Abstract
Identification of protein targets of post-translational modification is an important analytical problem in biology. Protein microarrays exposed to cellular extracts could offer a rapid and convenient means of identifying modified proteins, but this kind of biochemical assay, unlike DNA microarrays, depends on a faithful reconstruction of in vivo conditions. Over several years, concentrated cellular extracts have been developed, principally for cell cycle studies that reproduce very complex cellular states. We have used extracts that replicate the mitotic checkpoint and anaphase release to identify differentially regulated poyubiquitination. Protein microarrays were exposed to these complex extracts, and the polyubiquitinated products were detected by specific antibodies. We expected that among the substrates revealed by the microarray should be substrates of the anaphase promoting complex (APC). Among 8,000 proteins on the chip, 10% were polyubiquitinated. Among those, we found 11 known APC substrates (out of 16 present on the chip) to be polyubiquitinated. Interestingly, only 1.5% of the proteins were differentially ubiquitinated on exit from the checkpoint. When we arbitrarily chose 6 proteins thought to be involved in mitosis from the group of differentially modified proteins, all registered as putative substrates of the APC, and among 4 arbitrarily chosen non-mitotic proteins picked from the same list, 2 were ubiquitinated in an APC-dependent manner. The striking yield of potential APC substrates from a simple assay with concentrated cell extracts suggests that combining microarray analysis of the products of post-translational modifications with extracts that preserve the physiological state of the cell can yield information on protein modification under various in vivo conditions.
Keywords: anaphase promoting complex, EFA, post-translational modification, proteomics
Protein post-translational modifications have been implicated in virtually every aspect of cell regulation. Yet, work in this area, particularly on a genome-wide scale, has been hampered by a lack of suitable analytical methods. This is especially true for polyubiquitination where the products are heterogeneous and intermediates are found at low abundance in the cell. Hence, identifying polyubquitinated proteins in complex mixtures is very difficult. One simple way of identifying proteins is to array them on a solid support where the position on the array serves as a means of identification, thus obviating problems of analyzing heterogeneous mixtures of products. However, to use a protein array to generate post-translational modifications on specific proteins relies on in vitro system for carrying out the chemical reactions, in contrast to relying on a more physiological in vivo analysis. Concentrated functional mammalian cell extracts have been shown to recapitulate complex events, such as the ordered degradation of mitotic substrates, and, therefore, for many purposes, bridge the gap between in vivo and in vitro conditions (1). Thus, we wished to see whether we could use these special cell-free systems and combine them with protein microarrays to identify targets of post-translational modification in a physiological context. A few groups have pioneered protein microarrays in which full-length proteins are arrayed to identify the substrates of purified kinases (2, 3) or have used microarrays to study binding interactions with small molecules or other proteins (4, 5). These assays can produce useful biochemical information even though there is usually no attempt to reproduce exact cellular conditions. However, it is also likely that the use of purified enzymes is too permissive as it is well-known, for example, that purified kinases will label many proteins that are not important in vivo targets. Furthermore, these in vitro reactions lack many cellular components, such as adapter and scaffold proteins, competing substrates, phosphatases, and inhibitors, that help specify targets. To ascertain whether concentrated and functional in vitro extracts can generate physiological responses on solid-state arrays of full-length proteins, we studied the well characterized ubiquitination of proteins at the metaphase-anaphase transition. Using what we would like to call an extract-based functional assay (EFA), we have asked whether we can identify new targets of ubiquitination at the metaphase-anaphase transition in mammalian cells.
