Abstract
Bacteriophage T4 is a large-tailed E. coli virus whose capsid is 120 × 86 nm. ATP-driven DNA packaging of the T4 capsid results in the loading of a 171-kb genome in less than 5 minutes during viral infection. We have isolated 50-mg quantities of uniform 15N and [ε-15N]lysine-labeled bacteriophage T4. We have also introduced 15NH4+ into filled, unlabeled capsids from synthetic medium by exchange. We have examined lyo- and cryoprotected lyophilized T4 using 15N{31P} and 31P{15N} rotational-echo double resonance. The results of these experiments have shown that: (i) packaged DNA is in an unperturbed duplex B-form conformation; (ii) the DNA phosphate negative charge is balanced by lysyl amines (3.2%), polyamines (5.8%), and monovalent cations (40%); and (iii) 11% of lysyl amines, 40% of –NH2 groups of polyamines, and 80% of monovalent cations within the lyophilized T4 capsid, are involved in the DNA charge balance. The NMR evidence suggests that DNA enters the T4 capsid in a charge-unbalanced state. We propose that electrostatic interactions may provide free energy to supplement the nanomotor-driven T4 DNA packaging.
Keywords: DNA conformation, isotopic labeling, phage nanomotors, solid-state NMR
Introduction
Bacteriophage T4
Much attention has been given to phage T4, a large double-stranded DNA (dsDNA) tailed bacteriophage specific for E. coli, because its DNA polymerase, ligase, and kinase are widely used as reagents for DNA recombination.1 Consequently, the proteins involved in phage T4 assembly are well studied and the T4 nucleotide sequence has been completely determined.2 Moreover, in vitro assemblies of phage T4 and various mutants are possible.1 Although some basic mechanisms of phage T4 assembly have been established, there remain several unanswered challenging questions. We are most interested in the mechanism of DNA packaging, the crucial step of T4 assembly detailed below. T4 packages DNA faster than any other known bacteriophage3 and is an obvious choice for study.
Lytic life cycle of double-stranded DNA tailed bacteriophage T4
The lytic life cycle of phage T4 consists of four steps:1,2 (i) Adsorption and penetration. The tail fibers bind to receptors on the bacterial cell surface, the phage sheath contracts, and the phage tail penetrates the outer membrane of the bacterial cell wall, leading to the injection of phage DNA into the E. coli host cell. The DNA injection rate for phage T4 is about 4000 bp/s. (ii) Synthesis of early proteins. A set of early proteins is synthesized immediately after the injection of viral DNA. The proteins include an enzyme that degrades the host DNA, and a virus-specific DNA polymerase that replicates phage DNA. (iii) Synthesis of late proteins. Each of the many copies of phage DNA is used for transcription and translation of a set of late proteins, including lysozyme and the structural proteins used to make up the phage capsid and tail. (iv) Assembly process, DNA packaging, and release of the mature phage. In the late lytic life cycle of phage T4, the procapsid and tail are formed via a self-assembly process inside the host. The phage T4 DNA, 171 kb in length, is packaged into the procapsid by an ATP-driven motor resulting in a mature capsid. The tail and accessory proteins are then attached to the capsid to form a mature phage particle. Finally, lysozyme accomplishes lysis of the host cell to release mature phages. There are usually 50 to 200 phage particles produced per infected host cell.
DNA packaging in bacteriophage T4
The tightly packaged phage dsDNA (~500 mg/ml) exerts an internal pressure of tens of atmospheres on the capsid due to electrostatic repulsion and the stress from bending the DNA duplex.4 Compared to classical motors such as myosin II which generate only ~3 pN of force,5 phage DNA motors generate larger forces to overcome high internal pressure during packaging.3,6–9 For example, the phage φ29 packaging motor exerts forces up to 80 pN,9 while phage λ and phage T4 DNA packaging motors generate forces greater than 50 pN and 60 pN, respectively.3,7 The DNA packaging rate for phage T4 (with an average of 690 bp/s and ranging up to 1,840 bp/s) is at least 4 times faster than that of phage φ29 (about 165 bp/s).3
The phage T4 DNA packaging motor, a terminase holoenzyme, consists of two components, gp16 and gp17. The large subunit gp17 is a 70-kDa ATPase and the small subunit gp16 is an 18-kDa protein. The exact stoichiometery of the subunits in the holoenzyme is unknown.10 Gp17 alone is sufficient to package DNA in vitro.11,12
Single-molecule experiments have shown that the phage φ29 DNA motor translocates DNA by 2 base pairs per ATP hydrolysis.6 The estimated free energy release per ATP hydrolysis is sufficient to translocate DNA by as much as 6 base pairs,3 assuming the motor generates a force of 60 pN. To account for the fastest observed T4 DNA packaging rate of 1,840 bp/s,3 an ATP-driven phage T4 motor would have to hydrolyze 18,400 ATPs per minute, if each ATP hydrolysis translocates DNA by 6 bp. To hydrolyze this much ATP, each motor must either contain an active 46-mer of gp17 ATPase (a 5-mer is the current best estimate),10 or the in vivo hydrolysis rate for gp17 must be about 9 times the in vitro rate of 400 ATPs/gp17/min.11 Neither explanation seems plausible. Another possibility is that the step size, the number of base pairs translocated per ATP hydrolysis, is much greater than 6 bp. However, this can only be true if there is an additional packaging force. Electrostatic interactions could potentially provide the extra free energy needed and therefore the charge neutralization scheme in phage T4 is relevant to an understanding of the DNA packaging mechanism.
Charge balance within the T4 capsid
By analyzing 31P{15N} and 15N{31P} rotational-echo double-resonance13 (REDOR) dephasing of uniform 15N-labeled T4, specific [ε-15N]lysine-labeled T4, and 15NH4+-exchanged unlabeled phage T4, we determined that packaged T4 DNA has a B-form conformation, and that the DNA phosphate negative charges are partially balanced by lysyl amines (3.2%), polyamines (5.8%), and monovalent cations (40%). Presumably, the remaining phosphate charge is balanced by divalent cations. The N-P distance between nitrogens of the lysyl sidechains and the nearest-neighbor DNA phosphates is 3.5 Å, consistent with N-H…O-P hydrogen bonding.14 In addition, the N-P distances between the amine nitrogens of polyamines and nearby DNA phosphate groups are 3.5 Å and 5.5 Å, which means that some amine groups of polyamines provide full charge balance while others are more distant from phospates and provide only partial charge balance. The fact that some of the phosphate charge balance is due to amine groups that are inside the capsid before the DNA is loaded, leads to the suggestion that DNA may be charged unbalanced during entry, and electrostatics may therefore play a role in packaging.
Results
Conformation of DNA in bacteriophage T4
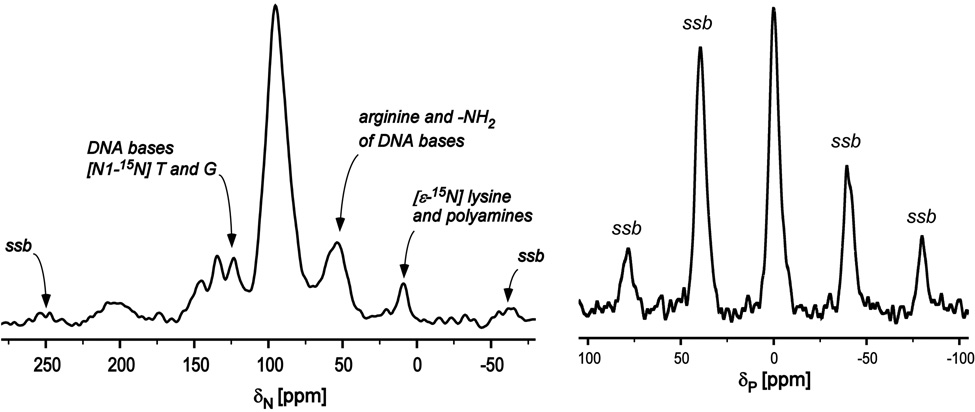
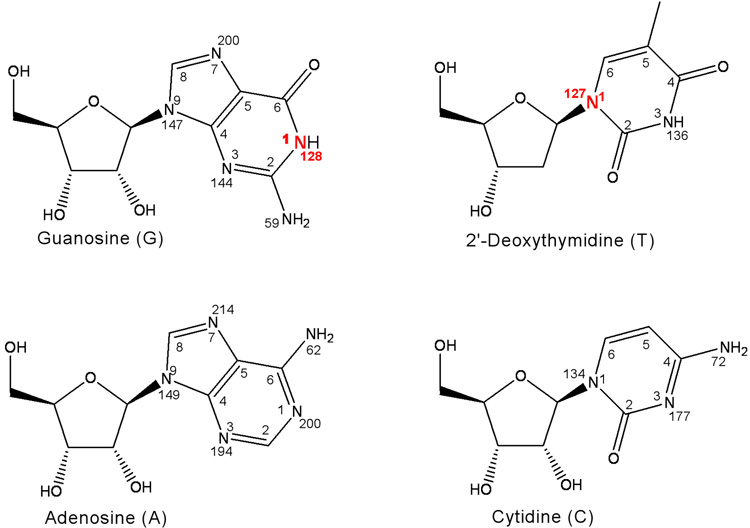
A 15N CPMAS spectrum of uniform 15N labeled phage T4 (Figure 1, left) has an intense peak at 95 ppm due to the peptide backbone, and a weaker peak at 9 ppm from both polyamines and lysyl amines. The peaks from 40 to 75 ppm are due to the sidechain nitrogens of arginine and the –NH2 moeity of DNA bases (Figure 2). The peaks between 125 and 150 ppm have contributions from three types of nitrogens: (i) the N1 nitrogens of DNA bases guanosine and thymidine at 127 ppm, (ii) the N1 nitrogens of cytidine and the N3 nitrogens of thymidine at 135 ppm, and (iii) the N9 nitrogens of adenosine, guanosine, and the N3 nitrogens of guanosine and histidine sidechains, all at 145 ppm (Figure 2).15
Figure 1.
