Abstract
We have identified new genomic alterations in the breast cancer cell line HCC1954, using high-throughput transcriptome sequencing. With 120 Mb of cDNA sequences, we were able to identify genomic rearrangement events leading to fusions or truncations of genes including MRE11 and NSD1, genes already implicated in oncogenesis, and 7 rearrangements involving other additional genes. This approach demonstrates that high-throughput transcriptome sequencing is an effective strategy for the characterization of genomic rearrangements in cancers.
Keywords: cancer genome, transcriptome sequencing
An achievable goal of the oncology community is to perform comprehensive sequence analysis of the cancer genome and its transcripts toward the identification of new detection, diagnostic and intervention strategies. The onset of cancer results from genomic alterations in precursor cells, and changes in the surrounding microenvironment (1) including the immune system (2). Comprehensive analysis of the active genes comprising the transcriptome has resulted in advances in our ability to understand the pathways involved in the progression of cancer, serves as a tool to delineate molecular differences in cancers, even among those that are from the same body site and appear similar by traditional approaches. Large-scale Sanger-based cDNA sequencing approaches contributed significantly to deciphering transcriptome complexity (3–5). However, cost has been a significant limitation. To address that issue, and to attain deep coverage of the transcriptome such that rare transcripts could be identified, tagging approaches such as SAGE (6), CAGE (7), and MPSS (8) have been used. However, the short tags that are used give a very limited view of the complete transcript set and variations therein such as through alternative splicing, translocations and point mutations.
Here, we use 454 Life Sciences pyrosequencing technology that enables relatively long sequence reads and deep transcriptome coverage. Our primary interest was to determine whether we could use this technology to identify genomic alterations, specifically gene fusions, and thus contribute to an integrated view of the genome and transcriptome alterations within the breast cancer cell line HCC1954. This cell line has been the subject of several large-scale genomic analyses including comprehensive exome sequencing to detect somatic point mutations and BAC sequencing to identify chromosome translocations, which thus allows direct comparison between the approaches (9–11). It is evident that the generation of sufficient depth of transcript coverage from a tumor and corresponding normal tissue will identify all expressed genes and the point mutations they contain. The focus of this current study is how translocations that result in chimeric genes can be elucidated and interpreted from transcript sequencing.
Results
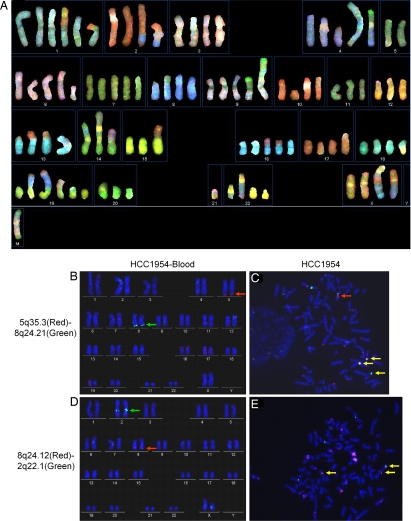
As shown by the spectral karyotyping (SKY), HCC1954 has a pseudotetraploid karyotype with an average of 92 chromosomes per cell (see Fig. 2A). The SKY analysis also reveals a large number of translocations involving most or all chromosomes. Using 454-FLX pyrosequencing we generated 510,703 cDNA sequences of average length 245 bp from the HCC1954 cell line. (See Methods and Fig. S1). We then initially aligned all cDNA sequences to RefSeq mRNAs (GenBank dataset available on March 28, 2008), revealing that >384,900 reads were uniquely associated well with 9,221 RefSeq genes.
Fig. 2.
SKY analysis of HCC1954 karyotype and FISH confirmation of interchromosomal translocations. (A) SKY picture of HCC1954 genome. (M) chromosome is too complicated to be assigned. (B and C) FISH using BACs adjacent to the break point CTC-1286C20 (5q35.3, labeled in red) and RP11–17E16 (8q24.21, labeled in green) generated fusion signals (yellow) in HCC1954. (D and E) FISH using BACs adjacent to the break point RP11–72M5 (8q24.12, labeled in red) and RP11–12M21 (2q22.1, labeled in green) generated fusion signals in HCC1954 (yellow). The red arrow in C indicates a possible event of additional duplication of 5q35.3 region.