Results
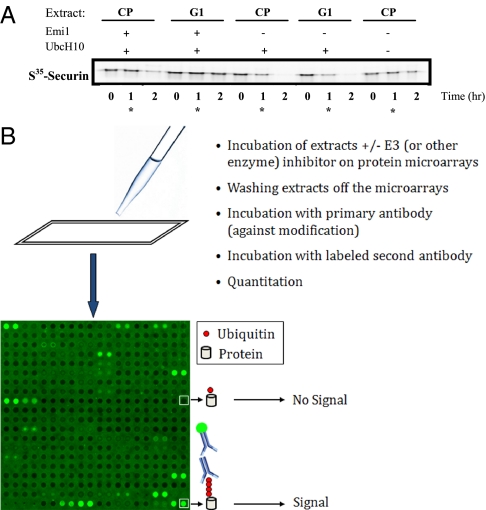
It was shown previously that the mitotic checkpoint (CP) system can be reproduced in extracts from nocodazole-arrested mammalian cells (1, 6, 7). Nocodazole prevents the formation of the mitotic spindle and thus produces a strong checkpoint signal that inhibits the anaphase promoting complex (APC) which is the major ubiquitin ligase in mitosis and G1. APC inhibition can be overcome by the addition of UbcH10 (7), an E2 that regulates mitotic cyclin degradation (8). Thus, there is a well-characterized system for detecting APC-dependent polyubiquitination events that occur when the checkpoint is released and the cells enter anaphase. Checkpoint extracts from HeLa S3 cells arrested with nocodazole were divided into 3 aliquots, one was retained and denoted as the checkpoint extract (CP-extract), one was supplemented with UbcH10 to relieve the checkpoint arrest (CP-released), and the third also received UbcH0 along with a specific inhibitor of APC, emi1(APC-inhibited). To ascertain the functionality of the extracts, an aliquot of each sample was removed and S35 labeled-securin, a well-characterized mitotic substrate, was added (Fig. 1A). Securin remained stable in CP-extracts, consistent with the inhibition of APC by the spindle checkpoint. Extracts supplemented with UbcH10 (CP-released) degraded securin rapidly while the addition of the APC inhibitor emi1 (APC-inhibited) stabilized securin for at least 60 minutes. The same 3 types of extracts were added to the protein microarrays for 60 minutes at room temperature. The microarray, produced by Invitrogen contained approximately 8,000 proteins spotted in duplicate at a reported level of around 10 pg per spot (median diameter = ≈150 μm). We used an anti-polyubiquitin (FK1) antibody (see supporting information (SI) Fig. S1) to detect ubiquitinated proteins on the microarray. Microarrays were scanned and the median signal intensity and local background of each spot was measured. Fig. 1B illustrates the process and depicts one representative scanned subarray (out of 48 that are on each microarray) and its signal.
Fig. 1.
(A) Assaying the biological state and competence of the extracts. To evaluate the activity of the extracts, S35-labeled securin was added to extract samples to follow its degradation. The reactions were stopped at the indicted times by the addition of sample buffer and were then analyzed by on SDS/PAGE and autoradiography. The star (*) labeled lanes reflect the state of the extracts at the time when incubation on the protein microarrays were stopped. (B) Signal on microarrays for the detection of polyubiqutination extracts with an active E3 (e.g., checkpoint released) or with an inhibitor or lacking the activity (e.g., checkpoint extracts or extracts inhibited with emi1) were incubated on protein microarrays and detected (Human ProtoArray, Invitrogen) as described. An example of one block/subarray out of the 48 on each microarray is given (16 rows × 16 columns).
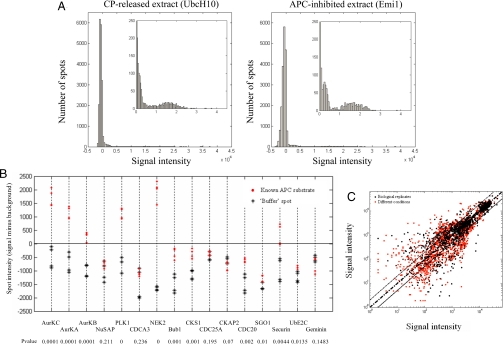
The vast majority of the spots on each microarray were either lower in intensity or similar to the background level (see Fig. 1B). As an example we present the data from two microarrays incubated with either CP-released (Fig. 2A, left panel) or APC-inhibited extracts (Fig. 2A, right panel). The inset depicts the net positive signal that was detected on each of these microarrays. Both microarrays show a very similar pattern of reactivity. The differences are restricted to a minor subset of proteins. To determine a threshold value for identifying polyubiquitin signals, we compared the reactivity level of the sixteen known APC substrates that were spotted on each microarray with the reactivity level of the ‘buffer’ spots located adjacent to them (in the same subarray). As shown in Fig. 2B, 11 of these substrates appeared to have a signal that was significantly higher than the buffer spots (P < 0.05). However, only 6 proteins of the 11 gave a positive signal (when considering the spot intensity minus its local background). Although 11 of the 16 spots were significantly modified, for the analysis presented here we filtered out 5 of these that were negative. Thus, in the following analysis, we consider only net positive signals. This applies a perhaps overly strict standard but we do this to reduce the potential false-positive rate.
Fig. 2.
(A) The distribution of the signal intensity minus background of all of the spots on a microarray. Reactivates were divided into 100 equally sized bins and the number of spots (y axis) at different intensity levels (x axis) of CP-released (Left) and APC-inhibited (Right) extracts were plotted. The inset represents a 20× magnification of the positive signals where the y axis ranges between 0 and 250 and the x axis ranges between 0 and 45,000. (B) The reactivity level of 16 known APC substrates (red dots) compared with the reactivity level of the ‘buffer’ spots located in the same subarray (black stars). These reactivities were then compared using two-sample t test to determine their significance and the P-values were labeled below each substrate. (C) Scatter plots of the positive signal intensities on each microarray were plotted to compare the variability between 2 biological replicates (black dots; x axis, CP-released; y axis, CP-released) vs. the variability between signals from two different conditions (red spots; x axis, APC-inhibited; y axis, CP-released).
