Abstract
The complex microbiome of the rumen functions as an effective system for the conversion of plant cell wall biomass to microbial protein, short chain fatty acids, and gases. As such, it provides a unique genetic resource for plant cell wall degrading microbial enzymes that could be used in the production of biofuels. The rumen and gastrointestinal tract harbor a dense and complex microbiome. To gain a greater understanding of the ecology and metabolic potential of this microbiome, we used comparative metagenomics (phylotype analysis and SEED subsystems-based annotations) to examine randomly sampled pyrosequence data from 3 fiber-adherent microbiomes and 1 pooled liquid sample (a mixture of the liquid microbiome fractions from the same bovine rumens). Even though the 3 animals were fed the same diet, the community structure, predicted phylotype, and metabolic potentials in the rumen were markedly different with respect to nutrient utilization. A comparison of the glycoside hydrolase and cellulosome functional genes revealed that in the rumen microbiome, initial colonization of fiber appears to be by organisms possessing enzymes that attack the easily available side chains of complex plant polysaccharides and not the more recalcitrant main chains, especially cellulose. Furthermore, when compared with the termite hindgut microbiome, there are fundamental differences in the glycoside hydrolase content that appear to be diet driven for either the bovine rumen (forages and legumes) or the termite hindgut (wood).
Keywords: CAZymes, cellulases, plant cell wall, pyrosequencing
Herbivores carry out a foregut fermentation that digests plant cell wall materials by a complex and efficient microbial process. The microbiome inhabiting the rumen is characterized by its high population density, wide diversity, and complexity of interactions. Bacteria predominate the rumen, with a variety of anaerobic protozoa and fungi (1), and the associated occurrence of bacteriophage is well documented (2). The use of small subunit (SSU) rRNA sequence analysis has allowed for a more complete description of the rumen microbiome and these inventories have demonstrated that a large microbial component remains uncultured (3–12) and that a high proportion of the fibrolytic population has not been thoroughly described (7, 8, 13, 14). The rumen habitat contains a consortium of microbes that harbor the complex lignocellulosic degradation system for the microbial attachment and digestion of plant biomass. However, the complex chemical processes required to break down the plant cell wall are rarely carried out by a single species. Evidence also suggests that the most important organisms and gene sets involved in the most efficient hydrolysis of plant cell walls are associated with the fiber portion of the rumen digesta (15). Because we continue to investigate the community structure of the rumen, it is also clear that the system is not fully characterized with respect to the metabolic potential, especially as the system relates to plant cell wall degradation. In this regard, our laboratory compared the microbial metagenomes from the rumen by using suppressive subtractive hybridization and detected an unexpectedly large difference in archaeal community composition between 2 steers fed identical diets and housed together (16), but we did not detect any functional differences related to plant cell wall hydrolysis. In contrast, metagenomic analysis of a bovine rumen expression library (17) identified 22 glycoside hydrolase (GH) clones of which 4 potentially represent undescribed families of GHs. A polyphenol oxidase (laccase) from this expression library has also been characterized and might play a role in ryegrass lignin digestion (18).
The rumen provides a unique genetic resource for the discovery of plant cell wall-degrading microbial enzymes for use in biofuel production, presumably because of coevolution of microbes and plant cell wall types. There are, however, limitations to metagenome mining (19), and the number of clones needed to represent the entire metagenome is staggering (20). Nonetheless, this approach does allow one to begin to harvest the remarkable and vast diversity present in a given metagenome (21). In this regard, next generation sequencing technologies, primarily the massively parallel DNA sequencing capacity of the 454 Genome Sequencer (Roche) (22), are now being applied to complex environmental samples (23–29). A comparison of the metabolic potential and functional genes in 45 distinct microbiomes showed that the 454 technology could describe the metabolic profiles of the microbial communities across 9 discrete environments and these could be related to biogeochemical characteristics of the environment (24). Indeed, a recent review of metagenomics (28) notes that approximately one third of published metagenomic studies have used pyrosequencing as a platform.
Massive depth metagenomic sequencing is an invaluable complement to what has already been learned about lignocellulose degradation in the rumen. The ease with which bovine ruminants can be fistulated allows for simple and rapid sampling strategies, and changes in diet and management can be easily implemented for metagenomic investigations on the microbial community and metabolic potential. Presented in this study is a comparative metagenome analysis of a gastrointestinal microbiome from a foregut-fermenting mammal by using the inexpensive, massively parallel, and rapid method of pyrosequencing.
Results and Discussion
Sequencing of the Rumen Microbiome.
Our goal was to provide a detailed characterization of the metabolic potential and GH content of the fiber-adherent (FA) microbiome, the location of plant cell wall hydrolysis, and to compare how microbiomes differ between different animals fed the same diet. We compared both phylotype and functional gene content in the FA microbiomes of different bovine animals fed the same diet (bovines 8, 64, and 71), and a pooled liquid (PL) microbiome from these same animals. Similar relationships were seen between rRNA gene sequence hits for bacterial SSU against the Ribosomal Database Project, and archaeal and eukaryotic SSU against European Ribosomal RNA databases (Table 1). The number of 16S rRNA gene hits in the metagenomic libraries (Table 1) is similar (228 to 722) to that found for our analysis of the chicken cecum where 401–510 SSU rRNA gene sequences per sample were observed (29). Rarefaction analysis (supporting information (SI) Fig. S1) of the 4 near full-length 16S rRNA gene sequence libraries that were sequenced in this study revealed a total of 161–259 operational taxonomic units (OTUs) (97% identity level) per library, which is similar to the estimate of Edwards et al. of 177 OTUs (10, 30). Considering all libraries together, a total of 510 unique OTUs were seen; however, nonparametric estimators [ACE and Chao1 (31)] suggested that as many as 771 OTUs (671–857 OTUs confidence interval) may have been present (Fig. S2). A majority of sequences (64%) belonged to 59 OTUs, which were shared between all libraries, contrasting to the 273 OTUs (containing 10% of sequences) that were restricted to a single library. These results suggest that the most populous organisms were present in all 4 libraries. Our random sample pyrosequencing was not amenable to rarefaction analysis because any single organism would be represented by multiple “hits” across the genome, making it impossible to establish a consistent OTU definition.