Detection of Chimeric Genes.
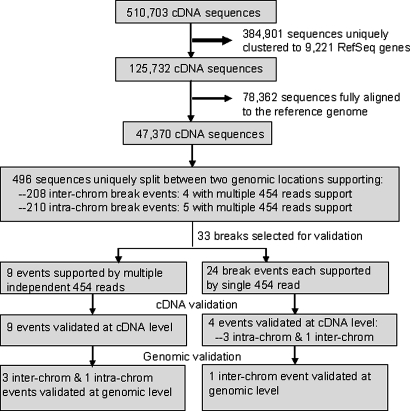
The set of sequences that did not cluster with RefSeq mRNAs were aligned to the human reference genome. The remaining 47,370 sequences including those that were neither aligned to RefSeq mRNA entries nor to the human genome at a full-length coverage were then pooled together and submitted to a computational analysis pipeline for the detection of chimeric transcripts, as revealed by cDNA that can be uniquely split between two genomic locations. Chimeric transcripts thus identified might result from genomic rearrangements. From the set of 47,370 nonalignable sequences, we identified 496 sequences that could be uniquely mapped to 2 distinct genomic locations that suggested genomic break points. The results of this analysis are detailed in Fig. 1.
Fig. 1.
Schematic diagram of chimeric transcript detection and validation.
Approximately half of the putative chimeric sequences described 208 interchromosomal rearrangement events (243/496) and the other half represented 210 intrachromosomal rearrangement events (253/496). We performed experimental validation for 33 putative chimeras including 9 with more than 1 supporting 454 read and 24 with only 1 supporting 454 read (Fig. 1).
Chimeric transcripts were experimentally first validated at the transcript level. Reverse transcriptase-PCR (RT-PCR) with a primer pair flanking the break junction of a chimeric transcript was performed on an independently prepared genomic DNA-free total RNA sample of the HCC1954. The chimeras were subsequently confirmed by Sanger sequencing of RT-PCR amplified bands. All 9 chimeric cDNA with multiple 454 read support and 4 chimeric cDNAs with single 454 read support were thus verified in the HCC1954 transcriptome. We also tested the existence of chimeric cDNAs in a matched blood cell line (HCC1954BL). Surprisingly, most chimeric transcripts that supported an intrachromosomal rearrangement were amplified from both HCC1954 and HCC1954BL (Table S1). However, all chimeric transcripts that suggested an interchromosomal rearrangement were only detected in HCC1954 but not in the control cell line (Fig. S2).
Validation of Genomic Rearrangements.
Exon repetition and gene fusion events have been reported to be present in normal cell lines as a result of trans-splicing rather than of de novo genomic rearrangements (12, 13). Thus, for those chimeras that were verified at the cDNA level we carried validation further to the genomic level, using a combination of long-range PCR (LR-PCR) and FISH experiments. For LR-PCR, we first tested the same set of primer pairs used in the chimeric cDNA amplification. If this approach was unsuccessful, then PCR-walking was performed with new primer sets along the proposed break junction regions (Fig. S3). LR-PCR amplified genomic fragments were confirmed by Sanger end-sequencing. For translocations in which the chromosomal break points were not observable by LR-PCR, we performed fluorescence in situ hybridization (FISH) with paired BAC probes flanking the break junction. Translocations t(5;8)(q35.3;q24.21) and t(8;2)(q24.12;q22) were clearly shown by the merge of fluorescent signals in HCC1954 only (Fig. 2).
Four interchromosomal translocations were confirmed at the genomic level, 3 supported by multiple 454 reads and 1 supported by single 454 read (Fig. 1, Table 1, and Table S2). Only 1 of 5 putative intrachromosomal rearrangements supported by multiple 454 reads was confirmed at the genomic level. Because most intrachromosomal chimeric transcripts with exon duplications exist in both HCC1954 and HCC1954BL cell lines, we suggest that those transcripts are likely to result from trans-splicing events. All confirmed genomic rearrangements were detected only in HCC1954.
