Abstract
Direct interaction between bacteria and epithelial cells may initiate or amplify the airway response through induction of epithelial defense gene expression by nuclear factor-κB (NF-κB). However, multiple signaling pathways modify NF-κB effects to modulate gene expression. In this study, the effects of CCAAT/enhancer binding protein (C/EBP) family members on induction of the leukocyte adhesion glycoprotein intercellular adhesion molecule-1 (ICAM-1) was examined in primary cultures of human tracheobronchial epithelial cells incubated with nontypeable Haemophilus influenzae. Increased ICAM-1 gene transcription in response to H. influenzae required gene sequences located at −200 to −135 in the 5′-flanking region that contain a C/EBP-binding sequence immediately upstream of the NF-κB enhancer site. Constitutive C/EBPβ was found to have an important role in epithelial cell ICAM-1 regulation, while the adjacent NF-κB sequence binds the RelA/p65 and NF-κB1/p50 members of the NF-κB family to induce ICAM-1 expression in response to H. influenzae. The expression of C/EBP proteins is not regulated by p38 mitogen-activated protein kinase activation, but p38 affects gene transcription by increasing the binding of TATA-binding protein to TATA-box–containing gene sequences. Epithelial cell ICAM-1 expression in response to H. influenzae was decreased by expressing dominant-negative protein or RNA interference against C/EBPβ, confirming its role in ICAM-1 regulation. Although airway epithelial cells express multiple constitutive and inducible C/EBP family members that bind C/EBP sequences, the results indicate that C/EBPβ plays a central role in modulation of NF-κB–dependent defense gene expression in human airway epithelial cells after exposure to H. influenzae.
Keywords: inflammation, transcription factors, NF-κB, mitogen-activated protein kinase
CLINICAL RELEVANCE
Haemophilus influenzae activates intercellular adhesion molecule-1 gene transcription in primary human airway epithelial cells. This work defines the importance of specific C/EBP family members and a mechanism for p38 mitogen-activated kinase modulation of defense gene expression.
Nontypeable Haemophilus influenzae frequently colonize respiratory mucosa and can produce respiratory tract infections that include otitis media, sinusitis, bronchitis, and pneumonia, particularly in patients with underlying pulmonary diseases such as chronic obstructive pulmonary disease, bronchiectasis, or cystic fibrosis (1, 2). When innate defense mechanisms in airway epithelia are overwhelmed by H. influenzae, an inflammatory response is initiated that recruits leukocytes, particularly neutrophils, to sites of infection (3–6). Intercellular adhesion molecule-1 (ICAM-1) is a cell adhesion molecule member of the immunoglobulin gene superfamily that is an important component of this response in airway epithelia (5). ICAM-1 normally has a restricted distribution, but cytokines and/or bacteria induce ICAM-1 expression on many different cell types (7, 8). ICAM-1 participates in the inflammatory response through interaction with its β2-integrin–containing counter-receptors LFA-1 (αLβ2, CD11a/CD18) and Mac-1 (αMβ2, CD11b/CD18) on leukocytes (9). Expression of ICAM-1 on airway epithelial cells is critical for airway defense through its capacity to modulate leukocyte recruitment and bacterial killing (4, 5).
Increased inflammatory gene expression (including ICAM-1) in epithelial cells in response to H. influenzae has been demonstrated in vitro and in vivo, and several signaling pathways that control this response have been identified. The transcription factor nuclear factor-κB (NF-κB) is clearly central to the regulation of inflammatory gene expression in response to bacteria, including H. influenzae (6, 10). Members of the mitogen-activated protein (MAP) kinase family appear to modulate ICAM-1 and other inflammatory genes in response to H. influenzae (6, 10, 11). In addition, phosphatidylinositol 3-kinase (PI 3-kinase) may alter inflammatory gene expression through effects on NF-κB, MAP kinases, and/or other mechanisms (12, 13). The CCAAT/enhancer-binding protein (C/EBP) family of transcription factors regulate many cellular processes, including inflammation (14). The six known members (α, β, γ, δ, ɛ, ζ) of this family of proteins contain a conserved basic leucine zipper (bZIP) domain at the carboxyl-terminus that is involved in dimerization and DNA binding, as well as activation and regulation domains (15). Three C/EBP genes (α, β, ɛ) express multiple functionally active polypeptides that are produced primarily by alternative translation initiation site utilization, regulated proteolysis, or differential splicing. C/EBP family members may participate in inflammatory gene activation, sometimes through cooperative interaction with NF-κB, providing precedent for the possibility of their involvement of ICAM-1 regulation in response to bacteria (16, 17). Although many reports in this area focus on regulation of chemokine expression in response to isolated bacterial components, the role that each pathway plays appears to be cell-, gene-, and stimulus-dependent. Furthermore, the molecular mechanisms through which these pathways control inflammatory gene expression are incompletely understood. Accordingly, we hypothesized that H. influenzae would modulate specific C/EBP family members to control the activation of ICAM-1 and other defense genes in human airway epithelial cells.
In this article, we describe experiments that assess specific C/EBP proteins in human airway epithelial cells in response to interaction with H. influenzae. We demonstrate that increased ICAM-1 expression is mediated, at least in part, through an increase in gene transcription rate that is controlled by an H. influenzae response element (HFRE) located at −200 to −135 in the 5′-flanking region of the ICAM-1 gene. Both C/EBPβ and NF-κB transcription factors interact with the HFRE to control ICAM-1 gene expression. Although p38 MAP kinases are activated and modulate ICAM-1 expression in epithelial cells in response to H. influenzae, p38 alters DNA binding of the basal transcription factor TATA-binding protein (TBP), but does not affect C/EBP expression or DNA binding. Our results support the concept that C/EBPβ plays an important role in modulation of NF-κB–dependent defense gene expression in human airway epithelial cells after exposure to H. influenzae and allows for precise control of inflammatory gene expression and rapid and efficient airway defense.
MATERIALS AND METHODS
Airway Epithelial Cell Isolation, Culture, and Bacterial Treatment
Human tracheal and bronchial samples from multiple individuals without lung disease were obtained under a protocol approved by the University of Iowa Institutional Review Board. Airways were dissected from lung tissue, and primary human tracheobronchial epithelial (hTBE) cells from the surface of airway mucosa were isolated by enzymatic dissociation. Cells were cultured in Laboratory of Human Carcinogenesis (LHC)-8e medium on plates coated with collagen and albumin as described previously (8, 18, 19). Aerated, log-phase cultures of H. influenzae strain 12 were prepared and quantitated as described previously (4, 6, 8). Bacteria were incubated in 100 μg/ml gentamicin for 30 minutes, and then 108 to 1010 colony-forming units (CFU)/ml (500–50,000 CFU/epithelial cell) of killed bacteria was incubated with epithelial cells in culture media for 0.5 to 24 hours. In some experiments, hTBE cells were pretreated for 1 hour with either vehicle control (DMSO) or the p38 MAP kinase inhibitor SB203580 (Calbiochem, La Jolla, CA). To assure reproducible and generalizable results, key experiments were repeated at least three times and this study used epithelial cells from 11 different individuals.
Nuclear Runoff Analysis
Relative gene transcription rates were assessed using nuclear runoff analysis as described previously (18, 20). Plasmids containing target cDNAs that were tested include: (1) pBluescriptIISK as a negative control vector (Stratagene, La Jolla, CA); (2) pCD1.8 containing human ICAM-1 cDNA in pCDM8 (a gift from D. Staunton, Harvard University) (21); (3) pHMαA-PX containing human skeletal α-actin cDNA in pBR322 as a low level positive control (a gift from P. Gunning, Children's Medical Research Institute, Wentworthville, NSW, Australia) (22); (4) pC4-c-rel containing human Rel/c-Rel cDNA in pCMV4 as a high-level positive control (a gift from W. Greene, University of California-San Francisco) (23); and (5) pSKAlu containing Alu repetitive sequence cDNA in pBluescriptSK as a high-level positive control (a gift from T. Ley, Washington University) (24).
Reporter and Expression Plasmids
Luciferase reporter plasmids driven by ICAM-1 5′-flanking sequences from −1294 or −134 to +7 (numbered from the downstream transcription initiation site) were constructed using the pBH-luc vector (a gift from C. Stratowa, Ernst Bohringer Institute, Vienna, Austria) as previously described (20, 25). Nested 5′-deletions of ICAM-1 sequence from −553, −294, −200, and −192 to +7 were generated by PCR using the full-length sequence plasmid as template and primers with 5′ restriction enzyme sites. PCR products were initially ligated into the TOPO TA cloning vector (Invitrogen, Carlsbad, CA), and then subcloned into pBH-luc. Double-stranded oligonucleotides containing ICAM-1 sequence from −200 to −135 with restriction enzyme sites on the ends were ligated upstream of the TA minimal promoter (nucleotides −36 to +8 of the fibronectin gene with the TATAA replaced by the SV40 early-gene TATA box equivalent TATTTAT) in pBH-TA-luc or the TK minimal promoter (consisting of nucleotides −33 to +9 of the herpes simplex virus thymidine kinase gene) in pBH-TK-luc (20). Putative transcription factor binding sites in the ICAM-1 sequence of pBH-1294+7ICAM-1-luc were mutated using a commercial site-directed mutagenesis kit (Stratagene). Mutations include: AP1 at −283 from 5′-TGATTCAA-3′(wild-type) to 5′-TGGTGCCA-3′(mutated), C/EBP at −198 from 5′-CGATTGC-3′(wild-type) to 5′-CGATATC-3′(mutated), and NF-κB at −187 from 5′-TTGGAAATTCC-3′(wild-type) to 5′-TTCCAAATTGC-3′(mutated). A plasmid that expressed a dominant-negative form of C/EBPβ was generated by PCR amplification of cDNA sequence corresponding to amino acids 201 to 346 of full-length human C/EBPβ cDNA, followed by ligation into the mammalian expression vector pCMV-Tag (Stratagene). This plasmid expressed methionine plus a 146–amino acid naturally occurring product also known as liver-enriched inhibitory protein (LIP) (15). An inactive form of this dominant-negative C/EBPβ was generated by mutation of amino acids 28 to 31 and 35 to 38 (numbered per the 146–amino acid sequence) to alanines as previously described (26) using the site-directed mutation kit. Insert orientation and sequence integrity were verified in all plasmid constructs by DNA sequencing. The pCMV-p38(agf) plasmid expressing dominant-negative p38 was a gift from R. Davis (University of Massachusetts) (27).
