Abstract
To cleave DNA, Type III restriction enzymes must communicate the relative orientation of two asymmetric recognition sites over hundreds of base pairs. The basis of this long-distance communication, for which ATP hydrolysis by their helicase domains is required, is poorly understood. Several conflicting DNA-looping mechanisms have been proposed, driven either by active DNA translocation or passive 3D diffusion. Using single-molecule DNA stretching in combination with bulk-solution assays, we provide evidence that looping is both highly unlikely and unnecessary, and that communication is strictly confined to a 1D route. Integrating our results with previous data, a simple communication scheme is concluded based on 1D diffusion along DNA.
Keywords: 1D diffusion, ATPase, helicase, single molecule, motor protein
The ability of enzymes bound at distant DNA sites to communicate with each other via long-range interactions is an important biological theme. Very often the underlying genetic processes, such as gene silencing, site-specific recombination, restriction, etc., rely on energy-independent DNA looping (1). For many other processes, such as in mismatch repair (2) and for both Type I and III restriction enzymes (REs) (3, 4), the long-range interaction relies on ATP hydrolysis and in these cases the contribution of general passive three-dimensional (3D) looping to communication remains controversial.
Restriction enzymes are a model family for studying long-range communication because the majority (and in particular all Type I and III REs) need to interact with two separate DNA sequences before cutting DNA. For the Type II REs, there is growing evidence that passive 3D DNA looping (Fig. 1A) is frequently used (5). In contrast, the Type I and III REs contain protein domains that are classified as Superfamily 2 (SF2) DNA helicases (6), and these domains are required for ATP-dependent DNA communication (7, 8). The role of the helicase domains in the Type I REs has been clearly defined (Fig. 1A); communication involves DNA loop extrusion driven by directional dsDNA translocation (9, 10), without DNA unwinding (11), with the motor making steps along the DNA of <2 bp and consuming on average one ATP for each bp moved (12). Cleavage occurs upon collision with a second translocating motor at random positions distant from the binding site (3). This is therefore a pure 1D directional communication process. In comparison, the communication mechanism for Type III REs has not been accurately defined and conflicting models have been proposed (4, 13, 14).
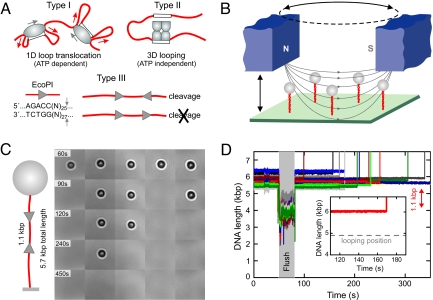
Fig. 1.
Modes of communication between two distant enzyme sites and highly parallel single DNA molecule detection assay. (A) Communication between distant DNA target sites by restriction enzymes. Type I REs use ATP-driven translocation to pull in large DNA loops (red arrows). Establishment of intersite contacts by Type II REs occurs mainly by passive, diffusive 3D looping. Type III REs require two asymmetric sites in an indirectly repeated orientation to cleave DNA approximately 25 bp downstream at one of the two sites (small gray arrows). (B) Sketch of the parallel magnetic tweezers setup. (C) Real-time DNA cleavage experiment with EcoPI on 5 DNA molecules (sketch on the Left) where the two 1.1-kb spaced enzyme sites are oriented in a HtH fashion (F = 1.5 pN). DNA cleavage is seen as disappearance of the 1-μm sized magnetic microspheres (black/white circles). (D) Simultaneous tracking of 15 DNA molecules during cleavage by EcoPI. DNA contour lengths have been corrected to account for incomplete stretching at F = 1.5 pN. During the period marked “flush”, enzyme is introduced into the flow cell. Subsequently, DNA molecules are cleaved and the microsphere lost, as seen by the apparent rapid DNA lengthening. Inset: Enlarged view of a time trace for a single DNA, showing the constant length throughout the reaction.
Type III REs require two copies of their asymmetric recognition site in an indirectly repeated, head-to-head (HtH) orientation (15) (Fig. 1A) cleaving the DNA 25–27 bp downstream of only one of the two sites. Given that Type III REs also require the ATPase activity of their SF2 helicase domains [albeit unrelated to Type I REs (6)], a Type I-like DNA loop translocation model was proposed in which translocation was unidirectional, accounting for the site-orientation preference (4). In support of this model, apparent DNA looping activity has been observed in atomic force microscope (AFM) studies (14, 16, 17). However, compared with Type I REs (12), Type III REs have a greatly reduced ATPase activity (4, 18), making a pure translocation-driven communication much less likely. To cope with this discrepancy, recent studies have elaborated on the original model by including several (up to five) passive 3D-looping steps before 1D translocation (14, 17). The looping is thought to occur with nonspecific DNA sites and shortens the intersite distance for the final translocation step. Although compelling, there are new problems associated with both model and experiments: First, 3D looping can only sense specific site orientations over long distance under special circumstances of DNA topology; and second, confining the DNA on mica during AFM measurements imposes a 2D geometry that can bias conformational flexibility.