Table 1.
Validated Genomic Rearrangements in HCC1954 Initially Detected by 454 Chimeric Transcripts
| Genomic changes | Chromosomes locations | Genes affected | Effect on coding | Genetic or somatic changes reported in cancers | Validation method |
|---|---|---|---|---|---|
| Interchrom | |||||
| intragenic to intergenic | t(5;8)(q35.3;q24.21) | NSD1 | truncation | Fusion protein in acute myeloid leukemia | cDNA and FISH |
| intragenic to intragenic | t(5;8)(p15.33;q24.21)* | CLPTM1L and PVT1 | truncation | Amplification of PVT1 linked to pathophysiology of ovarian and breast cancer | cDNA and Genomic |
| intragenic to intergenic | t(5;8)(q23.1;q23.1)* | EIF3E | truncation | Truncated form is tumorigenic in vivo. Decreased expression found in one third of all human breast carcinomas | cDNA and Genomic |
| intragenic to intergenic | t(4;11)(q32;q21) | MRE11A | truncation | Mutations found in many types of cancers including breast cancer. | cDNA and Genomic |
| intragenic to intragenic | t(9;18)(p24.1;q12.2) | PDCD1LG2 and C18orf10 | chimeric protein | Expression level of PDCD1LG2 is linked to tumor immune invasion. | cDNA and Genomic |
| Intragenic to intergenic | t(8;2)(q24.12;q22.1) | SAMD12 | chimeric protein | SAM domain assists protein dimerization. Chromosomal translocation of another SAM domain protein TEL linked to leukemia. | cDNA and FISH |
| Intrachrom | |||||
| inversion | 8q24.12: 8q24.22 | PHF20L1 and SAMD12 | truncation | N/A | cDNA and Genomic |
*t(5;8)(p15.33;q24.21) and t(5;8)(q23.1;q23.1) were identified by the additional approach as described in the text.
Our initial chimeric cDNA detection pipeline required that a chimeric cDNA uniquely maps to distinct locations. This conservative approach was designed to limit the identification of false positives, but would miss chimeric cDNAs that could be mapped to multiple genomic locations, such as would occur with the presence of pseudogenes. Therefore, to identify more potential genomic rearrangements we used an additional approach to identify chimeric transcripts that map to more than 1 genomic region. Resulting candidates with either multiple or only 1 supporting 454 reads were selected and subjected to verification. A total of 36 candidates events were identified through this approach. Of these, 6 were supported by multiple independent 454 reads. Two of these 6 chimeric events were confirmed by RT-PCR at the cDNA level and LR-PCR at genomic DNA level (Table 1, Table S1, and Figs. S2 and S3). Both resulted from interchromosomal rearrangements.
In total, 7 chimeric cDNAs resulting from 6 interchromosomal break events and 1 intrachromosomal break event were confirmed at both the transcriptional and genomic levels through validation assays, all present only in HCC1954 (the tumor cell line) (Table 1). Single 454 read provided supportive evidence for only 1 of them (Table S2). These chimeras could affect the transcription and protein products of at least 9 genes. Wild-type transcripts were detected in parallel with the chimeric transcript to different extents in the cell line (Table S2).
All verified break points tended to occur in either an intron or an intergenic region (Fig. 3). To produce the chimeric transcript, the transcription machinery either switched to the downstream exon (Fig. 3 A and C) or acquired a novel splicing acceptor in the intergenic sequence (Fig. 3B). In either break type, the consensus splicing sites were always observed on either side of the break junction of a chimeric cDNA when aligned to the genome. Although the mechanism is unknown, it is possible that the false positive chimeric cDNAs could be products of the cDNA preparation process.