Cell Transfection and Luciferase Reporter Gene Assay
Plasmid DNA was purified by two successive centrifugations through cesium chloride and then used to transfect hTBE cell cultures using a commercial liposomal DNA packaging system as described previously (18, 20, 28). Cell monolayers at 80 to 100% confluence in 22-mm tissue culture wells were treated with 1.6 μg of plasmid DNA and 4.8 μl GeneFECTOR reagent (VennNova, Pompano Beach, FL) in 400 μl of antibiotic-free medium for 2 to 4 hours at 37°C. Transfected cells were washed with complete medium containing 0.5% bovine serum albumin (BSA), and then incubated with bacteria for 18 hours. Where indicated, cells were pretreated for 1 hour before bacterial treatment with chemical inhibitors. In other samples, plasmids expressing control or dominant-negative proteins were included in the transfection mixture. Photinus pyralis luciferase activity was determined as previously described or using a commercial luciferase reporter assay kit (Promega, Madison, WI) (18, 20, 28). To allow normalization for transfection efficiency, 5 ng of pRL-TK (Promega) was also included in the transfection reaction and Renilla reniformis luciferase activity was determined.
Electrophoretic Mobility-Shift Assay
Nuclear extracts were prepared by cell membrane lysis in 10 mM Hepes, pH 7.8, 10 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 0.5 mM phenylmethysulfonylfluoride, 1 mM N-α-p-tosyl-L-lysine chloromethyl ketone, 5 μg/ml leupeptin, 2 μg/ml pepstatin, and 10% Nonidet P-40. Nuclei were isolated by centrifugation at 10,000 × g, and nuclear proteins extracted in 50 mM Hepes, pH 7.8, 300 mM NaCl, 50 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, 0.5 mM phenylmethysulfonylfluoride, 1 mM N-α-p-tosyl-L-lysine chloromethyl ketone, 5 μg/ml leupeptin, 2 μg/ml pepstatin, and 10% glycerol. Supernatants containing nuclear proteins were isolated by centrifugation at 10,000 × g, and protein concentration were determined using a Coomassie brilliant blue G-250 binding assay (Bio-Rad Laboratories, Hercules, CA). Double-stranded, synthetic oligonucleotides corresponding to ICAM-1, C/EBP consensus, NF-κB consensus, TFIID/TATA consensus (Santa Cruz Biotechnology, Santa Cruz, CA), or control cAMP response element (CRE) sequences were end-labeled with 32P and used as probes in electrophoretic mobility shift assays (EMSAs). Binding reactions were performed at 25°C for 20 minutes, followed by electrophoresis through 6% acrylamide. For ICAM-1, C/EBP, NF-κB, and CRE-binding reactions, the buffer contained 25 mM Tris, pH 7.5, 25 mM KCl, 12.5 mM MgCl2, 0.1 mM ZnSO4, 1 mM dithiothreitol, 0.1% Nonidet P-40, 20% glycerol, and 2 μg poly[d(I-C)], and electrophoresis was performed in 1× TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA, pH 8.4). For TFIID/TATA-binding reactions, the buffer contained 20 mM Tris, pH 8.0, 80 mM KCl, 10 mM MgCl2, 2 mM dithiothreitol, 10% glycerol, and 1 μg poly[d(G-C)], and electrophoresis was performed in 0.5× TBE buffer with 0.02% Nonidet P-40. Specificity of oligonucleotide binding was demonstrated both by competition for binding by unlabeled nucleotide of identical sequence and by lack of competition by a control oligonucleotide. Proteins in DNA-binding complexes were identified by supershift analysis using 2 to 5 μg of the following specific antibodies: rabbit polyclonal IgG sc-109 against human RelA/p65, rabbit polyclonal IgG sc-7178 against human NF-κB1/p50, rabbit polyclonal IgG sc-150 against human C/EBPβ, rabbit polyclonal IgG sc-636 against human C/EBPδ, goat polyclonal IgG sc-7658 against human C/EBPγ, rabbit polyclonal IgG sc-25770 against human C/EBPɛ, goat polyclonal IgG sc-9315 against human C/EBPα, and mouse IgG2b mAb clone 58C9 against human TBP from Santa Cruz Biotechnology.
Immunoblot Analysis
Epithelial expression levels of specific cellular and nuclear proteins were assessed by immunoblot analysis as described previously (19, 29–31). Whole cell protein extracts were prepared by lysis of cell monolayers in 50 mM Tris, pH 7.4, 150 mM NaCl, 1% Nonidet P-40, a protease inhibitor cocktail (Roche Bioscience, Palo Alto, CA), and a phosphatase inhibitor panel (Calbiochem, San Diego, CA). Nuclear extracts were prepared and protein concentrations were determined as outlined for EMSA. Equal amounts of protein were subjected to SDS-PAGE in 7.5 to 12% polyacrylamide, and electrophoretically transferred to polyvinylidene fluoride membranes (Bio-Rad Laboratories). Membranes were exposed to either 5% nonfat milk or 5% BSA in Tris-buffered saline with 0.1% Tween-20 to block nonspecific antigens, and then incubated with antibodies against a specific cellular protein. Primary antibodies against transcription factors were the same as for EMSA except: rabbit polyclonal IgG 39306 against human C/EBPα, and rabbit polyclonal IgG 39006 against human C/EBPδ from Active Motif (Carlsbad, CA). Other primary antibodies included: goat polyclonal IgG sc-59-G against human Sp1 from Santa Cruz Biotechnology; rabbit polyclonal IgG 4915 against human ICAM-1, rabbit polyclonal IgG 3084 against Thr235-phosphorylated human C/EBPβ, and rabbit polyclonal IgG 2406 against Ser82-phosphorylated human heat shock protein(HSP)-27 from Cell Signaling Technology (Beverly, MA); mouse IgG2a mAb clone AC-74 against human β-actin from Sigma-Aldrich (St. Louis, MO). Primary antibody binding was detected using goat antirabbit, goat antimouse, or donkey antigoat IgG conjugated to horseradish peroxidase (Santa Cruz Biotechnology) and an enhanced chemiluminescence detection system (Amersham Biosciences, Buckinghamshire, UK). Reprobing of membranes was done after washing in Restore buffer (Pierce, Rockford, IL) for 15 minutes at 25°C.
Real-Time Reverse Transcription PCR mRNA Analysis
Total cellular RNA was isolated using a commercial spin column isolation kit (Stratagene), and 1 μg was reverse transcribed using a commercial kit (Ambion, Austin, TX). Equal aliquots of the resulting cDNA were subjected to PCR using an iCycler iQ Fluorescence Thermocycler (Bio-Rad Laboratories) with SYBR Green I DNA dye (Molecular Probes, Eugene, OR), iTaq DNA Polymerase (Bio-Rad Laboratories), and the following primers designed with software by S. Rozen and H. Skaletsky (http://www.genome.wi.mit.edu/genome_software/other/primer3.html): (1) human ICAM-1, sense 5′-CATAGAGACCCCGTTGCCTA-3′ and antisense 5′-GAAATTGGCTCCATGGTGAT-3′; and (2) human hypoxanthine phosphoribosyltransferase (HPRT), sense 5′-TTGGAAAGGGTGTTTATTCTTC-3′ and antisense 5′-TCCCCTGTTGACTGGTCATT-3′. PCR conditions included denaturation at 95°C for 3 minutes, and then 45 cycles of 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds, followed by melting curve analysis. Fluorescence data was captured during annealing reactions, and specificity of the amplification was confirmed using melting curve analysis. Data were collected and recorded by iCycler iQ software (Bio-Rad Laboratories) and initially determined as a function of threshold cycle (Ct). Ct was defined as the cycle at which the fluorescence intensity in a given reaction tube rose above background, which was calculated as 10 times the mean standard deviation (SD) of fluorescence in all wells over the baseline cycles. Levels of mRNA are expressed relative to control HPRT levels, calculated as 2ΔCt.
RNA Interference
Control RNA and sequence-specific 25- to 27-nucleotide double-stranded RNA molecules that target human C/EBP mRNA sequences were synthesized (Integrated DNA Technologies, Coralville, IA) and transfected into hTBE cells at a final concentration of 150 or 300 nM using Lipofectamine 2000 (Invitrogen), using a modification of a protocol described previously (30). The following dsRNA sequences with the forward sequence containing a 5′-phosphorylation and two 2′-deoxynucleotides at the 3′-end and the reverse sequence containing a two-nucleotide 3′-overhang were used: C/EBPβ gene, forward 5′-pCCCGCCCGUGGUGUUAUUUAAAGaa-3′ and reverse 5′-UUCUUUAAAUAACACCACGGGCGGGAG-3′; C/EBPδ gene, forward 5′-pUGGACUUACCACCACUAAACUGCga-3′ and reverse 5′-UCGCAGUUUAGUGGUGGUAAGUCCAGG-3′; and C/EBPɛ gene, forward 5′-pGGGCAAGAAGGCAGUGAACAAAGat-3′ and reverse 5′-AUCUUUGUUCACUGCCUUCUUGCCCUU-3′. Scrambled sequences at the same concentration were used as controls. Transfection mixtures were assembled by mixing the Dicer-substrate small interfering (si)RNA in 100 μl of OptiMem medium with 4 μl Lipofectamine 2000 in 100 μl of OptiMem medium for 15 minutes at 25°C. Epithelial cells in 35-mm tissue culture plates were incubated with the 200 μl mixture in 2 ml of antibiotic-free LHC-8e medium for 16 to 18 hours, followed by treatment with bacteria (C/EBPδ) or incubation in fresh medium for an additional 48 hours before treatment with bacteria (C/EBPβ and C/EBPɛ).
