obtained by laser capture microdissection (LCM) of human bronchus specimens. We found that SMG expressed high levels of many transcripts encoding known or putative innate immune factors, including lactoferrin, zinc α-2 glycoprotein, and proline-rich protein 4. By contrast, surface epithelial cells expressed high levels of genes involved in basic nutrient catabolism, xenobiotic clearance, and ciliated structure assembly. Selected confirmation of differentially expressed genes in surface and SMG epithelia demonstrated the predictive power of this approach in identifying genes with localized tissue expression. To characterize metabolic differences between surface epithelial cells and SMG, immunostaining for a mitochondrial marker (isocitrate dehydrogenase) was performed. Because greater staining was observed in the surface compartment, we predict that these cells use significantly more energy than SMG cells. This study illustrates the power of LCM in defining the roles of specific anatomic features in airway biology and may be useful in examining how disease states alter transcriptional programs in the conducting airways.
Keywords: airway epithelia, microarray, submucosal gland
CLINICAL RELEVANCE
Unique transcriptional differences between airway surface and submucosal gland epithelia were identified in this study, expanding our knowledge of airway physiology. Many genes identified are clinically relevant because they metabolize cigarette smoke.
The airways maintain a dynamic interface with the environment. The continuity of the airway epithelium serves as a physical barrier to inhaled pathogens, while the secreted protein and peptide products of the tissue, in concert with ciliary function, support mucociliary clearance and host defense. The epithelium of the intrapulmonary airways contributes to pulmonary innate immunity as a first responder and as an important site of signal amplification. Through a variety of receptor systems, the surface epithelial cells sense and respond to inhaled chemical, microbial, thermal, and particulate stimuli to maintain health. Perturbation of the function of this tissue is involved in the pathogenesis of several diseases, including asthma, chronic obstructive pulmonary disease (COPD), and cystic fibrosis (CF).
The epithelial tissue of the conducting airways includes many specialized cell types and the spatial division of functions. Two anatomically distinct conducting airway compartments are the airway surface epithelium and submucosal glands (SMG). The surface epithelium of the cartilaginous conducting airways is composed of ciliated, nonciliated, goblet, and basal cell types. SMG consist of four histologically distinct regions: ciliated ducts, collecting ducts, mucous tubules, and serous acini and tubules (1, 2). Increasing evidence indicates that secretions from the submucosal glands are a major source of the liquid and mucins that line the airway surface (1, 2).
We hypothesized that the specialized functions of these epithelial regions are, in part, transcriptionally regulated. To test this hypothesis, we harvested the two distinct histologic compartments using laser capture microdissection (LCM). Large-scale expression profiling was then performed using high-density custom Affymetrix genechips (Affymetrix, Santa Clara, CA). This approach revealed a number of differentially expressed gene products that may contribute to the unique functions of these anatomical regions.
MATERIALS AND METHODS
Tissue Isolation
Primary and secondary bronchi were dissected within 12 to 14 hours after death from human donor lung tissues. The bronchi were opened by making a longitudinal incision through the posterior membranous region of the airway. A scalpel was used to make shallow cuts through the epithelium, and fine forceps were then used to strip the surface epithelium from underlying submucosal tissues. These surface epithelium strips were placed in a microfuge tube, frozen in liquid nitrogen, and stored at −80°C. Approximately 1-mm cubes of submucosal tissues (which contained the submucosal glands) were dissected from the posterior membranous region of the airways, placed in microfuge tubes, frozen in liquid nitrogen, and stored at −80°C. Multiple surface epithelium and submucosal tissue samples were collected from each specimen. Total RNA was isolated from representative epithelial and submucosal tissue samples and electrophoresed to verify RNA integrity. Only samples with intact RNA were used for further analysis. This study was approved by the institutional review board of the University of Iowa.
LCM
Frozen human bronchus tissues (stripped surface epithelium and submucosal tissues) were obtained from six individuals postmortem. The subject demographics are shown in Table 1. Five-micrometer thick frozen sections were prepared and thaw-mounted onto glass slides. Slides were stained with aqueous Mayer's hematoxylin, dehydrated through an alcohol series, and air-dried. Sections used for collecting surface epithelial cells and submucosal gland cells were processed within 1 hour of LCM. Prep Strips (Arcturus, Mountain View, CA) were used to remove debris from the sections before LCM. Surface epithelial cells and SMG were collected with a Pixcell II LCM system and CapSure Macro LCM caps (Arcturus). LCM was performed at ×40 magnification using a 7.5-μm laser beam, laser power of 75 mV, and a laser duration of 1.5 ms. Typically, approximately 12,000 pulses were collected per specimen on four to five different caps. The microdissected samples were obtained from about 10 to 15 sequential tissue sections for each specimen. After tissue collection was completed for each cap, the film was peeled from the cap and placed in a microfuge tube that contained 100 μl lysis buffer. The tube, usually containing five films from five caps, was heated at 42°C for 30 minutes, then stored at −80°C. Tubes containing the LCM sample extracts were stored until specimens from all of the donors were collected.
TABLE 1.
SUBJECT DEMOGRAPHICS
| Sample # | Age (yr) | Ethnicity | Sex | Cause of Death |
|---|---|---|---|---|
| A | 11 | White | M | Respiratory arrest, asthma |
| B | 44 | White | F | Intracranial hemorrhage; smoker |
| C | 17 | White | M | Head trauma from motor vehicle accident |
| D | 36 | White | M | Head trauma from motor vehicle accident; smoker |
| E | 46 | African American | M | Anoxia after seizure |
| F | 48 | White | F | Intracranial hemorrhage; smoker |
RNA Sample Preparation and cDNA Amplification
Total RNA was simultaneously isolated from all LCM preparations using the RNAqueous-4 PCR kit (Ambion, Austin, TX). Thirty nanograms of total RNA template were used to produce amplified cDNA using the Ovation Biotin RNA Amplification System, v2 (#3100; NuGEN Technologies, Inc., San Carlos, CA) following the manufacturer's protocol. Amplified cDNA product was purified with Zymo Research DNA Clean and Concentrator-25 (Zymo Research, Orange, CA). Amplified cDNA (3.75 μg) were processed using the FL-Ovation cDNA Biotin Module v2 (NuGEN Technologies #4200) to produce biotin-labeled antisense cDNA in 50- to 100-bp fragments.
After denaturation at 99°C for 2 minutes, 2.2 μg of fragmented, labeled cDNA were combined with hybridization control oligomer (b2) and control cRNAs (BioB, BioC, BioD, and CreX) in hybridization buffer and hybridized to a custom Affymetrix microarray. The microarray used in this study (HsAirway) included 23,002 probe sets derived from sequencing of cDNA libraries prepared from human lung, primary airway epithelial cells, and human alveolar macrophages (3). After an 18-hour incubation at 45°C, the arrays were washed, stained with streptavidin-phycoerythrin (Molecular Probes, Inc., Eugene, OR), and then amplified with an anti-streptavidin antibody (Vector Laboratories, Inc., Burlingame, CA) using the Fluidics Station 450 (Affymetrix). Arrays were scanned with the Affymetrix Model 3000 scanner and data collected using GeneChip operating software (GCOS) v1.4. Each sample and hybridization underwent a quality control evaluation, including percentage of probe sets reliably detecting between 40 and 60% present call and 3′-5′ ratio of the GAPDH gene less than 3.
Evaluation and Normalization of Affymetrix GeneChip Data
Microarray hybridizations were normalized using the RMA (robust multi-chip averaging) (4) method from Bioconductor (http://www.bioconductor.org; 5) to obtain summary expression values for each probe set. Gene expression levels were analyzed on a logarithmic scale. Additional quality assessment was performed using methods provided in Bioconductor. Specifically, an RNA degradation plot was constructed, and any samples with significant degradation were excluded from analysis. Similarly, each hybridization image was inspected for gross errors or corruption. Finally, a sample-level hierarchical clustering plot was constructed and assessed for outliers. The ANOVA statistical test was used to identify differentially expressed genes. Gene Set Enrichment Analysis (GSEA) (6, 7) was used to identify global changes in expression level for gene sets from MSigDB (Molecular Signatures Database, www.broad.mit.edu/gsea/msigdb/index.jsp) as well as several custom gene sets.
Confirmation of Array Data
Immunohistochemistry.