(Left) 15N CPMAS spectrum for uniform 15N labeled phage T4. (Right) 31P CPMAS spectrum for uniform 15N labeled phage T4. Both spectra resulted from 2048 scans. Spinning sidebands are identified as “ssb.” Magic angle spinning for both experiments was at 8,000 Hz.
Figure 2.
Structures, atom-numbering schemes and 15N isotropic chemical shifts for adenosine, guanosine, 2’-deoxythymidine, and cytidine.
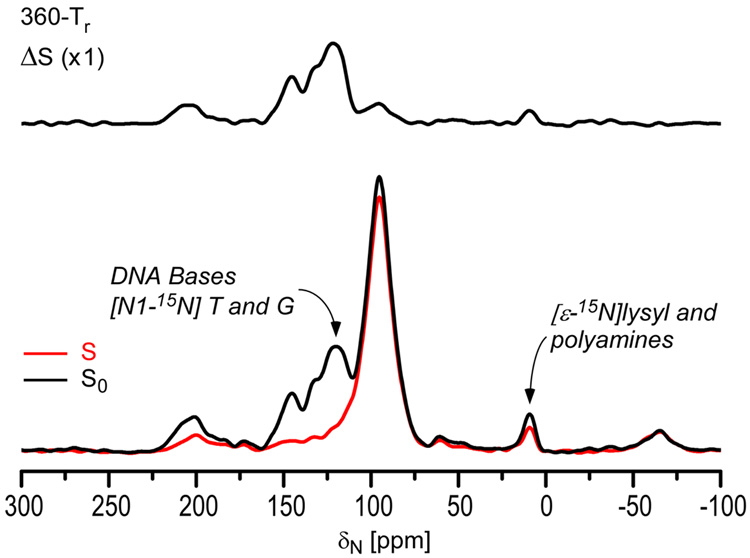
Figure 1 (right) shows a 31P CPMAS spectrum of uniform 15N labeled phage T4. The spinning sideband pattern, resulting from the large chemical shift anisotropy (CSA) of 31P of DNA phosphates, indicates that there is no large-amplitude motion of the DNA phosphate backbone. The minor dephasing at 95 ppm indicates DNA contact with capsid or internal proteins (cf, below). We assign the substantial 15N{31P} REDOR dephasing of uniform 15N labeled T4 for the peaks between 125 and 150 ppm (Figure 3) to nitrogens of DNA bases that are in proximity to DNA phosphates.
Figure 3.
15N{31P} REDOR spectra of uniform 15N labeled phage T4 after dipolar evolution for 360 Tr (45 ms). The REDOR difference is shown at the top and the normal full-echo spectrum (black) and diminished echo spectrum (red) at the bottom. The spectra were the result of the accumulation of 13,752 scans.
The C3’-endo sugar ring pucker is characteristic of A-DNA, while B-DNA has a C2’-endo sugar-ring pucker.16,17 This difference in sugar conformation between A-DNA and B-DNA results in different distances between nitrogens of DNA bases and phosphorus of the DNA backbone. For example, as shown in Figure 4, the average distance between N1 of guanosine and the two nearest phosphorus atoms is 5.78 Å for B-DNA (1d56.pdb), and 5.35 Å for A-DNA (1vj4.pdb). Thus, it appears that the 15N{31P} REDOR dephasing of the peak at 127 ppm might be used to determine the conformation of encapsulated phage T4 DNA.
Figure 4.
Conformations of B-DNA (1d56.pdb) and A-DNA (1vj4.pdb) of Table 1. Phosphorus atoms (blue) and nitrogen atoms (red) are highlighted.
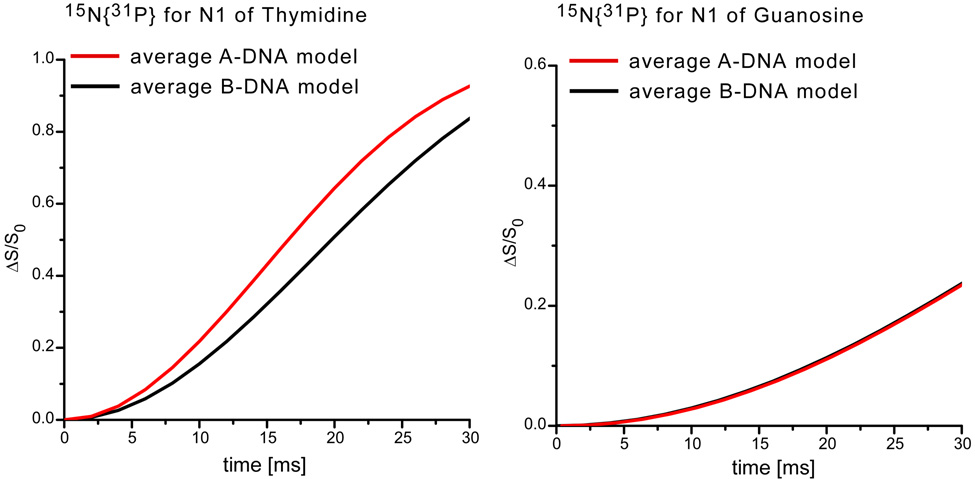
To explore this possibility, eleven A-DNAs and twelve B-DNAs were selected (Table 1) from the Nucleic Acid Database (NDB) to calculate a representative average 15N{31P} REDOR dephasing as a function of dipolar evolution time for the N1 nitrogens of thymine and guanosine in T4 DNA. REDOR dephasing for an N1 nitrogen of one of the DNAs, coupled to all phosphorus atoms with coordinates specified by the specific DNA structure, was calculated using standard methods.13,18 Individual contributions were summed and normalized. The results are shown in Figure 5. The calculated dephasing for the N1 nitrogens of thymine in B-DNA deviates significantly from that calculated for the corresponding nitrogens in A-DNA (Figure 5, left). However, there is no distinguishable difference in dephasing for the N1 nitrogens of guanosine (Figure 5, right). Fortunately, T4 DNA has twice as much thymine as guanosine.2
Table 1.
