Abstract
The aim of this study was to find conserved motifs in specific T cell receptor (TCR) α- and β-chains, and to analyse the association between complementarity determining region 3 (CDR3) spectratype and systemic lupus erythematosus (SLE) activity. TCR α- and β-chain CDR3 spectratypes were analysed in 20 SLE patients. The CDR3 spectratypes of three patients were monitored over time, and the CDR3 regions of clonally expanded T cells were sequenced. CDR3 spectratype analysis showed prominent usage of TCR AV8, AV14, AV23, AV30, AV31, BV2, BV8, BV11, BV14, BV16, BV19 and BV24 families in SLE patients. The CDR3 spectratype showed dynamic change correlating with SLE activity. The sequence of the CDR3 region in clonally expanded T cells suggested a conserved GGX amino acid motif in both α- and β-chains. The Ja34 and Jb2s1 region genes were found in high frequency. Both TCR Vα and Vβ gene usage is highly restricted in SLE, suggesting that the TCRs recognize a limited number of antigenic epitopes. The conserved motifs and limited use of joining region genes may indicate the recognition of similar antigenic epitopes in multiple individuals.
Keywords: autoimmunity, CDR3, Genescan, SLE, TCR
Introduction
Systemic lupus erythematosus (SLE) is a prototypic autoimmune disease characterized by an extensive diversity of clinical features, excessive autoantibody production, immune complex formation and immunologically mediated tissue injury [1]. T cells play an important role in the occurrence and development of SLE. The production of autoantibodies such as anti-double-stranded DNA (dsDNA), anti-ribonucleoprotein or anti-Smith antigen is facilitated by helper T cells. In addition, T cell infiltrates can be detected in various organs, including kidney, skin and salivary glands, where they may function as autoreactive T cells [2]. T cells recognize antigen specifically through the membrane protein T cell receptor (TCR). The complementarity determining region 3 (CDR3) of the TCR is a highly variable region that interacts directly with the antigen peptide. Each CDR3 sequence determines a unique T cell clonotype. After responding to autoantigen, specific T cells proliferate and form clonal populations. Thus, analysis of the frequency of use, variation in length and sequences of different TCR CDR3s (the spectratype) can reflect the heterogeneity of the T cell compartment and the functional status of particular T cells.
Based on a number of observations in human disease and in murine models of lupus, clonal expansion of T cells appears to play a major role in the immune pathogenesis of SLE [3–5]. Murata et al. found, based upon TCR usage, that intrarenal T cells in SLE showed an oligoclonal expansion [6], and TCR analysis of restricted T cells demonstrated an oligoclonal T cell expansion in the peripheral blood of SLE patients [7]. The accumulating T cell clones were relevant to disease activity and to the inflammatory lesions, and sequencing of selected TCR CDR3 regions indicated a high level of sequence conservation [8,9]. Despite demonstration of the relevance in SLE of the TCR CDR3 of expanded T cells, the nature of the conserved amino acids in the TCR CDR3 region have not been studied in a large sample of patients. In addition, most previous studies in human SLE focused on the TCR β-chain, involving little analysis of the TCR α-chain [10], although the α-chain can also bind to cationic residues in nucleosomal autoantigens [11]. The molecular basis of human leucocyte antigen (HLA)–peptide complex recognition by the TCR is more complex than described by our current understanding, which emphasizes only TCR β gene usage [12]. Furthermore, dynamic monitoring of the CDR3 spectratype and how this corresponds to different manifestations of SLE disease is rarely reported, and little is known about the association between changes in CDR3 spectratype and disease activity. Most previous investigators have analysed the TCR repertoire in SLE by indirect and labour-intensive methods such as quantitative polymerase chain reaction (PCR), Southern blotting and single-strand conformation polymorphism (SSCP), which cannot separate all genetically diverse TCR α- and β-chain genes [13]. Recently, the immunoscope spectratyping technique has been used increasingly for its advantages of convenience and high resolution. The technique allows information to be obtained about the composition, frequency of expression and length of the CDR3 region for each TCR family, to permit analysis of a global picture of the T cell repertoire. It is also useful in dynamic monitoring of the changes in CDR3 spectratype with disease activity.
In this study, we analysed the peripheral blood mononuclear cells (PBMC) of 20 SLE patients and six healthy controls by immunoscope spectratyping (GeneScan, Applied Biosystems, USA), in order to identify the CDR3 spectratypes of 32 TCR AV families and 24 BV families. In recent years, immunoscope spectratyping as a higher sensitive and accurate method has been used widely to detect the clonal of T cell and to analyse the TCR CDR3 gene repertoire. The principle of this technique is to design specific forward AV (BV) family and reverse AC (BC) primers, TCR gene segments (including complete CDR3 region) of each family can be amplified, and scanning the fluorescent PCR product can reflect the composition and expression frequency of each family. The TCR CDR3 regions of detected clonal expansions of T cells were sequenced, and the CDR3 spectratype was monitored at different disease states in three patients. Our data might assist in defining the immune mechanism and pathological role of T cells in SLE, and in revealing the relationship between changes in CDR3 spectratype and disease activity. Furthermore, it may shed light on strategies for further development of DNA vaccines and the study of individualized immunotherapy of SLE.