Human trachea and bronchus tissue was fixed in 4% paraformaldehyde and embedded in paraffin. Tissues were sectioned at 8 μm thickness and mounted on glass slides. Paraffin sections were deparaffinized, rehydrated, and washed with PBS. High temperature antigen retrieval was performed on sections (Antigen Unmasking Solution, cat. # H-3300; Vector Laboratories). After washing in PBS, endogenous peroxidases were inactivated, and sections were washed again in PBS. To block nonspecific antigen binding, the sections were pre-incubated for 30 minutes with normal serum obtained from the species in which the secondary antibody was raised. The sections were then incubated overnight at 4°C with selected primary antibodies diluted in blocking buffer, washed in PBS, and incubated with a biotinylated secondary antibody (Elite ABC kit, Vectastain; Vector Laboratories). Binding of the secondary antibody was detected with a peroxidase-avidin-biotin mixture (Elite ABC kit, Vectastain; Vector Laboratories). Sections were then incubated with a working solution of 3,3′-diaminobenzidine (DAB) and hydrogen peroxide. Finally, sections were counterstained with hematoxylin, mounted, viewed by light microscopy (Olympus BX60; Olympus, Center Valley, PA), and photographed (Olympus DP70 digital camera; Olympus). Primary antibodies specific for epithelial staining included mouse mAb to glutathione S-transferase α [38H11] (GSTα, ab49483; Abcam, Cambridge, MA), mouse mAb to NAD(P)H quinone oxidoreductase 1 [A180] (NQO1, ab28947; Abcam), rabbit polyclonal Ab to Cyp4B1 (HPA004331; Atlas Antibodies, Stockholm, Sweden), and rabbit polyclonal Ab to NAD+-dependent isocitrate dehydrogenase subunit γ (IDH3G, HPA002017; Atlas Antibodies). Primary antibodies specific for submucosal gland staining included rabbit polyclonal Ab to zinc-α-2-glycoprotein (ZAG, sc-13585; Santa Cruz Biotechnology), and rabbit polyclonal Ab to α-lactoferrin (ab24264–100; Abcam). Control primary antibodies included mouse IgG (MAB002; R&D Systems, Minneapolis, MN) and rabbit IgG (AB-105-C; R&D Systems).
RESULTS
LCM Facilitates Separate Analysis of Surface Epithelial Cell and SMG Gene Expression Profiles
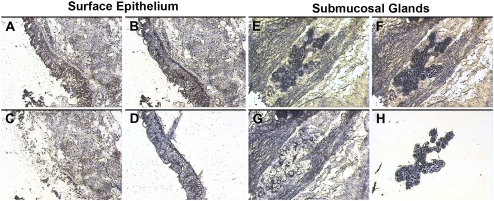
The capture of the targeted bronchial surface epithelial and submucosal gland cell populations was monitored and documented by photomicroscopy (Figure 1). The procedure yielded relatively pure populations of surface epithelia (Figure 1D) and submucosal glands (Figure 1H). The harvested tissues yielded RNA with sufficient quality and quantity for amplification, cRNA synthesis, and microarray hybridization procedures. While not the goal of the present study, it is clear from these data that LCM could also be adapted to harvest more selective cell populations from these samples for study, such as ciliated surface epithelial cells, ciliated duct epithelial cells, or populations of submucosal gland serous or mucous cells.
Figure 1.
Laser capture microdissection (LCM) of surface epithelia and submucosal glands (SMG). Representative images from the tissue isolation process are depicted for surface epithelia (left, A–D) and submucosal glands (right, E–H). A and E represent respective tissues before dissection. Captured cells attached to the LCM cap film are demonstrated for surface epithelia (B) and submucosal glands (F). After removal of the LCM cap, unselected tissue remains behind (C and G). The LCM process yields populations of surface epithelial cells (D) and submucosal gland cells (H) on the LCM cap film.
Microarray Hybridization Reveals Differential Gene Expression in Distinct Anatomical Regions
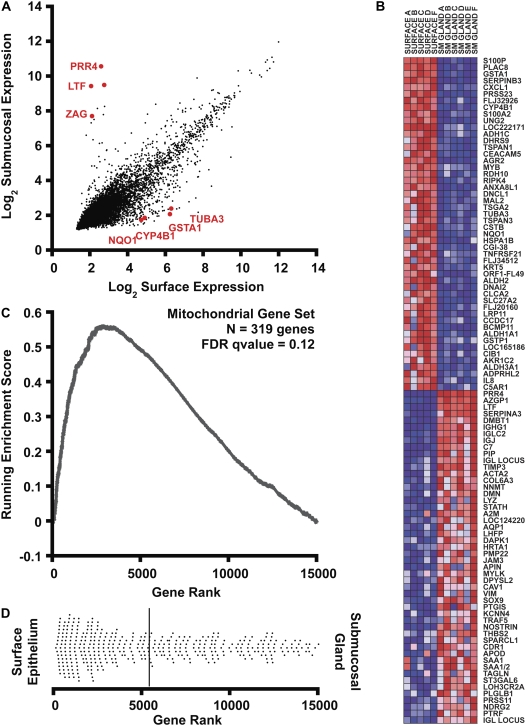
A total of six paired surface and submucosal gland samples were processed and hybridized to the HsAirway genechip. With the exception of one surface epithelium sample (Sample E) that showed signs of RNA degradation in the analysis of hybridization, all remaining samples passed quality control and were included in our analysis. After data collection, microarray data were normalized by RMA (4). In Figure 2A, we show that the level of expression in the two tissue compartments is similar for most of the individual genes studied after normalization and log2 transformation. However, there were many genes that deviated significantly from this trend. We separately list individual transcripts with statistically significant (P < 0.001) differences in expression and at least 4-fold enrichment in either surface epithelium (Table 2) or in SMG (Table 3).
Figure 2.
Bioinformatic analysis of gene expression. (A) Correlation of gene expression levels in surface epithelia (x axis) with submucosal glands (y axis). Expression signals were log2 transformed and normalized by the RMA procedure. Some genes selected for further analysis are displayed in red. (B) Gene set enrichment analysis (GSEA) heatmap output for the 100 most differentially expressed transcripts in human bronchus surface epithelium and submucosal glands. The columns represent unique array hybridizations using either surface epithelial RNA (SURFACE) or submucosal gland RNA (SM GLAND). The donor number is also indicated after the tissue type. Red indicates high expression and blue low expression. Probe IDs are listed on the right of the figure. (C) Metabolic pathway upregulation (mitochondrial genes) in surface epithelium assessed by gene set enrichment analysis. A total of 14,989 genes were rank ordered by relative expression in surface epithelium, and an enrichment score (ES) was calculated using these rank order data weighted by a correlation coefficient using GSEA software (7). The upward deviation of the ES curve demonstrates the positive enrichment of the gene set in surface epithelial cells. (D) The dot plot represents the ranks of individual genes belonging to the mitochondrial gene set (n = 319) within the cumulative list of genes. The distribution is skewed to the left, indicating enrichment of mitochondrial transcripts in surface epithelial cells. The solid bar represents the mean rank of this gene set.
TABLE 2.