To test the reproducibility of the assay and its ability to detect differential modification under different conditions, we compared spot intensities of microarrays that were incubated with different extract preparations (biological replicates) and with extracts under different conditions (CP released vs. APC-inhibited). Fig. 2C shows scatter plots of the positive spot reactivities in each comparison (log scale). Visually, the different conditions (red dots) produced a signal that was more spread and variable compared with the biological replicates that are closer to the diagonal (black dots). As expected, when we averaged the reactivity of spots per each protein, the variability between two biological replicate chips decreases in each of the conditions (Fig. S2).
To determine which proteins are modified at release from the mitotic checkpoint, we compared the signals from the CP-released and the APC-inhibited extracts. Two microarrays from each condition were examined and a two-tailed t test was used to identify differentially modified proteins. To determine significance we used a permutation-based P value calculation (9–11) and corrected for false discovery rate (FDR) using Storey's (12, 13) method (see Materials and Methods for details of the data analysis and functions that were used). Over 132 proteins were differentially modified (q-value <0.096) and these proteins are listed in Table S1. A list of the top 20 proteins in our list (sorted by q-value) is presented in Table 1. Using an algorithm for clustering by functional annotation [DAVID (14)], we identified which biological terms/functions are specifically enriched in our gene list compared to the background list of proteins that were spotted on the microarray. Fig. 3 presents a summary of the enriched functional terms (P < 0.05) and the proteins that were classified to each GO annotation (black squares). The percentage (%) of proteins that correspond to each GO category is presented. Interestingly, among the most enriched biological terms in our predicted list were ‘post-translational protein modification’ (GO:0043687, P value = 4.02E−10) and the ‘mitotic cell cycle’ (GO:0007049, P value = 0.01). A full table of the analysis is provided in Table S2.
Table 1.
Top 20 differentially modified proteins (see Table S1 for the full list) between CP-released and APC-inhibited extracts
| Name | Protein ID | P | q | pFDR value |
|---|---|---|---|---|
| Additional sex combs like 1 (Drosophila) | BC064984.1 | 0.001 | 0.040 | 0.048 |
| Ankyrin repeat domain 13 | BC032833.2 | 0.000 | 0.040 | 0.067 |
| Aurora kinase A (AURKA), | NM_003600.2 | 0.001 | 0.040 | 0.060 |
| Aurora kinase B (AURKB) | NM_004217.2 | 0.001 | 0.040 | 0.044 |
| cDNA clone MGC:39273 | BC024289.1 | 0.001 | 0.040 | 0.046 |
| Cytochrome P450, family 26, subfamily A, polypeptide 1 (CYP26A1) | NM_057157.1 | 0.001 | 0.040 | 0.053 |
| Dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit (DPM2) | NM_152690.1 | 0.001 | 0.040 | 0.055 |
| EGF-like repeats and discoidin I-like domains 3 | BC053656.1 | 0.002 | 0.040 | 0.041 |
| Ems1 sequence, mammary tumor and squamous cell carcinoma-associated (EMS1) | NM_138565.1 | 0.001 | 0.040 | 0.052 |
| Erythrocyte membrane protein band 4.1 like 5 | BC054508.1 | 0.000 | 0.040 | 0.093 |
| Feline sarcoma oncogene (FES) | NM_002005.2 | 0.002 | 0.040 | 0.040 |
| HTGN29 protein (HTGN29) | NM_020199.1 | 0.000 | 0.040 | 0.080 |
| Hypothetical protein DKFZp762O076 (DKFZp762O076) | NM_018710.1 | 0.001 | 0.040 | 0.042 |
| Hypothetical protein FLJ11184 | BC011842.2 | 0.001 | 0.040 | 0.070 |
| Hypothetical protein FLJ36175 | BC029520.1 | 0.001 | 0.040 | 0.040 |
| Hypothetical protein LOC143458 (LOC143458) | NM_174902.2 | 0.001 | 0.040 | 0.044 |
| Hypothetical protein MGC4618 (MGC4618) | NM_032326.1 | 0.002 | 0.040 | 0.042 |
| KIAA0157 protein (KIAA0157) | NM_032182.2 | 0.001 | 0.040 | 0.051 |
| MAX interacting protein 1 (MXI1), | NM_130439.1 | 0.001 | 0.040 | 0.044 |
| Nedd4 family interacting protein 1 (NDFIP1) | NM_030571.2 | 0.001 | 0.040 | 0.040 |
Protein reactivities of two microarrays in each condition were compared using 2-sided t test. Permutation-based P values were calculated and the false discovery rate was calculated for each gene based on Storey's q values estimation (see Materials and Methods).