DNA structures selected from the Nucleic Acid Database (NDB) for the calculation of 15N{31P} REDOR dephasing of N1 nitrogens of thymine and guanosine. The selected thymine and guanosine for each DNA structure are bolded. All the phosphorus atoms in each DNA structure were used in the calculation.
| B-DNA | A-DNA | ||
|---|---|---|---|
| NDB ID | Sequence | NDB ID | Sequence |
| BDJ036 | CGATATATCG | ADH006 | GGGGCCCC |
| GCTATATAGC | CCCCGGGG | ||
| BDJ037 | CGATATATCG | ADH010 | GGTATACC |
| GCTATATAGC | CCATATGG | ||
| BDJB48 | CGATCGATCG | ADH029 | GGGCGCCC |
| GCTAGCTAGC | CCCGCGGG | ||
| BDJB49 | CCAGGCCTGG | ADH030 | GGGTACCC |
| GGTCCGGACC | CCCATGGG | ||
| BDJB50 | CCAGGCCTGG | ADH031 | GGGTACCC |
| GGTCCGGACC | CCCATGGG | ||
| BDL001 | CGCGAATTCGCG | ADH054 | GAAGCTTC |
| GCGCTTAAGCGC | CTTCGAAG | ||
| BDL015 | CGCAAAAATGCG | AD1009 | GGATGGGAG |
| GCGTTTTTTACGC | CCTACCCTC | ||
| BDL020 | CGCGAATTCGCG | ADJ049 | CCCGGCCGGG |
| GCGCTTAAGCGC | GGGCCGGCCC | ||
| BDL029 | CGTGAATTCACG | ADJ050 | GCGGGCCCGC |
| GCACTTAAGTGC | CGCCCGGGCG | ||
| BDL038 | CGCAAATTTGCG | ADL025 | CCCCCGCGGGGG |
| GCGTTTAAACGC | GGGGGCGCCCCC | ||
| BDL042 | CGTAGATCTACG | ADL046 | GCGTACGTACGC |
| GCATCTAGATGC | CGCATGCATGCG | ||
| BDLBO3 | CGCGAATTCGCG | ||
| GCGCTTAAGCGC | |||
Figure 5.
Calculated 15N{31P} REDOR dephasing as a function of dipolar evolution time for the N1 of thymine (left) and the N1 of guanosine (right) for an A-DNA model (red) and a B-DNA model (black).
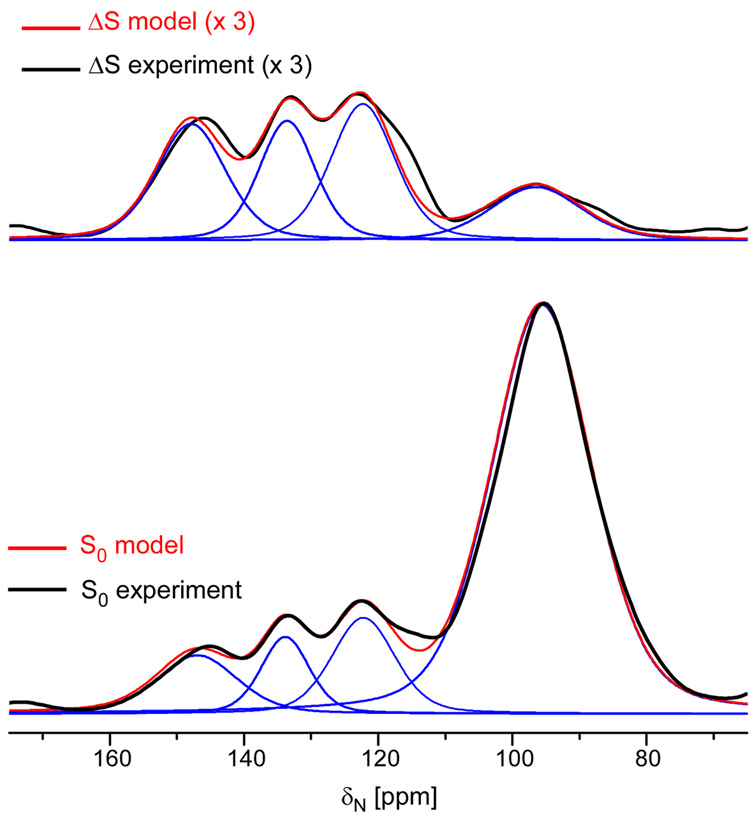
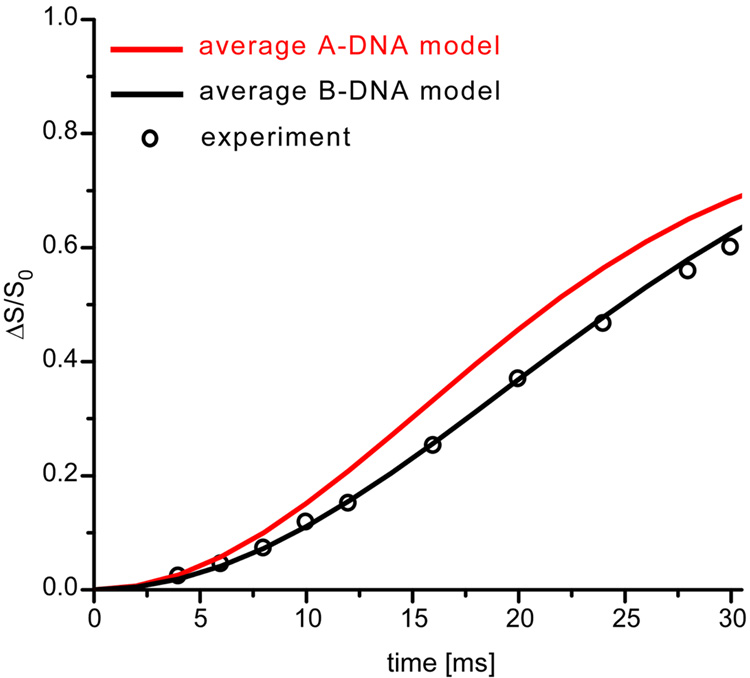
Deconvolution was used19 to analyze the experimental 15N{31P} REDOR dephasing of the N1 peak at 127 ppm. The spectrum between 75 and 160 ppm (Figure 6), was deconvoluted into four peaks at 95, 127, 135 and 145 ppm, based on the chemical shift values of DNA base nitrogens (Figure 2). The same lineshapes were used to analyze both the REDOR difference spectrum (ΔS) and the full-echo spectrum (S0). Comparison of the experimental 15N{31P} REDOR dephasing at 127 ppm to the calculated values assuming either A-DNA or B-DNA models (Figure 7), shows unambiguously that encapsulated T4 dsDNA is in an unperturbed B-DNA conformation.
Figure 6.
Expanded view of experimental (black line) and model (red line) 15N{31P} REDOR difference spectrum (top) and the full-echo spectrum (bottom) after 192 Tr. Deconvolution allows an estimate of the intensity of the N1 peak of DNA bases T and G (blue line).
Figure 7.
Experimental (circles) and calculated (solid lines) 15N{31P} REDOR dephasing (ΔS/S0) for the 127-ppm N1 peak of thymine and guanosine of phage T4 DNA, assuming an A-DNA model (red) or a B-DNA model (black). Estimated errors are within the symbols.
Contact between amines and DNA phosphates
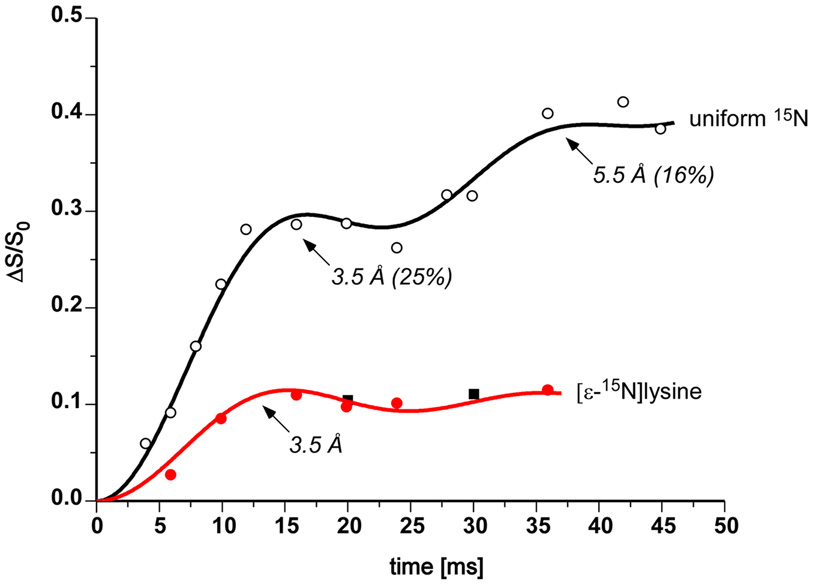
A substantial number of amines (lysyl amines and polyamines) are proximate to phage DNA phosphates because significant 15N{31P} REDOR dephasing is observed at 9 ppm for uniform 15N-labeled T4 (Figure 3, top). Similar contact for arginine and histidine sidechains is not observed. The analysis of the amine dephasing as a function of dipolar evolution time (Figure 8, open circles) shows that 25% of the amines have an N-P separation of just 3.5 Å and are therefore likely a part of N-H…O-P hydrogen bonds. These amines are primary charge-balance contributors. An additional 16% of the amines have a 5.5-Å N-P separation and we classify these amines as secondary charge-balance contributors.