Statistical Analysis
Reporter gene assays and realtime RT-PCR mRNA analyses were analyzed for statistical significance using a one-way ANOVA for a factorial experimental design. The multicomparison significance level for the one-way ANOVA was 0.05. If significance was achieved by one-way analysis, post-ANOVA comparison of means was performed using Scheffe F-tests (32).
RESULTS
The ICAM-1 HFRE Is Restricted to Specific C/EBP and NF-κB Sites
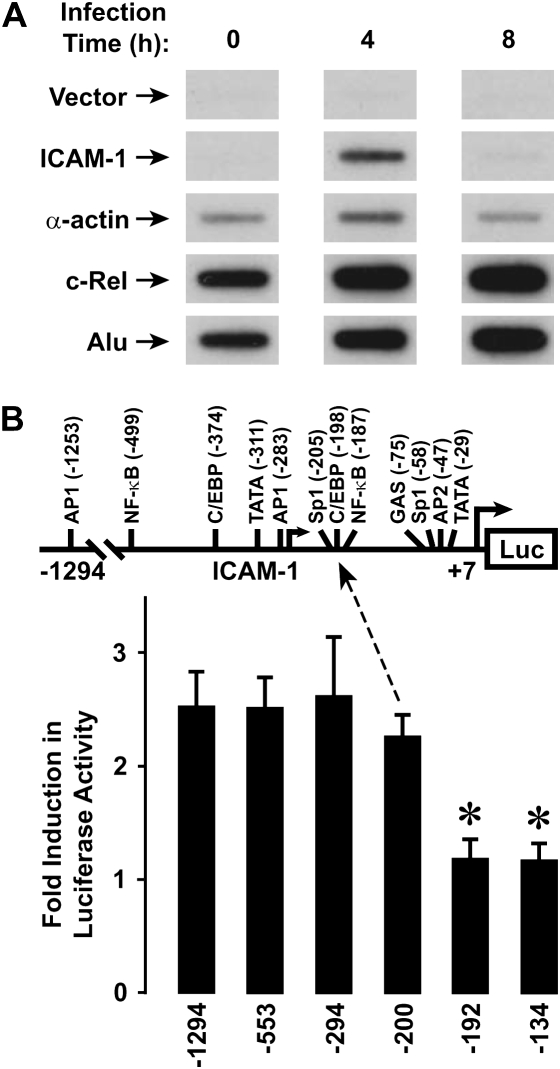
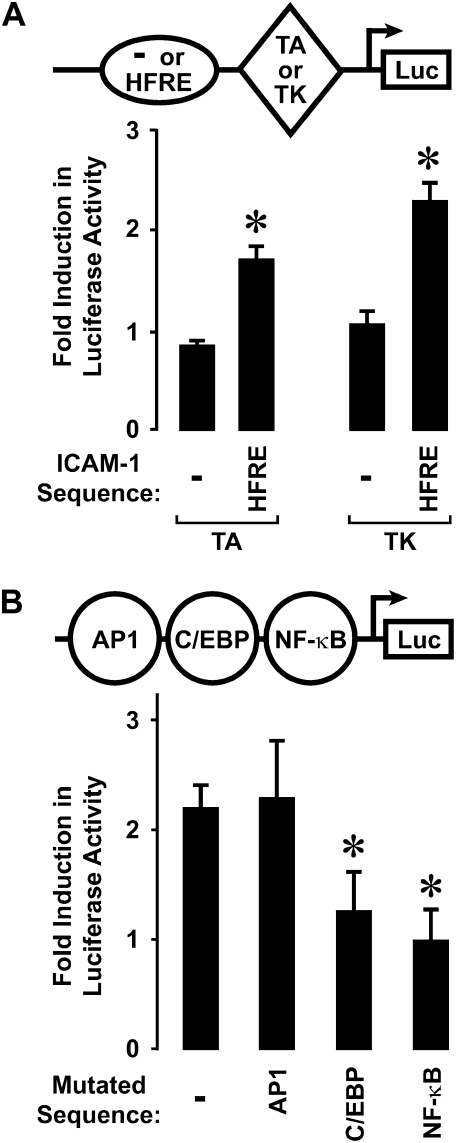
We previously reported that human airway epithelial cells responded to direct interaction with H. influenzae by increasing ICAM-1 mRNA and cell surface protein expression (4). Nuclear runoff assays demonstrated an increase in ICAM-1 gene transcription rate after hTBE cell exposure to H. influenzae for 4 hours (Figure 1A), confirming that regulation of this airway defense gene is mediated, at least in part, by bacterial induction of gene transcription. Accordingly, our next set of experiments focused on identifying transcriptional controls for ICAM-1 gene expression in epithelial cells exposed to H. influenzae. Experiments with reporter plasmids in which a luciferase gene is driven by ICAM-1 5′-flanking sequence indicated that 5′ deletion from −200 to −192 results in loss of promoter responsiveness to H. influenzae in hTBE cells (Figure 1B). Immediately downstream from this 5′ limit of the HFRE of the ICAM-1 gene are C/EBP and NF-κB enhancer sequences that participate in gene activation in response to other inflammatory stimuli (16). ICAM-1 gene sequence starting at this 5′ limit and containing these enhancers conferred reporter gene responsiveness to H. influenzae in plasmids driven by heterologous minimal promoters and transfected into hTBE cells (Figure 2A). Mutation of either the C/EBP or NF-κB sequences in the ICAM-1 HFRE sequence abrogated promoter responsiveness to H. influenzae (Figure 2B). However, mutation of the closest AP1 site that has been reported to modulate ICAM-1 in response to other stimuli had little effect (16). Taken together, our analysis indicates that specific C/EBP and NF-κB enhancer sequences in the ICAM-1 gene mediate gene activation in response to human airway epithelial cell interaction with H. influenzae.
Figure 1.
Haemophilus influenzae increases intercellular adhesion molecule (ICAM)-1 gene transcription. (A) Relative gene transcription rates were assessed using nuclear runoff assays with human tracheobronchial epithelial (hTBE) cells incubated in media without or with H. influenzae for the indicated durations. The positions of 32P-labeled transcripts that hybridized with immobilized vector without cDNA or vector containing ICAM-1, α-actin, c-Rel, and Alu repeat sequence cDNAs are indicated by arrows. Results are representative of three experiments. (B) ICAM-1 gene promoter activity was assessed using reporter gene assays with hTBE that were transfected with a plasmid containing a luciferase gene driven by ICAM-1 gene sequence from −1294, −553, −294, −200, −192, or −134 to +7 and then incubated without or with H. influenzae for 18 hours. Values are expressed as mean fold induction in luciferase activity relative to uninfected cells ± S.D. (n = 4–7 experiments, each with duplicate samples), and a significant difference from levels with constructs driven by the ICAM-1 gene sequence from −1294 to +7 is indicated by an asterisk.
Figure 2.
H. influenzae activates a response element in the ICAM-1 gene. (A) Gene promoter activity was assessed using plasmids containing a luciferase gene driven by a heterologous minimal promoter (either TA or TK). The luciferase reporter plasmid without or with the H. influenzae response element (HFRE) (nucleotides −200 to −135 of the ICAM-1 gene sequence) placed 5′ to the minimal promoter was transfected into hTBE cells. Cells were then incubated without or with H. influenzae for 18 hours and luciferase activity was measured. (B) Gene promoter activity was assessed by transfecting hTBE cells with a plasmid containing a luciferase gene driven by ICAM-1 gene sequence from −1294 to +7 without or with mutation of the AP1-, C/EBP-, or NF-κB–binding sites in or nearest to the HFRE. Cells were then incubated without or with H. influenzae for 18 hours and luciferase activity was measured. Values in both panels are expressed as mean fold induction in luciferase activity relative to uninfected cells ± SD (n = 3–6 experiments, each with duplicate samples), and a significant difference from levels with constructs driven by TA or TK heterologous promoters alone in A and the unmutated construct in B is indicated by an asterisk.
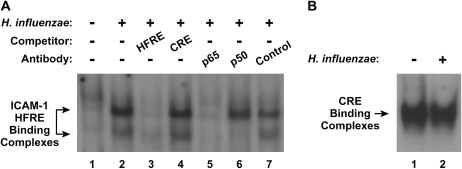
Previous reports indicate that NF-κB is required for induction of ICAM-1 expression in hTBE cells by H. influenzae (6, 10). To assess transcription factor binding to enhancer sequences identified in our promoter analysis, we performed EMSA with nuclear protein extracts from hTBE cells and oligonucleotides containing the HFRE from the ICAM-1 gene. These experiments identified specific protein complexes that bound to the ICAM-1 HFRE in extracts from hTBE cells incubated with H. influenzae, but not in extracts from uninfected cells (Figure 3A). The ICAM-1 HFRE-binding complexes were found to contain the p65 and p50 NF-κB subunits using antibody supershift analysis. Similar levels of nuclear protein binding to a cAMP response element indicated that differences in HFRE binding were not due to differences in nuclear protein isolation (Figure 3B). NF-κB–dependent induction of ICAM-1 expression in airway epithelial cells in response to bacteria was predictable based on other reports (6, 10, 16), but confirmation using primary airway epithelial cells and intact H. influenzae in our system was important for subsequent work directed at understanding C/EBP regulation of the ICAM-1 gene.