Fig. 3.
The Biological terms/functions specifically enriched in our gene list compared to the background list of proteins that were spotted on the microarray. We used functional annotation clustering in the Database for Annotation, Visualization, and Integrated Discovery. The proteins that were classified to each of the most enriched (P value <0.05) GO annotation terms are labeled in black. The percentage (%) of proteins that correspond to each GO category is also presented (see Table S2 for a full report of the results).
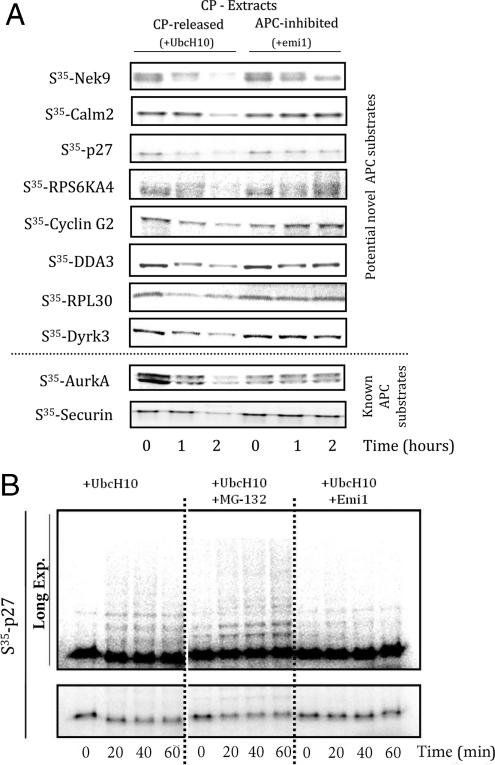
As discussed above, known APC substrates were found in this list. We suspected that the EFA procedure may have identified new ones as well. As a preliminary test, we chose 6 proteins from the predicted list that were suggested by other data to play a role in mitosis but for which there was no known APC connection (Nek9, Calm2, p27, RPS6KA4, cyclin G2, and DDA3). Four other proteins on this list (Zap-70, MAP3K11, RPL30, and Dyrk3) were not known to be involved in mitosis and were chosen arbitrarily with no expectation as to whether they were substrates. The standard assay for APC substrates involves expression by in vitro translation, using S35-labeled methionine, addition to CP-released and APC-inhibited extracts, and comparison of the results. Of the 4 control proteins, 2 (Zap-70 and MAP3K11) showed no degradation (Fig. S3). Surprisingly, the 2 other proteins, RPL30 and Dyrk3, which had no known mitotic connection but which were differentially labeled on the arrays, were degraded via APC (Fig. 4A). These proteins were clearly degraded in the CP extracts that were released into anaphase by UbcH10, but their degradation was inhibited by the addition of emi1. Of the 6 mitotic proteins, 5 (Nek9, Calm2, RPS6KA4, cyclin G2, and DDA3) were clearly degraded in an APC-dependent manner (Fig. 4A). The sixth, p27, appeared to be degraded slowly in the CP-released extracts as well. However, a longer exposure (Fig. 4B and Fig. S4) revealed that p27 accumulated polyubiquitin chains (that caused a gel shift) rather than being rapidly degraded. The addition of emi1 only partially inhibited the formation of ubiquitin chains. To examine whether endogenous proteins were also degraded, we blotted calm2 and p27 (for which there were good antibodies) in CP-released and APC-inhibited extracts. The signal for both endogenous p27 and calm2 declined in the extracts released from the checkpoint block. This could be due either to degradation of the substrate or to ubiquitination and the dispersal substrate-ubiquitin conjugates in a haze of high molecular-weight products. The loss of signals for both substrates was inhibited by emi1 (see Fig. S5).
Fig. 4.
(A) Soluble degradation assays for identified substrates. S35-labeled putative substrates (Nek9, Calm2, RPS6KA4, cyclin G2, RPL30, and Dyrk3) were added to CP-synchronized HeLa S3 extracts with and without the addition of the APC-inhibitor emi1. Reactions were stopped at 0, 60, and 120 min, and analyzed by SDS/PAGE (4–15%) and autoradiography. (B) Ubiqutination and limited degradation of p27. S35-labeled p27 was added to CP-synchronized HeLa S3 extracts with the addition of UbcH10 (1 μL; 1 mg/mL), UbcH10 and MG-132 (200 μM), or UbcH10 and Emi1 (5 mg/mL). The bottom panel shows the change in stability of p27 under these conditions. The top panel is the same gel overexposed to detect p27-conjugated ubiquitin chains.