Figure 8.
15N{31P} REDOR dephasing (ΔS/S0) for the 9-ppm amine peak as a function of dipolar evolution time for uniform 15N and [ε-15N]lysine-labeled bacteriophage T4. The observed dephasing (solid circles and squares) of [ε-15N]lysine-labeled phage T4 (100 mg lysine/L media, red; and 200 mg lysine/L, black) is consistent with that calculated (red line) assuming a single 15N-31P distance of 3.5 Å. The observed dephasing (open circles) of uniform 15N-labeled phage T4 is consistent with that calculated assuming an 15N-31P distance of 3.5 (± 0.2) Å for 25 (± 3) % of the amines, and 5.5 Å for 16%.
The biosynthesis of polyamines starts from arginine, which belongs to a different biosynthetic family than does lysine.20,21 Thus, only lysyl amines contribute to the 9-ppm peak of 15N spectra of [ε-15N]lysine-labeled T4. The 15N{31P} REDOR dephasing for this peak (Figure 8, solid red circles) shows that the N-P distance between lysyl amines and their nearest-neighbor DNA phosphates is also 3.5 Å. The dephasing maximum, which occurs after 16 ms of dipolar evolution, is 11%; that is, 11% of all lysyl amines are hydrogen bonded to phage T4 DNA phosphates.
Neither arginine sidechain nor histidine sidechain nitrogens have significant contact with DNA phosphates, based on the absence of dephasing for the 15N{31P} REDOR spectra of uniform 15N labeled T4 in the region between 40 and 75 ppm, and at 150 ppm (Figure 3, top). The dephasing for the peak at 145 ppm is mainly from the –NH2 moeity of DNA bases. These results suggest that arginine and histidine are not actively involved in the phage T4 DNA charge neutralization. In contrast, all three positively charged amino acid residues (lysine, arginine, and histidine residues) are involved in DNAhistone interactions to fold DNA into the nucleosome core particle.22,23 Why phage T4 DNA only interacts with lysyl amine but not the other two positively charged sidechain nitrogens remains an open question.
DNA phosphate charge balanced by lysyl amines and polyamines
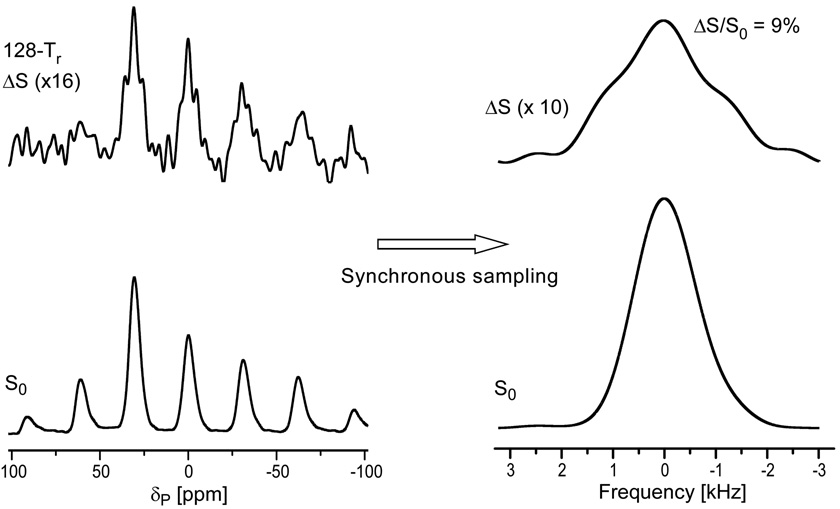
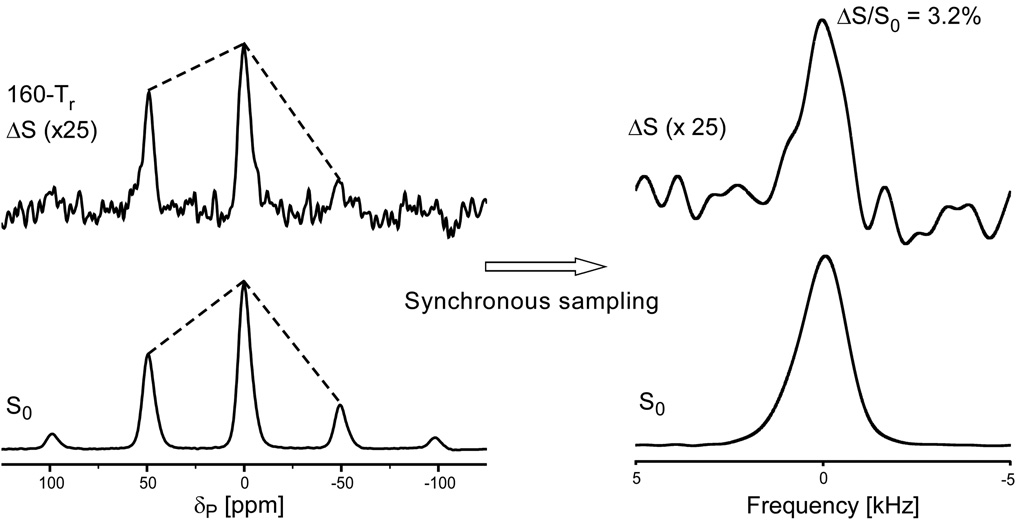
As described in the previous section, substantial numbers of polyamines and lysyl amines contribute to the T4 DNA charge balance. To determine quantitatively the percentage of T4 DNA phosphate charge that is balanced by amines, 31P{15N} DANTE-based frequency-selective REDOR24 (dbFSR, see Methods) was performed on uniform 15N-labeled phage T4. With the frequency selectivity provided by DANTE pulses,25 31P{15N} dbFSR reintroduces the dipolar interaction between DNA 31P and amine 15N only. Thus, we determined that 9% of the DNA phosphate charge is balanced by the combination of lysyl amines and polyamines (Figure 9).
Figure 9.
31P{15N} dbFSR spectra of uniform 15N-labeled phage T4 after 128 Tr of dipolar evolution. The full-echo spectrum (S0) is at the bottom of the figure and the REDOR difference spectrum (ΔS) is at the top. The DANTE carrier frequency was centered at the 9-ppm amine peak. The spectra on the left show sidebands resulting from magic-angle spinning at 6250 Hz. The NMR signal was digitized at 64 times the spinning speed so that the Fourier transform of every 64th time-domain data point resulted in the rotor-synchronized spectra on the right. The spectra were the result of the accumulation of 316,968 scans.
We also performed 31P{15N} REDOR NMR for [ε-15N]lysine-labeled phage T4 to determine that the percentage of T4 DNA charge that is balanced by lysyl amines is 3.2% (Figure 10). The dephasing maximum did not change when the concentration of 15N-labeled lysine in the growth medium was doubled, consistent with an isotopic enrichment of the ε-nitrogen of 95% or more.26 By comparison of this value to the 9% 31P{15N} dbFSR dephasing maximum of uniform 15N-labeled phage T4 (Figure 9), we conclude from the difference that the percentage of DNA charge that is balanced by polyamines is 5.8%.
Figure 10.
31P{15N} REDOR spectra of [ε-15N]lysine-labeled phage T4 after 160 Tr of dipolar evolution. The full-echo spectrum (S0) is at the bottom of the figure and the REDOR difference spectrum (ΔS) is at the top. The dissimilarity of REDOR difference and full-echo spectra (dotted lines) indicates an orientational preference for the 15N-31P internuclear vector relative to the 31P chemical shift tensor. The NMR signal was digitized at 40 times the spinning speed (10 kHz) so that the Fourier transform of every 40th time-domain data point resulted in the rotor-synchronized spectra on the right. The spectra were the result of the accumulation of 373,792 scans.