Figure 3.
H. influenzae induces NF-κB binding to the ICAM-1 gene. (A) Transcription factor binding to ICAM-1 HFRE gene sequence from −200 to −135 was assessed using electrophoretic mobility shift assay (EMSA) with nuclear extracts from hTBE cells that were incubated without or with H. influenzae for 4 hours. Specificity of protein binding was assessed by competition with unlabeled oligonucleotides containing the HFRE, but not an unrelated CRE. Protein components of the DNA-binding complexes were identified using supershift analysis by addition of antibodies against p65, p50, or control. The position of ICAM-1 HFRE-binding complexes is indicated by arrows. (B) Transcription factor binding to the CRE was assessed using EMSA with the same nuclear extracts as in A to verify equivalency of nuclear protein isolation. The position of CRE-binding complexes is indicated by an arrow. Results in both panels are representative of five experiments.
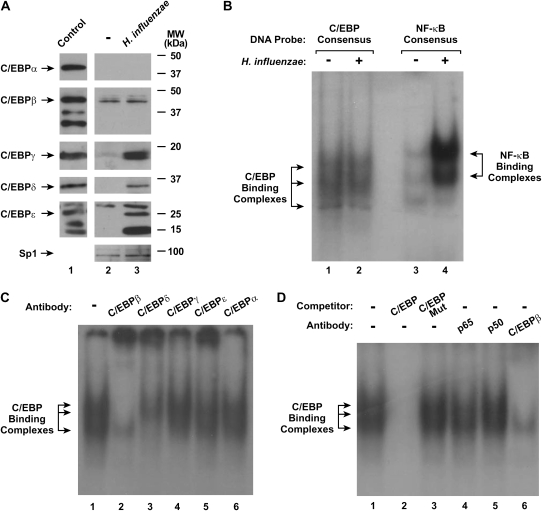
H. influenzae Induces Multiple C/EBP Family Members
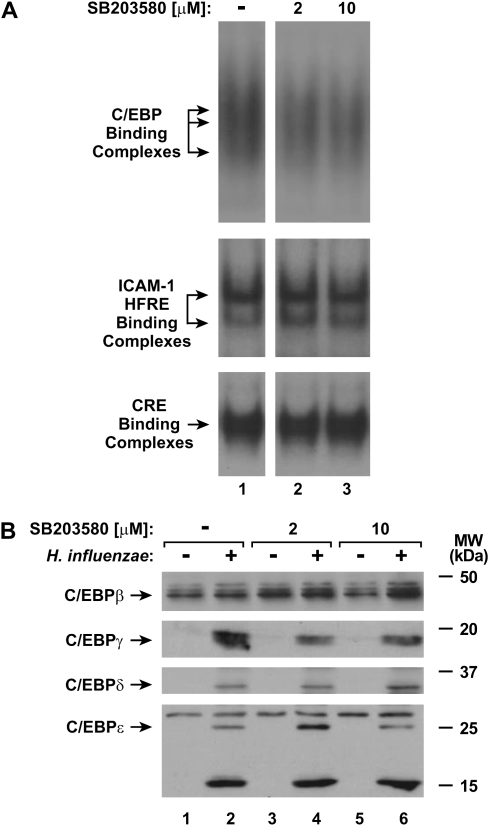
Under untreated conditions, hTBE cells expressed one form of both C/EBPβ and C/EBPɛ (Figure 4A). Treatment of cells with H. influenzae induced nuclear expression of C/EBPγ, C/EBPδ, and two smaller variants of C/EBPɛ. Attempts were made to discern specific C/EBP isoform binding to the ICAM-1 HFRE using EMSA and antibody supershift analysis, but these experiments were difficult to interpret (unpublished data). This was due to the high level of NF-κB compared with C/EBP-binding complexes generated in hTBE after exposure to H. influenzae, combined with the observation that both complexes migrated to similar gel positions in our EMSA analysis (Figure 4B). Supershift analysis using a C/EBP consensus sequence and extracts from hTBE cells treated with H. influenzae suggested that DNA-binding complexes were composed of C/EBPβ and C/EBPδ (Figure 4C). Antibodies against the C/EBPγ and C/EBPɛ (that are also expressed in H. influenzae–treated cells) had some minor effects on the complexes, suggesting that these isoforms (that are also expressed in H-influenzae–treated hTBE cells) may also participate in DNA binding. H. influenzae treatment of hTBE cells generated DNA-binding complexes that are specific for C/EBP, as EMSA with antibody supershift analysis indicated that the C/EBP-binding complex did not contain the NF-κB p65 and p50 subunits (Figure 4D). The results indicate that specific C/EBP isoforms and polypeptide variants are induced in hTBE cells treated with H. influenzae.
Figure 4.
H. influenzae affects nuclear expression of C/EBP family members. (A) C/EBP family and Sp1 (to verify equivalent isolation and loading) nuclear protein levels were assessed using immunoblot analysis of nuclear protein extracts from hTBE cell monolayers that were incubated without or with H. influenzae for 4 hours. Control samples were rat liver nuclear extract (Active Motif, Carlsbad, CA) for C/EBPα, THP-1 cell nuclear extract for C/EBPβ and C/EBPδ, purified C/EBPγ amino acids 39–147 linked to a His-tag (US Biological, Swampscott, MA), and HL-60 nuclear extract (Abcam, Cambridge, MA) for C/EBPɛ. The positions of C/EBP family members and control Sp1 are indicated by arrows. (B) Transcription factor binding to C/EBP and NF-κB consensus sequences that were similar in nucleotide number was assessed using EMSA with nuclear extracts from hTBE cell monolayers that were incubated without or with H. influenzae for 4 hours. The position of C/EBP- and NF-κB–binding complexes are indicated by arrows. (C) Transcription factor binding to a C/EBP consensus sequence was assessed using EMSA with nuclear extracts from hTBE cells that were incubated with H. influenzae for 4 hours. Supershift analysis was performed by addition of antibodies against C/EBPβ, C/EBPδ, C/EBPγ, C/EBPɛ, or C/EBPα. (D) Transcription factor binding to a C/EBP consensus sequence was assessed using EMSA with nuclear extracts from hTBE cells that were incubated with H. influenzae for 4 hours. Specificity of protein binding was assessed by competition with unlabeled oligonucleotides containing the C/EBP consensus sequence without (C/EBP) or with (C/EBP Mut) mutation. Supershift analysis was performed by addition of antibodies against NF-κB (p65 or p50) or C/EBPβ. Results in all panels are representative of three to five experiments.
H. influenzae Effects on C/EBP Family Members Are Not Regulated by p38 MAP Kinases
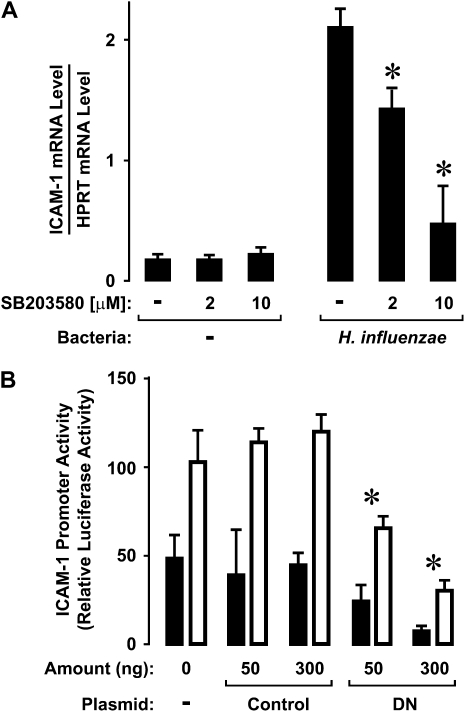
The MAP kinase family of evolutionarily conserved enzymes are activated by phosphorylation of specific threonine and tyrosine residues in response to cell surface events (33). Both extracellular signal–regulated kinase (ERK) and p38 MAP kinases are phosphorylated in hTBE cells after interaction with isolates of H. influenzae (6). However, inhibitors of ERK activation had little effect on ICAM-1 expression induced by H. influenzae (unpublished data). In contrast, pretreatment of hTBE cells with a small molecule inhibitor of the kinase function of p38 at concentrations previously demonstrated to inhibit epithelial cell ICAM-1 protein expression in response to H. influenzae also resulted in significant and dose-dependent inhibition of ICAM-1 mRNA levels (Figure 5A) (6). Similarly, ICAM-1 promoter activity assessed using a luciferase reporter plasmid driven by ICAM-1 gene sequence was significantly diminished by cotransfection of a plasmid expressing a dominant-negative form of p38 (Figure 5B). ICAM-1 promoter function appeared to require activated p38 under both basal conditions and after hTBE cell interaction with H. influenzae, suggesting involvement with a constitutive factor in ICAM-1 gene expression.
Figure 5.
H. influenzae induction of ICAM-1 expression requires p38 MAP kinase. (A) ICAM-1 mRNA levels were determined using realtime RT-PCR analysis of total RNA from hTBE cells monolayers that were left untreated or were pretreated with a p38 inhibitor at the indicated concentrations. Cells were then incubated without or with H. influenzae for 8 hours. Values are expressed as mean mRNA level compared with control HPRT mRNA ± S.D. (n = 3), and a significant difference in mRNA levels compared with cells not treated with the inhibitor is indicated by an asterisk. Results are representative of three experiments. (B) ICAM-1 gene promoter activity was assessed using reporter gene assays with hTBE cells that were cotransfected with a reporter plasmid containing a luciferase gene driven by ICAM-1 gene sequence from −1294 to +7 and either no expression plasmid, a control expression plasmid, or plasmid that expressed dominant-negative p38 (DN) in the indicated amount. Cells were then incubated without or with H. influenzae for 18 hours. Values are expressed as mean luciferase level relative to uninfected cells ± SD (n = 4 experiments, each with duplicate samples), and a significant difference between levels in cells transfected with dominant-negative p38 compared with control is indicated with an asterisk.