Materials and methods
Patient selection and disease activity assessment
Twenty patients, all meeting the criteria for the classification of SLE revised in 1997 [14], were referred to Nan fang Hospital and First Affiliated Hospital of Guangzhou University of Traditional Chinese Medicine. None of the patients showed symptoms of other connective tissue diseases. A signed consent form was obtained from each patient. Six healthy blood donors with no clinical or laboratory evidence of connective tissue diseases or immunological disorders were included in the study, which had approval from the appropriate ethics committee. To define the level of disease activity in the SLE patients, a standard scoring system, the SLE disease activity index (SLEDAI), was used [7], with results shown in Table 1. It is generally accepted that a SLEDAI score < 9 indicates non-active disease, a SLEDAI score > 9 indicates activity disease and a SLEDAI score > 20 indicates highly active disease. Immunological indices such as anti-dsDNA and serum complement levels were determined as markers of SLE activity. Of the 20 patients studied, four were in a period of inactive disease, while 16 had active SLE, and exhibited typical clinical and serological manifestation such as high fever, leucopenia, extremely reduced complement level and high titres of anti-dsDNA antibody. Five of these were patients with acute and severe manifestations of clinically active lupus. Among these five patients, one with autoimmune haemolytic anaemia was associated with cerebral infarction and deterioration of renal function, two had rapidly progressive lupus nephritis, with laboratory studies revealing massive proteinuria, thrombocytopenia and azotaemia, and another two had thrombocytopenia and mononeuritis multiplex. None of the patients took any anti-malarial drugs. All the patients were treated successfully with oral prednisolone therapy without other immunosuppressive agents, the dose of prednisolone being reduced with decreasing disease activity. Low-dose oral therapy was given to patients with inactive disease (Table 1). For two patients (LYJ and LSB), we detected a change in the spectratype of TCR α- and β-chain CDR3 between the active phase of the disease before treatment and while they were in clinical remission after treatment. One patient (LYS) had disease recurrence after leaving hospital and was hospitalized again, enabling us to follow her disease progress through occurrence, remission, recurrence and deterioration. Unfortunately, we failed to detect any α-chain CDR3 spectratype changes in this patient.
Table 1.
Clinical characteristic of patients and the number of T cell receptor (TCR) V gene families showing evidence of clonal expansion for each.
| Case | Age | C3† (g/l) | C4‡ (g/l) | Ds-DNA§ (U/ml) | SLEDAI | Prednisolone (mg/day) | Abnormal AV¶ | Abnormal BV¶ |
|---|---|---|---|---|---|---|---|---|
| LYJ1 | 24 | 0·18 | 0·05 | 324 | 24 | 80 | 30 | 22 |
| LYJ2 | 24 | 0·72 | 0·16 | 72 | 8 | 30 | 14 | 12 |
| ZHJ | 62 | 0·17 | 0·05 | +†† | 20 | 80 | 29 | 20 |
| DWN | 26 | 0·19 | 0·05 | 142·2 | 20 | 80 | 28 | 19 |
| ZTX | 58 | 0·26 | 0·08 | + | 20 | 80 | 29 | 18 |
| LSB1 | 17 | 0·32 | 0·089 | 198 | 19 | 60 | 24 | 18 |
| LSB2 | 17 | 0·98 | 0·18 | 26 | 5 | 20 | 11 | 10 |
| CYP | 55 | 0·28 | 0·07 | + | 14 | 50 | 20 | 16 |
| SWT | 20 | 0·51 | 0·06 | + | 13 | 50 | 15 | 18 |
| YXX | 17 | 0·36 | 0·08 | + | 13 | 50 | 16 | 16 |
| GX | 19 | 0·42 | 0·09 | + | 12 | 45 | 18 | 16 |
| JY | 18 | 0·52 | 0·083 | + | 12 | 40 | 16 | 13 |
| LYS1 | 23 | 0·644 | 0·092 | 132 | 13 | 55 | −‡‡ | 16 |
| LYS2 | 23 | 0·686 | 0·106 | 34·8 | 8 | 30 | − | 11 |
| LYS3 | 23 | 0·473 | 0·08 | 146 | 15 | 80 | − | 20 |
| LYS4 | 23 | 0·452 | 0·073 | 237 | 20 | 80 | − | 23 |
| HJM | 17 | 0·58 | 0·09 | + | 12 | 45 | 15 | 12 |
| LXM | 26 | 0·49 | 0·062 | + | 10 | 50 | 13 | 11 |
| ZXF | 36 | 0·38 | 0·093 | + | 10 | 60 | 15 | 14 |
| FSF | 28 | 0·62 | 0·12 | 72 | 10 | 45 | 12 | 10 |
| ZSF | 17 | 0·38 | 0·06 | + | 9 | 80 | 10 | 8 |
| CYX | 54 | 0·9 | 0·17 | 13 | 6 | 15 | 5 | 3 |
| LHJ | 36 | 1·28 | 0·28 | 15 | 5 | 15 | 4 | 2 |
| ZCX | 59 | 1·44 | 0·32 | 10 | 3 | 10 | 3 | 2 |
| LOP | 21 | 1·56 | 0·36 | 8·6 | 2 | 10 | 2 | 2 |
Reference: (0·93–1·88);
reference: (0·15–0·48),
reference: (0–25);
the number of clonal expansion families;
positive results;
missing data. SLEDAI: systemic lupus erythematosus disease activity index.