If we assume that the fraction of –NH2 groups of polyamines that have an N-P amine-phosphate separation of 3.5 Å is α, and a separation of 5.5 Å is 1-α, and if the number of lysyl amines is X and that of polyamine –NH2 groups is Y, then the two 15N{31P} dephasing plateaus of Figure 8 are: (0.11X + αY)/(X + Y) = 0.25 and (0.11X + Y)/(X + Y) = 0.41. In addition, 0.11X/αY = 3.2/5.8 = 0.55, which is the percentage of DNA charge balanced by lysyl amines relative to that charge balanced by amines in polyamines (Figure 9 and Figure 10). Because these three experimental values are ratios, there are only two parameters, X/Y and α, and the system is overdetermined. A reasonable fit gives X = 2Y and α = 0.40; that is, there are half as many amine groups in polyamines as in lysyl sidechains, and 40% of the –NH2 groups of polyamines are hydrogen bonded to T4 DNA phosphate (N-P distance of 3.5 Å). The remaining 60% are involved in a partial charge balance (N-P distance of 5.5 Å). The three experimental ratios of 0.25, 0.41, and 0.55 are estimated as 0.20, 0.40, and 0.55, respectively, by X = 2Y and α = 0.40. A better match could be obtained by assuming a slightly higher total concentration of polyamines and a smaller value for α, and allowing some of the amine groups of the polyamines to be remote from phosphates and so not contributing to the 15N{31P} dephasing.
DNA phosphate charge balance by monovalent cations
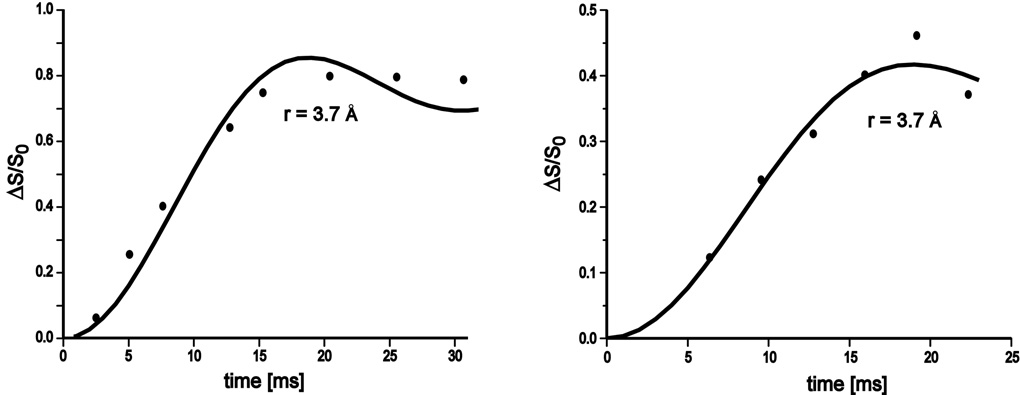
We prepared 15NH4+-exchanged phage T4 to determine the charge-screening effect of monovalent cations. Exchange did not occur at 37 °C but required an elevated temperature of 45 °C. The 15N{31P} and 31P{15N} REDOR dephasing as function of dipolar evolution time (Figure 11) establish that 80% of encapsulated ammonium cations account for 40% of the T4 DNA charge balance, assuming NH4+ replaces monovalent cations completely but cannot replace multivalent cations in bacteriophage T4.
Figure 11.
Experimental (closed circles) and calculated (solid lines) 15N{31P} (left) and 31P{15N} (right) REDOR dephasing (ΔS/S0) as a function of dipolar evolution time for 15NH4+-exchanged phage T4. The calculations assumed a single 15N-31P distance of 3.7 (± 0.2) Å.
Discussion
Local conformation of packaged DNA in bacteriophage T4
The REDOR analysis of intact phage T4 (Figure 5–Figure 9) shows quantitatively that the encapsidated dsDNA is predominately in a B-form conformation. Previously, the presence of a B-form conformation for dsDNA (in phage T7) had been inferred qualitatively from Raman difference spectra.27 The B-DNA conformation is considered the lowest energy state of double-helical DNA under physiological conditions.17 Its presence as the dominant form for T4 DNA indicates that phage DNA packaging does not significantly perturb the local B-DNA conformation.
Mullaney & Black showed that in vivo staphylococcal nuclease digestion of encapsidated DNA gives an interesting pattern by gel analysis, predominately a 160-bp repeat.28 This result supports a spiral-fold model where the encapsidated phage T4 dsDNA folds into about 700 to 800 segments of 160 to 300 bp of B-form DNA duplexes, arranged in 15 or 16 shells.29 This model is consistent with the present REDOR analysis. The number of bends or turns is small and would not be detected in the results of Figure 5–Figure 9. Other packing models28 are also possible, as long as the deviations from the B-form conformation are minor.
Balance of DNA phosphate charge
It has been suggested that a small number of basic internal proteins could account for the neutralization of about 5% of the DNA charge.30 Phage T4 contains some 1,100 molecules of three basic, lysine-rich internal proteins (iP1, iP2, and iP3).1 These three internal proteins are packaged with the DNA and contain about a quarter of the total lysine content of phage T4. REDOR analysis shows that 11% of lysyl amines account for 3.2% of the DNA charge balance (Figure 9 and Figure 10), consistent with the idea that the internal proteins are indeed used for T4 DNA charge balance.
The role of polyamines in the charge neutralization of phage T4 is somewhat controversial.4,30,31 Polyamine-deficient E. coli mutants can serve as hosts for the production of phage T4 (although the rate of production is slower than with wild type E. coli30), leading Earnshaw et al.4 to propose that polyamines are not directly involved in DNA packaging. In addition, even though intracapsid polyamines are injected into bacteria with the DNA during phage T2 infection,31 a subsequent function has not been established. Nevertheless, the titration curves of polyamines show that all of the amino groups are protonated at pH 7,30 and extraction experiments suggest that phage T4 contains enough intracapsid putrescine and spermidine to neutralize as much as one-third to one-half of the phage DNA charge30,32 REDOR analysis of phage T4 grown with wild-type E. coli confirms that intracapsid polyamines play a role in balancing T4 DNA charge (Figure 9), not at a 50% level, but at a more modest 5–10% level.
Smith et al. observed that both the DNA packaging velocity and the internal pressure of phage φ29 vary significantly with a change in ionic condition.8 They concluded that ionic screening strongly affects motor function and the packaging force. The primary cations found in phage T4 are Mg2+, Ca2+, Na+, K+, and NH4+.30 Isotopically labeled ammonium ion (15NH4+) is a proven probe of alkali ion-binding sites on DNA and in enzymes.33,34 We believe that the combination of 15NH4+ exchange with REDOR detection is well suited to study the location and charge screening of monovalent cations for packaged phage DNA. Thus, the REDOR-determined N-P distance between 15N-labeled ammonium cations and DNA phosphates in T4 is 3.7 Å (Figure 11), indicating that the ammonium cations are hydrogen bonded to T4 DNA phosphates. The 15N{31P} and 31P{15N} dephasing maxima establish quantitatively that 80% of monovalent cations in the capsid balance 40% of the total DNA charge.
Orientation of lysyl sidechains bound to T4 DNA phosphate
REDOR NMR measures dipolar coupling between pairs of spins and yields not only internuclear distances but also relative orientations that correlate the arrangement of dipolar and CSA tensors. Both O’Connor & Schaefer and Stueber et al. have shown that any difference in the REDOR dephasing rates of spinning sidebands is proof of a preferred CSA-dipolar orientation.35,36 Thus, the implication of the observed difference in sideband dephasing rates in the 31P{15N} REDOR spectra of [ε-15N]lysine-labeled T4 (Figure 10, left) is the existence of a preferred average orientation for the 15N-31P internuclear vector relative to the 31P chemical shift tensor.
We have suggested that lysine-rich internal proteins (iP1, iP2, and iP3) account for the primary lysine-phosphate contact in T4. We now argue that lysyl sidechains bind to phosphates with a preferred distance and orientation, which implies the existence of specific internal protein-DNA binding structures. The three-dimensional structure of internal protein 1 has been solved by solution-state NMR.37 All of the lysine sidechains are on one face of the structure, with the lysine C1 carbons separated from one another by just over 7 Å. The lysine sidechains therefore appear to be positioned to bind to packaged DNA, possibly stabilizing the turns in Black’s spiral-fold capsid model.28
Electrostatic forces in phage T4 DNA packaging
It seems reasonable to assume that the DNA charge is completely screened by cations and a DNA duplex and T4 capsid are neutral at the start of the packaging process. Nevertheless, we hypothesize that the DNA charge is only partially screened when DNA is in the entry channel of the capsid. We argue that negatively charged DNA in the entry channel could experience both attractive and repulsive forces assisting packaging. The attractive force would be due to the electrostatic interaction between negatively charged DNA and lysines and polyamines that had become positively charged inside the capsid.1 The repulsive force would be due to the electrostatic interaction between negatively charged DNA and the negatively charged DNA entry channel.38 For the repulsive force to assist in packaging however, the portal must be more negatively charged than the channel; that is, there must be a negative-charge gradient from portal to channel. Presumably, neutral DNA could pass through the portal driven by the ATP-powered nanomotor, become charge unbalanced possibly by the competition of nearby inorganic phosphate for cations, and then be accelerated away from the portal and through the channel by electrostatic repulsions.