TRANSCRIPTS ENRICHED IN SURFACE EPITHELIAL CELLS
| Probe Name | Gene | Gene Title | Max Fold Change | P Value |
|---|---|---|---|---|
| 204351_at | S100P | S100 calcium binding protein P | 41.0 | 8.42×10−8 |
| CD365047_s_at, 204268_at | S100A2 | S100 calcium binding protein A2 | 21.2 | 6.67×10−5 |
| 233157_x_at | FLJ32926 | Hypothetical protein FLJ32926 | 17.3 | 3.94×10−5 |
| 204470_at | CXCL1 | Chemokine (C-X-C motif) ligand 1 | 16.3 | 2.89×10−5 |
| 222271_at | — | Transcribed locus | 15.6 | 2.20×10−4 |
| 203924_at | GSTA1 | Glutathione S-transferase A1 | 15.6 | 1.26×10−5 |
| 226961_at | LOC222171 | Hypothetical protein LOC222171 | 15.2 | 1.28×10−6 |
| 209118_s_at, 202154_x_at | TUBA3 | Tubulin, alpha 3 | 13.2 | 6.00×10−4 |
| 206262_at | ADH1C | Alcohol dehydrogenase 1C, gamma polypeptide | 12.1 | 3.51×10−5 |
| 201884_at | CEACAM5 | Carcinoembryonic antigen-related cell adhesion molecule 5 | 11.6 | 3.82×10−4 |
| 202458_at | PRSS23 | Protease, serine, 23 | 11.2 | 2.91×10−6 |
| BU677128_x_at | ALOX15 | Arachidonate 15-lipoxygenase | 11.0 | 5.57×10−8 |
| 209720_s_at | SERPINB3 | Serine (or cysteine) proteinase inhibitor, clade B, member 3 | 10.6 | 1.32×10−8 |
| 230093_at | TSGA2 | Testis specific A2 homolog (mouse) | 10.3 | 3.80×10−4 |
| 201005_at | CD9 | CD9 antigen (p24) | 10.0 | 3.33×10−5 |
| 208680_at | PRDX1 | Peroxiredoxin 1 | 9.9 | 2.85×10−4 |
| 200824_at | GSTP1 | Glutathione S-transferase pi | 8.8 | 1.39×10−4 |
| 204798_at | MYB | v-myb myeloblastosis viral oncogene homolog (avian) | 8.8 | 5.25×10−5 |
| 219014_at | PLAC8 | Placenta-specific 8 | 8.7 | 1.87×10−7 |
| 200878_at | EPAS1 | Endothelial PAS domain protein 1 | 8.5 | 2.83×10−5 |
| 201650_at | KRT19 | Keratin 19 | 8.0 | 3.53×10−4 |
| CB853184_s_at | MUC16 | Mucin 16, cell surface associated | 7.7 | 8.51×10−4 |
| 200703_at | DNCL1 | Dynein, cytoplasmic, light polypeptide 1 | 7.6 | 4.17×10−4 |
| 212224_at | ALDH1A1 | Aldehyde dehydrogenase 1 family, member A1 | 7.4 | 3.28×10−4 |
| 208659_at | CLIC1 | Chloride intracellular channel 1 | 7.3 | 1.54×10−4 |
| CA944500_at | TNFRSF21 | Tumor necrosis factor receptor superfamily, member 21 | 7.2 | 1.47×10−4 |
| 201201_at | CSTB | Cystatin B (stefin B) | 7.2 | 9.73×10−5 |
| 210096_at | CYP4B1 | Cytochrome P450, family 4, subfamily B, polypeptide 1 | 7.1 | 7.01×10−7 |
| 201468_s_at | NQO1 | NAD(P)H dehydrogenase, quinone 1 | 6.9 | 7.47×10−4 |
| 226021_at | RDH10 | Retinol dehydrogenase 10 (all-trans) | 6.8 | 7.22×10−5 |
| 201012_at | ANXA1 | Annexin A1 | 6.7 | 9.76×10−6 |
| 225325_at | FLJ20160 | FLJ20160 protein | 6.6 | 1.76×10−4 |
| CB528441_at | TSC22D1 | TSC22 domain family, member 1 | 6.6 | 8.80×10−4 |
| 219795_at | SLC6A14 | Solute carrier family 6 (amino acid transporter), member 14 | 6.2 | 4.45×10−5 |
| 201820_at | KRT5 | Keratin 5 | 6.2 | 1.12×10−4 |
| 201563_at | SORD | Sorbitol dehydrogenase | 6.0 | 2.02×10−5 |
| 210021_s_at | UNG2 | Uracil-DNA glycosylase 2 | 6.0 | 3.64×10−6 |
| 221484_at | B4GALT5 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 | 6.0 | 2.79×10−4 |
| 209114_at | TSPAN1 | Tetraspanin 1 | 5.9 | 3.09×10−5 |
| 200627_at | PTGES3 | Prostaglandin E synthase 3 (cytosolic) | 5.8 | 9.26×10−4 |
| 211657_at | CEACAM6 | Carcinoembryonic antigen-related cell adhesion molecule 6 | 5.7 | 8.42×10−5 |
| 232968_at | FANK1 | Fibronectin type III and ankyrin repeat domains 1 | 5.6 | 9.36×10−4 |
| 210272_at | CYP2B7P1 | Cytochrome P450, family 2, subfamily B, polypeptide 7 pseudogene 1 | 5.6 | 1.29×10−6 |
| CA310634_s_at, 209186_at | ATP2A2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | 5.6 | 5.79×10−5 |
| 223000_s_at | F11R | F11 receptor | 5.6 | 6.38×10−4 |
| 201425_at | ALDH2 | Aldehyde dehydrogenase 2 family (mitochondrial) | 5.5 | 7.77×10−7 |
| 225060_at | LRP11 | Low-density lipoprotein receptor–related protein 11 | 5.4 | 3.81×10−4 |
| 221215_s_at | RIPK4 | Receptor-interacting serine-threonine kinase 4 | 5.4 | 1.05×10−5 |
| 216609_at | TXN | Thioredoxin | 5.2 | 9.57×10−5 |
| 200614_at | CLTC | Clathrin, heavy polypeptide (Hc) | 5.2 | 6.70×10−4 |
| 200642_at | SOD1 | Superoxide dismutase 1, soluble | 5.2 | 9.78×10−4 |
| 1562921_at | EP300 | E1A binding protein p300 | 5.1 | 4.12×10−4 |
| BU727905_s_at | COX6A1 | Cytochrome c oxidase subunit VIa polypeptide 1 | 5.1 | 5.42×10−4 |
| 228969_at | AGR2 | Anterior gradient 2 homolog (Xenopus laevis) | 5.1 | 9.35×10−7 |
| 201266_at | TXNRD1 | Thioredoxin reductase 1 | 5.1 | 4.15×10−4 |
| 224650_at | MAL2 | Mal, T-cell differentiation protein 2 | 5.0 | 3.04×10−5 |
| BM975341_s_at | CTNNA1 | Catenin (cadherin-associated protein), alpha 1 | 4.9 | 1.96×10−4 |
| 204734_at | KRT15 | Keratin 15 | 4.8 | 3.91×10−4 |
| 201030_x_at | LDHB | Lactate dehydrogenase B | 4.8 | 5.88×10−4 |
| 200972_at | TSPAN3 | Tetraspanin 3 | 4.7 | 2.61×10−4 |
| 204363_at | F3 | Coagulation factor III (thromboplastin) | 4.7 | 5.04×10−6 |
| 209154_at | TAX1BP3 | Tax1 binding protein 3 | 4.7 | 4.99×10−4 |
| 202581_at | HSPA1B | Heat shock 70-kD protein 1B | 4.7 | 2.06×10−4 |
| 219799_s_at | DHRS9 | Dehydrogenase/reductase (SDR family) member 9 | 4.6 | 1.10×10−4 |
| BM989324_at | IK | IK cytokine, down-regulator of HLA II | 4.6 | 8.36×10−4 |
| CA450425_x_at | AKR1C3 | Aldo-keto reductase family 1, member C3 | 4.6 | 5.79×10−4 |
| CB529214_at | HNRPC | Heterogeneous nuclear ribonucleoprotein C (C1/C2) | 4.5 | 7.55×10−4 |
| BU683199_s_at | DHCR24 | 24-dehydrocholesterol reductase | 4.5 | 4.65×10−5 |
| 212876_at | B4GALT4 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 | 4.4 | 2.12×10−5 |
| 201951_at | ALCAM | Activated leukocyte cell adhesion molecule | 4.4 | 1.47×10−4 |
| CA450425_at | AKR1C2 | Aldo-keto reductase family 1, member C2 | 4.4 | 9.14×10−4 |
| 1555961_a_at | HINT1 | Histidine triad nucleotide binding protein 1 | 4.3 | 1.71×10−4 |
| 201487_at | CTSC | Cathepsin C | 4.3 | 2.52×10−4 |
| 202804_at | ABCC1 | ATP-binding cassette, sub-family C, member 1 | 4.3 | 1.79×10−4 |
| 224707_at | ORF1-FL49 | Putative nuclear protein ORF1-FL49 | 4.3 | 4.78×10−4 |
| 209211_at | KLF5 | Kruppel-like factor 5 (intestinal) | 4.2 | 2.94×10−4 |
| 217728_at | S100A6 | S100 calcium binding protein A6 (calcyclin) | 4.2 | 7.65×10−5 |
| 217528_at | CLCA2 | Chloride channel, calcium activated, family member 2 | 4.1 | 9.17×10−4 |
| 202071_at | SDC4 | Syndecan 4 (amphiglycan, ryudocan) | 4.1 | 8.41×10−5 |
TABLE 3.
TRANSCRIPTS ENRICHED IN SUBMUCOSAL GLANDS
| Probe Name | Gene | Description | Maximum Fold Change | P Value |
|---|---|---|---|---|
| 204919_at | PRR4 | Proline rich 4 (lacrimal) | 288.3 | 7.53×10−11 |
| 202018_s_at, CB306796_at | LTF | Lactotransferrin | 189.5 | 5.55×10−9 |
| BU684206_s_at | SERPINA3 | Serpin peptidase inhibitor, clade A, member 3 | 61.8 | 6.43×10−6 |
| 209309_at | AZGP1 | α-2-glycoprotein 1, zinc | 55.5 | 2.70×10−8 |
| 209138_x_at | IGLC2 | Immunoglobulin lambda variable 3-21 | 51.3 | 1.27×10−5 |
| 202376_at | SERPINA3 | Serine (or cysteine) proteinase inhibitor, clade A, member 3 | 45.0 | 5.13×10−6 |
| 212592_at | IGJ | Immunoglobulin J polypeptide | 43.7 | 1.95×10−5 |
| CA447563_s_at | IGHG1 | Immunoglobulin heavy constant gamma 1 | 38.8 | 1.31×10−5 |
| 208250_s_at | DMBT1 | Deleted in malignant brain tumors 1 | 20.8 | 1.88×10−5 |
| 215121_x_at | IGL | Immunoglobulin lambda locus | 14.2 | 3.71×10−4 |
| 214836_x_at | — | HRV Fab N8-VL | 8.4 | 2.93×10−4 |
| 217757_at | A2M | α-2-macroglobulin | 8.3 | 8.26×10−4 |
| 202992_at | C7 | Complement component 7 | 6.9 | 1.59×10−4 |
| 201150_s_at | TIMP3 | Tissue inhibitor of metalloproteinase 3 | 6.1 | 8.18×10−5 |
Gene Set Enrichment Analysis
Because the surface epithelium and submucosal glands have different environmental exposures and functional roles, we hypothesized that the transcriptional profiles would reflect differential regulation of cellular and biochemical pathways in the two compartments. To test this hypothesis, we employed Gene Set Enrichment Analysis (GSEA), a technique that compares microarray data for curated sets of genes (7). Gene sets are classified on the basis of cytogenetic, functional, or regulatory motif similarity.