Table 1.
Summary of pyrosequence data obtained from different bovine rumen samples
| Parameters | Fiber-adherent fractions |
Liquid fraction |
||
|---|---|---|---|---|
| 8 | 64 | 71 | PL | |
| No. of sequences | 178,713 | 264,849 | 345,317 | 236,830 |
| Total length of sequences | 18,153,371 | 26,644,817 | 35,115,534 | 24,016,021 |
| Avg. length of sequences, bp | 101.7 | 101.6 | 100.6 | 101.4 |
| Avg. quality score* | 26.4 | 26.8 | 26.1 | 26.4 |
| Total coding sequences (EGTs)† (% of total sequences) | 47,885 (26.8) | 62,531 (23.6) | 160,698 (46.5) | 60,955 (25.7) |
| Archaea EGTs (% of total EGTs) | 1,067 (2.2) | 1,464 (2.3) | 1,098 (0.68) | 2,586 (4.2) |
| Bacteria EGTs (% of total EGTs) | 45,142 (94.3) | 59,162 (94.6) | 152,910 (95.2) | 55,634 (91.3) |
| Eukarya EGTs (% of total EGTs) | 726 (1.5) | 1,058 (1.7) | 980 (0.61) | 899 (1.5) |
| Virus EGTs (% of total EGTs) | 60 (0.12) | 89 (0.14) | 204 (0.13) | 87 (0.14) |
| Hypothetical sequences (% of total EGTs) | 4,500 (9.4) | 5,415 (8.7) | 14,751 (9.2) | 5,450 (8.9) |
| Nonhypothetical sequences (% of total EGTs) | 40,310 (84.2) | 52,470 (83.9) | 123,740 (77.0) | 51,369 (84.3) |
| Noncoding sequences (% of total sequences) | 130,595 (73.1) | 201,950 (76.3) | 183,894 (53.3) | 175,569 (74.1) |
| Number of SSU rDNA hits‡ (% of total sequences) | ||||
| Bacteria (Ribosomal Database Project) | 228 (0.13) | 362 (0.14) | 722 (0.21) | 291 (0.13) |
| Archaea (European Ribosomal Database) | 0 | 2 (8−4) | 2 (6−4) | 14 (6−3) |
| Eukarya (European Ribosomal Database) | 5 (3−3) | 4 (2−3) | 1 (3−4) | 1 (4−3) |
*The quality score of each base was provided by 454 Life Sciences and is analogous to the Phred score of Sanger Sequencing methods (53). The value cited here is the mean quality score per sequence. Avg., average.
†The BLASTX cutoff for environmental gene tags is 1 × 10−5.
‡The E value cutoff for SSU rDNA hits for all databases used is 1 × 10−5 with a minimum length of 50 bp.
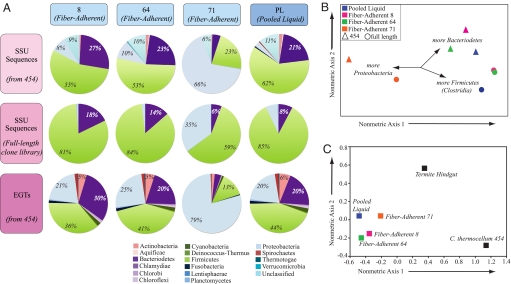
In theory, 454 pyrosequencing should randomly sample the whole metagenome. From our analysis, there does not appear to be a bias with respect to location on the 16S rRNA molecule for the sequences retrieved from these 4 microbiome samples (Fig. S3). The phylogenetic composition [bacterial phyla (32, 33)] for both 16S rRNA gene and environmental gene tags (EGTs) (34) followed a similar distribution for metagenomic libraries (Fig. 1A), again highlighting the randomness of the sequenced libraries, as expected for a random sampling of genes. However, nonmetric multidimensional scaling (NM-MDS) revealed that the phylogenetic make-up of near full-length 16S rRNA gene libraries was markedly different from that of the metagenomic 16S rRNA gene sequences (Fig. 1B), with the former being dominated by Firmicutes (particularly of the class Clostridia) and tending to be deficient in Proteobacteria and Bacteroidetes sequences. Additionally, NM-MDS analysis (Fig. 1B) shows that phylogenetic composition of microbiome PL is resolved in both full-length 16S rRNA gene libraries and pyrosequenced samples, with an equidistant distribution from fiber-adherent microbiomes 8 and 64 in the latter case.
Fig. 1.
Comparison of taxonomic and functional variation in the rumen microbiome. (A) Phylogenetic composition of bacterial phyla from pyrosequence 16S rDNA sequences, 16S rDNA from full length clone libraries and EGTs from 4 pyrosequenced bovine rumen samples. The number of sequences in each of the bacterial phyla from the FA (bovines 8, 64, and 71) and PL rumen microbiomes is shown. (B) Multidimensional scaling plots comparing phylogenetic composition from pyrosequence and full-length 16S rRNA gene sequences. (C) Nonmetric multidimensional scaling plots comparing plant polysaccharide degrading components between Clostridium thermocellum genome and other metagenomic microbiomes. A hierarchal clustering analysis was conducted on the frequency distribution of sequences showing similarity to each gene within 3 gene groups using a χ2 analysis (Table S3).