Isolation of mononuclear cells
Blood samples were obtained from patients with SLE and six healthy volunteers. Mononuclear cells were isolated from heparinized whole blood by Ficoll-Hypaque gradient centrifugation.
RNA extraction and cDNA synthesis
Total RNA was extracted from 2 × 106 PBMC using the total RNA extraction kit (R6834-01 EZNA TM Total RNA Kit I; Omega Biotechnology Companies, USA), according to the manufacturer's instructions, and quantified by spectrophotometry. One microgram of total RNA was transcribed to first-strand cDNA by incubation at 42°C for 1 h with 250 pm oligo-dT primer, 200 U Moloney murine leukaemia virus (M-MuLV) reverse transcriptase and 250 uM of each deoxyribonucleoside triphosphate (dNTP) in a total volume of 20 µl.
Primers
The primers used for TCR BV and TCR AV family-specific amplification were those described by Yao et al.[15].
The PCR amplification of cDNA
CDR3 size analysis within the TCR α-chain was performed by semi-nested PCR. Separate first-round PCR amplification reactions were performed for each of the 32 human AV gene families. These were carried out in a volume of 50 µl containing 2 µl each of the forward AV primer and the reverse AC primer, 2·0 mM MgCl2, 10 mM Tris-HCL, 200 mM of each dNTP and 1 µl of cDNA. Following an initial denaturing step at 95°C for 5 min, PCR was carried out with 35 cycles of denaturing at 95°C for 30 s, annealing at 60°C for 30 s and extension at 72°C for 90 s, with a final extension at 72°C for 10 min. An aliquot of 8 µl of each PCR product was electrophoresis on a 1·5% agarose gel and analysed by ethidium bromide staining. Separate second-round PCR amplifications for each of the 32 human AV gene families were carried out in 25 µl volume containing 2 µl each of the forward AV primer and the reverse AC-FAM primer, 2·0 mM MgCl2, 10 mM Tris-HCL and 200 mM of each dNTP, with 4 µl of the first-round products as the template. Four cycles of denaturing at 95°C for 2 min, annealing at 60°C for 2 min and 72°C for 2 min, were performed, followed by a final extension at 72°C for 10 min. An aliquot of 8 µl of each PCR product was electrophoresis on a 2% agarose gel, stained with ethidium bromide and visualized under ultraviolet light [16].
The PCR amplification of TCR β-chain CDR3 cDNA for CDR3 spectratype analysis of the Vβ gene was performed as described previously [17].
Analysis of CDR3 spectratype by GeneScan
An aliquot of 2 µl of the fluorescent PCR products was mixed with 2 µl of formamide and 0·5 µl of loading dye [25 mM ethylenediamine tetraacetic acid, 50 ng/ml dextran blue]. The mixture was denatured at 95°C for 2 min, and 2 µl was loaded onto a prewarmed 6% acrylamide sequencing gel and run for 2 h on a 50-lane Applied Biosystems model 373A DNA sequencer (Applied Biosystems). The data were analysed by GeneScan software version 672 [18]. The evaluation of relative fluorescence intensity (RI) was described as: RI (%) = 100 × (clonal peak area)/(total peak area). The following criteria were used to determine whether a clonal T cell expansion had occurred: a single peak with an RI greater than 35%, twin peaks with each peak having an RI greater than 25%, a skewed distribution with the peak RI of the skewing family greater than 25% or a skewed distribution with some families being expressed at very low levels or not at all.
Sequencing the CDR3 of TCR AV and BV families showing restricted usage
The PCR products of TCR AV or TCR BV families showing clonal expansion were purified by gel electrophoresis and the purified products were amplified using the same TCR AV or TCR BV family sense primers and TCR AC or BC anti-sense primers (not FAM-labelled), under the same PCR conditions as for the first amplification. Nucleotide sequences of the amplified products were determined on an ABI 377 DNA sequencer [18].
Results
Peripheral T cell clonality in SLE patients
When visualized in agarose gels, the PCR product of every TCR AV and TCR BV family in healthy controls has a common specific band of approximately 250 base pairs (bp) (Fig. 1). However, this specific band could not be seen for several V gene families in the PBMC of SLE patients. This single band actually consists of several bands differing in length by 3 bp that can be separated in a sequencing gel. In healthy controls, approximately eight bands can be seen for each TCR V gene family. However, in the samples of PBMC in SLE patients, fewer than eight bands or even a single band were observed for some TCR AV or BV families. Figure 2 shows, for a representative patient (SWT), the fluorescent reverse transcription (RT)–PCR products of 32 TCR AV families, 24 TCR BV families and the BC controls. The RT–PCR products were analysed on agarose gel electrophoresis by ethidium bromide staining and on acrylamide sequencing gel electrophoresis by fluorescence. The PCR products of AV families were amplified with semi-nested PCR. In the agarose gel, the lowermost band is the PCR product of the TCR α-chain.