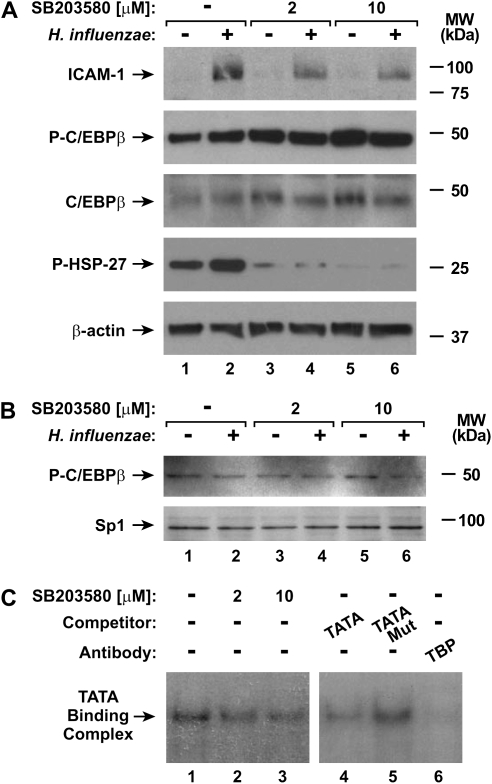
To identify mechanisms for p38 MAP kinase regulation of ICAM-1 expression in airway epithelial cells in response to H. influenzae, we first focused on potential interactions with transcription factors that regulate ICAM-1. Inhibition of p38 phosphorylation did not clearly affect C/EBP or NF-κB binding complex formation in EMSA experiments using nuclear extracts from hTBE cells pretreated with the p38 chemical inhibitor and then treated with H. influenzae (Figure 6A). Furthermore, no change in constitutive or induced expression of C/EBP isoforms or polypeptide variants was seen in cells pretreated with the p38 inhibitor (Figure 6B). The phosphorylation state of C/EBPβ also regulates its DNA-binding and gene transactivation capacity (15, 34). However, pretreatment of hTBE cells with the p38 inhibitor did not consistently affect the phosphorylation of cellular C/EBPβ, despite clearly inhibiting constitutive phosphorylation of HSP-27 and H. influenzae induction of ICAM-1 expression (Figure 7A). The level of C/EBPβ phosphorylation was also examined in nuclear extracts from hTBE cells. Similar to cellular extracts, there was no evidence of altered C/EBPβ phosphorylation in the nuclei of cells pretreated with the p38 inhibitor (Figure 7B). Taken together, these results indicate that p38 does not modulate the level or phosphorylation of C/EBPβ.
Figure 6.
Inhibiting p38 MAP kinase phosphorylation does not alter C/EBP levels. (A) Transcription factor binding to C/EBP consensus sequence, ICAM-1 gene HFRE sequence from −200 to −135, or control CRE sequence was assessed using EMSAs with nuclear extracts from hTBE cell monolayers that were left untreated or were pretreated with a p38 inhibitor at the indicated concentrations, and then incubated with H. influenzae for 4 hours. (B) C/EBP family nuclear protein levels were assessed using immunoblot analysis of nuclear protein extracts from hTBE cell monolayers that were left untreated or were pretreated with a p38 inhibitor at the indicated concentrations. Cells were then incubated without or with H. influenzae for 4 hours. Results in both panels are representative of three experiments.
Figure 7.
Inhibiting p38 MAP kinase phosphorylation alters TFIID function. (A) ICAM-1, phosphorylated and total C/EBPβ, HSP27, and β-actin protein levels were assessed using immunoblot analysis of extracts from hTBE cell monolayers that were left untreated or were pretreated with a p38 inhibitor at the indicated concentrations, and then incubated without or with H. influenzae for 24 hours. (B) Phosphorylated C/EBPβ and Sp1 nuclear protein levels were assessed using immunoblot analysis of nuclear protein extracts from hTBE cell monolayers that were left untreated or were pretreated with a p38 inhibitor at the indicated concentrations, and then incubated without or with H. influenzae for 4 hours. (C) Transcription factor binding to a TFIID consensus sequence were assessed using EMSA with nuclear extracts from hTBE cell monolayers that were left untreated or were pretreated with a p38 inhibitor at the indicated concentrations, and then incubated with H. influenzae for 4 hours. Specificity of protein binding was assessed by competition with unlabeled oligonucleotides containing the TFIID consensus sequence (TATA), but not a mutated sequence (TATA Mut). TFIID participation in the binding complex was identified using supershift analysis by addition of antibody against TBP. The position of TATA-binding complexes is indicated by an arrow. Results in all panels are representative of three experiments.
Based on a previous report indicating that p38 modulates basal transcription complex function under certain conditions through phosphorylation of TBP and affecting its capacity to bind DNA containing the TATA box sequence (35), we focused on the TBP component of the basal transcription complex. Extracts from hTBE cells pretreated with the p38 inhibitor before exposure to H. influenzae demonstrated decreased TBP binding to a TATA box–containing TFIID-binding protein consensus sequence in EMSA analysis (Figure 7C). Specificity for the TATA complexes was demonstrated by cold oligonucleotide competition and antibody supershift analysis. These findings indicate that p38 MAP kinases do not regulate ICAM-1 gene expression through direct effects on C/EBP or NF-κB, but one mechanism for p38 effects is through regulation of TBP binding to TATA box–containing promoters.
Specific C/EBP Family Members Regulate ICAM-1
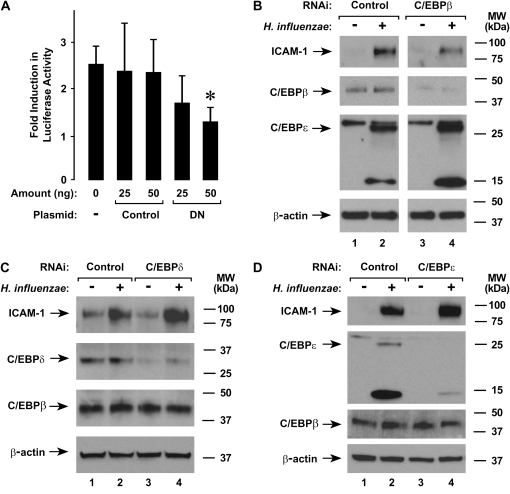
To confirm a role for C/EBP family members in regulation of ICAM-1 gene expression in hTBE cells by H. influenzae, we used two experimental approaches to inhibit the expression or function of C/EBP family members. In the first experimental system, reporter plasmids with a luciferase gene driven by ICAM-1 gene sequence were cotransfected with a plasmid that expressed a dominant-negative form of C/EBPβ without or with a mutation that affects inhibitory function. The dominant-negative form of C/EBPβ expressed by this plasmid consisted of the C-terminal 20-kD form of C/EBPβ, which occurs naturally in some cells and serves a dominant-negative function against the larger, activating forms of C/EBPβ (15). Expression of dominant-negative C/EBPβ significantly decreased luciferase activity produced by the ICAM-1 promoter reporter construct in response to H. influenzae, while the mutated dominant-negative control had little effect (Figure 8A). For the second approach, the expression of specific C/EBP proteins was decreased using RNA interference. When hTBE cells were transfected with siRNA specific for C/EBPβ and then treated with H. influenzae, ICAM-1 expression was decreased compared with cells transfected with control siRNA (Figure 8B). Specificity of the siRNA was demonstrated by showing that C/EBPβ, but not C/EBPɛ, expression was decreased. In contrast, siRNA that decreased C/EBPδ (Figure 8C) or C/EBPɛ (Figure 8D) levels had little effect on ICAM-1 expression. Similar experiments were performed using RNA interference targeted at C/EBPγ isoforms, but the siRNA tested suffered from lack of specificity (unpublished data). Taken together, these experiments indicate that C/EBPβ is required for full activation of the ICAM-1 gene promoter in hTBE cells in response to H. influenzae.
Figure 8.
H. influenzae induction of ICAM-1 expression requires C/EBPβ. (A) ICAM-1 gene promoter activity was assessed using reporter gene assays with hTBE cells that were cotransfected with a reporter plasmid containing a luciferase gene driven by ICAM-1 gene sequence from −1294 to +7 and either no expression plasmid or a plasmid that expressed either dominant-negative C/EBPβ with (Control) or without mutation (DN) in the indicated amount. Cells were then incubated without or with H. influenzae for 18 hours. Values are expressed as mean fold induction in luciferase activity relative to uninfected cells ± SD (n = 3 experiments, each with duplicate samples), and a significant difference between levels in cells transfected without and with dominant-negative C/EBPβ is indicated with an asterisk. (B–D) ICAM-1, C/EBPβ, C/EBPδ, C/EBPɛ, and β-actin protein levels were assessed using immunoblot analysis of extracts from hTBE cell monolayers that were transfected with control siRNA or siRNA against (B) C/EBPβ, (C) C/EBPδ, or (D) C/EBPɛ. Cells were then incubated without or with H. influenzae for 18 hours. Results are representative of three to five experiments.