Further insight into the diversity within the 4-metagenomic samples was obtained by comparing the number of SSU rRNA gene sequences and EGTs in different bacterial phyla in the FA and the PL rumen microbiomes (Fig. 1A). Whereas classifying EGTs from short pyrosequencing reads has been challenging, a recent report demonstrates that EGTs as short as 27 aa can accurately be classified with an average specificity ranging from 97% for Superkingdom to 93% for Order (33). Bacterial specific EGTs represented from 91.3% to 95.2% of total EGTs and the distribution of phylotypes fell predominantly into the Bacteriodetes, Firmicutes, and Proteobacteria phyla, regardless of the microbiome analyzed. Additionally, the taxonomic profiles of the top BLAST hits were consistent across all microbiomes (Table S1). Further resolution at the taxonomic levels of both Class and Order reveals that the phylogenetic distribution of 16S rRNA gene sequences are similar for major taxa between FA-microbiomes 8 and 64 and the PL microbiome (Fig. S4). SSU 16S rRNA gene sequences matching the genus Ruminococcus, a fibrolytic rumen bacterium, were rare in all libraries (e.g., 7 SSU rRNA gene pyrosequences total), which agrees with previous molecular studies of the rumen (4, 8, 11, 13, 32, 35, 36). Additionally, the distribution of EGTs from the Bacteria is remarkably congruent with the distribution of 16S rRNA phylotypes. However, the phylogenetic distribution of 16S rRNA phylotypes was strikingly different for the FA-microbiome of bovine 71 (Fig. 1A), wherein two-thirds of the 16S rRNA gene sequences (463 of 722) fell into the Gammaproteobacteria class, whereas in the other FA samples, the Gammaproteobacteria class represented only 3.5% to 8% of the sequences the FA-microbiomes from bovines 8 and 64, respectively. This enrichment in Gammaproteobacteria appeared to be at the expense of Firmicutes and Bacteroidetes (both underrepresented in bovine 71) and this shift in phyla enrichment was associated with the primary nonmetric axis of Fig. 1B. The distribution of significant EGTs within orders in Proteobacteria was reasonably consistent between the metagenomes (Fig. S5).
Archaeal EGTs constituted ≈2.3% of EGTs in metagenome libraries (Table 1), matching well with previous estimates placing Archaea at 0.5% to 3.0% of the microbiome (5, 37) with the majority corresponding to methanogenic classes (Fig. S6). Few Eukarya phylotypes (≈1.3%) were identified in these microbiomes (Table 1), and were most similar to the Viridiplantae (i.e., feed), Metazoa (i.e., host), and Fungi. Fungal rRNA gene sequences were not identified in any of these samples; however, 19% of eukaryotic EGTs appear to be fungal (Fig. S6). These distributions are consistent with the environment that was sampled, with the exception that members of the Chytridiomycota were not detected. The anaerobic rumen fungi fall into this phylogenetic group and are thought to be associated with plant cell wall hydrolysis (38). Three sequences with similarity to the rumen protozoa were found in the samples from bovine 8 (Dasytricha ruminantium U57769) and bovine 64 (Polyplastron multivesiculatum U27815 and Dasytricha ruminantium U57769). Virus EGTs were rare (0.1%) (Table 1) and were composed primarily of dsDNA viruses (Fig. S7). The lack of viral sequences may be because of their extensive diversity and limited representation in public databases. Additionally, viral sequences may have been overlooked because they were not enriched for during cellular fractionation procedures. These EGT proportions match our current knowledge of the rumen microbiome community structure.
Subsystems-based annotations (SEED database) were used to gain a better understanding of how these phylogenetic trends could be used to predict the metabolic potential of these microbiomes. To extend this analysis, we applied statistical methods (39), which compare those subsystems that are more, or less, represented in the different microbiomes. Table S2 shows the subsystems that are significantly over-represented in each sample when compared with the others at P < 0.02. Again, the distribution of subsystems is strikingly different for the FA-microbiome from the rumen of bovine 71, which is predominated with metabolisms consistent with a community that has shifted away from a soluble, more easily fermentable carbohydrate-based metabolism. It appears that bovine 71 had not adapted to the higher-fiber diet. This is consistent with the metabolism represented by the Gammaproteobacteria. Whereas most of the Gammaproteobacteria sequences were most similar to sequences from Psychrobacter-like organisms (from arctic samples), they probably are not from this genus, but rather from a close relative (40).
Plant Polysaccharide Degradative Enzymes.
We have chosen the rumen as the microbiome for studying lignocellulosic degradation as it is obvious that this natural habitat has selected for and evolved a complex lignocellulosic degradation system (1). We compared the plant polysaccharidase EGTs in the different microbiomes (Fig. S8). All of the microbiomes have the metabolic potential to hydrolyze carbohydrate components of the plant cell wall including cellulose, xylan components such as acetylxylan, arabinosides, and xylosides, and fructosides, fucosides galactosides, glucosides, mannosides and rhamnosides. Again, EGTs for these plant cell wall hydrolyzing enzymes are in lower abundance in the FA-microbiome from the rumen of bovine 71, which is congruent with the apparent shift in metabolism seen in the subsystem and the phylogenetic analyses.