Fig. 1.
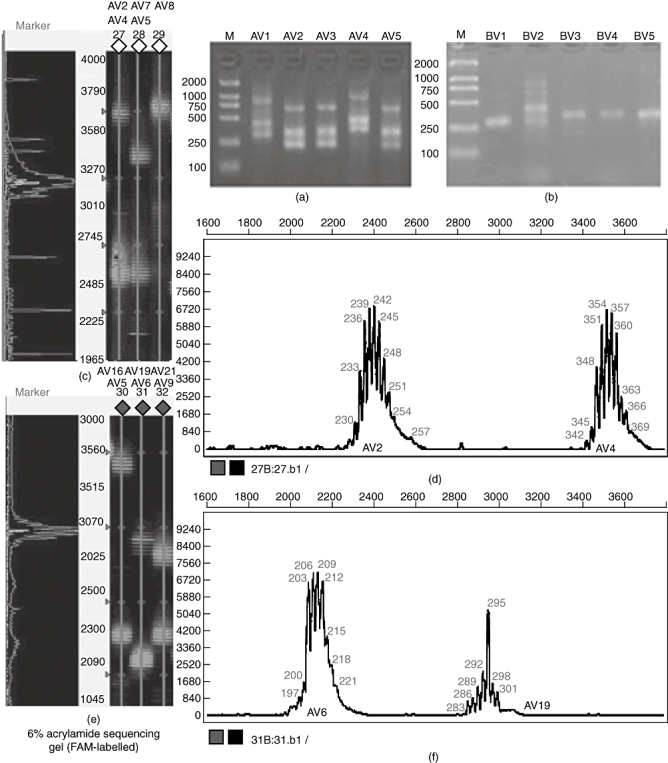
The GeneScan results of size and peak intensity of partial T cell receptor (TCR) AV and BV CDR3 products of healthy control-1 and patient SWT. (a) The polymerase chain reaction (PCR) products of TCR AV1-AV5 of healthy control-1 analysed on a 1·5% agarose gel by ethidium bromide staining. (b) The PCR products of TCR BV1-BV5 of healthy control-1 analysed on 1·5% agarose gels. (c) Partially fluorescent PCR products of TCR AV of healthy control-1 analysed on a 6% acrylamide sequencing gel. (d) CDR3 size and fluorescence intensity analysis of AV2 and AV4 of healthy control-1. (e) Partially fluorescent PCR products of TCR AV of patient SWT analysed on a 6% acrylamide sequencing gel. (f) CDR3 size and fluorescence intensity analysis of AV6 and AV19 of patient SWT.
Fig. 2.
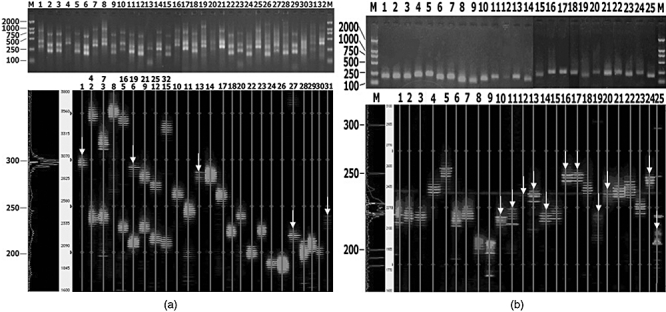
The profiles of CDR3 length distribution for 32 T cell receptor (TCR) AV families, 24 TCR BV families and the BC controls for patient SWT. (a) Numbers 1–32 denote the corresponding polymerase chain reaction (PCR) products for TCR families AV1-AV32. In the sequencing gel, lanes 2, 3 and 5–9 include two TCR AV families in one line. (b) Numbers 1–24 denote the corresponding PCR products of TCR families BV1-BV24; 25 indicates the BC control. Bands fewer than eight are marked with a white arrow.
We compared the CDR3 length distribution of healthy controls and SLE patients. In healthy control donors, the peak map of the CDR3 length for all the TCR AV and TCR BV families displayed the Gaussian distribution expected of polyclonal T cell populations (Fig. 1). CDR3 distribution of healthy individuals shows high polymorphism. In SLE patients, however, some families showed an abnormal distribution characteristic of oligoclonal T cell expansion lower than eight peaks; as few as two or three peaks were even observed, and a few families with monoclonal T cell expansion showed a single peak. The distribution of peaks appeared frequently skewed to some extent. CDR3 distribution of SLE patients shows varying degrees of restriction. Analysis of the CDR3 length distribution of T cell populations in SLE patients revealed prominent usage of TCR AV8 (76% of patients), AV14 (71%), AV23 (81%), AV30 (76%), AV31 (76%), BV2 (80%), BV8 (72%), BV11 (72%), BV14 (72%), BV16 (84%), BV19 (84%) and BV24 (96%). The profiles of CDR3 length distribution for 32 TCR AV families, 24 TCR BV families and the BC controls for patient SWT are shown in Fig. 3. The number of V gene families with abnormal peaks in different patients, and even within one patient at different stages of disease, correlated with the SLEDAI score of the patients (Table 1).