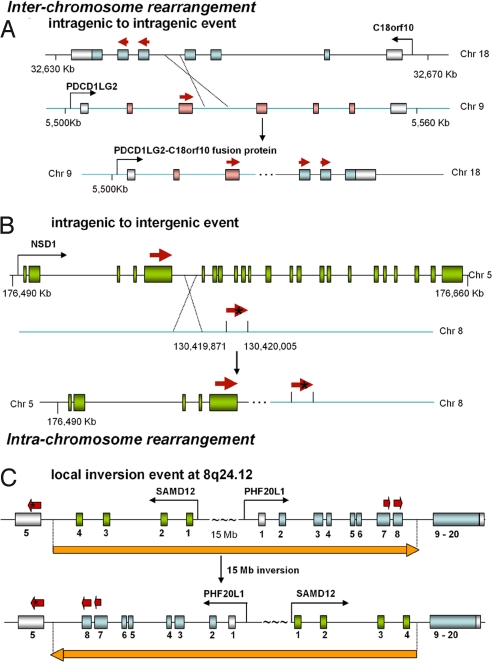
Fig. 3.
Schematic diagram of genomic rearrangement events captured by 454 transcriptome sequencing. Upper shows wild-type structures and Lower shows the rearranged structure. Red thick arrows are chimeric cDNAs captured by 454 reads. Untranslated exons are shown in gray bars, whereas translated exons are shown in colored bars. (A) Translocation between chromosomes 9 and 18 created an in-frame chimeric product proposed to be composed of 120 aa from 5′ terminus of PDCD1LG2 and 172 aa from 3′ terminus of C18orf10. (B) Transcription of the chimeric transcript involving NSD1 continued for another 134 bp on chromosome 8 before poly(A) tail was added to the mRNA. Translation of the chimeric protein contained 1,265 aa from the 5′end of NSD1 plus 19 aa from the intergenic region on chromosome 8 before stopped by an in-frame stop codon marked by an asterisk. (C) An ≈15-Mb genomic fragment was flipped as shown by the orange arrow. The PHF20L1 gene is truncated by a stop codon marked by an asterisk.
There are 2 types of breaks for the 6 verified interchromosomal translocations: intragenic to intragenic and intragenic to intergenic (Table 1 and Fig. 3 A and B). The one verified intrachromosomal rearrangement is a local intronic inversion (Table 1 and Fig. 3C). We were unable to detect the alternate broken chromosome for all rearrangement events that involved 2 genes, suggesting that alternate transcripts were rarely produced. We also observed that in most cases (5 of 7) the rearrangement events resulted in protein truncation rather than extension of the ORF, suggesting loss-of-function of critical genes (Table 1). We noted that chromosome 8 seemed to be very frequently involved (5 of 7 events) in genomic rearrangements in HCC1954. In particular, 8q24 seemed a recombination hot spot (4 break events), which coincides with the findings that polymorphic loci at 8q24 are risk factors for breast and prostate cancer from genome wide studies (14, 15).
We identified chromosomal events that led to truncation of proteins derived from genes shown to have oncogenic roles. For example, MRE11A, a key component of the DNA mismatch repair pathway, functions in DNA double-strand break repair. It has been reported to accumulate somatic mutations that lead to truncations of its protein in many types of tumors, including breast cancer (16–24). In the HCC1954 cancer cell line, this gene is truncated at its DNA binding domain because of a translocation event between chromosomes 11 and 4 (Fig. 4A). For chimeras involving the gene NSD1, transcription after exon 5 of NSD1 shifted to an intergenic sequence on chromosome 8 (Fig. 3B). NSD1 fusion protein has been detected in acute myeloid leukemia (25). SAMD12 gene, which is involved in both interchromosomal and intrachromosomal rearrangement events in HCC1954, has not been linked to oncogenesis. However, its frequent disruption in HCC1954 implicates that it might have a role in the cancer cell's phenotype.
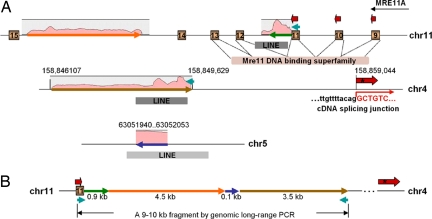
Fig. 4.