An example of the GSEA method is illustrated in Figures 2B to 2D, with special emphasis on enrichment data for the mitochondrial metabolism gene set (Figures 2C and 2D). Briefly, the 23,002 probe sets that belong to the microarray were reduced to eliminate all unknown or redundant genes. For the remaining genes (n = 14,989), a correlation score was determined based on the relative expression in surface epithelial cells versus SMG. The remaining genes were subsequently rank ordered by their correlation score into a gene list (L). Within L, surface epithelial–enriched transcripts appeared first and SMG-enriched transcripts were last. The heatmap (Figure 2B) displays the 100 individual genes exhibiting the greatest correlation with either biological state. Red and blue colors represent high and low expression, respectively.
To compare the expression of a predefined gene set between the two biological states, the positions of all individual genes comprising the gene set were located within the ranked list L. For example, the mitochondrial gene set (S) contained 391 genes within L. The locations of these genes within L are presented as a dot plot in Figure 2D. Although the mitochondrial genes were distributed widely throughout the ranked list, most genes ranked highly for surface epithelial enrichment. To quantify how well the mitochondrial gene set was enriched in the surface epithelial cells, an enrichment score (ES) was calculated. The computation and statistical validation of ES were fully discussed previously (7). The ES is a running sum that starts and ends at zero, by definition. A large deviation of the ES above zero at positions between 1 and N would represent gene set enrichment in the epithelium, whereas a negative deviation would reflect enrichment in the SMG. Statistical significance was assessed by permutation of the biological sample labels and estimating how frequently an ES more extreme than the observed maximal ES would occur by random chance. Figure 2C shows the running ES for mitochondrial genes plotted versus gene rank. This curve shows significant deviation above zero, consistent with pathway enrichment in surface epithelia. Using the GSEA analysis package (available at www.broad.mit.edu/gsea/), we identified 76 gene sets enriched in surface epithelial cells at a false discovery rate (FDR) less than 25%. Table 4 presents the most highly enriched functional gene sets correlating with the surface epithelium. Among the most enriched gene sets in surface epithelia were catabolic pathways, electron transport, and xenobiotic detoxification. By contrast, no gene sets were enriched in submucosal gland epithelia at an FDR less than 25%.
TABLE 4.
GENE SETS WITH SIGNIFICANT ENRICHMENT IN SURFACE EPITHELIUM SAMPLES
| Gene Set | N | Enrichment Score | Nominal P Value | FDR q- Value | FWER P Value |
|---|---|---|---|---|---|
| Urea cycle and metabolism of amino groups | 15 | 0.820 | 0.000 | 0.115 | 0.046 |
| Arginine and proline metabolism | 34 | 0.756 | 0.000 | 0.074 | 0.065 |
| Glutathione metabolism | 42 | 0.625 | 0.000 | 0.173 | 0.276 |
| Glycolysis and gluconeogenesis | 48 | 0.693 | 0.000 | 0.136 | 0.359 |
| Tryptophan metabolism | 39 | 0.613 | 0.002 | 0.123 | 0.37 |
| Mitochondria | 391 | 0.561 | 0.000 | 0.122 | 0.395 |
| Glycerolipid metabolism | 44 | 0.561 | 0.000 | 0.120 | 0.447 |
| Bile acid biosynthesis | 19 | 0.792 | 0.000 | 0.108 | 0.565 |
| Phenylalanine metabolism | 24 | 0.782 | 0.000 | 0.104 | 0.565 |
| Stilbene, coumarine, and lignin biosynthesis | 24 | 0.782 | 0.000 | 0.099 | 0.565 |
| Drug resistance and metabolism | 92 | 0.464 | 0.000 | 0.094 | 0.573 |
| Down-regulated by glutamine | 262 | 0.540 | 0.000 | 0.087 | 0.578 |
| Fatty acid metabolism | 40 | 0.641 | 0.004 | 0.086 | 0.584 |
Definition of abbreviations: FDR, false discovery rate; FWER, familywise error rate.
Transcripts Enriched in Surface Epithelial Cells
A structural difference between the two tissue compartments included in this analysis is the abundance of ciliated cells in the surface epithelium. We predicted that transcripts encoding components of cilia would be enriched in the surface epithelium samples. Cilia are built upon a 9+2 microtubular structure of α- and β-tubulin heterodimers (8). Dyneins, tubulin-dependent ATPases, are ciliary motor proteins. Several mRNAs encoding dynein isoforms were enriched in the surface epithelium samples, including DNAH5, DNCL1, DNAI2, DNCL2B, DNAI1, and DNALI1. Two tubulin isoforms, TUBA3 and TUBB3, were also differentially expressed. We observed that expression of testis-specific A2 homolog TSGA2 (meichroacidin) was highly enriched in the surface epithelium. This gene was recently found to encode a radial spoke protein in the axoneme of airway cilia (9).
Gene products that metabolize xenobiotic molecules were highly enriched in the surface epithelium. These included two that encode aldehyde dehydrogenases, ALDH2 and ALDH3A1, a gene encoding a quinone reductase (NQO1), and a cytochrome p450 isoform (CYP4B1). In addition, a number of cell surface receptors were up-regulated in surface epithelia, including the CXCL1 and C5AR1 chemokine receptors, CEACAM5, and two tetraspanin isoforms (TSPAN1 and TSPAN3). We also observed a large number of gene products with surface enrichment that are either novel or have no known role in airway surface epithelium function.
Transcripts Enriched in Submucosal Glands
Submucosal glands produce an array of secreted proteins that kill invading pathogens, yet minimize damage to host tissue (10). We found that submucosal glands expressed several genes encoding secreted proteins at levels that were orders of magnitude higher than in the surface epithelium. Importantly, we observed enrichment of transcripts for well-known submucosal gland gene products (e.g., lactoferrin, lysozyme, and α1-antitrypsin), thus confirming the effectiveness of LCM in recovering populations of SMG epithelia. We postulate that many of the other transcripts enriched in SMG also play roles in innate immunity. For instance, lacrimal proline rich protein (also known as LPRP, PROL4, PRP-4, and PRR4) was highly enriched in SMG, and was previously reported to be expressed at high levels in glands present in oral and conjunctival mucosa (11, 12).
In addition to secreting innate immune effectors, a growing body of literature now recognizes that a major function of SMG is secretion of approximately 95% of the fluid and electrolytes in airway surface liquid (1). Disruption of fluid and electrolyte transport in SMG (as in cystic fibrosis) can result in changes in airway surface liquid composition and the rheologic properties of airway mucus (13). Because SMG play such an important role in fluid secretion, we were interested in identifying which fluid and electrolyte transporters were highly expressed in this tissue fraction. Differentially expressed genes of this class included aquaporin 1 (AQP1) (14), and a calcium activated potassium channel (KCNN4) (15). Transcripts for enzymes involved in post-translational modification and transport, including caveolin and a sialyltransferase enzyme, ST3 β-galactoside α-2,3 sialyl transferase 6 (ST3GAL6), were enriched in the SMG cells. ST3GAL6 may play a role in synthesis of mucins, an important function of SMG.
The acini of SMG are surrounded by myoepithelial cells, which can be stimulated to contract and release fluid and secretions into the airway (16). Three genes known to be expressed in smooth muscle were highly enriched in the SMG samples, including myosin light chain kinase (MYLK), α-2 actin (ACTA2), and transgelin (TAGLN). SMG preparations also had significant enrichments of various cytoskeletal and extracellular matrix proteins, including desmuslin (DMN), vimentin (VIM), and collagen type VI α-3 (COL6A3). An important nonepithelial resident of respiratory submucosal tissue is mucosal-associated lymphoid tissue (MALT), including lymphocytes and plasma cells that closely neighbor SMG (17). We observed that several immunoglobulin genes were enriched in the SMG preparations. For instance, IGLC2, the immunoglobulin lambda variable chain, was among the highest expressed transcripts in the SMG. This trend was also observed for the immunoglobulin heavy chain IGHG1.