DISCUSSION
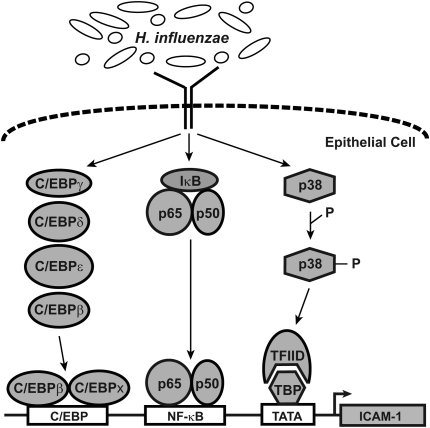
Expression of ICAM-1 and other defense genes in airway epithelial cells in response to bacterial infection is mediated directly by epithelial cell interaction with bacteria or their products and/or indirectly through communication with other cells via soluble mediators (4, 36, 37). Direct contact between H. influenzae and airway epithelial cells has been demonstrated during airway infection with attachment being mediated by specific bacterial cell surface molecules such as adhesins (38, 39). Other bacterial surface molecules (e.g., lipooligosaccharide, P6 outer membrane protein) as well as soluble bacterial factors have been demonstrated to initiate epithelial cell gene expression (10, 11, 40). Thus, epithelial cells are activated by multiple bacterial molecules during airway infection with H. influenzae, and the coordination of responses to these stimuli results in activation of multiple signaling pathways that control ICAM-1 expression (Figure 9). Binding of C/EBPβ and NF-κB to enhancer sequences close to the transcription start site is critical in regulation of the ICAM-1 gene in airway epithelial cells. Phosphorylation of p38 MAP kinases is also important in ICAM-1 gene regulation through effects on TBP binding to the TATA box and initiation of transcription. This complex, multitiered system of signaling pathways allows for modulation of both enhancesome and basal transcription complex function in the ICAM-1 gene, thereby permitting rapid and precise control of expression levels.
Figure 9.
Pathways that regulate ICAM-1 expression. Model of signaling pathways that interact in airway epithelial cells to regulate ICAM-1 expression in response to H. influenzae.
The ICAM-1 gene HFRE contains a C/EBP-binding sequence that is required for maximal gene expression in response to H. influenzae, as well as specific inflammatory stimuli (41, 42). Airway epithelial cells express multiple C/EBP family members, either constitutively or after interaction with H. influenzae. Formation of homo- or hetero-dimers is required for DNA binding and gene regulation by these transcription factors, and thus multiple C/EBP dimer combinations may form that have different effects on gene expression (15). Our results demonstrated that C/EBPβ and C/EBPɛ are expressed under baseline conditions in hTBE cells, while exposure to H. influenzae induced the expression of C/EBPγ and C/EBPδ. In addition, two additional variants of C/EBPɛ were detected after epithelial cell exposure to this bacterium, providing additional diversity for possible dimers. This finding is interesting in light of reports that C/EBPɛ expression is thought to be restricted primarily to myeloid and lymphoid cells (15). Using dominant-negative and RNA interference strategies, the participation of C/EBPβ in H. influenzae–induced ICAM-1 expression was confirmed. Using specific siRNA, we found no evidence that C/EBPδ or C/EBPɛ have a primary role in induction of ICAM-1 in hTBE cells in response to H. influenzae. As some of the smaller C/EBP family members (e.g., C/EBPγ, the liver inhibitory protein variant of C/EBPβ, and the 14-kD form of C/EBPɛ) lack an intact activation domain and thus can serve dominant-negative functions, it seems unlikely that these peptides mediate ICAM-1 gene activation (43). C/EBPα was not expressed in hTBE cells, and C/EBPζ was not investigated as it has been shown to not bind the C/EBP consensus DNA sequence, which is similar to the sequence in the ICAM-1 gene HFRE (44). Taken together, it appears that C/EBPβ has a primary role in ICAM-1 gene regulation in airway epithelial cells. C/EBPβ can function as a homodimer and may also undergo a cooperative interaction with NF-κB to regulate gene expression (17, 45).
Although C/EBPβ and C/EBPɛ are constitutively expressed in hTBE cells, it is possible that additional C/EBP activation is required for full H. influenzae induction of ICAM-1 expression. Phosphorylation of C/EBPβ affects its DNA-binding and transactivation capacity, and can be induced through one of multiple signaling pathways present in epithelial cells (15, 34). Signaling pathways reported to regulate C/EBPβ phosphorylation at Thr235 include p38 and ERK MAP kinases, and phosphatidylinositol-3 (PI3) kinase (34, 46, 47). However, we found no clear evidence that C/EBPβ phosphorylation at Thr235 was affected by epithelial cell interaction with H. influenzae or by p38 MAP kinase inhibition. This correlated with results showing that the p38 inhibitor had no effect on C/EBP binding to the ICAM-1 HFRE. Chemical inhibitors of ERK and PI3 kinases also did not affect levels of total or phosphorylated C/EBPβ in hTBE cells (unpublished data). These findings indicate that C/EBPβ (likely phosphorylated) is constitutively bound to the ICAM-1 HFRE, and suggest that other post-translational modifications of C/EBPβ or the level of other C/EBP family members control ICAM-1 expression, or that C/EBP proteins directly facilitate NF-κB DNA binding or effects on gene activation (17, 45, 48).
The final common pathway from detection of bacteria (including H. influenzae) to increased expression of many inflammatory genes is through induction of the transcription-activating complex NF-κB (6, 10). Activated NF-κB has been shown to regulate ICAM-1 expression in several cell types (41, 49–53). Of the NF-κB/Rel family, which includes p65 (RelA), p50 (NF-κB1), p52 (NF-κB2), RelB, and Rel (c-Rel), the ICAM-1 gene HFRE in human airway epithelial cells binds p65 and p50, likely as a heterodimer that is the most prevalent transactivating form of NF-κB (54). It has been demonstrated that this enhancer site can also bind (and may have a preference for) p65 homodimers, and our results support this possibility in epithelial cells after interaction with H. influenzae (49–52).
There appears to be cell type and stimulus specificity in transcription factor control of ICAM-1 expression. Much of the previous work on NF-κB–dependent regulation of ICAM-1 focused on TNF-α–induced activation. Similar to H. influenzae effects on primary hTBE cells, induction of ICAM-1 expression in A549 cells in response to TNF-α is regulated by NF-κB and C/EBPβ (55). However, although NF-κB and C/EBPβ binding to the ICAM-1 gene HFRE has been demonstrated using nuclear extracts from TNF-α–treated A549 and HeLa cells, and the endothelial cell line EVC304, this was not seen with Jurkat cells (55). In addition, there appears to be cell type and stimulus specificity for C/EBP family member participation in ICAM-1 expression, as C/EBPδ has been shown to function with C/EBPβ to control ICAM-1 in astrocytes in response to TNF-α and interleukin-1β (56). However, C/EBPα and C/EBPβ bind ICAM-1 gene sequence using TNF-α–treated Hep G2 cells extracts, while C/EBPδ does not (41). Other reports indicate that activator protein-1 (AP-1) is involved in TNF-α induction of ICAM-1 expression in A549 and ECV304 cells (57, 58), which is also different than hTBE cells. Although some differences in regulation could be due to culture or other experimental conditions, the different patterns of transcription factors that control ICAM-1 expression demonstrates the complexity of regulation of this and other airway defense genes in response to inflammatory stimuli. Thus, assumptions that similar cells types respond to stimuli in the same way may not be valid, particularly when complex stimuli (such as bacteria) are involved.
Our studies indicate that ICAM-1 expression is also modulated by p38 MAP kinases, similar to many genes that participate in the inflammatory response. H. influenzae has been shown to modulate expression of other genes using p38, but these kinases can increase or decrease expression depending on the gene and conditions tested (10–12, 59). In contrast, despite activation of ERK kinases and reports that these kinases participate in epithelial cell IL-8 expression in response to H. influenzae and other mediators, there was no evidence in our studies that ERK kinases control bacterial induction of ICAM-1 expression in hTBE cells (11, 60, 61). These findings are in light of conflicting reports on the role of MAP kinases in epithelial cell ICAM-1 expression, in which discrepancies in results likely reflect differences in cell models, stimuli, and inhibitory strategy (53, 62, 63). Furthermore, mechanisms for p38 effects on epithelial defense gene expression in response to H. influenzae have not been well defined. MAP kinases phosphorylate nuclear and cytoplasmic proteins, thereby regulating cellular gene expression by modulating transcription factor or basal transcription complex function, mRNA stability, and/or translation efficiency (64). Interaction between MAP kinases and NF-κB–dependent gene expression has been identified in the regulation of several genes that mediate inflammation (35, 61). Our results indicate that one mechanism for p38 MAP kinase effects is through control of TBP binding to the ICAM-1 gene promoter. This p38 effect likely affects the function of many epithelial genes up-regulated by H. influenzae.
Epithelial cells are often the first to encounter bacteria in the airway, and can respond to intact bacteria or bacterial products. Many pathogen-associated molecular patterns are recognized by members of the Toll-like receptor (TLR) family of surface proteins on cells in the airway (65, 66). One important example is detection of a component of the outer membrane of gram-negative bacteria called lipopolysaccharide (LPS) by host cell TLR4 resulting in inflammatory gene activation. Haemophilus species synthesize a form of LPS, referred to as lipooligosaccharide (LOS), that contains an oligosaccharide linked to lipid A without repeating subunit O-antigen polysaccharide chains. However, airway epithelial cells are poorly responsive to LOS and LPS, suggesting that these bacterial molecules are not directly responsible for defense gene activation after epithelial cell interaction with H. influenzae (4). In contrast, the P6 outer membrane lipoprotein of H. influenzae has been shown to activate NF-κB and other signaling pathways through binding to TLR2, and this bacterial molecule may regulate epithelial cell responses (10). However, we have found that purified H. influenzae P6 is a poor inducer of epithelial cell ICAM-1 expression (unpublished data). Other H. influenzae factors that could control epithelial cell gene activation include LOS glycoforms containing phosphorylcholine that bind to the platelet-activating factor receptor, and small cytoplasmic molecules that activate multiple epithelial cell signaling pathways (11, 40). Although intact bacteria were used for our studies, bacterial products were likely also present in this model system. This parallels the situation in the airway, where epithelial cells are exposed to multiple bacterial factors, as well as the bacteria themselves, and therefore must coordinate all of the induced signals into an appropriate defense response.