Detailed analysis of t(4;11)(q32;q21). (A) Local genomic features of chromosomes involved. The interchromosome translocation between chromosomes 11q21 and 4q32 truncates MRE11A at its DNA binding domain. Chimeric cDNAs span exon 9, 10 and 11 (brown bars) of MRE11A and intergenic sequences on chromosome 4 as shown by thick red arrows. A 9- to 10-kb genomic fragment containing the break junction was amplified with primers on chromosome 11 and chromosome 4 as shown in light blue arrows. Consensus splice acceptor sequences used for transcription of the chimeric cDNA on chromosome 4 are shown. LINE repeats are shown in shaded gray bars. The transcription orientation of MRE11A gene is marked by a black arrow. An in-frame stop codon in the chimeric 454 cDNA is marked by an asterisk. Coverage of 454 reads (shown by shaded pink areas) mapped to the 9- to 10-kb fragment was determined from an assembly output graph from CLC Bio. Coverage of the genome at the break junction is between 500× and 3,200×. (B) Final fine assembly of the break junction of t(4;11)(q32;q21) by mapping and assembly of 454 sequences on the 9- to 10-kb genomic fragment.
We also compared our validated candidates with a study that used FISH and BAC sequencing analyses for the same cell line (10), and in which 13 gene loci were reported to be affected by somatic chromosomal rearrangements. The events affecting genes PHF20L1 and NSD1 were identified in both the previous study and our reports here.
To determine the specific molecular rearrangements of the breakpoints occurring at t(4;11)(q32;q21) and t(5;8)(p15.33;q24.21) we undertook PCR walking and additional Sanger and 454 DNA sequencing. Our initial results demonstrated that the t(4;11)(q32;q21) translocation joins the MRE11A gene immediately after exon 11 on chromosome 11 with an intergenic region on chromosome 4 (Fig. 4A). PCR walking was able to amplify a genomic fragment ≈9 kb that traversed the break junction (Fig. S4). The 9-kb fragment was first end-sequenced using Sanger chemistry, then subjected to a half plate of 454 shotgun sequencing, which generated >60,000 reads with an average read length of 220 bp. Combining the results from de novo assembly and mapping to the reference genome of the 454 shotgun sequences (see Methods), we assembled the break junction of t(4;11)(q32;q21) to base pair resolution (Fig. 4B). Surprisingly, t(4;11)(q32;q21) is a complex event that involves both intrachromosomal and interchromosomal rearrangement events. A portion of the MRE11A intron 14 was found inverted and joined with intron 11 directly before the break point on chromosome 11. In addition, a 114 base pair sequence, which is a less conserved LINE repeat element exactly matching to chromosome 5 between 63,051,940 and 63,052,053, bridges chromosomes 11 and 4.
The t(5;8)(p15.33;q24.21) break junction, of which the genomic LR-PCR product was <1 kb, was fully determined by Sanger sequencing. Sequences at both break points for t(4;11)(q32;q21) and t(5;8)(p15.33;q24.21) are rather unique. Recently identified DNA motifs in nonallelic homologous recombination hot spot (26, 27) were not observed at either of these break junctions.
Assembly of the 454 cDNA sequences reveals that the translational products of chimeric transcripts include chimeric and truncated proteins. The t(9;18)(p24.1;q12.2) translocation is predicted to result in a fusion protein in which the 5′end of PDCD1LG2 encoding 120 aa and containing an Ig subtype domain (IPR003599) is fused in frame with the 3′end of C18orf10 encoding 172 aa. In the case of the t(8;2)(q24.12;q22.1) translocation, the predicted protein product is a fusion protein in which 7 aa at C terminus of the SAMD12 gene product is replaced by 45 aa encoded by the intergenic sequence on chromosome 2. However, the entire functional SAM domain in SAMD12 is predicted to be preserved in the chimeric protein.