Whether such an hypothesis is correct is an open question. The following five observations support that the notion that charge balance and electrostatic interactions somehow affect the phage DNA packaging rate: (i) Smith et al. observed that both DNA packaging speed and the internal pressure of phage φ29 vary significantly with a change in ionic conditions.8 They concluded that ionic screening strongly affects the DNA motor function and the packaging force.8 (ii) Hafner et al. grew phage λ and phage T4 with a polyamine-deficient E. coli strain.20 They observed that phage λ production was only 1% of the expected value, and that the phage T4 production rate had decreased by one-third compared to growth with wild type E.coli.20 (iii) Rao & Black reported that spermidine was required for efficient DNA packaging in vitro.39 (iv) Black has reported a ~2 minute lag at 30 °C in formation of phage was observed for mutants of T4 lacking the internal proteins.40 (v) Simpson et al. showed that the DNA entry channel of phage φ29 is 75 Å in length and 36 Å in diameter, and is lined with helices with exposed negative charges.40 These authors proposed that the DNA entry channel might repel DNA, permitting smooth passage during packaging and ejection.38 We argue that the repulsive force due to the negatively charged channel acts more directly by providing free energy to help package DNA rapidly. This hypothesis could be tested by optical-tweezer measurements of DNA packaging rates into prohead mutants lacking internal proteins and polyamines.9,40
Materials and Methods
Bacteriophage T4 growth and purification
Bacteriophage T4 (ATCC 11303-B4) and E.coli B (ATCC 11303) were both purchased from American Type Culture Collection. Ultrafiltered nuclease-free grade 1 water (Solution 2000 water purifier, model 2002BL; Aqua Solution, Inc., Jasper, GA) was used in all preparations.
E. coli B cells in suspension were grown in a defined medium, a modification of M9 at pH 7.2, which contained the following on a per liter basis: 7.5 g of Na2HPO4·2H2O, 3 g of KH2PO4, 0.5 g of NaCl, 1 g of NH4Cl, 5 g of D-glucose, 11.5 µg of ZnSO4·7H2O, 19.5 µg of Na2MoSO4, 22.5 µg of CuSO4·5H2O, 13.5 µg of MnSO4· H2O, 19 µg of CoCl2·6H2O, 1 mg of Thiamine-HCl, 1mM of MgSO4, 100 µM of CaCl2 and 10 µM of FeCl2.
Typically, phage T4 lysate was prepared by infecting an E. coli B culture at 37 °C, shaking at 200 rpm in an Environ-Shaker (Lab-lines Instruments, Inc., Melrose Park IL). The initial infection was at a multiplicity of infection (MOI) equal to 3 followed by the superinfection at the same MOI ten minutes later. The MOI at which infection occurs is crucial to all experiments. The culture was incubated at 37 °C with vigorous shaking until cell lysis occurred. To complete cell lysis, 10 ml chloroform per liter of lysate was added. All lysate were centrifuged at 5,000 r.p.m. in a Sorvall GS-3 rotor (4,225 g) for 20 minutes to remove debris. Precipitation was then performed by adding NaCl and polyethylene glycol (PEG) 8000 to a final concentration of 0.5 M and 10%, respectively. The lysate was stirred at 4 °C for 12 hours and spun in a Sorvall GS-3 rotor at 7000 r.p.m. (8,281 g) for 30 minutes. The phage pellet was resuspended in TSG buffer, containing 0.01 M Tris-HCl (pH 7.5) and 0.15 M NaCl, followed by pancreatic DNase I (Sigma-Aldrich, St. Louis, MO) treatment according to Carlson et. al.1 PEG precipitation was repeated and the phage T4 pellet was resuspended in the TSG buffer. DEAE-cellulose anion-exchange chromatography was used to further purify the phage particles and the purity of each collected fraction was examined according to Reddy et al.41 The purified phage containing fractions were finally ultracentrifuged in a Beckman Ti 50.2 rotor at 50,000 g for 20 minutes to obtain the phage T4 pellet.
Uniform 15N labeling and [ε-15N]lysine labeling
To prepare uniform 15N labeled phage T4 samples, the cultures were grown in modified M9 in which the ammonium chloride was replaced by 15NH4Cl (Sigma-Aldrich, St. Louis, MO). For the [ε-15N]lysine labeling scheme, the culture was grown with modified M9 medium supplemented with L-[ε-15N]lysine at a concentration of either 100 or 200 mg/L (Cambridge Isotope Laboratory Inc. Andover MA). All the phage T4 samples were lyophilized for NMR measurements. Prior to lyophilization, each T4 pellet was resuspended in 3 ml of buffer containing 5 mM HEPES, 10 mM trehalose as a lyoprotectant, and 0.5% PEG 8000 as a cryoprotectant. Samples were frozen with liquid nitrogen and then lyophilized for 72 hours. A plaque assay performed on a phage suspension from a small portion of a lyophilized sample determined that the viability of lyophilized phage T4 to be 14%.
15NH4+-exchanged bacteriophage T4
The purified, unlabeled phage pellet was resuspended in a modified TSG buffer containing 0.01 M Tris-HCl and 0.15 M 15NH4Cl (the pH was adjusted to 7.5 with Tris-Base). The phage suspension was incubated at 37 °C for 16 hours and then heated to 45 °C for 2 hours to facilitate 15N-ammonium ion exchange. Cation exchange in T4 capsids does not occur readily at 37 °C. Phage T4 particles were then spun in a Beckman Ti 50.2 rotor at 50,000 g for 20 minutes and the pellet was rinsed and resuspended with cryo- and lyoprotected buffer followed by centrifugation (at 50,000 g) and lyophilization.
REDOR and DNA packaging
Rotational-echo double resonance (REDOR) NMR, a distance measurement, was used to study the conformation of intracapsid dsDNA and to estimate the amount of DNA phosphate charge balanced by couterions (monovalent cations, lysyl amines and polyamines…etc) in proximity to DNA phosphates.
REDOR NMR measures heteronuclear dipolar coupling between pairs of spins. REDOR is performed in two parts and both parts start with the establishment of S-spin magnetization, the observed spin, via a cross-polarization transfer from protons. In the first half, no dephasing pulses are applied and full-intensity rotational echoes are formed at the end of each rotor period. In the second half, additional rotor-synchronized π pulses are applied to the I spin, the dephasing spin, in the center of the rotor period, and the diminished rotational echoes are observed. The spectrum acquired in the first half is designated as the S0 spectrum, while that acquired in the second half is the S spectrum. The difference in signal intensity between the two spectra (ΔS = S0 - S) depends on the dipolar coupling between I spin and S spin and hence the corresponding I-S internuclear distance.
Spectrometer
All NMR experiments were performed using a spectrometer equipped with a four-frequency (HFPN) transmission-line probe.36 The probe has a 10.55-mm long, 5.12-mm i.d. analytical coil, and a Chemagnetics/Varian magic-angle spinning stator. The spectrometer was controlled by a Tecmag pulse programmer. Radio frequency (rf) pulses for 1H (499.882 MHz) and 19F (470.274MHz) were amplified, for the first stage, by 50-W American Microwave Technology power amplifiers (AMT 3137), followed by second-stage amplification using 2-kW and 1-kW Creative Electronics (CE) tube amplifiers, respectively. For 31P(202.355 MHz) and 15N (50.654 MHz), rf pulses were amplified by 1-kW and 2-kW American Microwave Technology power amplifiers, respectively. The amplifiers were under active control. A 12-Tesla static magnetic field was provided by an 89-mm bore Magnex superconducting solenoid.