When the number of EGTs within GH families and cellulosome modules (dockerins, cohesins, etc.) are compared (Table 2), within the bovine rumen samples and compared with the termite hindgut microbiome (41), several interesting observations can be made. First, in the bovine microbiomes there is a wide diversity of GH catalytic modules with >3,800 sequences belonging to 35 GH families. This abundance is in striking contrast with the finding of only 9 carbohydrate-binding modules from 3 families (CBM6, CBM13 and CBM32), only one of which (CBM6) is likely to bind cellulose. Only 3 dockerin modules and no cohesins or CBM3 modules were detected, even though these modules are common in cellulosomes (42). This suggests that cellulosome-based systems for plant cell wall hydrolysis are rare in this community. Microbial cellulases fall into GH families 5, 6, 7, 8, 9, 12, 44, 45, 48 and 74, and whereas a few members of families GH5 and GH9 were found in all of the bovine metagenomes, only a single family GH48 (cellobiohydrolase) was detected in the FA-microbiome from the rumen of bovine 64. Additionally, no cellulases from families 6, 7, 12, 44, 45 and 74 could be found in the FA-microbiome. Surprising was the finding of a reduced set of enzymes for the digestion of xylan main chains (only 3 GH11 and 18 GH10 xylanases detected) and of enzymes for the hydrolysis of the main chain of pectin (9 GH28 members and a single pectate lyase from family PL9). In contrast with the small number of enzymes devoted to the hydrolysis of the main chain of cellulose, hemicelluloses, and pectins, the metagenomes displayed a diversity of enzymes that digest the side chains of these polymers, and oligosaccharides thereof. Families GH2 and GH3, which contain a large range of glycosidases cleaving nonreducing carbohydrates in oligosaccharides and the side chains of hemicelluloses and pectins, were particularly abundant, with >700 members in each family. This is ≈30 times more abundant than the ≈25 members found in cellulase families GH5 and GH9. Other abundant glycosyl hydrolase families, which contain enzymes active on side chains of hemicelluloses and pectins, include family GH31 (295 members), GH43 (244 members) and GH51 (257 members). We also note that there appear to be few of the several families of GHs and CBMs that are found in large numbers in fungi (GH6, GH7, GH61 and CBM1). None of these are found in the bovine microbiome, as are other typical fungal GH families such as GH72 (fungal cell wall remodeling) and GH47 (accessory to N-glycosylation). This is consistent with the low numbers of fungal SSU rRNA gene and EGTs detected for this low abundance microbial group.
Table 2.
Comparative analysis of cellulosome-based systems between the Clostridium thermocellum (C.T.) genome and other metagenomic microbiomes
| CAZy family* | C.T. genome† | C.T. 454 model | Pooled liquid | Fiber-8 | Fiber-64 | Fiber-71 | Termite hindgut‡ |
|---|---|---|---|---|---|---|---|
| GH1 | 2 | 21 | 7 | 4 | 7 | 20 | 22 |
| GH2 | 1 | 15 | 218 | 185 | 228 | 114 | 23 |
| GH3 | 3 | 10 | 207 | 194 | 207 | 96 | 69 |
| GH4 | 0 | 0 | 16 | 9 | 7 | 2 | 14 |
| GH5 | 11 | 165 | 7 | 11 | 5 | 4 | 56 |
| GH8 | 1 | 19 | 8 | 3 | 4 | ND | 5 |
| GH9 | 16 | 355 | 7 | 6 | 6 | 5 | 9 |
| GH10 | 6 | 99 | 10 | 5 | 7 | 4 | 46 |
| GH11 | 1 | 10 | 2 | ND | 1 | ND | 14 |
| GH13 | 2 | 30 | 47 | 36 | 37 | 39 | 48 |
| GH15 | 1 | 9 | ND | ND | ND | 1 | 0 |
| GH16 | 2 | 15 | ND | ND | ND | 1 | 1 |
| GH18 | 3 | 20 | 2 | ND | 3 | 1 | 17 |
| GH23 | 2 | 7 | ND | ND | ND | ND | 52§ |
| GH25 | 0 | 0 | 1 | 1 | ND | ND | 1 |
| GH26 | 3 | 41 | 2 | 5 | 6 | 5 | 15 |
| GH27 | 0 | 0 | 16 | 21 | 23 | 5 | 4 |
| GH28 | 0 | 0 | 9 | 9 | ND | ND | 6 |
| GH29 | 0 | 0 | 31‖ | 34‖ | 29‖ | 16‖ | 0 |
| GH30 | 0 | 0 | 3** | 3** | 2** | 1** | 0 |
| GH31 | 0 | 0 | 101 | 72 | 80 | 42 | 26 |
| GH32 | 0 | 0 | 12** | 8** | 5** | 2** | 0 |
| GH33 | 0 | 0 | 2 | ND | 1 | 1 | 0 |
| GH35 | 0 | 0 | 21 | 8 | 9 | 10 | 3 |
| GH36 | 0 | 0 | 47 | 43 | 48 | 48 | 5–7 |
| GH38 | 0 | 0 | 22 | 16 | 19 | 11 | 11 |
| GH39 | 0 | 0 | 2 | 3 | 3 | 1 | 3 |
| GH42 | 0 | 0 | 10 | 7 | 15 | 13 | 24 |
| GH43 | 6 | 72 | 68 | 72 | 69 | 35 | 16 |
| GH44 | 1 | 28 | ND | ND | ND | ND | 6¶ |
| GH48 | 2 | 60 | ND | ND | 1 | ND | 0¶ |
| GH51 | 1 | 10 | 73 | 54 | 86 | 44 | 18–19 |
| GH53 | 1 | 13 | 15 | 16 | 18 | 17 | 12 |
| GH54 | 0 | 0 | ND | ND | 3 | 1 | 0 |
| GH57 | 0 | 0 | 2 | ND | ND | 1 | 17 |
| GH74 | 1 | 26 | ND | ND | ND | ND | 7¶ |
| GH77 | 0 | 0 | ND | ND | 2 | ND | 14 |
| GH78 | 0 | 0 | 41‖ | 37‖ | 38‖ | 18‖ | 0 |
| GH81 | 1 | 8 | ND | ND | ND | ND | 0 |
| GH92 | 0 | 0 | 43 | 67 | 66 | 28 | 2 |
| GH94 | 3 | 66 | ND | ND | ND | ND | 68–132§ |
| GH97 | 0 | 0 | 47‖ | 67‖ | 59‖ | 20‖ | 0 |