The t(4;11)(q32;q21) translocation encodes a truncated form of the MRE11A protein. Transcription of the chimeric MRE11A gene extends for only 281 base pairs on chromosome 4 before the poly(A) tail. Translation of the chimeric transcript was predicted to be truncated with 308 aa through exon11 from MRE11A plus 31 aa encoded by the intergenic sequence acquired from chromosome 4, then halted by an in-frame stop codon. The MRE11A DNA binding domain was disrupted by this translocation event.
Discussion
In this proof-of-principle study we sought to identify somatic genome rearrangements based on DNA sequencing of a cell's transcriptome. Through this approach we specifically looked for genomic rearrangements that are of greatest interest—those that are expressed in the cancer cell and that might direct phenotypes associated with cancer development and progression. The results reported here substantiate the notion that deep analysis of the transcriptome can reveal such genomic changes, thereby highlighting active genomic regions that might contribute to the breast cancer phenotype. This study builds upon previous studies, using the 454 Life Sciences sequencing technology that were focused on deep sequencing of cancer cell transcriptomes to identify alternative transcript splice forms and point mutations (28, 29). Our results suggest that with a deeper coverage of transcriptome sequence, genomic rearrangements that result in chimeric genes will be identified in an even more efficient and comprehensive manner.
Our study also complements that of Ng et al. (30) that used an alternative technology, GIS-PET, to explore new chimeric transcripts within 2 tumor cell lines. Together the previous studies and the findings described here show that the deep sequencing of a tumor transcriptome provides important new opportunities to identify changes in the transcriptome that reflect the underlying cancer genome.
Chromosome rearrangements can lead to gene fusions that result in gain or loss of function. Gene fusion has been shown to be the critical factor in oncogenesis in both leukemias and solid tumors. For example, gene fusion between TMPRSS2 and ETS family of genes (ERG and ETV) has been reported in more than half of prostate cancer patients (31, 32). Although no definitive fusion protein structure is generated in the case of TMPRSS2-ETS, overexpression of this chimeric transcript is associated with the cancer prognosis (33, 34). However, the BCR-ABL1 fusion gene produces an overactive fusion protein that carries tyrosine kinase activity in leukemias (35, 36). In our study, each of the verified breakpoints and fusions detected involves genes that function in cell growth, apoptosis or DNA repair. Some of these genes have been reported to be directly linked to cell malignancy at different stages and types of cancer. One translocation identified by this study occurs between chromosome 11 and chromosome 4 and severely truncates the MRE11A gene (Fig. 4A). MRE11A may function as a tumor suppressor gene by forming a complex with RAD50 to mediate DNA mismatch repair. Point mutations that lead to truncated proteins have been reported in breast cancer patients by various groups. Our results suggest that genomic translocations may be an additional mechanism of inactivation of this gene in breast cancer, and that it may be another frequent mechanism in parallel with point mutations.
Although the analysis reported here is focused toward identification of genomic changes expressed in the transcriptome, it is also clear that the 454 sequencing approach can successfully identify numerous alternative (or aberrant) splice forms that might also play a role in oncogenesis. An achievable goal will be the integration of genome-specific and transcript-specific events toward the goal of understanding the pathways and networks that contribute to oncogenesis, and that might suggest new detection, diagnostic, and therapeutic strategies. Databases that provide the ability to compare and contrast these changes across a broad range of cancers will be essential for identifying features that might be shared across cancers, thereby affording opportunities to apply new intervention approaches effectively to all cancers for which patient outcome might be improved.
Methods
454 Sequencing of the Transcriptome.
Two micrograms of total RNA was extracted from HCC1954 tumor cell line by TRIzol. cDNA was prepared using the SMART technology (37) (service was provided by Evrogen). For the first stand cDNA, oligo(dT) and TS oligo were used. To enrich rare messenger RNAs, a cDNA normalization step was introduced using DSN (duplex specific nuclease) technology (service was provided by Evrogen Inc.). Further cDNA amplification was carried out to increase the cDNA amount required in the 454 sequencing step by using SMART primers.