Cross-polarization magic-angle spinning (CPMAS) and REDOR NMR
The phage T4 samples were packed in a 4-mm ceramic rotor and spun at 6,250, 8,000 or 10,000 Hz. The spinning speed was under active control to within ±3 Hz. Rotors were spun at 8,000 Hz (Tr = 125 µs) for 15N-observe CPMAS, 31P-observe CPMAS, and 15N{31P} REDOR NMR for uniform 15N labeled and [ε-15N]lysine-labeled phage T4. Unless otherwise noted, parameters included: 2-ms 55-kHz 1H-15N Hartmann-Hahn CP match, 1-ms 55-kHz 1H-31P Hartmann-Hahn CP match, 6-µs 31P π pulse, 7-µs 15N π pulse, and proton decoupling at 100 kHz. For 31P{15N} REDOR NMR of [ε-15N]lysine-labeled phage T4, the magic-angle spinning was at 10,000 Hz and the π pulse length was 6.8 µs for 31P. For 15NH4+-exchanged phage T4, spinning was at 6,250 Hz and 10,000 Hz for 15N{31P} REDOR and 31P{15N} REDOR, respectively.
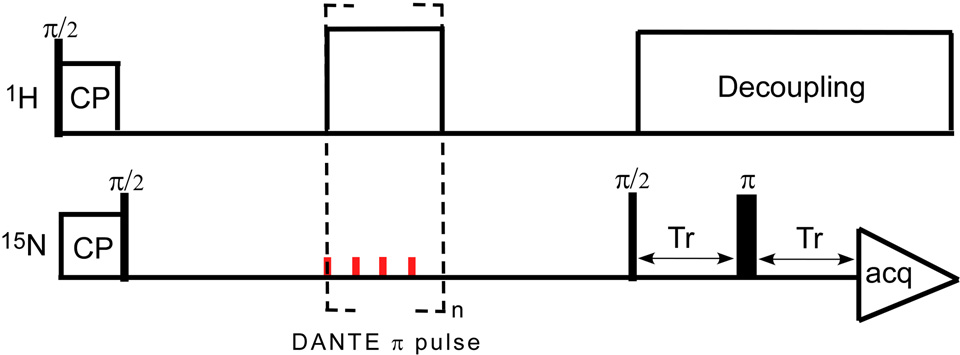
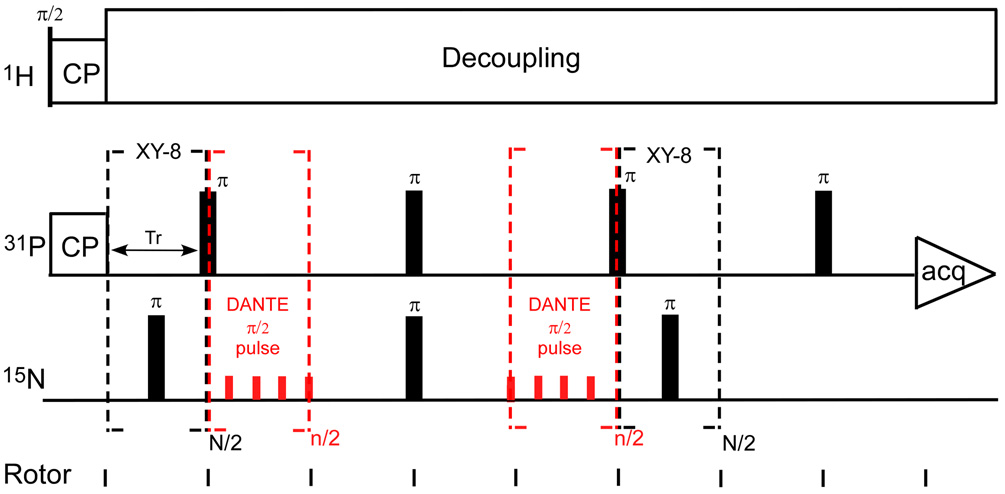
DANTE-based, frequency-selective REDOR (dbFSR)
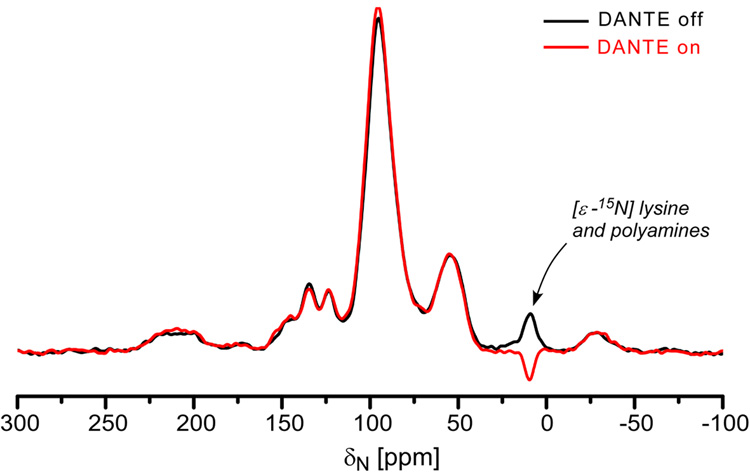
A dbFSR experiment24 combines a REDOR pulse sequence with DANTE inversion to reintroduce selectively the dipolar interaction between observed spins within a narrow frequency range (S) and the dephased nuclei (I). A DANTE-based, frequency-selective CPMAS pulse sequence (Figure 12) was used to calibrate the DANTE pulses by selectively inverting the amine peak of the 15N CPMAS spectrum (Figure 13).
Figure 12.
DANTE-based frequency-selective CPMAS pulse sequence.
Figure 13.
15N observed DANTE-based CPMAS spectra of uniform 15N-labeled phage T4 obtained with (red) and without (black) the application of a DANTE π pulse at the 9-ppm amine peak.
The dbFSR pulse sequence is shown in Figure 14. A full-echo signal (S0) is obtained without the DANTE pulses (red) while the frequency-selective recoupled signal (S) was obtained by applying two DANTE π/2 pulses. The parameters of 31P{15N} dbFSR for uniform 15N-labeled phage T4 were as follows: magic-angle spinning at 6,250 Hz, 1.1-ms 50-kHz 1H-31P Hartmann-Hahn CP match, 6-µs 31P π pulse, 7-µs 15N π pulse, and proton decoupling at 100 kHz. Each DANTE π/2 pulse was composed of eight 1- µs DANTE pulses with rf amplitude of 15.625 kHz and separation of a quarter of one rotor period (39 µs).
Figure 14.
DANTE-based, frequency-selective REDOR (dbFSR) pulse sequence developed by Kaustov et al. (reference 24). N/2 is an integral multiple of eight and the phase cycling for each eight-pulse unit is xy8. Each DANTE π/2 pulse is composed of n/2 rotor-synchronized π/n RF pulses. The small CSA of the –NH2 moeity allows multiple pulses per rotor period.