| GH106 | 0 | 0 | 9 | 9 | 11 | 4 | 2 |
| CBM3 | 23 | 134 | ND | ND | ND | ND | 9 |
| CBM4 | 4 | 44 | ND | ND | ND | ND | 5¶ |
| CBM6 | 10 | 53 | ND | 1 | ND | ND | 13 |
| CBM9 | 1 | 0 | ND | ND | ND | ND | 5 |
| CBM11 | 1 | 14 | ND | ND | ND | ND | 3¶ |
| CBM13 | 2 | 9 | 1 | ND | 1 | 2 | 0 |
| CBM22 | 4 | 32 | ND | ND | ND | ND | 5¶ |
| CBM25 | 2 | 2 | ND | ND | ND | ND | 0 |
| CBM30 | 1 | 9 | ND | ND | ND | ND | 1–8¶ |
| CBM32 | 1 | 2 | ND | 3 | ND | 1 | 4 |
| CBM35 | 7 | 7 | ND | ND | ND | ND | 0–1¶ |
| CBM42 | 4 | 5 | ND | ND | ND | ND | 0 |
| CBM44 | 1 | 9 | ND | ND | ND | ND | 0 |
| CBM48 | 1 | 11 | ND | ND | ND | ND | 0–1 |
| CE1 | 3 | 36 | 5 | 10 | 22 | 8 | NR |
| CE2 | 1 | 14 | 1 | 1 | 1 | ND | 4–34 |
| CE3 | 1 | 5 | ND | ND | ND | ND | NR |
| CE4 | 3 | 17 | 6 | 2 | 5 | 4 | 4–34 |
| CE6 | 0 | 0 | ND | ND | ND | 1 | NR |
| CE7 | 1 | 3 | ND | 2 | 3 | 1 | NR |
| CE8 | 1 | 6 | ND | ND | ND | ND | NR |
| CE9 | 2 | 13 | ND | ND | ND | ND | NR |
| CE12 | 1 | 14 | ND | ND | ND | ND | NR |
| COH | 29 | 85 | ND | ND | ND | ND | NR |
| DOC1 | 76 | 128 | 2 | ND | 1 | ND | NR |
| PL1 | 2 | 5 | ND | ND | ND | ND | 5 |
| PL9 | 0 | 0 | ND | 1 | ND | ND | NR |
| PL11 | 1 | 25 | ND | ND | ND | ND | 5¶ |
| Total CBM | 62 | 331 | 1 | 4 | 1 | 3 | 36–45 |
| Total CE | 13 | 108 | 12 | 15 | 31 | 14 | 8–68 |
| Total GH | 70 | 1099 | 1108 | 1005 | 1105 | 610 | 636–703 |
A 454 simulation of the C. thermocellum genome generated 291,058 sequences resulting in 56,142 pegs. The number of sequences corresponding to each CAZy family (E value cutoff is 1 × 10−5) from the C. thermocellum genome, a 454 short fragment simulation of GH detection in the C. thermocellum genome, fiber-adherent microbiomes, pooled liquid microbiome, and the termite hindgut microbiome is shown (ND, not detected; NR, not reported).
*CAZy database (www.cazy.org).
†JGI link to C. thermocellum genome (http://genome.jgi-psf.org/mic_home.html).
‡Derived from supplemental of Warnecke et al. (46).
§Genes overrepresented in termite hindgut microbiome.
¶Genes underrepresented in termite hindgut microbiome.
‖Genes overrepresented in bovine rumen microbiomes.
**Genes underrepresented in bovine rumen microbiomes.
When compared with the termite hindgut metagenome study (41), the termite hindgut microbiome contains more representatives of protein modules that are involved in degradation of the main chain of cellulose (CBM6 and GH5, 9, 44, and 74) and xylan (GH10 and 11), and GH94 (cellobiose phosphorylase) compared with the bovine microbiomes. This could be because of the diet differences (wood versus forages and legumes), or it could be that the short reads from pyrosequencing do not detect CBMs or dockerins. To address this question we performed a 454 pyrosequencing short read simulation for the detection of these protein motifs in the Clostridium thermocellum genome, a model cellulolytic microbe (Table 2 and Fig. S9). From this analysis, dockerins appear more difficult to detect in the simulated 454 short read query than are CBMs and GHs. We found 5 CBM “454 fragments” per CBM in the genome, ≈15.7 GH “454 fragments” per GH in the genome, and only ≈1.7 dockerin “454 fragments” per dockerin in the genome. Dockerins possess a shorter length than the average GH, whereas CBMs are intermediate in size. The detection of CBMs is approximately one-third of that of GHs and this is likely because of the relative average sizes of the modules. So, although the 454 short reads do detect dockerins and CBMs when they are present, their under-representation may be because of the inability to detect them by using pyrosequence short reads. Nonetheless, when one focuses on GH and CBM content, an analysis of 71 genes highlights fundamental differences in the GH content of these 2 microbiomes (Tables 2, Fig. 1C, and Table S3).
The differences detected in GH content between the termite hindgut and rumen microbiomes are consistent with an interpretation that diet, wood, or forages, is a contributing factor to the metabolic potentials that have been detected. This can also be seen by the different mechanisms by which these biomes deal with nitrogen metabolism. In the termite hindgut, there appears to be an abundance of genes involved in nitrogen fixation, presumably because of a diet of wood that is depleted in nitrogen (41). For the most part, ruminant diets are rich in nitrogen containing compounds (43), and so there should be little need for nitrogen fixation in the rumen microbiome. This is reflected in the EGT content for the Nitrogen Metabolism subsystem (Table S4) in our rumen samples. The most abundant EGTs detected are those involved in the uptake of nitrogenous compounds, with very few EGTs for enzymes involved in nitrogen fixation.