An aliquot of ≈5 ng of cDNA sample was evaluated on a gel to determine the size range for the cDNA preparation. Random shearing was achieved by nebulization that was tightly controlled to ensure an average final length of ≈500bp. The removal of small fragments (<300 bp) was done with SPRI beads. Two full sequencing runs were conducted using the 454 GS FLX platform.
Bioinformatics Algorithms for Chimeras.
454 sequences were clustered to 38,015 RefSeq mRNAs (queried from National Center for Biotechnology Information on March 28, 2008), using PASA software (www.sourceforge.org) with 96% identity and 95% coverage cutoffs. All cDNAs that failed PASA were subjected to blat search against the human reference genome (National Center for Biotechnology Information build 36). cDNAs with >90% length coverage mapped to a human chromosomal location were filtered out. The remaining cDNAs were screened for chimeras based on each cDNA being uniquely split between 2 distinct chromosomal locations by blat, with each alignment at least 25 bp long and with a combined length coverage of at least 90%.
For additional algorithm to identify chimeric cDNAs, 454 sequences were located in the human genome, using megablast and local alignments were produced with SIBsim4 (http://sibsim4.sourceforge.net). Sequences that mapped to different chromosomes or to the opposite strands of the same chromosome were investigated further. Putative chimeras that were supported by at least 2 sequences and/or where the breakpoint was not traversed by nonchimeric transcripts were tested by RT-PCR.
Validation of Chimeric cDNAs and Genomic Breaks.
For transcript level confirmation, candidate genomic alternations were verified by RT-PCR in independently prepared DNA-free RNA samples made from the tumor cell line and the corresponding blood cell line as control. Primer pairs flanking the chimeric transcript were used. Sanger sequencing of the PCR products was performed to confirm the amplified bands.
For genomic level confirmation, long range PCR (LR-PCR) was performed using the TakaRa LA PCR kit. LR-PCR products were purified and subsequently sequenced either by Sanger sequencing or 454 sequencing. See Fig. S3 for primers used.
SKY.
Metaphase chromosomes were prepared from HCC1954 cells. SKY was done using the SkyPaint DNA kit (Applied Spectral Imaging) following standard manufacturer's protocols. Spectral images were captured using a microscope (E800; Nikon) equipped with ASI spectral cube, 60× objective, and analyzed using SKYView software from Applied Spectral Imaging. At least 7 metaphases were analyzed per sample.
FISH.
FISH experiments were done using manufacture's standard protocol. Briefly, BAC clones were amplified and purified. BAC DNA was labeled by nick translation with Spectrum Red and Green. Labeled BAC DNA was hybridized to metaphase spreads from the cell lines.
454 Assembly and Mapping.
Our 454 reads of the 9- to 10-kb fragment were assembled de novo and mapped to the reference genome by using Newbler and CLC Genomics Workbench analysis tools for the break junction of t(4;11)(q32;q21). Results were combined with chimeric cDNA and end sequences to permit the fine mapping of the final break point.
Supplementary Material
Acknowledgments.
We thank Dr. Jiaqi Huang and Dr. Pauline Ng for their critical advice and review of this manuscript and Dr. William C. Nierman for helping us with cDNA amplification. This work was conducted as part of the Hilton-Ludwig Cancer Metastasis Initiative, funded by the Conrad N. Hilton Foundation and the Ludwig Institute for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812945106/DCSupplemental.