To probe the intersite communication by Type III REs in an environment that allows 3D motion of the DNA, we used single molecule magnetic tweezers in combination with bulk solution biochemical techniques. We found that for the two Type III REs, EcoPI and EcoP15I, specific DNA cleavage was both efficient and rapid when DNA looping was suppressed. Moreover, the cleavage kinetics were unaffected by stretching forces covering the full range of DNA conformations, from almost completely elongated to randomly coiled. Irrespective of the applied force, active or passive DNA loops were never detected. These results strictly limit the possible intersite communication mechanisms. They show that the 3D looping is not a prerequisite of, and is most likely not even a consequence of, long-range communication. Given the low ATPase rate of Type III REs, we instead propose 1D diffusion along the DNA contour as the driving mechanism.
Results
Highly Parallel Magnetic Tweezers to Study DNA Cleavage.
To monitor active or passive DNA looping in a 3D rather than a 2D environment, we used magnetic tweezers (19). This technique allows stretching and twisting of single DNA molecules whereas simultaneously recording the DNA end-to-end distance in the presence or absence of a DNA-binding protein. In brief, this is achieved by attaching DNA at one end to the surface of a glass flow cell and at the other end to a magnetic microsphere (Fig. 1B). Permanent magnets placed above the flow cell generate a magnetic force on the microsphere and, in turn, on the DNA molecule. The DNA end-to-end distance is determined from images of the microspheres recorded by microscopy. Magnetic tweezers have been applied to a variety of DNA-interacting enzymes (20). In particular, they have been used to monitor the translocation driven DNA looping by Type I REs (10). Similarly, applying this technique to Type III REs should allow unambiguous discrimination between active translocation or passive 3D diffusion as the driving force for any loop formation.
To obtain strong statistics more efficiently, we improved the magnetic tweezers set-up and software by enabling simultaneous tracking and manipulation of multiple DNA molecules. For a sufficiently dense coverage of the flow cell, between 10 to 30 microsphere-DNA constructs could be found per 140 × 110 μm2 field-of-view (Fig. 1 B and C). Because the magnetic field gradient is essentially constant over this area (≈0.2% deviation) and the microsphere-to-microsphere variability is approximately 10%, all imaged microsphere-DNA constructs experienced, within 10% error, the same force. Unwanted multiple DNA tethers were distinguished from single nicked and unnicked DNA tethers by their characteristic supercoiling behavior (19). Experiments were then initiated by introducing enzyme into the cell.
EcoPI Can Cleave DNA Specifically Without Looping.
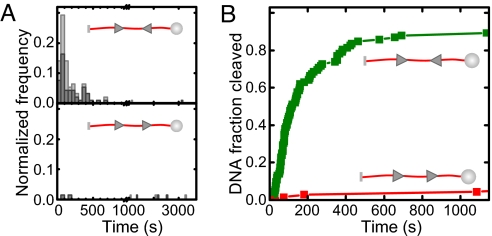
Using the highly parallel magnetic tweezers we monitored the DNA looping and cleavage activity of EcoPI on a substrate with two HtH oriented sites separated by 1.1 kb (Fig. 1 C and D). A force of 1.5 pN was chosen in this experiment, where the DNA was sufficiently stretched (89% of its contour length) to prevent diffusive looping between the two EcoPI sites (Fig. 1C, see supporting information (SI) Fig. S1). Relatively rapid DNA cleavage was observed for the majority (≈90%) of DNA molecules, identified by disappearance of the magnetic microspheres from the field-of-view (Fig. 1C). For all of the molecules tested (n = 92), DNA cleavage was never preceded by any loop formation between the two sites (Fig. 1C). Because EcoPI, in the absence of the cofactor S-adenosyl methionine (AdoMet), is known to exhibit moderate nonspecific cleavage activity, that is, which does not rely on communication between two sites (21), we tested EcoPI under identical conditions on a head-to-tail (HtT) substrate, which differs only in the orientation of the second EcoPI site (Fig. 2). Although cleavage can be observed, it is much slower and is confined to <10% of the molecules, in agreement with previous data (21). Therefore, the rapid DNA cleavage seen on the HtH substrate can be attributed in the vast majority of cases to specific cleavage events dependent on intersite communication. We also tested cleavage of both substrates in the presence of AdoMet. As expected (21), whereas cleavage of the HtT substrate was never observed (n = 37), rapid cleavage of the HtH substrate independent of DNA looping was observed (n = 62), albeit with a lower efficiency (≈60%, data not shown).
Fig. 2.