We can conclude from these data, that in the rumen microbiome, initial colonization of fiber appears to be by organisms that have enzymes that attack the easily available side chains of complex plant polysaccharides and not the more recalcitrant main chains, especially cellulose. So, we propose that whereas the previous evidence suggested that the most important organisms and gene sets for efficient hydrolysis of plant cell walls were associated with the fiber portion of the rumen digesta (15), there appears to be a dynamic process with initial colonization by one subset of organisms, which is probably later replaced by another subset of organisms that degrade the main chains of cellulose and xylan. Furthermore, when compared with the termite hindgut microbiome, there are fundamental differences in the GH content that appear to be diet driven for either the bovine rumen (forages and legumes) or the termite hindgut (wood). This interpretation, based on the gene-centric approach presented herein, is consistent with the recent SSU rRNA gene surveys of 60 different vertebrate animal gastrointestinal microbiomes (44–46), which concluded that these microbiomes are similar at the phylum level, and phylogenetic composition is influenced by diet, morphology, and phylogeny.
Over the past decade, extensive suites of molecular based approaches have been developed that have enabled the study of uncultured microorganisms. These studies have been the direct outcome of the use of SSU rRNA targets, which have been widely used to study the bacterial and archaeal diversity, community structure, and microbial interactions within these microbial ecosystems (47–49). The gene-centric pyrosequence approach and subsystems-based annotations allowed us to gain an understanding of the metabolic potential of these microbiomes that could not have been achieved using clone library approaches that are not random and have both a PCR and cloning basis. The extended uses of increased depth sequencing technologies will not only allow for the discovery of unique or additional diversity, but also has the potential to detect minor species below the current sequencing depth of most studies. The use of pyrosequencing and a systems approach generates sequence information in a comparative context based on the ecology of the microbial communities that inhabit the rumen. This allows us to simultaneously analyze both the metabolic potential and diversity of the rumen microbiome.
Materials and Methods
Rumen Sampling.
Samples of whole rumen contents were obtained from 3 fistulated 5-yr old Angus Simmental Cross steers (samples 8, 64, and 71) averaging ≈500 kg of that were housed at Illinois State University and maintained on a NRC restricted diet of medium-quality grass-legume hay at maintenance intake based on 2001 Dairy National Research Council for grass-legume hay (50). The animals were fed once a day for the entire length of the study (a total of 8 weeks including 2 weeks before sampling). ≈3 l of whole rumen digesta (fiber-adherent (FA) and liquid associated microbes) was collected from the dorsal third rumen 6 weeks after the beginning of the study, 1 hour after the morning feeding. Samples were then partitioned into FA fractions and liquid fractions before DNA extraction (14, 51). The pooled sample (PL) (derived from a mixture of liquid fractions from each of the 3 steers) was thoroughly mixed before the DNA extraction (14, 52). Samples were stored at −80 °C until DNA extraction.
DNA Extraction and Purification.
Genomic DNA was extracted by using a protocol similar to the extraction of high molecular weight DNA for rumen and fecal contents (51). Deviation from this protocol included proceeding with the Qiagen DNA Stool Kit manufacturer's protocol (Qiagen) after the addition of 960 μl of ASL buffer to the samples. DNA purity and concentration was analyzed by spectrophotometric quantification and gel electrophoresis.
Pyrosequencing and Sequence Analysis.
Three FA samples and 1 PL sample were subjected to a single pyrosequence run by 454 Life Sciences by using a 454 Life Sciences Genome Sequencer GS20 (454 Life Sciences) and analyzed using the SEED Annotation Engine (http://seed.sdsu.edu/FIG/index.cgi) (53). The sequences were compared using the BLASTX algorithm with an expected cutoff of 1 × 10−5 (27). The detection and assignment of sequences to families of carbohydrate-active enzymes was done by adapting BLAST-based procedures against the libraries of catalytic and noncatalytic modules generated from the CAZy database (www.cazy.org). The BLASTN algorithm (E <1 × 10−5 and a sequence length hit >50 nt) was used to identify SSU rRNA genes from release 9.3.3 of the RDP database [(54); http://rdp.cme.msu.edu/], and the European Ribosomal RNA database (http://www.psb.ugent.be/rRNA/index.html). RDP was used for robust Bacterial classification and the European Ribosomal RNA database was used to classify eukaryal and archaeal sequences. The metagenomes used in this article are freely available from the SEED platform and are being made accessible from CAMERA and the National Center for Biotechnology Information Short Read Archive. The National Center for Biotechnology Information genome project IDs used in this study are: 28605, 28607, 28609, and 286011.
Construction and Phylogenetic Analysis of 16S rRNA Gene Clone Libraries.
Universal prokaryotic primers 8FPL (AGTTTGATCCTGGCTCAG) and 1492RPL (GGYTACCTTGTTACGACTT) were used to amplify the 16S rRNA gene in a 30-cycle PCR (details provided in SI Text). 16S rRNA gene PCR products were cloned into pCR-4-TOPO vectors (Invitrogen) and electroporated into electrocompetent E. coli DH10B (Invitrogen). Transformants were selected by plating onto selective SOB/amp agar plates and plasmid template DNA from each transformant was prepared by a modified alkaline lysis method. The nucleotide sequences were determined by cycle sequencing using BigDye Terminator (Applied Biosystems) and run on ABI 3730xl sequencers (Applied Biosystems). Sequences were then trimmed to remove the vector sequence. A total of 3,617 16S rRNA gene sequences were aligned in Greengenes. The BLASTN algorithm (E <1 × 10−5) was used to identify sequences by using the RDP database [(54); http://rdp.cme.msu.edu/] whereas rarefaction analysis was performed by using DOTUR (http://www.plantpath.wisc.edu/fac/joh/dotur.html) (31), with the distance matrices analyses calculated in ARB (http://www.arb-home.de/). Near full-length 16S rRNA gene sequences of microbiomes PL, FA-8, FA-64, and FA-71 have been deposited in GenBank under accession numbers EU842125-EU842929, EU842930-EU843925, EU843926-EU844782 and EU844783-EU845741, respectively.
Statistical Analyses.