The results from our studies indicate that multiple signaling pathways that are active in airway epithelial cells after exposure to H. influenzae regulate ICAM-1 gene transcription by differential modulation of enhancesome and basal transcription complex function. To accumulate these findings, multiple strategies for signaling pathway inhibition were used to modulate ICAM-1 gene expression in hTBE cells in response to H. influenzae. These strategies include use of small molecule inhibitors, dominant-negative protein expression, and RNA interference. Although specificity or inefficient delivery currently limit use of these approaches in vivo, these inhibitory strategies could be improved in the future to alter inflammation that is detrimental to host airway function. Although airway epithelial cells express multiple C/EBP family members, C/EBPβ appears to play a primary role in ICAM-1 gene regulation and targeting its expression or function may be one approach to modulate ICAM-1 expression and airway inflammation. A better understanding of cellular mechanisms that control airway defense responses may allow for the development of therapeutic strategies that selectively modify inflammation that causes decreased pulmonary function without decreasing beneficial defense functions inherent in airway epithelium.
Acknowledgments
The authors gratefully acknowledge R. Davis, W. Greene, P. Gunning, T. Ley, C. Stratowa, D. Staunton, and the Cell Culture and Tissue Core Facility of the University of Iowa Center for Gene Therapy for generous gifts of cells and reagents; B. Carter, M. Monick, and G. Hunninghake for helpful discussion; and B. Nardy and N. Laurie for technical assistance.
This research was supported by grants from the National Heart, Lung, and Blood Institute (grant numbers R01HL065752 and R01HL082505 to D.C.L.) and Cystic Fibrosis Foundation. The University of Iowa Center for Gene Therapy is supported by grants from the National Institutes of Health and Cystic Fibrosis Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
Originally Published in Press as DOI: 10.1165/rcmb.2008-0104OC on August 14, 2008
Conflict of Interest Statement: M.A.B. is employed by Integrated DNA Technologies (IDT), Inc., which offers oligonucleotides for sale similar to some of the compounds described in this manuscript. However, IDT is not a publicly traded company and he does not own shares or equity in IDT. None of the other authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.van Alphen L, Caugant DA, Duim B, O'Rourke M, Bowler LD. Differences in genetic diversity of nonencapsulated Haemophilus influenzae from various diseases. Microbiology 1997;143:1423–1431. [DOI] [PubMed] [Google Scholar]
- 2.Muhlebach MS, Stewart PW, Leigh MW, Noah TL. Quantitation of inflammatory responses to bacteria in young cystic fibrosis and control patients. Am J Respir Crit Care Med 1999;160:186–191. [DOI] [PubMed] [Google Scholar]
- 3.Hill AT, Campbell EJ, Hill SL, Bayley DL, Stockley RA. Association between airway bacterial load and markers of airway inflammation in patients with stable chronic bronchitis. Am J Med 2000;109:288–295. [DOI] [PubMed] [Google Scholar]
- 4.Frick AG, Joseph TD, Pang L, Rabe AM, St. Geme JW III, Look DC. Haemophilus influenzae stimulates ICAM-1 expression on respiratory epithelial cells. J Immunol 2000;164:4185–4196. [DOI] [PubMed] [Google Scholar]
- 5.Humlicek AL, Pang L, Look DC. Modulation of airway inflammation and bacterial clearance by epithelial cell ICAM-1. Am J Physiol 2004;287:L598–L607. [DOI] [PubMed] [Google Scholar]
- 6.Chin CL, Manzel LJ, Lehman EE, Humlicek AL, Shi L, Starner TD, Denning GM, Murphy TF, Sethi S, Look DC. Haemophilus influenzae from patients with chronic obstructive pulmonary disease exacerbation induce more inflammation than colonizers. Am J Respir Crit Care Med 2005;172:85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dustin ML, Rothlein R, Bhan AK, Dinarello CA, Springer TA. Induction by IL 1 and interferon-γ: tissue distribution, biochemistry, and function of a natural adherence molecule (ICAM-1). J Immunol 1986;137:245–254. [PubMed] [Google Scholar]
- 8.Aldallal N, McNaughton EE, Manzel LJ, Richards AM, Zabner J, Ferkol TW, Look DC. Inflammatory response in airway epithelial cells isolated from patients with cystic fibrosis. Am J Respir Crit Care Med 2002;166:1248–1256. [DOI] [PubMed] [Google Scholar]
- 9.Albelda SM, Smith CW, Ward P. Adhesion molecules and inflammatory injury. FASEB J 1994;8:504–512. [PubMed] [Google Scholar]
- 10.Shuto T, Xu H, Wang B, Han J, Kai H, Gu X, Murphy TF, Lim DJ, Li J. Activation of NF-κB by nontypeable Hemophilus influenzae is mediated by toll-like receptor 2–TAK1-dependent NIK-IKK α/β-IκBα and MKK3/6-p38 MAP kinase signaling pathway in epithelial cells. Proc Natl Acad Sci USA 2001;98:8774–8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang B, Cleary PP, Xu H, Li J. Up-regulation of interleukin-8 by novel small cytoplasmic molecules of nontypeable Haemophilus influenzae via p38 and extracellular signal-regulated kinase pathways. Infect Immun 2003;71:5523–5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B, Lim DJ, Han J, Kim YS, Basbaum CB, Li J. Novel cytoplasmic proteins of nontypeable Haemophilus influenzae up-regulate human MUC5ac mucin transcription via a positive p38 mitogen-activated protein kinase pathway and a negative phosphoinositide 3-kinase-AKT pathway. J Biol Chem 2002;277:949–957. [DOI] [PubMed] [Google Scholar]
- 13.Newcomb DC, Sajjan U, Nanua S, Jia Y, Goldsmith AM, Bentley JK, Hershenson MB. Phosphatidylinositol 3-kinase is required for rhinovirus-induced airway epithelial cell interleukin-8 expression. J Biol Chem 2005;280:36952–36961. [DOI] [PubMed] [Google Scholar]
- 14.Poli V. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J Biol Chem 1998;273:29279–29282. [DOI] [PubMed] [Google Scholar]
- 15.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function, and regulation. Biochem J 2002;365:561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roebuck KA, Finnegan A. Regulation of intercellular adhesion molecule-1 (CD54) gene expression. J Leukoc Biol 1999;66:876–888. [DOI] [PubMed] [Google Scholar]
- 17.Stein B, Baldwin ASJ. Distinct mechanisms for regulation of the interleukin-8 gene involves synergism and cooperativity between C/EBP and NF-κB. Mol Cell Biol 1993;13:7191–7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Look DC, Roswit WT, Frick AG, Gris-Alevy Y, Dickhaus DM, Walter MJ, Holtzman MJ. Direct suppression of Stat1 function during adenoviral infection. Immunity 1998;9:871–880. [DOI] [PubMed] [Google Scholar]
- 19.Joseph TD, Look DC. Specific inhibition of interferon signal transduction pathways by adenoviral infection. J Biol Chem 2001;276:47136–47142. [DOI] [PubMed] [Google Scholar]
- 20.Look DC, Pelletier MR, Holtzman MJ. Selective interaction of a subset of interferon-γ response element binding proteins with the intercellular adhesion molecule-1 (ICAM-1) gene promoter controls the pattern of expression on epithelial cells. J Biol Chem 1994;269:8952–8958. [PubMed] [Google Scholar]
- 21.Staunton DE, Merluzzi VJ, Rothlein R, Barton R, Marlin SD, Springer TA. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell 1989;56:849–853. [DOI] [PubMed] [Google Scholar]
- 22.Erba HP, Gunning P, Kedes L. Nucleotide sequence of the human gamma cytoskeletal actin mRNA: anomalous evolution of vertebrate non-muscle actin genes. Nucleic Acids Res 1986;14:5275–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ballard DW, Dixon EP, Peffer NJ, Bogerd H, Doerre S, Stein B, Greene WC. The 65-kDa subunit of human NF-kappa B functions as a potent transcriptional activator and a target for v-Rel-mediated repression. Proc Natl Acad Sci USA 1992;89:1875–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanson RD, Connolly NL, Burnett D, Campbell EJ, Senior RM, Ley TJ. Developmental regulation of the human cathepsin G gene in myelomonocytic cells. J Biol Chem 1990;265:1524–1530. [PubMed] [Google Scholar]
- 25.Voraberger G, Schafer R, Stratowa C. Cloning of the human gene for intercellular adhesion molecule 1 and analysis of its 5′-regulatory region. J Immunol 1991;147:2777–2786. [PubMed] [Google Scholar]
- 26.Williams SC, Baer M, Dillner AJ, Johnson PF. Crp2 (C/EBP beta) contains a bipartite regulatory domain that controls transcriptional activation, DNA binding and cell specificity. EMBO J 1995;14:3170–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davis RJ. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem 1995;270:7420–7426. [DOI] [PubMed] [Google Scholar]
- 28.Look DC, Pelletier MR, Tidwell RM, Roswit WT, Holtzman MJ. Stat1 depends on transcriptional synergy with Sp1. J Biol Chem 1995;270:30264–30267. [DOI] [PubMed] [Google Scholar]
- 29.Ramaswamy M, Shi L, Monick MM, Hunninghake GW, Look DC. Specific inhibition of type I interferon signal transduction by respiratory syncytial virus. Am J Respir Cell Mol Biol 2004;30:893–900. [DOI] [PubMed] [Google Scholar]
- 30.Ramaswamy M, Shi L, Varga SM, Barik S, Behlke MA, Look DC. Respiratory syncytial virus nonstructural protein 2 specifically inhibits type I interferon signal transduction. Virology 2006;344:328–339. [DOI] [PubMed] [Google Scholar]
- 31.Humlicek AL, Manzel LJ, Chin CL, Shi L, Excoffon KJ, Winter MC, Shasby DM, Look DC. Paracellular permeability restricts airway epithelial responses to selectively allow activation by mediators at the basolateral surface. J Immunol 2007;178:6395–6403. [DOI] [PubMed] [Google Scholar]
- 32.Zar JH. Biostatistical analysis. Englewood Cliffs, NJ: Prentice-Hall, Inc.; 1984.