GSEA Reveals Transcriptional Basis for Increased Energy Consumption by Surface Epithelial Cells
A critical role of the surface epithelium is the clearance of inhaled particles trapped in airway mucus, a function that requires the constitutive and coordinated activity of ciliated columnar epithelial cells (18). Because this activity requires energy, we hypothesized that surface epithelial cells transcribe genes involved in energy production more than submucosal cells. We found that gene sets encoding pathways involved in catabolism of basic nutrients (sugars, amino acids, and lipids) were highly enriched in the surface epithelium (consistent with data presented in Figures 2C and 2D). When we investigated a more focused catabolic pathway (glycolysis and the Krebs cycle), we observed that many genes with highest correlation to surface epithelia corresponded to known regulatory steps in these pathways. For instance, phosphofructokinase, pyruvate kinase, and pyruvate dehydrogenase were all enriched in the surface epithelium (see Figure E1 in the online supplement). By contrast, probes that correlated better with SMG included fructose-(1,6)-bisphosphatase and pyruvate dehydrogenase kinase, genes encoding enzymes that down-regulate glycolysis.
Drug and Xenobiotic Response
The surface epithelium performs several other important functions that require energy. Among these is the metabolism and clearance of inhaled xenobiotic molecules. We observed surface enrichment of many xenobiotic responsive genes, including xenobiotic-responsive transcription factors such as the aryl hydrocarbon receptor (AHR), phase I enzymes of the CYP450 class (CYP3A5 and CYP2E1), epoxide hydrolases, glutathione S-transferases, and drug exporters such as ABCC1.
Antioxidant Enzymes
Gene set enrichment analysis revealed that several enzymes involved in the clearance of reactive oxygen species (ROS) were expressed at higher levels in surface epithelial cells. ROS are produced as a consequence of a high metabolic rate and exposure to environmental toxins. In addition, the airway epithelia generate hydrogen peroxide at the apical surface as an innate immune mechanism (19–21). Consequently, we observed that surface epithelial cells express a battery of genes involved in the removal of peroxides and other ROS. The transcripts encoding peroxiredoxins 1, 2, 5, and 6 as well as their electron donor substrates (thioredoxins) were all highly enriched in the surface epithelium samples.
Mitochondrial Distribution Is Highly Skewed toward Surface Epithelial Cells
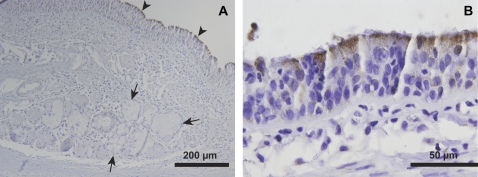
GSEA of our microarray data demonstrated that genes involved in mitochondrial metabolism are expressed at higher levels in surface epithelia compared with submucosal glands. To test whether mitochondria are more abundant in the surface epithelium, we performed immunohistochemical staining for the γ subunit of the NAD+-dependent isocitrate dehydrogenase complex (isocitrate dehydrogenase 3-γ, IDH3G). This enzyme is a ubiquitously expressed mitochondrial matrix protein in mammals (22, 23), and catalyzes the oxidative decarboxylation of isocitrate. This reaction is an irreversible and energy-yielding step in the Krebs cycle. Thus, IDH3G is not only a marker of mitochondria, but also an indicator of aerobic activity and energy production. We found that the tissue distribution of IDH3G is highly skewed, with significantly greater immunostaining observed in surface epithelial cells compared with submucosal gland cells (Figure 3A). IDH3G immunostaining was most robust in the apical cytoplasm of ciliated cells (Figures 3A and 3B).
Figure 3.
Mitochondrial staining is greater in conducting airway surface epithelial cells compared with submucosal gland cells. Immunohistochemical staining of bronchus tissue is presented for isocitrate dehydrogenase (IDH3G), a ubiquitous Krebs cycle enzyme with known localization to the mitochondrial matrix. (A) IDH3G immunostaining is highly enriched in surface epithelial cells (arrowheads), with less intense staining observed in SMG epithelial cells (arrows). (B) IDH3G staining was most intense immediately below the apical surface of ciliated epithelial cells. Representative of n = 3 human donor bronchi examined.
Gene Sets Enriched in the Submucosal Glands
While we did observe that a number of individual genes were highly expressed in the submucosal glands relative to the surface epithelium, GSEA did not reveal any known pathways or classes of transcripts that were expressed at significantly higher levels in the SMG. This could reflect the specialized nature of these cells, which may not be accounted for in the functional gene sets curated to date. Glandular cells have proved more difficult to culture than surface epithelial cells (2), and consequently less is known about them.
Confirmation of Specific Genes
A subset of differentially regulated gene products was selected to validate the results of the microarray study. Here we focused on a limited subset of genes that were (1) expressed at significant levels, with over 10-fold greater expression in either surface or submucosal gland epithelia; and (2) statistically reproducible differential expression (P < 0.001). To validate the results for these selected gene products, we performed immunohistochemical staining of human trachea or bronchus sections and compared the expression levels in the two compartments. Isotype control immunoglobulin (IgG) was used to control for the specificity of secondary antibody hybridization. We also noted expression in nonepithelial tissues, when applicable.
Surface Epithelial–Enriched Transcripts
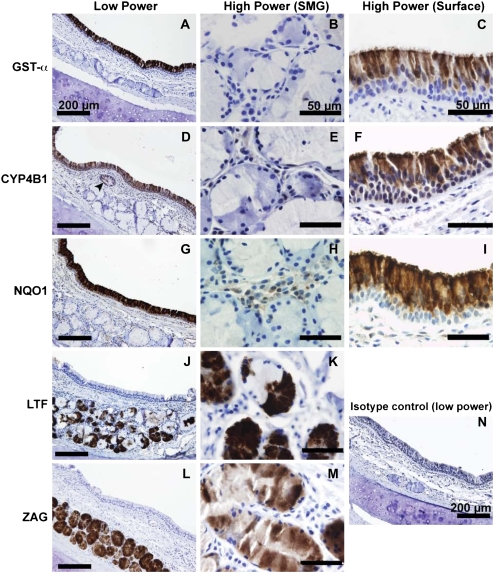
Glutathione S-transferase α (GST-α) is preferentially expressed in surface epithelial cells (Figure 4A), and was used as a positive control in this study. GST-α was not observed within the ducts or acini of the SMG, and was also absent in the glandular interstitium (Figure 4B). Within the surface epithelium, we observed GST-α is localized principally to ciliated columnar cells, but not the basal cell layer or goblet cells, consistent with earlier observations (Figure 4C) (24). Cytochrome P450 isoform 4B1 (CYP4B1) immunostaining was observed in the cytoplasm of surface epithelial cells (Figure 4D), with greatest intensity at the apical surface and on cilia (Figure 4F). We also observed considerable staining for CYP4B1 in ciliated gland ducts (Figure 4D, arrowhead). However, the tubular contents of the duct did not stain for CYP4B1. Acinar submucosal gland epithelial cells were uniformly negative for CYP4B1, as were cells within the interstitial spaces between acini (Figure 4E).
Figure 4.
Immunohistochemical localization of selected gene products confirms the microarray results. (A) GST-α is widely expressed in the surface epithelium. (B) Staining was observed very rarely in serous and mucus submucosal glands. (C) The most intense staining was observed at the apical surface of ciliated epithelial cells. (D) CYP4B1 was specifically observed on a subset of surface epithelial cells, with additional staining in cells lining the gland duct (arrowhead). (E) Under higher power, CYP4B1 was absent in the submucosal glands. (F) Surface CYP4B1 immunostaining was most intense near cilia, which could be consistent with the prediction that CYP4B1 is secreted. Minimal immunostaining was observed in the basal layer of surface epithelial cells. (G) Expression of CYP4B1 declined in the more distal portions of submucosal gland ducts (data not shown). NQO1 staining was widely expressed in the surface epithelium, consistent with previous studies (25). (H) NQO1 was not detectable in submucosal gland epithelia, but light staining was observed in interstitial cells. (I) The basal layer of surface airway epithelial cells was deficient in NQO1 expression. Surface epithelial staining within columnar cells appeared cytoplasmic, with a bias toward apical expression. NQO1 immunostaining was also present in vascular endothelial cells and in chondrocytes of bronchial cartilage (data not shown). (J and K) LTF is expressed highly in the serous demilunes, consistent with earlier observations (26). (L and M) ZAG expression was intense in the vast majority of acinar gland cells, and appeared to be present in both serous and mucus cell types. A representative tissue section stained with isotype control antibody is presented in panel N.