Specificity of DNA cleavage by EcoPI in the single-molecule assay. (A) Histograms of the cleavage times for a HtH substrate (Upper) and for a HtT substrate (Lower) recorded at 1.5 pN. Cleavage time is the period between start of the enzyme flush until cleavage was observed (Fig. 1D). Counts were normalized by the total number N of molecules investigated with n = 92 for the HtH and n = 68 for the HtT substrate. Light gray bars are for all molecules, gray bars for the intact fraction only. (B) Cleavage kinetics for the HtH and the HtT substrate obtained by integrating the histograms in A.
DNA Cleavage by EcoPI Is Independent of Force.
Although the above results are inconsistent with DNA looping, it could be argued that looping is suppressed by the applied stretching force. We therefore investigated DNA cleavage at three stretching forces that cover the full gamut of DNA conformations: approximately 0.01 pN (random coil), 0.1 pN (extended to 55%), and 5 pN (extended to 94%) (Fig. 3A). Similarly fast DNA cleavage was observed at all three forces (Fig. 3 A and B). Although we can exclude loop formation between the EcoPI sites at 5 pN (see Fig. S1b), loops could potentially have been formed at 0.01 pN and 0.1 pN (see arrows in Fig. 3A). The applied force reduces the looping probability by only 1.2-fold at 0.01 pN and by ≈10-fold at 0.1 pN (Fig. 4A) (22), whereas it hardly affects the loop life-time. At 0.1 pN we can, however, exclude loops with a life time larger than the approximate 1 s time resolution at this force (see Fig. S1a and gray curve in Fig. 3A), whereas at 0.01 pN the poor time resolution of approximately 50 s does not permit such conclusions given the time scale of the experiments.
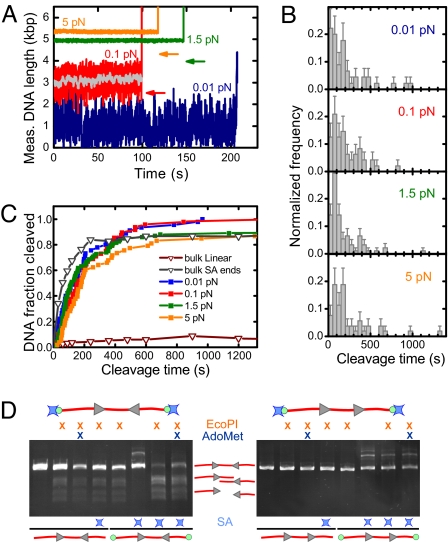
Fig. 3.
Force dependence of DNA cleavage by EcoPI. (A) Individual time traces for the HtH substrate taken at 0.01 pN, 0.1 pN, 1.5 pN, and 5 pN. The arrows indicate the expected apparent DNA length for the given force if a loop between the two EcoPI sites would have been formed. The gray trace was obtained by smoothing the 0.1 pN trace (red) with a 1-s sliding average window. (B) Histograms of the cleavage times for the HtH substrate measured at 0.01 pN (n = 45), 0.1 pN (n = 48), 1.5 pN (n = 92), and 5 pN (n = 53). (C) Cleavage kinetics for the HtH substrate at the four forces measured compared with the cleavage kinetics obtained in bulk experiments. For the bulk data the same substrate was used as in the tweezers experiments, which was either simply linearized or linearized with streptavidin (SA) attached to the ends (see below). (D) Bulk DNA cleavage measured after one hour using 15 nM EcoPI on 2 nM HtH (Left) or HtT (Right) linear DNA, with or without biotin ends (green circles). Where indicated, streptavidin (blue stars) and 100 μM AdoMet were included. DNA samples were separated by agarose gel electrophoresis.
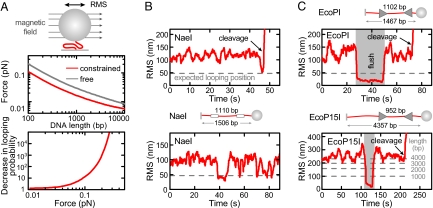
Fig. 4.
Absence of looping for DNA in random coil configuration. (A) Residual entropic force for a 1-μm microsphere as a function of DNA length (25) and probability decrease for formation of a 1,000-bp loop as a function of force (22). Calculations have been done for a microsphere which can either rotate in all directions (free) or only around one axis parallel to the surface (constrained), as achieved with an applied magnetic field (see sketch). Forces are obtained as described in ref. 25 using the DNA extension from simulations. (B) Detection of DNA looping for the Type II RE NaeI by TPM with a constrained microsphere. The sketch illustrates the DNA construct. DNA looping by NaeI is seen as a large decrease in the RMS amplitude of the lateral fluctuations. The gray dashed line shows the expected RMS amplitude after a loop is formed, as obtained from simulations. The graphs show looping just before DNA cleavage (Upper) and repetitive looping (Lower). The time resolution was approximately 2 s because of 2-s sliding window averaging. (C) DNA cleavage by EcoPI and EcoP15I on randomly coiled DNA. Constrained TPM measurements were carried out using either short 1.5-kb constructs or the longer constructs from the force experiments (see sketches; Fig. 1 and Fig. S3). Time resolution for the short constructs is approximately 2 s and for the long constructs is approximately 10 s.