Discussion
We developed an extract-based, large-scale profiling system for the detection of polyubiquitinated proteins using microarrays. To test this approach, we looked for APC-dependant polyubiquitination signals in extracts made from cells released from the mitotic checkpoint. The fraction of proteins that showed significant changes in ubiqutination after release of the mitotic checkpoint was small (9%). Significantly, this list included several known APC substrates that were present on the microarray, suggesting that there was specificity to the reaction. Of the known APC substrates, 16 were present on the microarray, and, of these, 11 were identified to be significantly polyubiquitinated and 6 had a net positive signal. Finally, we have been able to use this technique to identify 7 potential APC substrates (Nek9, Calm2, RPS6KA4, cyclin G2, RPL30, and Dyrk3) and validated their degradation/ubiquitination in the somatic extracts. We also found p27 to be significantly ubiquitinated on release from the mitotic checkpoint but only partially degraded. The yield of new potential APC substrates in this simple assay is remarkable when compared with other methods, yet we believe that although the results are impressive, the assay is far from optimized. The proteins in these commercial microarrays are all tagged at the N terminus with GST for ease in purification. N-terminal tags would interfere with some known substrates like cyclin and securin. Therefore, it would be important to have both N-terminal and C-terminal tags or no tags for each protein. If our goal were to identify all possible APC substrates in the cell cycle, we would have discriminated in our experimental design against Cdh1-APC substrates. The extracts we used reflected the cell cycle state corresponding to release from metaphase and this state would have been enriched in APC-cdc20. Wider coverage of Cdh1 could be achieved by using bona fide G1 extracts (15). As for false positives among the 132 proteins, we are not sure how many are truly false. There is no reason to expect that every ubiquitinated protein in mitosis should be an APC substrate. It would actually be very interesting to study further the differentially modified proteins that showed a clear ubiquitination signal but which were not inhibited by emi1; these might have been targets of other E3 ligases that were present in the extract.
A special feature of assaying for ubiquitination, rather than wholesale degradation in extracts, is that measurement of statistically valid changes in ubiquitination can reflect the degradation of a localized or minor subset of a protein. Bulk cellular assays for degradation may not be able to detect such events due to lack of sensitivity. For example, cyclin B degradation in Drosophila early embryos only happens near the nuclei in a large syncytial cytoplasm (16), and the loss of cyclin B in the total extract is undetectable. Had Tim Hunt used Drosophila eggs instead of sea urchin eggs (17), cyclin would not have been discovered. However, an elevated ubiqutination signal might have been easier to detect. We may have encountered a similar situation when we identified several proteins with EFA that were previously reported to localize to the spindle. Calmodulin has been reported to change its abundance only at the G1/S transition and maintain its level throughout the remainder of the cell cycle (18). However, calmodulin localizes to spindle poles during mitosis (19, 20) where it potentiates microtubule depolymerization (21). It was also shown that microinjection of calmodulin into cells prolongs the time from nuclear envelope breakdown to anaphase (22). Thus, it is quite possible that calmodulin activity is regulated by localized APC-mediated degradation. Interestingly, NimA-related protein kinase 9 (Nek9/Nercc1) activity was recently shown to specifically increase in mitosis and to accumulate a phosphorylated form at the spindle poles (23). Since Nek9 was shown to bind Ran-GTPase it was speculated that it might be involved in spindle organization. Xenopus Nek9 was detected on the pole of spindles assembled in vitro in frog egg extracts (24) and mammalian cells injected with anti-Nek9 antibodies after mitotic entry accumulated defects in mitotic spindle formation and chromosome segregation. Its activity might also be locally controlled by localized degradation. In addition, Cyclin G2 was shown to be a centrosome-associated nucleocytoplasmic shuttling protein where it was suggested to influence microtubule stability (25), and DDA3 was shown to be localized to spindle poles and to be involved in microtubule dynamics at metaphase (26). Thus, bulk changes in protein abundance may be small but the degradation of a subset of a protein might be very important. In fact, the centrosome generally might be a site for localized degradation in mitosis; proteasomal machinery was previously shown to be associated with centrosomes (27), and two main regulators of mitosis, the APC-activator cdc20 (28, 29) and cyclin B (30), have been shown to be regulated at the centrosomes. The role of APC-mediated degradation of RPS6KA4, Dyrk3, and RPL30 in mitosis or G1 now awaits further study.