References
- 1.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–150. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 2.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 3.Adams MD, et al. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 4.Strausberg RL, Buetow KH, Emmert-Buck MR, Klausner RD. The cancer genome anatomy project: Building an annotated gene index. Trends Genet. 2000;16:103–106. doi: 10.1016/s0168-9525(99)01937-x. [DOI] [PubMed] [Google Scholar]
- 5.Camargo AA, et al. The contribution of 700,000 ORF sequence tags to the definition of the human transcriptome. Proc Natl Acad Sci USA. 2001;98:12103–12108. doi: 10.1073/pnas.201182798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 7.Kodzius R, et al. CAGE: Cap analysis of gene expression. Nat Methods. 2006;3:211–222. doi: 10.1038/nmeth0306-211. [DOI] [PubMed] [Google Scholar]
- 8.Brenner S, et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 9.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 10.Bignell GR, et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–1303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 12.Frantz SA, et al. Exon repetition in mRNA. Proc Natl Acad Sci USA. 1999;96:5400–5405. doi: 10.1073/pnas.96.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Wang J, Mor G, Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:1357–1361. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- 14.Ghoussaini M, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer A, et al. Association of chromosomal locus 8q24 and risk of prostate cancer: A hospital-based study of German patients treated with brachytherapy. Urol Oncol. 2008 doi: 10.1016/j.urolonc.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 16.McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- 17.Hsu HM, et al. Breast cancer risk is associated with the genes encoding the DNA double-strand break repair Mre11/Rad50/Nbs1 complex. Cancer Epidemiol Biomarkers Prev. 2007;16:2024–2032. doi: 10.1158/1055-9965.EPI-07-0116. [DOI] [PubMed] [Google Scholar]
- 18.Barber TD, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci USA. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ottini L, et al. MRE11 expression is impaired in gastric cancer with microsatellite instability. Carcinogenesis. 2004;25:2337–2343. doi: 10.1093/carcin/bgh257. [DOI] [PubMed] [Google Scholar]
- 20.Donahue SL, Campbell C. A Rad50-dependent pathway of DNA repair is deficient in Fanconi anemia fibroblasts. Nucleic Acids Res. 2004;32:3248–3257. doi: 10.1093/nar/gkh649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, et al. Three classes of genes mutated in colorectal cancers with chromosomal instability. Cancer Res. 2004;64:2998–3001. doi: 10.1158/0008-5472.can-04-0587. [DOI] [PubMed] [Google Scholar]
- 22.Heikkinen K, Karppinen SM, Soini Y, Makinen M, Winqvist R. Mutation screening of Mre11 complex genes: Indication of RAD50 involvement in breast and ovarian cancer susceptibility. J Med Genet. 2003;40:e131. doi: 10.1136/jmg.40.12.e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giannini G, et al. Human MRE11 is inactivated in mismatch repair-deficient cancers. EMBO Rep. 2002;3:248–254. doi: 10.1093/embo-reports/kvf044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukuda T, et al. Alterations of the double-strand break repair gene MRE11 in cancer. Cancer Res. 2001;61:23–26. [PubMed] [Google Scholar]
- 25.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–812. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 26.Arnheim N, Calabrese P, Tiemann-Boege I. Mammalian meiotic recombination hot spots. Annu Rev Genet. 2007;41:369–399. doi: 10.1146/annurev.genet.41.110306.130301. [DOI] [PubMed] [Google Scholar]
- 27.Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 28.Sugarbaker DJ, et al. Transcriptome sequencing of malignant pleural mesothelioma tumors. Proc Natl Acad Sci USA. 2008;105:3521–3526. doi: 10.1073/pnas.0712399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bainbridge MN, et al. Analysis of the prostate cancer cell line LNCaP transcriptome using a sequencing-by-synthesis approach. BMC Genomics. 2006;7:246. doi: 10.1186/1471-2164-7-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng P, Wei CL, Ruan Y. Paired-end diTagging for transcriptome and genome analysis. Curr Protoc Mol Biol. 2007 doi: 10.1002/0471142727.mb2112s79. Chapter 21:Unit 21 12. [DOI] [PubMed] [Google Scholar]
- 31.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 33.Mehra R, et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007;20:538–544. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 34.Nam RK, et al. Expression of the TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. Br J Cancer. 2007;97:1690–1695. doi: 10.1038/sj.bjc.6604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rowley JD. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. lett. [DOI] [PubMed] [Google Scholar]
- 36.Rowley JD. Chromosome translocations: Dangerous liaisons revisited. Nat Rev Cancer. 2001;1:245–250. doi: 10.1038/35106108. [DOI] [PubMed] [Google Scholar]
- 37.Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. Reverse transcriptase template switching: A SMART approach for full-length cDNA library construction. Biotechniques. 2001;30:892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.