If DNA looping facilitates intersite communication (14), one would expect tremendously faster DNA cleavage at low forces, where DNA looping would be possible, compared with slower DNA cleavage at high forces, where DNA looping would be suppressed. No large change in the cleavage time distribution (Fig. 3B) or kinetics (Fig. 3C) were observed (see Fig. S2 for further analysis), irrespective of force, showing that even at the lowest forces possible, communication is not facilitated by looping.
DNA End Capping Stimulates Cleavage.
To rule out any constraints imposed by the single-molecule assay, for example, surface attachment, we compared our cleavage kinetics with those obtained in bulk using the same conditions and DNA substrates as above. Strikingly, hardly any (<10%) cleavage was observed even after 1 h of incubation (Fig. 3C). One potential difference is that the DNA ends were blocked at the attachment points in the tweezers assay. We therefore tested whether blocked DNA ends in bulk solution can restore efficient cleavage. As a block we used a single streptavidin attached to each end via a biotinylated nucleotide (Fig. 3D, Methods). Remarkably, the streptavidin attachment restored nearly 100% cleavage efficiency (Fig. 3D). This effect was only observed if the ends were biotinylated and streptavidin added, proving that the cleavage enhancement is solely because of blocking of the DNA ends. The same result was obtained with AdoMet. In contrast, cleavage was not observed on an end-blocked HtT substrate (Fig. 3D). The cleavage kinetics now provides good agreement with the single-molecule data. The slightly slower single-molecule cleavage is most likely because of time uncertainties in the addition of enzyme (Fig. 1D). These observations are inconsistent with existing directional models for Type III REs, in which communication is confined to the region between the sites.
EcoP15I Can Cleave DNA Without Looping.
The AFM studies which support DNA looping used EcoP15I exclusively (14, 16, 17). EcoP15I is very closely related to EcoPI and behaves similarly (23). We repeated our measurements using EcoP15I (Fig. S3). The results obtained using EcoP15I were, qualitatively, identical to those obtained with EcoPI, that is, rapid cleavage without DNA looping, similar cleavage kinetics at low (0.01 pN) and high (1.5 pN) forces and stimulation of cleavage by DNA end-capping (in a concentration dependent manner).
Absence of Stable Loops at Minimum Applied Force.
We also wanted to directly test DNA loop formation at the smallest possible forces within our setup, which was so far limited because of poor time resolution (see above). Better time resolution can be achieved by observing the lateral fluctuations of a (sub)-micrometer-sized particle tethered to a glass surface via the DNA molecule. The root-mean-square amplitude of these fluctuations is then a measure of the DNA length, both of which reduce upon loop formation. This technique, called tethered particle motion (TPM), has for example been used to detect 3D DNA looping by Type II REs (24). Despite the absence of external force, the DNA still experiences a residual entropic stretching force in these experiments because of volume exclusion (25). Depending on the parameters (microsphere size, DNA length), this entropic force can reach tens of fN (Fig. 4A). To reduce this force we performed our measurements by using magnetic microspheres in the presence of a weak magnetic field (residual force <5 fN) which limits microsphere rotations to those around the axis parallel to the field producing an approximate 2-fold force reduction (Fig. 4A). By using magnetic microspheres 1 μm in diameter we calculate the total force for an approximate 5-kb construct to be approximately 0.01 pN and for a 1.5-kb construct to be approximately 0.02 pN. At both forces the looping probability is only slightly changed (Fig. 4A).
We first tested our experimental system by using NaeI, for which 3D looping has been reported (24). We could readily detect NaeI-mediated DNA looping on a short 1.5-kb long substrate for an intersite distance of 1.1 kb and, notably, could resolve loops that precede DNA cleavage (Fig. 4B). We executed similar experiments by using EcoPI or EcoP15I on our long (5.7 kb and 4.3 kb, respectively) and on short 1.5-kb substrates (Fig. 4C). We did not observe loop formation on any of the substrates. For the short substrates, we can specifically rule out intersite looping with an approximate 2-s time resolution. Even the limited resolution with the long substrates still permits us to exclude extensive looping that includes upstream DNA as suggested (14).
Intersite Communication Requires Significantly less than 1 ATP per bp.