The ubiquitination system regulates some of the most important circuits in the cell, such as DNA damage control (31) and cell division (32), but we are only slowly identifying the targets. Recent discoveries stress that regulation is the product of a network of reactions that include a vast array of E3 and E2 enzymes, deubiquitinases, and other proteins that regulate ubiquitination and related modifications. Any of several activities might be regulated to change the stability of a particular substrate. Thus, it may not be easy to infer regulatory control on the specific substrates by assaying purified E3 enzymes alone. Simplified in vitro biochemical assays give some information but ultimately these targets need to be identified under physiological conditions, conditions that may only be truly obtainable within organisms. Thus, every putative substrate ultimately has to be confirmed by in vivo measurements. It is the task of biochemists, cell biologists, and geneticists to identify likely targets for further in vivo study. The use of concentrated functional extracts has proved very useful in this regard because it is both biochemically manipulatable and comes close, in some cases, to in vivo function. We have found that extracts can have the capacity to maintain the proper regulatory balance, and this has led to the identification of some of the most important regulatory circuits in the cell cycle.
In summary, the EFA we describe provides a proof-of-principle for the ability to screen post-translational modifications in different cellular conditions, using functional extracts and protein microarrays. The real challenge will be to make concentrated extracts that completely or nearly completely retain the specific functional properties of the cytosolic, nuclear, or other compartmental contents. Recent steps are encouraging. Frog egg extracts or marine egg extracts seemed at one time to be uniquely capable for in vitro reconstitution of complex cell cycle processes. Yet, in the last few years we have developed somatic cell systems that are as active and as stable as Xenopus egg extracts. With the proper extracts it should be possible to use protein microarrays for many purposes in the study of cell cycle progression, differentiation, signaling, and response to drugs and other forms of inhibitors, as well as extracts made from engineered cells or from certain disease conditions. Finally, the EFA method can be easily adapted to follow other modifications, such as sumoylation, neddylation, and phosphorylation, and may allow us to further explore the global modification state of many biological conditions. Thus, the combination of concentrated and functional cellular extracts with protein arrays should open up many questions in biology and medicine.
Materials and Methods
Tissue Culture and Cell Synchronization.
HeLa S3 cells were synchronized in prometaphase by treatment with nocodazole, in G1 by a release from nocodazole arrest. Cells were incubated in thymidine-containing (2 mM) medium for 24 h. Cells were released into fresh medium for 8 h, followed by a nocodazole arrest (0.1 μg/mL) for 12 h. For G1 cells, nocodazole-arrested cells were released into fresh medium for 4 h. Cells were harvested, washed with PBS, and processed for extraction.
Extract Preparation.
Extracts were prepared as previously described (15). Briefly, HeLa S3 cells were synchronized with thymidine for 20 h, released for 3 h, and then arrested in mitosis by the addition of nocodazole for an additional 11 h. Synchronized cells are then harvested, washed with phosphate buffer saline (PBS), lysed in swelling buffer (25 mM Hepes pH 7.5, 1.5 mM MgCl2, 5 mM KCl, 1 mM DTT, and a complete protease inhibitors (Roche), and homogenized by freeze-thawing and passage through a needle (CP-extracts). G1-extracts are prepared in the same manner with a 4-h release from nocodazole arrest. Extracts were cleared by subsequent centrifugations (5 min at 2,795 × g; 60 min at 21,920 × g). Extract (20 μl, 25 mg/mL) was supplemented with degradation mixture: 1.5 mg/mL ubiquitin (Boston Biochem), 150 mM creatine phosphate, 20 mM ATP; 2 mM EGTA; and 20 mM MgCl2 (pH 7.6).
Incubation of Extracts with Microarrays.
Human ProtoArray microarrays (Invitrogen) were washed three times (10 min each) with TBS containing 0.05% Tween 20 (TBS-T) in a plastic box containing 20 mL buffer (gently tilted) and then blocked for 4 h at 4 °C with Microarrays Blocking solution (ArrayIt). Extracts were preincubated with either Emi1 (c-terminal region; 5 μL, 1 mg/mL) or H20 for 30 min at room temperature. One hundred μL CP or G1 extracts (≈25 mg/mL) were then supplemented with UbcH10 (5 μM; Boston Biochem) and incubated under a coverslip on the microarrays for 1 h at RT. The arrays were then washed as above and incubated overnight with 100 μL anti-polyubiquitin antibody (FK1, 1 mg/mL; Biomol) diluted 1:250. To label modified proteins, an anti-mouse Cy3-conjugated secondary antibody (3 μL; 1 mg/mL, Jackson ImmunoResearch Laboratories, diluted 1:250) was incubated for 1 h at RT. The arrays were washed again, spin-dried (200 × g, 5 min), and scanned with a GenePix 4000B scanner.
Images and Data Processing.
Results were recorded as TIFF files and Images were quantified using GenePix Pro 5 feature extraction software (version 4000B). Scanning parameters were set so that none of the spots showed saturation: PMT gain value = 400 and laser power = 30%. For each spot, the local background intensity was subtracted from the median spot intensity.