NAD(P)H quinone oxidoreductase (NQO1) immunostaining was largely confined to surface epithelial cells (Figure 4G). There was no expression of NQO1 in the submucosa, except for light staining of interstitial cells between the glandular acini (Figure 4H). There was a noticeable expression gradient, with greatest staining at the apical surface of the pseudostratified epithelium, and a near absence of staining in the basal layer (Figure 4I). Both ciliated and nonciliated cells appeared positive for NQO1. In addition to surface epithelial expression, moderate immunostaining was also observed in various nonepithelial cell types, including vascular endothelial cells in arterioles and venules as reported previously (25) and chondrocytes of the tracheal cartilage (data not shown). Epithelial expression of NQO1 ended abruptly in submucosal gland ducts (data not shown).
Submucosal Gland–Enriched Transcripts
Abundant lactoferrin (LTF) staining was found in granules of serous submucosal cells as well as in the glandular lumen (Figures 4J and 4K) (26). Mucous cells of the SMG did not express LTF. Resident interstitial cells of the SMG did not stain positively for LTF expression, nor did cells of the glandular collecting ducts and surface epithelium. ZAG immunostaining was widely distributed within tracheal SMG (Figure 4L). Although a previous report indicated expression of ZAG was limited to the serous cells of SMG (27), our results (Figures 4L and 4M) show significant staining was found in both serous and mucous cell types. ZAG was not expressed elsewhere in the submucosa, and production disappears in the collecting duct cells. However, the lumenal contents of collecting ducts were highly positive, as was the fluid layer on top of the surface epithelium. Surface epithelial cells did not contain ZAG.
DISCUSSION
LCM is a powerful technique that can be used to isolate specific cell populations from solid organs. In this study, we have demonstrated that LCM can be used to characterize gene expression in surface versus SMG epithelia of human conducting airways. This technique could be adapted to study even more specialized cell types of the conducting airway or to analyze differences between normal and pathologic states. Because the tissues are sampled directly from an in vivo setting, LCM minimizes biological changes that can occur when cells adapt to tissue culture in vitro. LCM has been used effectively to characterize lung cancers, providing greater understanding of tumor gene expression in vivo (28–30). This new approach may help stratify disease prognosis based on individual tumor gene expression profiles (30, 31), that is, by comparing tumor cells with surrounding normal tissues (28), or by identifying new tumor markers (32). LCM can also be used to prepare specimens for other downstream applications such as comparative genomic hybridization (33). A previous LCM study focused on the transcriptome of the murine bronchus, and demonstrated that RNA amplification steps did not significantly alter the observed gene expression profile (34). Although LCM has been widely used to compare malignant cells to their normal counterparts, no published studies have characterized gene expression of normal tissues in separate anatomic components of the human airway. We used LCM to delineate the gene expression profiles of two unique airway tissues: the surface epithelium and SMG.
SMG and surface epithelia are derived from the same precursor cells, with glands arising from invaginations of the surface epithelium during fetal development (35). After submucosal invasion, the nascent glandular tubules branch and arborize to form acini, increasing their total surface area. Our study shows that the transcriptional profiles of SMG and airway epithelia are divergent and reflect their unique functions.
Recently, the contents of airway surface liquid were analyzed by high-throughput proteomic characterizations of salivary and bronchoalveolar lavage (BAL) fluid (36, 37). Our work confirms and complements these studies. Many of the differentially regulated genes identified in our study were among the most abundant proteins present in BAL fluid characterized in these proteomic studies. Our investigation provides information about the origin of many of these molecules. In addition, the results of progressive transcriptional analysis of polarizing airway epithelial cell cultures by microarray (38) are highly congruent with the transcriptional signature of surface epithelial cells described in our study. This positive correlation demonstrates the value of polarized cell culture models in mimicking the biological state of the surface epithelium in vivo.
An important distinction between surface epithelial cells and SMG is the robust difference in expression of energy yielding pathways. Previous studies of energy utilization by tracheal epithelial cells focused on culture models and have found that energy consumption increases during epithelial polarization (39). However, it is difficult to culture submucosal glands to reliably compare their energy usage to surface epithelia. Using LCM to isolate populations of SMG cells, we could directly compare energy-yielding pathways at the transcriptional level from cells in either compartment. GSEA revealed that the surface airway epithelial cells express genes involved in catabolism at higher levels than in SMG cells (Figures 2C and 2D). Thus, surface epithelial cells may be transcriptionally poised for greater energy consumption than SMG cells. By immunohistochemically probing for isocitrate dehydrogenase (IDH3G), a Krebs cycle enzyme residing within the mitochondrial matrix, we demonstrated that either mitochondria are more numerous in surface epithelial cells or that the mitochondria of surface epithelial cells contain greater levels of IDH3G compared with mitochondria in SMG cells (Figure 3).
These results provide evidence that aerobic metabolism is increased in surface epithelial cells, and indicate the unique metabolic demands of these cells. Two important functions of surface epithelial cells are electrolyte transport and mucociliary clearance. The lungs generate approximately 100 ml of mucus daily, much of which originates in submucosal glands. Approximately 10 ml of this mucus are raised to the glottis by surface epithelial cells (18). The remaining 90 ml of fluid are reabsorbed by surface epithelia. Fluid reabsorption requires transport of electrolytes against steep electrochemical gradients, and accounts for a significant fraction of energy expenditure by cultured epithelial cells (40). The physical work of mucus elevation out of the lungs requires the coordinated beating of cilia and represents another important energy expenditure. The localization of mitochondria near the apical pole of surface epithelial cells may be consistent with cilia movement as a major energy sink. We observed that many transcripts involved in cilia motility, such as DNAI and TSGA2, are highly enriched in surface epithelium samples. Although the major protein components of cilia are known, these are highly complex organelles and probably contain over 650 unique proteins (8). We predict that other gene products involved in cilia motion are also enriched in the surface epithelium, and that this approach may help identify presently unknown cilia-related gene products.
The difference in aerobic activity between surface epithelial cells and SMG could reflect the greater oxygen availability at the epithelial surface. However, the higher oxygen pressure at the epithelial surface, together with inhaled xenobiotics, could also contribute to oxidative stresses at the apical surface. We observe that enzymes that function as antioxidants or in the clearance of xenobiotics are highly expressed in surface epithelia compared with SMG. Interestingly, the up-regulation of transcripts involved in xenobiotic metabolism has also been described during epithelial differentiation in vitro (38) as well as in conducting airways of cigarette smokers in vivo (41). As examples of gene products used in xenobiotic clearance, we show immunohistochemical distribution of NQO1, CYP4B1, and GST-α. Each of these three gene products is expressed in the surface epithelium with an apical to basal expression gradient. All three of these enzymes require energy to function, either in the form of reduced NADH, NADPH, or reduced glutathione.
Enzymatic clearance of xenobiotic compounds by oxidation or conjugation appears to be an important defense mechanism of surface epithelial cells. However, these enzymes can also activate otherwise inert molecules to create carcinogens and mutagens. The ubiquitous pollutants benzene and polycyclic aromatic hydrocarbons (PAH) are hydroxylated by CYP450 enzyme systems and then further oxidized to yield quinones (42). Unsubstituted quinones form adducts with a variety of nucleophiles, including amino groups of nucleic acids (43). NAD(P)H quinone oxidoreductase 1 (NQO1) is a cytoplasmic enzyme that reduces quinones to hydroquinone forms to protect against DNA damage (43). NQO1 expression in bronchial epithelia was observed in a previous study (25), but the question of SMG expression was not addressed. We observed a similar distribution of surface epithelial staining, with no staining in the submucosal gland acini. We also found expression of NQO1 in the cytoplasm of chondrocytes, endothelial cells, and leukocytes of the submucosal space. Because NQO1 defends against xenobiotic toxicity (44) and it is highly expressed in the airway surface epithelia, its relationship to lung cancer remains an important question (45).
CYP4B1 is an extrahepatic CYP450 family member with high expression in the lung (46). This study shows that CYP4B1 is expressed in the ciliated submucosal gland ducts as well as in the surface epithelium, in concordance with the microarray results. Much of the interest in CYP4B1 surrounds the possible role of extrahepatic cytochromes P450 in the bioactivation of cigarette smoke compounds. Human CYP4B1 is thought to have diminished functionality due to a mutation in a conserved domain (47). However, its expression in the lung and other extrahepatic tissues (48–51) raises the possibility that it could modulate smoke bioactivation.