Fig. 3.
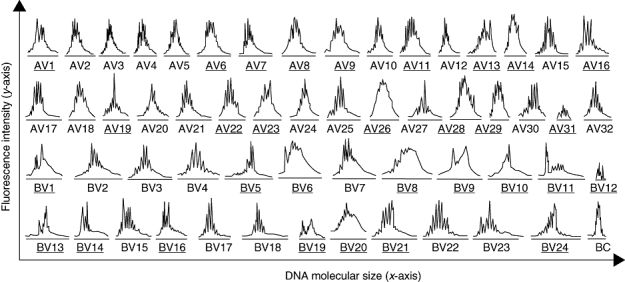
The CDR3 spectratype of T cell receptor 32 AV families and 24 BV families for one representative systemic lupus erythematosus patient SWT. Gene families showing evidence of clonal expansion are underlined.
Monitoring of the CDR3 spectratype over time in three patients
We compared the CDR3 spectratype of every V gene family in three patients before and after prednisolone treatment. We found that some V gene families, including AV1, BV13, BV20 and BV21, that showed evidence of oligoclonal expansion before treatment were restored after treatment to the normal Gaussian distribution, or to a polyclonal expansion with some skewed spectratypes. Other V gene families such as AV9, BV8, BV22 and BV23 in patient LYJ, which showed normal patterns or polyclonal expansion with skewed spectratypes before treatment, were replaced by oligoclonal expansion. Still other V gene families, such as AV32 and BV19 in patient LYJ, changed from being absent or expressed at low levels to showing evidence of polyclonal expansion (Fig. 4).
Fig. 4.
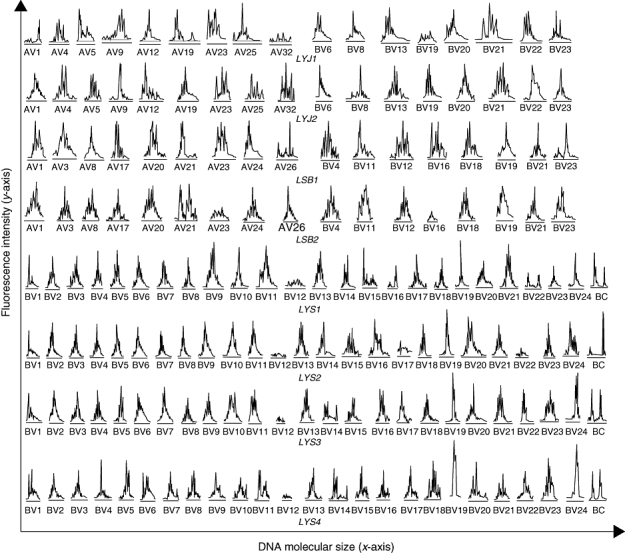
Partial analysis of changes in usage of T cell receptor AV and BV families in patients LYJ and LSB before and after treatment, and characterization of all BV families at four stages of disease for patient LYS.
Molecular analysis of the TCR α- and β-CDR3 region
Those PCR products showing evidence of clonal expansion by GeneScan analysis were selected for CDR3 sequencing. The results showed a high frequency of use of joining (J) gene segments Jα34, Jβ2-1 and Jβ2-7, and of N nucleotide insertion in both α- and β-chains, resulting in a high frequency of a GGX amino acid motif (Tables 2 and 3).
Table 2.
The partial sequences of T cell receptor (TCR) CDR3s of α-chain from T cell, showing evidence of monoclonal expansion.
| Case | AV | 3′Vα | N+5′Proximal Jα |
|---|---|---|---|
| LYJ1 | AV7 | CAVR | ATDGCNKLIFG |
| AV19 | CAVR | GRSARQLTFG | |
| AV31 | CLLG | NSGYALNFG | |
| ZHJ | AV7 | CAVR | VTGNQFYFG |
| AV12 | CVVN | DTGRRALTFG | |
| AV20 | CLVGD | GTAANNLFFG | |
| ZTX | AV3 | CATD | KSYNRGGKLIFG |
| AV18 | CAF | STTDSWGKLQFG | |
| LSB1 | AV8 | CAEN | AKEGGNKLIFG |
| AV18 | CAF | SGSRNSRYAVNFG | |
| CYP | AV7 | CAVR | FGTGGGRFNKLIFG |
| SWT | AV27 | CAVD | MGAISNKLIFG |
| AV19 | CAVR | ARGGGGRELTFG | |
| YXX | AV2 | CAVN | KSGGGADKLTFG |
| GX | AV5 | CALD | AANESGGNKLTFG |
| JY | AV20 | CLVGD | GTDASTNKLIFG |
| DWN | AV11 | CAVE | DNTGGFKTIFG |
| HJM | AV12 | CALSEA | GGTSYGKLTFG |
| LXM | AV12 | CALS | DTSSGSARQLTFG |
| ZXF | AV21 | CAAS | ASVGATNKLIFG |
| ZSF | AV14 | CVVS | GEDRDDKLIFG |
| CYX | AV7 | CAVR | DSKRELIFG |
Table 3.