Data Processing and Normalization.
The data set was organized in a matrix where each column contains the reactivities measured for a given array and each row contains the reactivities measured for a given protein over all arrays. The negative values were set to zero and the data were then normalized using the quantile normalization algorithm (33).
Data Analysis.
To determine for each protein the significance of the difference between different conditions, we used a 2-tailed t test. The null hypothesis was generated by permuting the values from all of the conditions and then separating the permuted samples arbitrarily into 2 groups. We used the “crossvalind” function in matlab to perform the permutations and the “mattest” function in matlab to perform the two-tailed t test. Specifically, for each protein we first calculated the observed t statistic. Then, for each protein separately (i.e., for each row) we randomly chose 4 values (out of possible 8) of that protein and recalculated the test statistic after separating the samples into 2 arbitrary groups of 4 samples. The entire data set was permutated 1,000 times and 10,000 times, and very little difference was observed in the t-score distributions, indicating that we have efficiently randomized our data set. We used 10,000 permutations to calculate the set of t statistics across all proteins as a null distribution. To estimate P-values for each protein we then count the number of permutations whose t statistics are greater than or equal to the observed (un-permutated) t statistics and divide it by the total number of permutations [see (9–11) for details on the method]. To estimate the false discovery rate (FDR) we have used Storey's method (32) and calculated the q-value for each protein using ‘mafdr’ function in matlab. The q-value is a measure of significance correcting for the false-discovery rate whereas the P value is a measure of significance correcting for the false-positive rate (12). Thus, the q-value for a certain protein is the fraction of times we would estimate that a protein is significant when it is actually not significant. We chose a q-value of 0.096 as the threshold since q-values higher than 0.096 varied greatly in P-values (see arrow in Fig. S6). We have listed the proteins that passed this threshold, their permutation-based P-values, and their corrected pFDR values in Table S1 in the SI Text.
Gene Functional Classification.
Using the Database for Annotation, Visualization, and Integrated Discovery [DAVID (34)], we identified which biological terms are specifically enriched in our gene list compared to the background list of proteins that were spotted on the microarray. The Functional Annotation Clustering tool was used with custom classification stringency and the Biological Process (BP_ALL) gene ontology term.
Degradation Assays.
Human cDNA clones were obtained from the National Institutes of Health (NIH) mammalian gene collection (MGC). Proteins Coupled in vitro transcription and translation were performed using a rabbit reticulocyte lysate system (TnT SP6, Promega) or Wheat germ (TnT Coupled Wheat Germ Extract systems, Promega) according to the manufacturer's instructions. S35-labeled substrates were added to G1 or CP extracts of synchronized HeLa S3 cells (see extract preparation). Aliquots were removed at 0, 30, 60, and 90 min, and analyzed by SDS/PAGE (4–15%) and autoradiography. Additionally, endogenous protein levels were determined in the extracts by Western blotting at the indicated times (actin, Sigma; securin, MBL; calmodulin, Upstate; p27, Upstate).
Supplementary Material
Acknowledgments.
We thank Einav Laser for her support, Tzachi Pilpel, Itai Yanai, Mike Springer, Shai Shen-Orr, Shay Tal, Ana Hernandez, Amit Tzur, Tao Wu, Martin Wuehr, and Francisco Quintana for helpful discussions and Becky Ward for critically reading this manuscript. This work was supported by the NIH grant GM039023.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812892106/DCSupplemental.