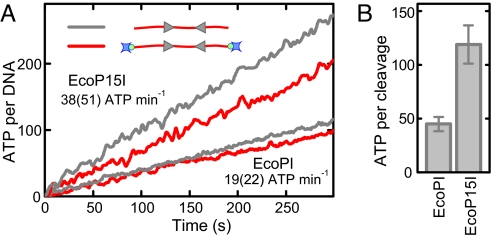
Pure translocation-driven communication requires significant ATP consumption. For example, Type I REs consume approximately 1 ATP per bp translocated with up to 60,000 ATP min−1 per site (12). To derive for our experiments an upper limit for such a coupling ratio, that is, the amount of ATP per communication distance, we measured the ATPase activity for EcoPI and EcoP15I under the same conditions and on the same linear DNA, as used above. We used a coumarin-labeled PBP sensor (26), which allows determination of the ATPase activity on a millisecond time scale. Our measurements show a constant ATPase rate over the time-course of cleavage with only 19 ± 2 (capped) and 22 ± 2 (uncapped) ATP min−1 per DNA for EcoPI and 38 ± 4 (capped) and 51 ± 5 (uncapped) ATP min−1 per DNA for EcoP15I (Fig. 5A) in agreement with previous reports for EcoP15I (4). We calculated the average number of ATP molecules hydrolyzed per DNA cleavage by dividing the ATPase rate (Fig. 5A) with the DNA cleavage rate for capped substrates (Fig. S4), which yielded 45 ± 7 and 119 ± 18 ATP per cleavage for EcoPI and EcoP15I, respectively (Fig. 5B). This equates to approximately 25 and 8 bp communicated per ATP hydrolyzed, respectively.
Fig. 5.
ATPase activity of Type III REs. (A) Kinetics of ATP hydrolysis (per DNA) for EcoPI and EcoP15I measured for capped and uncapped HtH substrates at 1 mM ATP. The ATPase rates indicated are for capped and uncapped (in brackets) substrates. (B) Average number of ATP molecules required to cleave a DNA molecule obtained by dividing the ATPase rate by the cleavage rate measured under the same conditions (Fig. S4).
Discussion
Type III REs Communicate in a 1D Manner Without Looping.
We show that Type III REs can communicate between their sites in a strict 1D fashion without recourse to loop formation. We can even rule out loops (minimum lifetime ≈1 s) at lower forces, conditions where looping is feasible (see Figs. 3A and 4). Additional evidence against DNA looping is the low force dependency of the cleavage which occurs on the same time scale as in bulk (Figs. 3C and S3b). In contrast, for Type II REs which use 3D diffusive looping, strongly force-dependent kinetics is observed (27). We also show that DNA ends upstream of the sites can influence the efficiency of communication. Our results set narrow constraints on the possible intersite communication mechanisms that could be used by Type III REs.
We note that whilst our results concur with other studies in bulk solution that argue against 3D DNA looping (23, 28), they are contradictory to recent AFM investigations where extensive 3D passive and active DNA looping was reported (14, 16, 17). However, the AFM studies could not show that DNA cleavage is directly dependent on the looped topology. Although structures found with AFM (17) have been interpreted as supercoiled DNA, established biochemical assays failed to detect supercoiling (28) more similar to this study. Also, during AFM investigations the DNA is bound to mica. This is typically achieved by using divalent ions such as Mg2+, which compete with monovalent ions such that DNA binding is only optimized at reduced monovalent ion concentrations (29). Thus, the AFM buffers were optimised for imaging but did not provide optimum conditions for cleavage (14). More critically, surface-bound DNA is kept in a semimobile 2D configuration. Confinement to a lower dimension will significantly enhance the intramolecular interactions seen as avoidance of self-crossing and volume exclusion (29). This potentially can favor interactions not seen in a 3D environment.
Our careful analysis under a variety of forces argues that DNA looping by EcoPI and EcoP15I is unlikely in a 3D environment and that communication occurs in 1D irrespective of force. Moreover, 3D looping is conceptually problematic, because the site orientation of one site needs to be communicated to the other (23). Although possible at short spacings, any topological bias vanishes for longer distances (30) and cannot reconcile differences in specificity of 10-fold or more.
1D Diffusion Model for Intersite Communication.