In larger mammals, submucosal glands are a prominent feature of cartilaginous airways. These structures supply the majority of airway surface liquid (1) and also secrete a myriad of antimicrobial compounds. Lysozyme and lactoferrin are important innate immune effectors that either exert direct antimicrobial activity or deprive pathogenic microbes of nutrients such as iron (52). Because host defenses are often functionally redundant, it remains likely that other secreted antimicrobial products of submucosal glands exist but are yet unknown. Lacrimal proline rich protein (PRR4) was among the most highly expressed genes in human conducting airway submucosal glands. It was previously identified as an abundant product of lacrimal and salivary glands (11, 12), and is thought to play a role in maintenance of dental health by altering oral flora (53). Given its coexpression with other innate immune effectors, we predict that it plays a similar role in the conducting airways.
Among the most highly enriched SMG products in this study was ZAG. ZAG was first purified from blood plasma in the early 1960s (54). It was named for its electrophoretic mobility in the same range as α-2 globulins and for its low solubility in dilute zinc solutions. Several glands abundantly secrete ZAG; however, its function is still poorly understood. Three interesting properties of ZAG may suggest a possible role in airway biology. First, ZAG expression in semen correlates well with sperm motility, suggesting the protein may act as a modulator of the viscoelastic properties of semen (55). It is plausible to view ZAG as a protein that helps liquify mucous secretions and allow them to flow through gland ducts. Second, ZAG is related structurally to the MHC class I molecule, with identical domain structure and a small binding cleft (56), suggesting a possible immunomodulatory role. Third, ZAG has been reported to possess RNase activity (57), and thus may play a role in antiviral defense against RNA viruses.
SMG play important roles in serious respiratory diseases such as CF, COPD, and asthma. However, many knowledge gaps exist in our understanding of these structures. Our results demonstrate that SMG and surface epithelial cells are transcriptionally dissimilar, and provide clues about their respective functions. Using rigorous tissue isolation methods such as LCM in concert with powerful genomic and proteomic techniques may yield much more information about the products and functions of glandular epithelial cells of the respiratory submucosa.
Supplementary Material
Acknowledgments
Kevin Knudtson, Gary Hauser, Phil Karp, and Ariadna Arias gave valuable technical assistance. Thomas Waldschmidt, Michael Welsh, and Jennifer Bartlett provided helpful discussions. Gary Hunninghake assisted in the development of the HsAirway Genechip. Technical services were provided by the University of Iowa DNA Sequencing Core, the Cell Morphology Core, the Gene Transfer Vector Core (partially supported by the Cystic Fibrosis Foundation and NHLBI PPG HL-51670), and the Center for Gene Therapy for Cystic Fibrosis (NIH P30 DK-54759).
This work was supported by the National Institutes of Health (N01 AI-30040, P.B.M., and RO1 HL-50050, J.M.S), the Cystic Fibrosis Foundation (MCCRAY00V0, P.B.M.), The Roy J. Carver Charitable Trust (P.B.M.), and a Research to Prevent Blindness Career Development Award (T.E.S.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2008-0240OC on August 14, 2008
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Wine JJ, Joo NS. Submucosal glands and airway defense. Proc Am Thorac Soc 2004;1:47–53. [DOI] [PubMed] [Google Scholar]
- 2.Ballard ST, Inglis SK. Liquid secretion properties of airway submucosal glands. J Physiol 2004;556:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheetz TE, Zabner J, Welsh MJ, Coco J, Eyestone Mde F, Bonaldo M, Kucaba T, Casavant TL, Soares MB, McCray PB Jr. Large-scale gene discovery in human airway epithelia reveals novel transcripts. Physiol Genomics 2004;17:69–77. [DOI] [PubMed] [Google Scholar]
- 4.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 2003;31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004;5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007;23:3251–3253. [DOI] [PubMed] [Google Scholar]
- 7.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol 2007;8:880–893. [DOI] [PubMed] [Google Scholar]
- 9.Shetty J, Klotz KL, Wolkowicz MJ, Flickinger CJ, Herr JC. Radial spoke protein 44 (human meichroacidin) is an axonemal alloantigen of sperm and cilia. Gene 2007;396:93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartlett JA, Fischer AJ, McCray PB. Innate Immune Functions of the Airway Epithelium. Contrib Microbiol 2008;15:147–163. [DOI] [PubMed] [Google Scholar]
- 11.Lamkin MS, Arancillo AA, Oppenheim FG. Temporal and compositional characteristics of salivary protein adsorption to hydroxyapatite. J Dent Res 1996;75:803–808. [DOI] [PubMed] [Google Scholar]
- 12.Dickinson DP, Thiesse M. A major human lacrimal gland mRNA encodes a new proline-rich protein family member. Invest Ophthalmol Vis Sci 1995;36:2020–2031. [PubMed] [Google Scholar]
- 13.Inglis SK, Corboz MR, Ballard ST. Effect of anion secretion inhibitors on mucin content of airway submucosal gland ducts. Am J Physiol 1998;274:L762–L766. [DOI] [PubMed] [Google Scholar]
- 14.Hasegawa H, Lian SC, Finkbeiner WE, Verkman AS. Extrarenal tissue distribution of CHIP28 water channels by in situ hybridization and antibody staining. Am J Physiol 1994;266:C893–C903. [DOI] [PubMed] [Google Scholar]
- 15.Jensen BS, Strobaek D, Christophersen P, Jorgensen TD, Hansen C, Silahtaroglu A, Olesen SP, Ahring PK. Characterization of the cloned human intermediate-conductance Ca2+-activated K+ channel. Am J Physiol 1998;275:C848–C856. [DOI] [PubMed] [Google Scholar]
- 16.Shimura S, Sasaki T, Sasaki H, Takishima T. Contractility of isolated single submucosal gland from trachea. J Appl Physiol 1986;60:1237–1247. [DOI] [PubMed] [Google Scholar]
- 17.Zhu J, Qiu Y, Valobra M, Qiu S, Majumdar S, Matin D, De Rose V, Jeffery PK. Plasma cells and IL-4 in chronic bronchitis and chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2007;175:1125–1133. [DOI] [PubMed] [Google Scholar]
- 18.Wanner A, Salathe M, O'Riordan TG. Mucociliary clearance in the airways. Am J Respir Crit Care Med 1996;154:1868–1902. [DOI] [PubMed] [Google Scholar]
- 19.Moskwa P, Lorentzen D, Excoffon KJ, Zabner J, McCray PB Jr, Nauseef WM, Dupuy C, Banfi B. A novel host defense system of airways is defective in cystic fibrosis. Am J Respir Crit Care Med 2007;175:174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geiszt M, Witta J, Baffi J, Lekstrom K, Leto TL. Dual oxidases represent novel hydrogen peroxide sources supporting mucosal surface host defense. FASEB J 2003;17:1502–1504. [DOI] [PubMed] [Google Scholar]
- 21.Conner GE, Wijkstrom-Frei C, Randell SH, Fernandez VE, Salathe M. The lactoperoxidase system links anion transport to host defense in cystic fibrosis. FEBS Lett 2007;581:271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenner V, Nyakatura G, Rosenthal A, Platzer M. Genomic organization of two novel genes on human Xq28: compact head to head arrangement of IDH gamma and TRAP delta is conserved in rat and mouse. Genomics 1997;44:8–14. [DOI] [PubMed] [Google Scholar]
- 23.Nichols BJ, Hall L, Perry AC, Denton RM. Molecular cloning and deduced amino acid sequences of the gamma-subunits of rat and monkey NAD(+)-isocitrate dehydrogenases. Biochem J 1993;295:347–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anttila S, Hirvonen A, Vainio H, Husgafvel-Pursiainen K, Hayes JD, Ketterer B. Immunohistochemical localization of glutathione S-transferases in human lung. Cancer Res 1993;53:5643–5648. [PubMed] [Google Scholar]
- 25.Siegel D, Franklin WA, Ross D. Immunohistochemical detection of NAD(P)H:quinone oxidoreductase in human lung and lung tumors. Clin Cancer Res 1998;4:2065–2070. [PubMed] [Google Scholar]