The partial sequences of T cell receptor (TCR) CDR3s of β-chain from T cell, showing evidence of monoclonal expansion.
| Case | BV | 3′Vβ | N-D-N | 5′ProximalJβ |
|---|---|---|---|---|
| ZTX | BV11 | CASS | SVNM | FFGP(J2S1) |
| LSB1 | BV3 | CASR | TGTGP | YEQYFGP(J2S7) |
| CYP | BV2 | CASR | GGGRH | EQYFGP(J2S7) |
| YXX | BV5 | CASSL | GLEQAAR | SNEQFFGP(J2S1) |
| LYS1 | BV1 | ALYF CASS | APN | SPLHFGN(J1S6) |
| BV4 | CSVE | AGGFG | EQFFGP(J2S1) | |
| BV19 | ALYL CASS | VNRGGT | SYEQYFGP(J2S7) |
Discussion
Normally, autoreactive T cells undergo clonal deletion or anergy. However, in SLE, exposure of cryptic epitopes of autoantigens during cell apoptosis, together with excess co-stimulation and abnormal antigen processing with inappropriate T cell help, could potentially contribute to the escape of T cells from immunological self-tolerance [1]. Identifying the characteristics of autoreactive T cells in SLE may help to clarify the role of T cells in the immune pathogenesis of the disease and provide guidance for clinical application of this information.
The TCR Vβ used by autoreactive T cells in SLE has been reported widely. Murata et al. showed that, based on PCR and Southern blot analysis, Vβ8 and Vβ20 were expressed highly in renal biopsies [6], and Kolowos et al. found that Vβ1, Vβ13·1, Vβ15 and Vβ16 families showed oligoclonal expansions in the PBMC of SLE patients [9]. Furukawa et al. reported restricted usage of Vβ8 and Vβ13 in SLE skin lesions [19]. In contrast, little is known about TCR Vα gene usage in SLE. Restriction fragment length polymorphism studies have revealed a link between one framework gene encoding the constant region of TCR Vα and SLE [20], and autoimmune T helper lines from four SLE patients showed a significant increase in the usage of the Vα8 gene family [21]. Little information is available about the restricted use of Vα genes in vivo, the relationship between highly expressed V families and SLE activity and conserved sequences of the TCR α-chain. To understand further the use of TCR Vα genes in SLE, and in an attempt to correlate the complete structure of the α/β TCR with antigenic specificity, we used the GeneScan technique to investigate the TCR CDR3 spectratype of 20 SLE patients and six healthy controls. The CDR3 spectratype of the α-chain in PBMC, like that of the β-chain, displayed a Gaussian distribution in healthy controls, and abnormal distribution of various Vα families in SLE patients. The T cells of patients with inactive disease and those in remission showed little evidence of clonal expansion compared with the T cells of patients with active disease. We also observed prominent use of some TCR V gene families in SLE patients. These data are consistent with parts of previous studies.
The restricted usage of distinctive TCR V gene families suggests common specificity and similar immunogenic agents in a number of SLE patients [22]. Datta's group have found the autoimmune T cells of lupus recognizing five major epitopes in nucleosomal histones, namely H1′(22–42), H2B (10–33), H3 (85–105), H4 (16–39) and H4 (71–94) [23]. Perhaps there are several major peptides driving oligoclonal T cell expansion in these SLE patients. To filter and confirm the major peptide-specific TCR, we should simulate the possible TCR structure according to the particular TCR α- and β-chain CDR3 sequence in SLE patients and analyse the specificity of these TCRs binding the major peptides in the background of specific HLA types using techniques including enzyme-linked immunospot, fluorescence activated cell sorter and tetramer to filter and confirm the TCR specific for the major epitopes.
To date, there is no broadly effective clinical method available for prevention and treatment of SLE. Some investigators have applied partial TCR gene sequences of pathogenic T cells to produce DNA-based vaccines for treatment of autoimmune diseases [24–26]. After being expressed in vivo, the DNA vaccine can result in selective inhibition or deletion of the pathogenic T cell clones. We intend to analyse a large number of samples to identify representative conserved CDR3 sequences. Based on the identification of these sequences, siRNA can be synthesized to interfere with the expression of TCRs related closely to disease. TCRs with the highest affinity for different types of HLA–peptide complexes can be screened by protein structure modelling and tetramer technology to produce either a protein vaccine or a DNA-based vaccine. In the present study, through sequencing the CDR3 of clonally expanded T cells in the PBMC of SLE patients, we found a highly conserved GGX amino acid motif in the CDR3 of both TCR α- and β-chains. Conserved motifs, such as LXG in the CDR3 β-chain of skin-specific T cells and SSG, GQG and VRG in the CDR3 β-chain of intrarenal T cells, have been reported [2,6]. These results suggest that the GGX motif may also be linked with common antigen epitopes. We also found an increased appearance of Ja34 and Jb2s1 in the proximal regions of the TCR CDR3 in our study. High-frequency usage of these two Jβ genes was also detected by Kolowos et al.[9]. Much larger numbers of patients are required in future studies to identify the conserved motif and to prove its association with disease.