If intersite communication is restricted to 1D, what can be the mechanism behind it? Two general suggestions are loop-independent 1D motion along DNA (13) or extension of a protein filament (2). Here, we will discard the latter possibility because the moderate excess of enzyme over DNA used in our studies does not provide sufficient molecules for extensive filamentous growth. However, 1D translocation models also have severe problems. (i) The imposed strict directionality fails to explain the cleavage stimulation by DNA end capping (see above). (ii) Type III REs fail to displace triplexes (Fig. S5), in contrast to all other bona fide dsDNA translocases tested. (iii) The ATP consumption required per cleavage event is much too low to allow significant translocation (Fig. 5B). For example, 45 ATP molecules are hydrolyzed by EcoPI until cleavage is achieved, providing an unrealistic minimum step size of 25 bp per ATP within a translocation-only model or a negligible length of DNA translocated within a partial-translocation model. Similar values have been obtained for the Type III RE PstII (18). The observation that end-capping greatly stimulates DNA cleavage indicates that communication is not just restricted to the intersite region, but extends over the entire DNA, which provides even more unrealistic coupling values within translocation models (e.g., >129 bp per ATP for EcoPI). Therefore, given the ATPase values obtained, any significant role for translocation can be excluded. The situation is similar for EcoP15I (119 ATP per cleavage or 8 bp communicated per ATP), although in theory it would be able to translocate a more significant proportion of the intersite DNA. However, the amounts of ATP consumed and apparent step sizes are extreme limits calculated under the unlikely assumptions that communication is restricted to the intersite area and always successful. Given the similarities of EcoP15I with EcoPI (>90% sequence identity between the ATPases) and the very similar observations made, we do not expect different mechanisms.
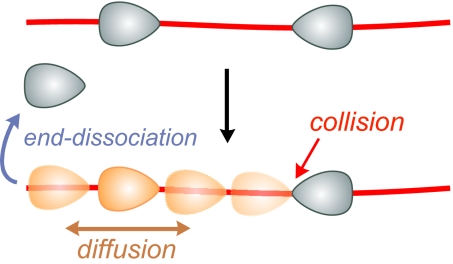
For these reasons and given our arguments against looping (above), we propose 1D diffusion to be the driving force for intersite communication by Type III REs (Fig. 6). In fact, 1D diffusion integrates 1D communication and low ATPase activity and can readily explain our data and all of the bulk solution data on Type III REs thus far, including the original roadblock experiments which were used to establish the dogma (4). The enzymes, which will associate with the DNA in an oriented fashion (Fig. 6), determined by their binding sites, only need to maintain their orientation during diffusion to convey information regarding the site orientation to another target site. Accordingly a site-orientation dependent cleavage complex can be formed (see SI Text and Figs. S6 and S7 for more detail on the diffusion model).
Fig. 6.
Model for the intersite communication by Type III REs based on 1D diffusion. The enzymes bind to DNA in an orientation determined by their binding site. After diffusion is triggered, one enzyme slides bidirectionally along the DNA and either falls off at a DNA end or encounters a second enzyme where, depending on its orientation, DNA cleavage may be triggered.
Our new model for intersite communication by Type III REs provides a fundamental basis for future work on these enzymes and allows making testable predictions. In fact, to further support 1D diffusion, we carried out cleavage experiments on linear, uncapped DNA with fixed intersite spacing but variable DNA end lengths (see SI Text and Fig. S8). This provided a faster cleavage with increasing end length, which can only be explained within the framework of a diffusion model.
Interestingly, 1D diffusion is now the widely accepted mechanism in mismatch repair for communication between mismatch and dam methylation sites, which mark the parental strand (2, 31). Beyond the overall communication task, many experimental observations are similar to Type III REs, for example, low ATPase rates, disruption of communication by protein roadblocks, trapping of diffusing complexes by capped DNA ends (2). It might be that both enzyme systems have independently developed a very similar 1D diffusion communication, making it a more wide-spread phenomenon.
Materials and Methods
DNA and Proteins.
EcoPI DNA substrates were based on either pAMS3 (5,711 bp, HtH-oriented sites with 1,102-bp spacing) or pAMS4 (5,709 bp, HtT-oriented sites with 1,079-bp spacing). 5.7-kb tweezers substrates were prepared by cutting with NotI and XbaI (17 bp downstream) to leave the EcoPI sites approximately 2.5 kb and 2.1 kb from the ends. Biotin- or digoxigen-modified attachment handles (≈0.6 kb) were prepared by cutting a 1.2-kb biotin- or digoxigen-dUTP labeled PCR fragment with XbaI or NotI, respectively, and subsequent ligation to the digested plasmid. The 1.5-kb substrate for the low force experiments was made by PCR from pAMS3 using primers 5′-modified with biotin or digoxigenin. For the bulk cleavage and ATPase experiments plasmids were cleaved using XbaI alone. The ends of the linearized plasmids were biotinylated by using Klenow polymerase and biotin-dUTP. The 1.5-kb NaeI substrate (Fig. 4B) was made by PCR analogously to the short EcoPI substrate. EcoPI and EcoP15I were produced as described in ref. 23.
Single-Molecule Cleavage Assays.
The basic magnetic tweezers protocol has been described (19). Images were acquired with a frequency of 60 Hz. In brief, DNA constructs were bound to 1-μm streptavidin-coated superparamagnetic microspheres (Invitrogen) and flushed into a flow cell, with a bottom cover slip coated with anti-digoxigenin. Subsequently, 15 nM EcoPI or EcoP15I were added in buffer R (50 mM Tris·HCl, pH 8.0, 50 mM KCl, 10 mM MgCl2, 1 mM DTT, and 100 μg ml−1 BSA, 20 ± 2°C) plus 4 mM ATP. Where indicated the buffer was supplemented with 100 μM AdoMet. For the NaeI experiments, 50 units ml−1 NaeI (NEB), were added in NEBuffer 2 (NEB) supplemented with 100 μg ml−1 BSA and 3.66 μg ml−1 unlabeled background dsDNA.