The comparison of 16S rRNA gene composition of the metagenomic libraries and the near full-length 16S rDNA libraries were conducted by using Nonmetric Multidimensional Scaling (NM-MDS) (55). The Bray-Curtis similarity coefficient [coefficient S17 of (56)] was calculated for each possible pair of libraries using a Phylum-level percentage contribution assessment of library composition (details provided in SI Text). The resulting 8-by-8 similarity matrix was used to conduct NM-MDS, graphically representing the ranked similarity between all pairs of samples (i.e., most similar samples are found closest together). NM-MDS was conducted with the software Primer 5 for Windows (57), using 20 random starting configurations, with the optimal two-dimensional solution having a final stress of 0.
A similar NM-MDS approach was taken to examine the similarity/differences in the abundances of CAZy proteins between the 4 bovine metagenomes and the microbial community from the termite hindguts (41), and a metagenome constructed from a bacterial species known to degrade cellulose, Clostridium thermocellum. The complete C. thermocellum genome (3,843,301 bp) was randomly sampled with replacement to generate a simulated metagenome. A hierarchical clustering analysis was conducted on the number of sequences showing similarity to each protein using a χ2 analysis (preformed on SPSS v15). The resultant 6-by-6 dissimilarity matrix was used to conduct a NM-MDS (SPSS v15) with a single random start and 100 iterations. The optimal two-dimensional solution is presented with a minimum stress set at 0.0001.
Supplementary Material
Acknowledgments.
The authors also thank Corinne Rancurel for her help with the CAZy computer routines developed specifically for this work, Larry Berger for his help with the animal study, the Ribosomal Database Project at Michigan State University, and James Cole and Benli Chai for their invaluable contributions. This project was supported by the United States Department of Agriculture (USDA) Cooperative State Research, Education, and Extension Service National Research Initiative Competitive Grant 2006–35206-16652 (to B.A.W. and K.E.N.) and the Finnish Cultural Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. EU842125-EU842929, EU842930-EU843925, EU843926-EU844782, and EU844783-EU845741).
This article contains supporting information online at www.pnas.org/cgi/content/full/0806191105/DCSupplemental.
References
- 1.Hespell RB, Akin DE, Dehority BA. In: Gastrointestinal Microbiology. Mackie RI, White BA, Isaacson R, editors. Vol 2. New York: Chapman and Hall; 1997. pp. 59–186. [Google Scholar]
- 2.Klieve AV, Bauchop T. Morphological diversity of ruminal bacteriophages from sheep and cattle. Appl Environ Microbiol. 1988;54:1637–1641. doi: 10.1128/aem.54.6.1637-1641.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kocherginskaya SA, Aminov RI, White BA. Analysis of the rumen bacterial diversity under two different diet conditions using denaturing gradient gel electrophoresis, random sequencing, and statistical ecology approaches. Anaerobe. 2001;7:119–134. [Google Scholar]
- 4.Lin C, et al. Taxon specific hybridization probes for fiber-digesting bacteria suggest novel gut-associated. Fibrobacter Syst Appl Microbiol. 1994;17:418–424. [Google Scholar]
- 5.Lin C, Raskin L, Stahl D. Microbial community structure in gastrointestinal tracts of domestic animals: Comparative analyses using rRNA-targeted oligonucleotide probes. FEMS Microbiol Ecol. 1997;22:281–294. [Google Scholar]
- 6.Lin C, Stahl DA. Taxon-specific probes for the cellulolytic genus Fibrobacter reveal abundant and novel equine-associated populations. Appl Environ Microbiol. 1995;61:1348–1351. doi: 10.1128/aem.61.4.1348-1351.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson KE, et al. Phylogenetic analysis of the microbial populations in the wild herbivore gastrointestinal tract: Insights into an unexplored niche. Environ Microbiol. 2003;5:1212–1220. doi: 10.1046/j.1462-2920.2003.00526.x. [DOI] [PubMed] [Google Scholar]
- 8.Stahl DA, Flesher B, Mansfield HR, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tajima K, et al. Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl Environ Microbiol. 2001;67:2766–2774. doi: 10.1128/AEM.67.6.2766-2774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tajima K, et al. Rumen bacterial community transition during adaptation to high-grain diet. Anaerobe. 2000;6:273–284. [Google Scholar]
- 11.Tajima K, et al. Phylogenetic analysis of archaeal 16S rRNA libraries from the rumen suggests the existence of a novel group of archaea not associated with known methanogens. FEMS Microbiol Lett. 2001;200:67–72. doi: 10.1111/j.1574-6968.2001.tb10694.x. [DOI] [PubMed] [Google Scholar]
- 12.Whitford MF, et al. Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe. 1998;4:153–163. doi: 10.1006/anae.1998.0155. [DOI] [PubMed] [Google Scholar]
- 13.Krause DO, et al. 16S rDNA sequencing of Ruminococcus albus and Ruminococcus flavefaciens: Design of a signature probe and its application in adult sheep. Microbiology. 1999;145:1797–1807. doi: 10.1099/13500872-145-7-1797. [DOI] [PubMed] [Google Scholar]
- 14.Larue R, et al. Novel microbial diversity adherent to plant biomass in the herbivore gastrointestinal tract, as revealed by ribosomal intergenic spacer analysis and rrs gene sequencing. Environ Microbiol. 2005;7:530–543. doi: 10.1111/j.1462-2920.2005.00721.x. [DOI] [PubMed] [Google Scholar]
- 15.Forsberg CW, Cheng K-J, White BA. In: Gastrointestinal Microbiology. Mackie RI, White BA, editors. Vol 1. New York: Chapman and Hall; 1997. pp. 319–379. [Google Scholar]
- 16.Galbraith EA, Antonopoulos DA, White BA. Suppressive subtractive hybridization as a tool for identifying genetic diversity in an environmental metagenome: The rumen as a model. Environ Microbiol. 2004;6:928–937. doi: 10.1111/j.1462-2920.2004.00575.x. [DOI] [PubMed] [Google Scholar]