There is an increasing body of evidence confirming the oligoclonal activation of T cells in PBMC of SLE patients. However, we still know little about how the clonotype of these oligoclonal T cells and the CDR3 spectratype changes in PBMC with the disease activity of human SLE. Takashi et al. observed that T cell BV family clonal expansions assessed by RT–PCR/SSCP analysis correlate with disease activities [7]. However, we compared the CDR3 spectratype of every AV and BV family in three patients during changes in disease activity status, and found that the CDR3 spectratype changed dynamically in accordance with disease activity. Some TCR V gene families showing a skewed spectratype in active disease were restored to a normal Gaussian distribution in remission. The number of V gene families showing evidence of clonal expansion reduced gradually as the SLEDAI score decreased, suggesting that this method has potential value for clinical diagnosis. In view of the variable course of SLE, analysis of the changes in clonal expansion of T cells and in CDR3 corresponding with SLE disease activity may be helpful for implementation of individual treatment.
This study found that the CDR3 of different self-reactive T cell clones in SLE patients was highly homologous. Because autoreactive T cells can identify several types of autoantigen with completely different structures [27], it is reasonable to speculate that conserved sequences identified in analysis of CDR3 sequences in a large number of SLE patients may represent TCRs specific for a particular autoantigen. We analysed part of the CDR3 sequence of 20 SLE patients and found a conserved amino acid motif GGX present in both TCR α- and β-chains. This motif might be involved in binding to a particular antigen common to a number of patients. For reasons beyond our control, we were not able to perform HLA-phenotype analysis or sorting of T lymphocyte subsets, which would have added considerable information to this study. Our further studies will be aimed at a detailed characterization of the relationship between the conserved motif and the corresponding antigen in a larger number of SLE patients with a clearly identified HLA background, as dynamic monitoring of more patients dynamically discloses the function of various TCR V gene families in the progress of SLE.
Acknowledgments
This work was supported financially by the National Natural Science Foundation of China (30771952, 30771971), the Program for New Century Excellent Talents in University (NCET-07-0410), the Natural Science Foundation of Guangdong Province (07117783) and the Major State Basic Research Development Program (973) (2007CB512405). We thank Ying-huan Chen and Yuan Hu for their critical roles in the collection of clinical samples.
References
- 1.Hoffman RW. T cells in the pathogenesis of systemic lupus erythematosus. Clin Immunol. 2004;113:4–13. doi: 10.1016/j.clim.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Kita Y, Kuroda K, Mimori T, et al. Receptor clonotypes in skin lesions from patients with systemic lupus erythematosus. J Invest Dermatol. 1998;110:41–6. doi: 10.1046/j.1523-1747.1998.00072.x. [DOI] [PubMed] [Google Scholar]
- 3.Holbrook MR, Tighe PJ, Powell RJ. Restrictions of T cell receptor beta chain repertoire in the peripheral blood of patients with systemic lupus erythematosus. Ann Rheum Dis. 1996;55:627–31. doi: 10.1136/ard.55.9.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutmuller M, Baelde HJ, Ouellette S, De Heer E, Bruijn JA. T-cell receptor Vbeta gene expression in experimental lupus nephritis. Immunology. 1998;95:18–25. doi: 10.1046/j.1365-2567.1998.00565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou G, Fujio K, Sadakata A, Okamoto A, Yu R, Yamamoto K. Identification of systemically expanded activated T cell clones in MRL/lpr and NZB/WF1 lupus model mice. Clin Exp Immunol. 2004;136:448–55. doi: 10.1111/j.1365-2249.2004.02473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murata H, Matsumura R, Koyama A, et al. T cell receptor repertoire of T cells in the kidneys of patients with lupus nephritis. Arthritis Rheum. 2002;46:2141–7. doi: 10.1002/art.10432. [DOI] [PubMed] [Google Scholar]
- 7.Mato T, Masuko K, Misaki Y, et al. Correlation of clonal T cell expansion with disease activity in systemic lupus erythematosus. Int Immunol. 1997;9:547–54. doi: 10.1093/intimm/9.4.547. [DOI] [PubMed] [Google Scholar]
- 8.Talken BL, Bailey CW, Reardon SL, Caldwell CW, Hoffman RW. Structural analysis of TCR alpha and beta chains from human T-cell clones specific for small nuclear ribonucleoprotein polypeptides Sm-D, Sm-B and U1-70 kDa: TCR complementarity determining region 3 usage appears highly conserved. Scand J Immunol. 2001;54:204–10. doi: 10.1046/j.1365-3083.2001.00930.x. [DOI] [PubMed] [Google Scholar]