References
- 1.Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 2.Ptacek J, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 3.Schnack C, Danzer KM, Hengerer B, Gillardon F. Protein array analysis of oligomerization-induced changes in alpha-synuclein protein-protein interactions points to an interference with Cdc42 effector proteins. Neuroscience. 2008;154:1450–1457. doi: 10.1016/j.neuroscience.2008.02.049. [DOI] [PubMed] [Google Scholar]
- 4.Gupta R, et al. Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol Syst Biol. 2007;3:116. doi: 10.1038/msb4100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emili AQ, Cagney G. Large-scale functional analysis using peptide or protein arrays. Nat Biotechnol. 2000;18:393–397. doi: 10.1038/74442. [DOI] [PubMed] [Google Scholar]
- 6.Braunstein I, Miniowitz S, Moshe Y, Hershko A. Inhibitory factors associated with anaphase-promoting complex/cylosome in mitotic checkpoint. Proc Natl Acad Sci USA. 2007;104:4870–4875. doi: 10.1073/pnas.0700523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 8.Yu H, King RW, Peters JM, Kirschner MW. Identification of a novel ubiquitin-conjugating enzyme involved in mitotic cyclin degradation. Curr Biol. 1996;6:455–466. doi: 10.1016/s0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
- 9.Pan W. On the use of permutation in and the performance of a class of nonparametric methods to detect differential gene expression. Bioinformatics. 2003;19:1333–1340. doi: 10.1093/bioinformatics/btg167. [DOI] [PubMed] [Google Scholar]
- 10.Xie Y, Pan W, Khodursky AB. A note on using permutation-based false discovery rate estimates to compare different analysis methods for microarray data. Bioinformatics. 2005;21:4280–4288. doi: 10.1093/bioinformatics/bti685. [DOI] [PubMed] [Google Scholar]
- 11.Yang H, Churchill G. Estimating p-values in small microarray experiments. Bioinformatics. 2007;23:38–43. doi: 10.1093/bioinformatics/btl548. [DOI] [PubMed] [Google Scholar]
- 12.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storey JD, Tibshirani R. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol Biol. 2003;224:149–157. doi: 10.1385/1-59259-364-X:149. [DOI] [PubMed] [Google Scholar]
- 14.Huang da W, et al. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- 16.Su TT, Sprenger F, DiGregorio PJ, Campbell SD, O'Farrell PH. Exit from mitosis in Drosophila syncytial embryos requires proteolysis and cyclin degradation, and is associated with localized dephosphorylation. Genes Dev. 1998;12:1495–1503. doi: 10.1101/gad.12.10.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans T, Rosenthal ET, Youngblom J, Distel D, Hunt T. Cyclin: A protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33:389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- 18.Chafouleas JG, Bolton WE, Hidaka H, Boyd AE, 3rd, Means AR. Calmodulin and the cell cycle: involvement in regulation of cell-cycle progression. Cell. 1982;28:41–50. doi: 10.1016/0092-8674(82)90373-7. [DOI] [PubMed] [Google Scholar]
- 19.Welsh MJ, Dedman JR, Brinkley BR, Means AR. Calcium-dependent regulator protein: localization in mitotic apparatus of eukaryotic cells. Proc Natl Acad Sci USA. 1978;75:1867–1871. doi: 10.1073/pnas.75.4.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moser MJ, Flory MR, Davis TN. Calmodulin localizes to the spindle pole body of Schizosaccharomyces pombe and performs an essential function in chromosome segregation. J Cell Sci. 1997;110:1805–1812. doi: 10.1242/jcs.110.15.1805. [DOI] [PubMed] [Google Scholar]
- 21.Keith C, DiPaola M, Maxfield FR, Shelanski ML. Microinjection of Ca++-calmodulin causes a localized depolymerization of microtubules. J Cell Biol. 1983;97:1918–1924. doi: 10.1083/jcb.97.6.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keith CH. Effect of microinjected calcium-calmodulin on mitosis in PtK2 cells. Cell Motil Cytoskeleton. 1987;7:1–9. doi: 10.1002/cm.970070102. [DOI] [PubMed] [Google Scholar]
- 23.Roig J, Mikhailov A, Belham C, Avruch J. Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev. 2002;16:1640–1658. doi: 10.1101/gad.972202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roig J, Groen A, Caldwell J, Avruch J. Active Nercc1 protein kinase concentrates at centrosomes early in mitosis and is necessary for proper spindle assembly. Mol Biol Cell. 2005;16:4827–4840. doi: 10.1091/mbc.E05-04-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arachchige Don AS, et al. Cyclin G2 is a centrosome-associated nucleocytoplasmic shuttling protein that influences microtubule stability and induces a p53-dependent cell cycle arrest. Exp Cell Res. 2006;312:4181–4204. doi: 10.1016/j.yexcr.2006.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jang CY, et al. DDA3 recruits microtubule depolymerase Kif2a to spindle poles and controls spindle dynamics and mitotic chromosome movement. J Cell Biol. 2008;181:255–267. doi: 10.1083/jcb.200711032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wigley WC, et al. Dynamic association of proteasomal machinery with the centrosome. J Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kallio MJ, Beardmore VA, Weinstein J, Gorbsky GJ. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J Cell Biol. 2002;158:841–847. doi: 10.1083/jcb.200201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raff JW, Jeffers K, Huang JY. The roles of Fzy/Cdc20 and Fzr/Cdh1 in regulating the destruction of cyclin B in space and time. J Cell Biol. 2002;157:1139–1149. doi: 10.1083/jcb.200203035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J, Raff JW. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 1999;18:2184–2195. doi: 10.1093/emboj/18.8.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooks CL, Gu W. Dynamics in the p53-Mdm2 ubiquitination pathway. Cell Cycle. 2004;3:895–899. [PubMed] [Google Scholar]
- 32.King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 33.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 34.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.