- 17.Ferrer M, et al. Novel hydrolase diversity retrieved from a metagenome library of bovine rumen microflora. Environ Microbiol. 2005;7:1996–2010. doi: 10.1111/j.1462-2920.2005.00920.x. [DOI] [PubMed] [Google Scholar]
- 18.Beloqui A, et al. Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen - Biochemical properties, structural analysis, and phylogenetic relationships. J Biol Chem. 2006;281:22933–22942. doi: 10.1074/jbc.M600577200. [DOI] [PubMed] [Google Scholar]
- 19.Henne A, Daniel R, Schmitz RA, Gottschalk G. Construction of environmental DNA libraries in Escherichia coli and screening for the presence of genes conferring utilization of 4-hydroxybutyrate. Appl Environ Microbiol. 1999;65:3901–3907. doi: 10.1128/aem.65.9.3901-3907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Handelsman J, et al. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem Biol. 1998;5:R245–249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- 21.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 24.Dinsdale EA, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 25.Roesch LFW, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sogin ML, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards RA, et al. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics. 2006;7:57–69. doi: 10.1186/1471-2164-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hugenholtz P, Tyson GW. Microbiology - Metagenomics. Nature. 2008;455:481–483. doi: 10.1038/455481a. [DOI] [PubMed] [Google Scholar]
- 29.Qu A, et al. Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome. PLoS ONE. 2008;3:e2945. doi: 10.1371/journal.pone.0002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edwards JE, Mcewan NR, Travis AJ, Wallace RJ. 16S rDNA library-based analysis of ruminal bacterial diversity. Antonie van Leeuwenhoek. 2004;86:263–281. doi: 10.1023/B:ANTO.0000047942.69033.24. [DOI] [PubMed] [Google Scholar]
- 31.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koike S, Yoshitani S, Kobayashi Y, Tanaka K. Phylogenetic analysis of fiber-associated rumen bacterial community and PCR detection of uncultured bacteria. FEMS Microbiol Lett. 2003;229:23–30. doi: 10.1016/S0378-1097(03)00760-2. [DOI] [PubMed] [Google Scholar]
- 33.Krause L, et al. Phylogenetic classification of short environmental DNA fragments. Nucleic Acids Res. 2008;36:2230–2239. doi: 10.1093/nar/gkn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 35.Briesacher SL, et al. Use of DNA probes to monitor nutritional effects on ruminal prokaryotes and Fibrobacter succinogenes S85. J Anim Sci. 1992;70:289–295. doi: 10.2527/1992.701289x. [DOI] [PubMed] [Google Scholar]
- 36.Denman SE, Mcsweeney CS. Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol Ecol. 2006;58:572–582. doi: 10.1111/j.1574-6941.2006.00190.x. [DOI] [PubMed] [Google Scholar]
- 37.Ziemer CJ, et al. Comparison of microbial populations in model and natural rumens using 16S ribosomal RNA-targeted probes. Environ Microbiol. 2000;2:632–643. doi: 10.1046/j.1462-2920.2000.00146.x. [DOI] [PubMed] [Google Scholar]
- 38.Trinci APJ, et al. Anaerobic Fungi in Herbivorous Animals. Mycol Res. 1994;98:129–152. [Google Scholar]
- 39.Rodriguez-Brito B, Rohwer F, Edwards RA. An application of statistics to comparative metagenomics. BMC Bioinformatics. 2006;7:162–173. doi: 10.1186/1471-2105-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bozal N, Montes MJ, Tudela E, Guinea J. Characterization of several Psychrobacter strains isolated from Antarctic environments and description of Psychrobacter luti sp nov and Psychrobacter fozii sp nov. Int J Syst Evol Microbiol. 2003;53:1093–1100. doi: 10.1099/ijs.0.02457-0. [DOI] [PubMed] [Google Scholar]
- 41.Warnecke F, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 42.Bayer EA, Shoham Y, Lamed R. In: Prokaryotes. Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. New York: Springer-Verlag; 2006. pp. 578–617. [Google Scholar]
- 43.Cotta MA, Russell JB. In: Gastrointestinal Microbiology. Mackie RI, White BA, editors. Vol 1. New York: Chapman and Hall; 1997. pp. 380–423. [Google Scholar]
- 44.Dethlefsen L, Mcfall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ley R, et al. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat Rev Micro. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 48.Theron J, Cloete TE. Molecular techniques for determining microbial diversity and community structure in natural environments. Crit Rev Microbiol. 2000;26:37–57. doi: 10.1080/10408410091154174. [DOI] [PubMed] [Google Scholar]
- 49.Torsvik V, Ovreas L. Microbial diversity and function in soil: From genes to ecosystems. Curr Opin Microbiol. 2002;5:240–245. doi: 10.1016/s1369-5274(02)00324-7. [DOI] [PubMed] [Google Scholar]
- 50.Council NR. Nutrient Requirements of Dairy Cattle. Washington, DC: Natl Acad Press; 2001. pp. 1–381. [Google Scholar]
- 51.Yu Z, Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques. 2004;36:808–812. doi: 10.2144/04365ST04. [DOI] [PubMed] [Google Scholar]
- 52.Dehority BA, Grubb JA. Effect of short-term chilling of rumen contents on viable bacterial numbers. Appl Environ Microbiol. 1980;39:376–381. doi: 10.1128/aem.39.2.376-381.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cole JR, et al. The Ribosomal Database Project (RDP-II): Sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kruskal JB. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika. 1964;29:1–27. [Google Scholar]
- 56.Legendre P, Legendre L. Numerical Ecology. 2nd English Ed. Amsterdam: Elsevier; 1998. pp. 1–853. [Google Scholar]
- 57.Clarke KR, Gorley RN. PRIMER-E. Plymouth, UK: PRIMER-E Ltd; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.