Abbreviations
- CPMAS
cross-polarization magic-angle spinning
- CSA
chemical shift anisotropy
- dbFSR
DANTE-based frequency-selective REDOR
- REDOR
rotational-echo double resonance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Karam JD. Molecular biology of bacteriophage T4. American Society For Microbiology Press; 1994. [Google Scholar]
- 2.Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Rüger W. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuller DN, Raymer D, Kottadiel V, Rao VB, Smith DW. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc. Natl. Acad. Sci. USA. 2007;104:16868–16873. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Earnshaw WC, Casjens SR. DNA packaging by the double-stranded DNA bacteriophages. Cell. 1980;21:319–331. doi: 10.1016/0092-8674(80)90468-7. [DOI] [PubMed] [Google Scholar]
- 5.Mehta AD, Rief M, Spudich JA, Smith DA, Simmons RM. Single-molecule biomechanics with optical methods. Science. 1999;283:1689–1695. doi: 10.1126/science.283.5408.1689. [DOI] [PubMed] [Google Scholar]
- 6.Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 7.Fuller DN, Raymer D, Rickgauer JP, Robertson RM, Catalano CE, Anderson DL, Grimes S, Smith DW. Measurements of single DNA molecule packaging dynamics in bacteriophage λ reveal high forces, high motor processivity, and capsid transformations. J. Mol. Biol. 2007;373:1113–1122. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuller DN, Rickgauer JP, Jardine PJ, Grimes S, Anderson DL, Smith DA. Ionic effects on viral DNA packaging and portal motor function in bacteriophage. Proc. Natl. Acad. Sci. USA. 2007;104:11245–11250. doi: 10.1073/pnas.0701323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage phi29 portal moter can package DNA against a large internal force. Nature. 2001;413:748–751. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 10.Rao VB, Black LW. DNA packaging in Bacteriophage T4. In: Catalano CE, editor. Viral Genome Packagin Machines: Genetics, Structures, and Mechanism. Klumer Academics/Plenum Publisher; 2005. [Google Scholar]
- 11.Baumann RG, Black LW. Isolation and characterization of T4 bacteriophage gp17 terminase, a large subunit multimer with enhanced ATPase activity. J. Biol. Chem. 2003;278:4618–4627. doi: 10.1074/jbc.M208574200. [DOI] [PubMed] [Google Scholar]
- 12.Oram M, Sabanyagam C, Black LW. Modulation of the packaging reaction of bacteriophage T4 terminase by DNA structure. J. Mol. Biol. 2008;381:61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gullion T, Schaefer J. Rotational-echo, double-resonance NMR. J. Magn. Reson. 1989;81:196–200. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]; Detection of Weak Heteronuclear Dipolar Coupling by Rotational-Echo Double-Resonance NMR. Adv. Magn. Reson. 1989;13:57–83. [Google Scholar]
- 14.McDowell LM, Schmidt A, Cohen ER, Studelska DR, Schaefer J. Structural constraints on the ternary complex of 5-enolpyruvylshikimate-3-phosphate synthase from rotational-echo double resonance NMR. J. Mol. Biol. 1996;256:160–171. doi: 10.1006/jmbi.1996.0074. [DOI] [PubMed] [Google Scholar]
- 15.Stueber D, Grant DM. 13C and 15N chemical shift tensors in adenosine, guanosine dihydrate, 2'-deoxythymidine, and cytidine. J. Am. Chem. Soc. 2002;124:10539–10551. doi: 10.1021/ja012485c. [DOI] [PubMed] [Google Scholar]
- 16.Berg JM, Tymoczko JL, Stryer L. Biochemistry. fifth edit. DNA replication, recombination and repair, W. H. Freeman and Company; 2002. [Google Scholar]
- 17.Saenger W. Principles of nucleic acid structure. New York: Springer-Verlag; 1984. pp. 51–104. [Google Scholar]
- 18.Goetz JM, Schaefer J. REDOR dephasing by multiple spins in the presence of molecular motion. J. Magn. Reson. 1997;127:147–154. doi: 10.1006/jmre.1997.1198. [DOI] [PubMed] [Google Scholar]
- 19.Massiot D, Fayon F, Capron M, King I, Le Calvé S, Alonso B, Durand J-O, Bujoli B, Gan Z, Hoatson G. Modelling one- and two-dimensional solid-state NMR spectra. Magn. Reson. Chem. 2001;40:70–76. [Google Scholar]
- 20.Hafner EW, Tabor CW, Tabor H. Mutants of Escherichia coli that do not contain 1.4-diaminobutane (putrescine) or spermidine. J. Biol. Chem. 1979;254:12419–12426. [PubMed] [Google Scholar]
- 21.Berg JM, Tymoczko JL, Stryer L. Biochemistry. Fifth edition. New York: W. H. Freeman and Company; 2002. pp. 665–692. [Google Scholar]
- 22.Saenger W. Principles of nucleic acid structure. New York: Springer-Verlag; 1984. pp. 432–458. [Google Scholar]
- 23.Widom J. Structure, dynamics, and function of chromatin in vitro. Annu. Rev. Biophys. Biomol. Struct. 1998;27:285–327. doi: 10.1146/annurev.biophys.27.1.285. [DOI] [PubMed] [Google Scholar]
- 24.Kaustov L, Kababya S, Belakhov V, Baasov T, Shoham Y, Schmidt A. Inhibition mode of a bisubstrate inhibitor of KDO8P synthase: a frequency-selective REDOR solid-state and solution NMR characterization. J. Am. Chem. Soc. 2003;125:4662–4669. doi: 10.1021/ja028688y. [DOI] [PubMed] [Google Scholar]
- 25.Morris GA, Freeman R. Selective excitation in Fourier transform nuclear magnetic resonance. J. Magn. Reson. 1978;29:433–462. doi: 10.1016/j.jmr.2011.08.031. [DOI] [PubMed] [Google Scholar]
- 26.Tong G, Pan Y, Dong H, Pryor R, Wilson GE, Schaefer J. Structure and dynamics of pentaglycyl bridges in the cell walls of Staphylococcus aureus by 13C{15N} REDOR NMR. Biochemistry. 1997;36:9859–9866. doi: 10.1021/bi970495d. [DOI] [PubMed] [Google Scholar]
- 27.Overman SA, Aubrey KL, Reilly KE, Osman O, Hayes SJ, Serwer P, Thomas GJJ. Conformation and interactions of the packaged doubled-stranded DNA genome of bacteriophage T7. Biospectroscopy. 1998;4:S47–S56. doi: 10.1002/(SICI)1520-6343(1998)4:5+<S47::AID-BSPY6>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 28.Mullaney JM, Black LW. Activity of foreign proteins targeted within the bacteriophage T4 head and prohead: implications for packaged DNA structure. J. Mol. Biol. 1998;283:913–929. doi: 10.1006/jmbi.1998.2126. [DOI] [PubMed] [Google Scholar]
- 29.Black LW, Newcomb WW, Boring JW, Brown J. Ion etching of bacteriophage T4: support for a spiral-fold model of packaged DNA. Proc. Natl. Acad. Sci. USA. 1985;82:7960–7964. doi: 10.1073/pnas.82.23.7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ames BN, Dubin DT. The role of polyamins in neutralization of bacteriophage deoxyribonucleic acid. J. Biol. Chem. 1960;235:769–775. [PubMed] [Google Scholar]
- 31.Hershey AD. Some minor components of bacteriophage T2 particles. Virology. 1957;4:237–264. doi: 10.1016/0042-6822(57)90061-2. [DOI] [PubMed] [Google Scholar]
- 32.Ames BN, Dubin DT, Rosenthal SM. Presence of polyamines in certain bacterial virus. Science. 1958;127:814–816. doi: 10.1126/science.127.3302.814-a. [DOI] [PubMed] [Google Scholar]
- 33.Hud NV, Schultze P, Sklenář V, Feigon J. Binding sites and dynamics of ammonium Ions in a telomere repeat DNA quadruplex. J. Mol. Biol. 1999;285:233–243. doi: 10.1006/jmbi.1998.2327. [DOI] [PubMed] [Google Scholar]
- 34.Hud NV, Sklenář V, Feigon J. Location of ammonium ions in the minor groove of DNA duplexes in solutions and origin of DNA A-tract bending. J. Mol. Biol. 1999;286:651–660. doi: 10.1006/jmbi.1998.2513. [DOI] [PubMed] [Google Scholar]
- 35.O'Connor RD, Schaefer J. Relative CSA-dipolar orientation from REDOR sidebands. J. Magn. Reson. 2002;154:46–52. doi: 10.1006/jmre.2001.2461. [DOI] [PubMed] [Google Scholar]
- 36.Stueber D, Mehta AK, Chen Z, Wooley KL, Schaefer J. Local order in polycarbonate glasses by 13C{19F} rotational-echo double-resonance NMR. J. Polym. Sci., Part B: Polym. Phys. 2006;44:2760–2775. [Google Scholar]
- 37.Rifat D, Wright NT, Varney KM, Weber DJ, Black LW. Restriction endonuclease Inhibitor IPI* of bacteriophage T4: a novel structure for a dedicated target. J. Mol. Biol. 2008;375:720–734. doi: 10.1016/j.jmb.2007.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage f29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao VB, Black LW. Cloning, overexpression and purification of the terminnase proteins gp16 and gp17 of bacteriophage T4. J. Mol. Biol. 1988;200:475–488. doi: 10.1016/0022-2836(88)90537-2. [DOI] [PubMed] [Google Scholar]
- 40.Black L. Bacteriophage T4 internal protein mutants: isolation and properties. Virology. 1974;60:166–179. doi: 10.1016/0042-6822(74)90374-2. [DOI] [PubMed] [Google Scholar]
- 41.Reddy KJ, Kuwabara T, Sherman LA. A simple and efficient procedure for the isolation of high-quality phage λ DNA using a DEAE-cellulose column. Anal. Biochem. 1988;168:324–331. doi: 10.1016/0003-2697(88)90325-9. [DOI] [PubMed] [Google Scholar]