- 26.Masson P, Heremans JF, Prignot J. Immunohistochemical localization of the iron-binding protein lactoferrin in human bronchial glands. Experientia 1965;21:604–605. [DOI] [PubMed] [Google Scholar]
- 27.Tada T, Ohkubo I, Niwa M, Sasaki M, Tateyama H, Eimoto T. Immunohistochemical localization of Zn-alpha 2-glycoprotein in normal human tissues. J Histochem Cytochem 1991;39:1221–1226. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi K, Nishioka M, Kohno T, Nakamoto M, Maeshima A, Aoyagi K, Sasaki H, Takenoshita S, Sugimura H, Yokota J. Identification of genes whose expression is upregulated in lung adenocarcinoma cells in comparison with type II alveolar cells and bronchiolar epithelial cells in vivo. Oncogene 2004;23:3089–3096. [DOI] [PubMed] [Google Scholar]
- 29.Kikuchi T, Daigo Y, Katagiri T, Tsunoda T, Okada K, Kakiuchi S, Zembutsu H, Furukawa Y, Kawamura M, Kobayashi K, et al. Expression profiles of non-small cell lung cancers on cDNA microarrays: identification of genes for prediction of lymph-node metastasis and sensitivity to anti-cancer drugs. Oncogene 2003;22:2192–2205. [DOI] [PubMed] [Google Scholar]
- 30.Miura K, Bowman ED, Simon R, Peng AC, Robles AI, Jones RT, Katagiri T, He P, Mizukami H, Charboneau L, et al. Laser capture microdissection and microarray expression analysis of lung adenocarcinoma reveals tobacco smoking- and prognosis-related molecular profiles. Cancer Res 2002;62:3244–3250. [PubMed] [Google Scholar]
- 31.Hoang CD, D'Cunha J, Tawfic SH, Gruessner AC, Kratzke RA, Maddaus MA. Expression profiling of non-small cell lung carcinoma identifies metastatic genotypes based on lymph node tumor burden. J Thorac Cardiovasc Surg 2004;127:1332–1341. (discussion 1342). [DOI] [PubMed] [Google Scholar]
- 32.He P, Varticovski L, Bowman ED, Fukuoka J, Welsh JA, Miura K, Jen J, Gabrielson E, Brambilla E, Travis WD, et al. Identification of carboxypeptidase E and gamma-glutamyl hydrolase as biomarkers for pulmonary neuroendocrine tumors by cDNA microarray. Hum Pathol 2004;35:1196–1209. [DOI] [PubMed] [Google Scholar]
- 33.Shibata T, Uryu S, Kokubu A, Hosoda F, Ohki M, Sakiyama T, Matsuno Y, Tsuchiya R, Kanai Y, Kondo T, et al. Genetic classification of lung adenocarcinoma based on array-based comparative genomic hybridization analysis: its association with clinicopathologic features. Clin Cancer Res 2005;11:6177–6185. [DOI] [PubMed] [Google Scholar]
- 34.Betsuyaku T, Senior RM. Laser capture microdissection and mRNA characterization of mouse airway epithelium: methodological considerations. Micron 2004;35:229–234. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Driskell RR, Engelhardt JF. Airway glandular development and stem cells. Curr Top Dev Biol 2004;64:33–56. [DOI] [PubMed] [Google Scholar]
- 36.Merkel D, Rist W, Seither P, Weith A, Lenter MC. Proteomic study of human bronchoalveolar lavage fluids from smokers with chronic obstructive pulmonary disease by combining surface-enhanced laser desorption/ionization-mass spectrometry profiling with mass spectrometric protein identification. Proteomics 2005;5:2972–2980. [DOI] [PubMed] [Google Scholar]
- 37.Plymoth A, Yang Z, Lofdahl CG, Ekberg-Jansson A, Dahlback M, Fehniger TE, Marko-Varga G, Hancock WS. Rapid proteome analysis of bronchoalveolar lavage samples of lifelong smokers and never-smokers by micro-scale liquid chromatography and mass spectrometry. Clin Chem 2006;52:671–679. [DOI] [PubMed] [Google Scholar]
- 38.Ross AJ, Dailey LA, Brighton LE, Devlin RB. Transcriptional profiling of mucociliary differentiation in human airway epithelial cells. Am J Respir Cell Mol Biol 2007;37:169–185. [DOI] [PubMed] [Google Scholar]
- 39.Kondo M, Tamaoki J, Sakai A, Kameyama S, Kanoh S, Konno K. Increased oxidative metabolism in cow tracheal epithelial cells cultured at air-liquid interface. Am J Respir Cell Mol Biol 1997;16:62–68. [DOI] [PubMed] [Google Scholar]
- 40.Welsh MJ. Energetics of chloride secretion in canine tracheal epithelium: comparison of the metabolic cost of chloride transport with the metabolic cost of sodium transport. J Clin Invest 1984;74:262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harvey BG, Heguy A, Leopold PL, Carolan BJ, Ferris B, Crystal RG. Modification of gene expression of the small airway epithelium in response to cigarette smoking. J Mol Med 2007;85:39–53. [DOI] [PubMed] [Google Scholar]
- 42.Rothman N, Smith MT, Hayes RB, Traver RD, Hoener B, Campleman S, Li GL, Dosemeci M, Linet M, Zhang L, et al. Benzene poisoning, a risk factor for hematological malignancy, is associated with the NQO1 609C→T mutation and rapid fractional excretion of chlorzoxazone. Cancer Res 1997;57:2839–2842. [PubMed] [Google Scholar]
- 43.Joseph P, Jaiswal AK. NAD(P)H:quinone oxidoreductase1 (DT diaphorase) specifically prevents the formation of benzo[a]pyrene quinone-DNA adducts generated by cytochrome P4501A1 and P450 reductase. Proc Natl Acad Sci USA 1994;91:8413–8417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lan Q, Zhang L, Li G, Vermeulen R, Weinberg RS, Dosemeci M, Rappaport SM, Shen M, Alter BP, Wu Y, et al. Hematotoxicity in workers exposed to low levels of benzene. Science 2004;306:1774–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiyohara C, Yoshimasu K, Takayama K, Nakanishi Y. NQO1, MPO, and the risk of lung cancer: a HuGE review. Genet Med 2005;7:463–478. [DOI] [PubMed] [Google Scholar]
- 46.Nhamburo PT, Gonzalez FJ, McBride OW, Gelboin HV, Kimura S. Identification of a new P450 expressed in human lung: complete cDNA sequence, cDNA-directed expression, and chromosome mapping. Biochemistry 1989;28:8060–8066. [DOI] [PubMed] [Google Scholar]
- 47.Zheng YM, Fisher MB, Yokotani N, Fujii-Kuriyama Y, Rettie AE. Identification of a meander region proline residue critical for heme binding to cytochrome P450: implications for the catalytic function of human CYP4B1. Biochemistry 1998;37:12847–12851. [DOI] [PubMed] [Google Scholar]
- 48.Hakkola J, Pasanen M, Hukkanen J, Pelkonen O, Maenpaa J, Edwards RJ, Boobis AR, Raunio H. Expression of xenobiotic-metabolizing cytochrome P450 forms in human full-term placenta. Biochem Pharmacol 1996;51:403–411. [DOI] [PubMed] [Google Scholar]
- 49.Roos PH, Belik R, Follmann W, Degen GH, Knopf HJ, Bolt HM, Golka K. Expression of cytochrome P450 enzymes CYP1A1, CYP1B1, CYP2E1 and CYP4B1 in cultured transitional cells from specimens of the human urinary tract and from urinary sediments. Arch Toxicol 2006;80:45–52. [DOI] [PubMed] [Google Scholar]
- 50.Czerwinski M, McLemore TL, Gelboin HV, Gonzalez FJ. Quantification of CYP2B7, CYP4B1, and CYPOR messenger RNAs in normal human lung and lung tumors. Cancer Res 1994;54:1085–1091. [PubMed] [Google Scholar]
- 51.Imaoka S, Yoneda Y, Matsuda T, Degawa M, Fukushima S, Funae Y. Mutagenic activation of urinary bladder carcinogens by CYP4B1 and the presence of CYP4B1 in bladder mucosa. Biochem Pharmacol 1997;54:677–683. [DOI] [PubMed] [Google Scholar]
- 52.Singh PK, Parsek MR, Greenberg EP, Welsh MJ. A component of innate immunity prevents bacterial biofilm development. Nature 2002;417:552–555. [DOI] [PubMed] [Google Scholar]
- 53.Li T, Bratt P, Jonsson AP, Ryberg M, Johansson I, Griffiths WJ, Bergman T, Stromberg N. Possible release of an ArgGlyArgProGln pentapeptide with innate immunity properties from acidic proline-rich proteins by proteolytic activity in commensal streptococcus and actinomyces species. Infect Immun 2000;68:5425–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burgi W, Schmid K. Preparation and properties of Zn-alpha 2-glycoprotein of normal human plasma. J Biol Chem 1961;236:1066–1074. [PubMed] [Google Scholar]
- 55.Ahlgren G, Rannevik G, Lilja H. Impaired secretory function of the prostate in men with oligo-asthenozoospermia. J Androl 1995;16:491–498. [PubMed] [Google Scholar]
- 56.Sanchez LM, Chirino AJ, Bjorkman P. Crystal structure of human ZAG, a fat-depleting factor related to MHC molecules. Science 1999;283:1914–1919. [DOI] [PubMed] [Google Scholar]
- 57.Lei G, Arany I, Tyring SK, Brysk H, Brysk MM. Zinc-alpha 2-glycoprotein has ribonuclease activity. Arch Biochem Biophys 1998;355:160–164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.