- 33.Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev 2001;81:807–869. [DOI] [PubMed] [Google Scholar]
- 34.Piwien-Pilipuk G, MacDougald O, Schwartz J. Dual regulation of phosphorylation and dephosphorylation of C/EBP modulate its transcriptional activation and DNA binding in response to growth hormone. J Biol Chem 2002;277:44557–44565. [DOI] [PubMed] [Google Scholar]
- 35.Carter AB, Knudtson KL, Monick MM, Hunninghake GW. The p38 mitogen-activated protein kinase is required for NF-kappaB-dependent gene expression. The role of TATA-binding protein (TBP). J Biol Chem 1999;274:30858–30863. [DOI] [PubMed] [Google Scholar]
- 36.Look DC, Keller BT, Rapp SR, Holtzman MJ. Selective induction of intercellular adhesion molecule-1 by interferon-γ in human airway epithelial cells. Am J Physiol 1992;263:L79–L87. [DOI] [PubMed] [Google Scholar]
- 37.DiMango E, Zar HJ, Bryan R, Prince A. Diverse Pseudomonas aeruginosa gene products stimulate respiratory epithelial cells to produce interleukin-8. J Clin Invest 1995;96:2204–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hers JFP, Mulder J. The mucosal epithelium of the respiratory tract in muco-purulent bronchitis caused by Haemophilus influenzae. J Pathol Bacteriol 1953;56:103–108. [DOI] [PubMed] [Google Scholar]
- 39.St. Geme JW III. Molecular determinants of the interaction between Haemophilus influenzae and human cells. Am J Respir Crit Care Med 1996;154:S192–S196. [DOI] [PubMed] [Google Scholar]
- 40.Swords WE, Ketterer MR, Shao J, Campbell CA, Weiser JN, Apicella MA. Binding of the non-typeable Haemophilus influenzae lipooligosaccharide to the PAF receptor initiates host cell signalling. Cell Microbiol 2001;3:525–536. [DOI] [PubMed] [Google Scholar]
- 41.Hou J, Baichwal V, Cao Z. Regulatory elements and transcription factors controlling basal and cytokine-induced expression of the gene encoding intercellular adhesion molecule 1. Proc Natl Acad Sci USA 1994;91:11641–11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chini BA, Fiedler MA, Milligan L, Hopkins T, Stark JM. Essential roles of NF-kappaB and C/EBP in the regulation of intercellular adhesion molecule-1 after respiratory syncytial virus infection of human respiratory epithelial cell cultures. J Virol 1998;72:1623–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooper C, Henderson A, Artandi S, Avitahl N, Calame K. Ig/EBP (C/EBP gamma) is a transdominant negative inhibitor of C/EBP family transcriptional activators. Nucleic Acids Res 1995;23:4371–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ron D, Habener JF. Chop, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev 1992;6:439–453. [DOI] [PubMed] [Google Scholar]
- 45.Matsusaka T, Fujikawa K, Nishio Y, Mukaida N, Matsushima K, Kishimoto T, Akira S. Transcription factors NF-IL6 and NF-kappa B synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc Natl Acad Sci USA 1993;90:10193–10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aouadi M, Jager J, Laurent K, Gonzalez T, Cormont M, Binétruy B, Le Marchand-Brustel Y, Tanti JF, Bost F. P38MAP kinase activity is required for human primary adipocyte differentiation. FEBS Lett 2007;581:5591–5596. [DOI] [PubMed] [Google Scholar]
- 47.Pham T, Langmann S, Schwarzfischer L, El Chartouni C, Lichtinger M, Klug M, Krause SW, Rehli M. CCAAT enhancer-binding protein β regulates constitutive gene expression during late stages of monocyte to macrophage differentiation. J Biol Chem 2007;282:21924–21933. [DOI] [PubMed] [Google Scholar]
- 48.Cui TX, Piwien-Pilipuk G, Huo JS, Kaplani J, Kwok R, Schwartz J. Endogenous CCAAT/enhancer binding protein beta and p300 are both regulated by growth hormone to mediate transcriptional activation. Mol Endocrinol 2005;19:2175–2186. [DOI] [PubMed] [Google Scholar]
- 49.Ledebur HC, Parks TP. Transcriptional regulation of the intercellular adhesion molecule-1 gene by inflammatory cytokines in human endothelial cells: essential roles of a variant NF-kappa B site and p65 homodimers. J Biol Chem 1995;270:933–943. [DOI] [PubMed] [Google Scholar]
- 50.Aoudjit F, Brochu N, Bélanger B, Stratowa C, Hiscott J, Audette M. Regulation of intercellular adhesion molecule-1 gene by tumor necrosis factor-alpha is mediated by the nuclear factor-kappaB heterodimers p65/p65 and p65/c-rel in the absence of p50. Cell Growth Differ 1997;8:335–342. [PubMed] [Google Scholar]
- 51.Paxton LL, Li LJ, Secor V, Duff JL, Naik SM, Shibagaki N, Caughman SW. Flanking sequences for the human intercellular adhesion molecule-1 NF-kappaB response element are necessary for tumor necrosis factor alpha-induced gene expression. J Biol Chem 1997;272:15928–15935. [DOI] [PubMed] [Google Scholar]
- 52.Rahman A, Anwar KN, True AL, Malik AB. Thrombin-induced p65 homodimer binding to downstream NF-κB site of the promoter mediates endothelial ICAM-1 expression and neutrophil adhesion. J Immunol 1999;162:5466–5476. [PubMed] [Google Scholar]
- 53.Holden NS, Catley MC, Cambridge LM, Barnes PJ, Newton R. ICAM-1 expression is highly NF-κB-dependent in A549 cells - no role for ERK and p38 MAPK. Eur J Biochem 2004;271:785–791. [DOI] [PubMed] [Google Scholar]
- 54.Tak PP, Firestein GS. NF-κB: a key role in inflammatory diseases. J Clin Invest 2001;107:7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Catron KM, Brickwood JR, Shang C, Li Y, Shannon MF, Parks TP. Cooperative binding and synergistic activation by RelA and C/EBPβ on the intercellular adhesion molecule-1 promoter. Cell Growth Differ 1998;9:949–959. [PubMed] [Google Scholar]
- 56.Lee SJ, Hou J, Benveniste EN. Transcriptional regulation of intercellular adhesion molecule-1 in astrocytes involves NF-kappaB and C/EBP isoforms. J Neuroimmunol 1998;92:192–207. [DOI] [PubMed] [Google Scholar]
- 57.Chen C, Chow M, Huang W, Lin Y, Chang Y. Flavonoids inhibit tumor necrosis factor-α-induced upregulation of intercellular adhesion molecule-1 (ICAM-1) in respiratory epithelial cells through activator protein-1 and nuclear factor-κB: structure-activity relationships. Mol Pharmacol 2004;66:683–693. [DOI] [PubMed] [Google Scholar]
- 58.Kobuchi H, Roy S, Sen CK, Nguyen HG, Packer L. Quercetin inhibits inducible ICAM-1 expression in human endothelial cells through the JNK pathway. Am J Physiol 1999;277:C403–C411. [DOI] [PubMed] [Google Scholar]
- 59.Shuto T, Imasato A, Juno H, Sakai A, Xu H, Watanabe T, Rixter DD, Kai H, Andalibi A, Linthicum F, et al. Glucocorticoids synergistically enhance nontypeable Haemophilus influenzae-induced toll-like receptor 2 expression via a negative cross-talk with p38 MAP kinase. J Biol Chem 2002;277:17263–17270. [DOI] [PubMed] [Google Scholar]
- 60.Li J, Johnston D, Iazvovskaia S, Tan A, Lin A, Hershenson MB. Signaling intermediates required for NF-κB activation and IL-8 expression in CF bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol 2003;284:L307–L315. [DOI] [PubMed] [Google Scholar]
- 61.Zhou L, Tan A, Iasvovskaia S, Li J, Lin A, Hershenson MB. Ras and mitogen-activated protein kinase kinase kinase-1 coregulate activator protein-1- and nuclear factor-κB-mediated gene expression in airway epithelial cells. Am J Respir Cell Mol Biol 2003;28:762–769. [DOI] [PubMed] [Google Scholar]
- 62.Tamanini A, Rolfini R, Nicolis E, Melotti P, Cabrini G. MAP kinases and NF-κB collaborate to induce ICAM-1 gene expression in the early phase of adenovirus infection. Virology 2003;307:228–242. [DOI] [PubMed] [Google Scholar]
- 63.Lin FS, Lin CC, Chien CS, Luo SF, Yang CM. Involvement of p42/p44 MAPK, JNK, and NF-κB in IL-1β-induced ICAM-1 expression in human pulmonary epithelial cells. J Cell Physiol 2005;202:464–473. [DOI] [PubMed] [Google Scholar]
- 64.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature 2001;410:37–40. [DOI] [PubMed] [Google Scholar]
- 65.Akira S. Toll-like receptor signaling. J Biol Chem 2003;278:38105–38108. [DOI] [PubMed] [Google Scholar]
- 66.Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science 2003;300:1524–1525. [DOI] [PubMed] [Google Scholar]