DNA Bulk Cleavage Assays.
DNA cleavage assays were carried out at 20 ± 2°C using buffer R, 15 nM EcoPI or EcoP15I and 2 nM DNA. Enzyme was premixed with buffer and cleavage was initiated by adding DNA. For the end-capping experiments, DNA with biotinylated ends was preincubated with a 50-fold molar excess of streptavidin. Reactions were incubated for the times indicated and subsequently stopped by adding 0.5 volumes of stop buffer (Blue-Orange loading buffer (Promega) diluted 1:1, 100 mM Tris·HCl, pH 8.0, and 100 mM EDTA) and placing them on ice. Reaction products were separated by agarose gel electrophoresis and stained with ethidium bromide. Cleavage kinetics was determined by quantification of the fluorescence.
ATPase Activity Assay.
ATPase activity was measured using coumarin-labeled PBP as described (12, 26). Reactions were initiated by mixing equal volumes of a DNA solution with an enzyme solution to give final conditions of 2 nM DNA, 15 nM enzyme, 1 mM ATP, 25 μM PBP, and Buffer R (without BSA plus 100 nM streptavidin) at 20 ± 0.1°C. Because of the slow ATPase rates, background Pi contamination from the ATP stocks could not be removed by purine nucleoside phosphorylase. Therefore, measurements could only be carried out at 1 mM ATP to be within the linear range of the PBP. To allow comparison, additional cleavage experiments at 1 mM were carried out (Fig. S4).
Supplementary Material
Acknowledgments.
This work was supported by the Deutsche Forschungsgemeinschaft (R.S.), by Wellcome Trust Grants 067439 and 084086 (to M.D.S.), the Biotechnology and Biological Sciences Research Council (M.D.S.), and the Marie Curie Research Training Network “DNA Enzymes”. M.D.S. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.H. is a guest editor invited by the Editorial Board. Freely available online through the PNAS open access option.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807193106/DCSupplemental.
References
- 1.Halford SE, Welsh AJ, Szczelkun MD. Enzyme-mediated DNA looping. Annu Rev Biophys Biomol Struct. 2004;33:1–24. doi: 10.1146/annurev.biophys.33.110502.132711. [DOI] [PubMed] [Google Scholar]
- 2.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: Functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 3.Studier FW, Bandyopadhyay PK. Model for how type I restriction enzymes select cleavage sites in DNA. Proc Natl Acad Sci USA. 1988;85:4677–4681. doi: 10.1073/pnas.85.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meisel A, Mackeldanz P, Bickle TA, Krüger DH, Schroeder C. Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis. EMBO J. 1995;14:2958–2966. doi: 10.1002/j.1460-2075.1995.tb07296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gowers DM, Bellamy SRW, Halford SE. One recognition sequence, seven restriction enzymes, five reaction mechanisms. Nucleic Acids Res. 2004;32:3469–3479. doi: 10.1093/nar/gkh685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McClelland SE, Szczelkun MD. Molecular motors that process DNA. In: Pingoud A, editor. Restriction Endonucleases, Nucleic Acids and Molecular Biology. Berlin: Springer Verlag; 2004. pp. 111–135. [Google Scholar]
- 7.Davies GP, Powell LM, Webb JL, Cooper LP, Murray NE. EcoKI with an amino acid substitution in any one of seven DEAD-box motifs has impaired ATPase and endonuclease activities. Nucleic Acids Res. 1998;26:4828–4836. doi: 10.1093/nar/26.21.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McClelland SE, Dryden DTF, Szczelkun MD. Continuous assays for DNA translocation using fluorescent triplex dissociation: Application to type I restriction endonucleases. J Mol Biol. 2005;348:895–915. doi: 10.1016/j.jmb.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Firman K, Szczelkun MD. Measuring motion on DNA by the type I restriction endonuclease EcoR124I using triplex displacement. EMBO J. 2000;19:2094–2102. doi: 10.1093/emboj/19.9.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seidel R, et al. Real-time observation of DNA translocation by the type I restriction modification enzyme EcoR124I. Nat Struct Mol Biol. 2004;11:838–843. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- 11.Stanley LK, et al. When a helicase is not a helicase: dsDNA tracking by the motor protein EcoR124l. EMBO J. 2006;25:2230–2239. doi: 10.1038/sj.emboj.7601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seidel R, Bloom JGP, Dekker C, Szczelkun MD. Motor step size and ATP coupling efficiency of the dsDNA translocase EcoR124I. EMBO J. 2008;27:1388–1398. doi: 10.1038/emboj.