- 9.Kolowos W, Gaipl US, Voll RE, et al. CD4+ peripheral T cells from patients with systemic lupus erythematosus (SLE) are clonally expanded. Lupus. 2001;10:321–31. doi: 10.1191/096120301671176280. [DOI] [PubMed] [Google Scholar]
- 10.Kurokawa M, Kato T, Masuko-Hongo K, et al. Characterisation of T cell clonotypes that accumulated in multiple joints of patients with rheumatoid arthritis. Ann Rheum Dis. 1999;58:546–53. doi: 10.1136/ard.58.9.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao C, Osman GE, Adams S, Datta SK. T cell receptor alpha-chain repertoire of pathogenic autoantibody-inducing T cells in lupus mice. J Immunol. 1994;152:1462–70. [PubMed] [Google Scholar]
- 12.Der Simonian H, Band H, Brenner MB. Increased frequency of T cell receptor Va12·1 expression on CD8+ T cells: evidence that Va participates in shaping the peripheral T cell repertoire. J Exp Med. 1991;174:639–48. doi: 10.1084/jem.174.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang C, Kanagawa O. Ordered and coordinated rearrangement of the TCR alpha locus: role of secondary rearrangement in thymic selection. J Immunol. 2001;166:2597–601. doi: 10.4049/jimmunol.166.4.2597. [DOI] [PubMed] [Google Scholar]
- 14.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 15.Yao XS, Xie ZJ, Ma L, et al. Analysis of the CDR3 region of alpha/beta T-cell receptors (TCRs) and TCR BD gene double-stranded recombination signal sequence breaks end in peripheral blood mononuclear cells of T-lineage acute lymphoblastic leukemia. Clin Lab Haematol. 2006;28:405–15. doi: 10.1111/j.1365-2257.2006.00827.x. [DOI] [PubMed] [Google Scholar]
- 16.Yao XS, Zhang GW, Ma L, et al. Analysis of the CDR3 length of TCRaβ T cells in the peripheral blood of patients with chronic hepatitis B. Hepatol Res. 2006;35:10–18. doi: 10.1016/j.hepres.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Luo W, Ma L, Yao XS, et al. Complementarity-determining region 3 analysis of T cell receptor beta chain variable region in peripheral blood mononuclear cells of patients with systemic lupus erythematosus. Nan Fang Yi Ke Da Xue Xue Bao. 2006;26:1128–31. [PubMed] [Google Scholar]
- 18.Assaf C, Hummel M, Dippel E, et al. High detection rate of T-cell receptor beta chain rearrangements in T-cell lymphoproliferations by family specific polymerase chain reaction in combination with the GeneScan technique and DNA sequencing. Blood. 2000;96:640–6. [PubMed] [Google Scholar]
- 19.Furukawa F, Tokura Y, Matsushita K, et al. Selective expansions of T cells expressing V beta 8 and V beta 13 in skin lesions of patients with chronic cutaneous lupus erythematosus. J Dermatol. 1996;23:670–6. doi: 10.1111/j.1346-8138.1996.tb02679.x. [DOI] [PubMed] [Google Scholar]
- 20.Tebib JG, Alcocer-Varela J, Alarcon Segovia D, Schur PH. Association between a T cell receptor restriction fragment length polymorphism and systemic lupus erythematosus. J Clin Invest. 1990;86:1961–7. doi: 10.1172/JCI114930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desai-Mehta A, Mao C, Rajagopalan S, Robinson T, Datta SK. Structure and specificity of T cell receptors expressed by potentially pathogenic anti-DNA autoantibody-inducing T cells in human lupus. J Clin Invest. 1995;95:531–41. doi: 10.1172/JCI117695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu L, Kaliyaperumal A, Boumpas DT, Datta SK. Major peptide autoepitopes for nucleosome-specific T cells of human lupus. J Clin Invest. 1999;104:345–55. doi: 10.1172/JCI6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datta SK. Major peptide autoepitopes for nucleosome-centered T and B cell interaction in human and murine lupus. Ann N Y Acad Sci. 2003;987:79–90. doi: 10.1111/j.1749-6632.2003.tb06035.x. [DOI] [PubMed] [Google Scholar]
- 24.VandenBark AA, Morgan E, Bartholomew R, et al. TCR peptide therapy in human autoimmune disease. Neurochem Res. 2001;26:713–30. doi: 10.1023/a:1010951706830. [DOI] [PubMed] [Google Scholar]
- 25.Matsumoto Y, Jee Y, Sugisaki M. Successful TCR-based immunotherapy for autoimmune myocarditis with DNA vaccines after rapid identification of pathogenic TCR. J Immunol. 2000;164:2248–54. doi: 10.4049/jimmunol.164.4.2248. [DOI] [PubMed] [Google Scholar]
- 26.Miyakoshi A, Yoon WK, Jee Y, Matsumoto Y. Characterization of the antigen specificity and TCR repertoire, and TCR-based DNA vaccine therapy in myelin basic protein-induced autoimmune encephalomyelitis in DA rats. J Immunol. 2003;170:6371–8. doi: 10.4049/jimmunol.170.12.6371. [DOI] [PubMed] [Google Scholar]
- 27.De Silva-Udawatta M, Kumar SR, Greidinger EL, Hoffman RW. Cloned human TCR from patients with autoimmune disease can respond to two structurally distinct autoantigens. J Immunol. 2004;172:3940–7. doi: 10.4049/jimmunol.172.6.3940. [DOI] [PubMed] [Google Scholar]