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raghavendra NK, Rao DN. Unidirectional translocation from recognition site and a necessary interaction with DNA end for cleavage by Type III restriction enzyme. Nucleic Acids Res. 2004;32:5703–5711. doi: 10.1093/nar/gkh899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crampton N, et al. DNA looping and translocation provide an optimal cleavage mechanism for the type III restriction enzymes. EMBO J. 2007;26:3815–3825. doi: 10.1038/sj.emboj.7601807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meisel A, Bickle TA, Krüger DH, Schroeder C. Type III restriction enzymes need two inversely oriented recognition sites for DNA cleavage. Nature. 1992;355:467–469. doi: 10.1038/355467a0. [DOI] [PubMed] [Google Scholar]
- 16.Reich S, Gössl I, Reuter M, Rabe JP, Krüger DH. Scanning force microscopy of DNA translocation by the Type III restriction enzyme EcoP15I. J Mol Biol. 2004;341:337–343. doi: 10.1016/j.jmb.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 17.Crampton N, et al. Fast-scan atomic force microscopy reveals that the type III restriction enzyme EcoP15I is capable of DNA translocation and looping. Proc Natl Acad Sci USA. 2007;104:12755–12760. doi: 10.1073/pnas.0700483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sears A, Peakman LJ, Wilson GG, Szczelkun MD. Characterization of the Type III restriction endonuclease PstII from Providencia stuartii. Nucleic Acids Res. 2005;33:4775–4787. doi: 10.1093/nar/gki787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Revyakin A, Ebright RH, Strick TR. Single-molecule DNA nanomanipulation: Improved resolution through use of shorter DNA fragments. Nat Methods. 2005;2:127–138. doi: 10.1038/nmeth0205-127. [DOI] [PubMed] [Google Scholar]
- 20.Seidel R, Dekker C. Single-molecule studies of nucleic acid motors. Curr Opin Struct Biol. 2007;17:80–86. doi: 10.1016/j.sbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Peakman LJ, Antognozzi M, Bickle TA, Janscak P, Szczelkun MD. S-adenosyl methionine prevents promiscuous DNA cleavage by the EcoP1I type III restriction enzyme. J Mol Biol. 2003;333:321–335. doi: 10.1016/j.jmb.2003.08.042. [DOI] [PubMed] [Google Scholar]
- 22.Sankararaman S, Marko JF. Formation of loops in DNA under tension. Phys Rev E. 2005;71 doi: 10.1103/PhysRevE.71.021911. 021911. [DOI] [PubMed] [Google Scholar]
- 23.Peakman LJ, Szczelkun MD. DNA communications by Type III restriction endonucleases–confirmation of 1D translocation over 3D looping. Nucleic Acids Res. 2004;32:4166–4174. doi: 10.1093/nar/gkh762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Broek B, Vanzi F, Normanno D, Pavone FS, Wuite GJL. Real-time observation of DNA looping dynamics of Type IIE restriction enzymes NaeI and NarI. Nucleic Acids Res. 2006;34:167–174. doi: 10.1093/nar/gkj432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segall DE, Nelson PC, Phillips R. Volume-exclusion effects in tethered-particle experiments: Bead size matters. Phys Rev Lett. 2006;96 doi: 10.1103/PhysRevLett.96.088306. 088306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webb MR. Development of fluorescent biosensors for probing the function of motor proteins. Mol Biosyst. 2007;3:249–256. doi: 10.1039/b614154d. [DOI] [PubMed] [Google Scholar]
- 27.Gemmen GJ, Millin R, Smith DE. Tension-dependent DNA cleavage by restriction endonucleases: Two-site enzymes are “switched off” at low force. Proc Natl Acad Sci USA. 2006;103:11555–11560. doi: 10.1073/pnas.0604463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janscak P, Sandmeier U, Szczelkun MD, Bickle TA. Subunit assembly and mode of DNA cleavage of the type III restriction endonucleases EcoP1I and EcoP15I. J Mol Biol. 2001;306:417–431. doi: 10.1006/jmbi.2000.4411. [DOI] [PubMed] [Google Scholar]
- 29.Rivetti C, Guthold M, Bustamante C. Scanning force microscopy of DNA deposited onto mica: Equilibration versus kinetic trapping studied by statistical polymer chain analysis. J Mol Biol. 1996;264:919–932. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- 30.Kingston IJ, Gormley NA, Halford SE. DNA supercoiling enables the type IIS restriction enzyme BspMI to recognise the relative orientation of two DNA sequences. Nucleic Acids Res. 2003;31:5221–5228. doi: 10.1093/nar/gkg743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorman J, et al. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol Cell. 2007;28:359–370. doi: 10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.