Abstract
Busulfan (Bu) resistance is a major obstacle to hematopoietic stem cell transplantation (HSCT) of patients with chronic or acute myelogenous leukemia (CML or AML). We used gene expression analysis to identify cellular factors underlying Bu resistance. Two Bu-resistant leukemia cell lines were established, characterized and analyzed for differentially expressed genes. The CML B5/Bu2506 cells are 4.5-fold more resistant to Bu than their parental B5 cells. The AML KBM3/Bu2506 cells are 4.0-fold more Bu-resistant than KBM3 parental cells. Both resistant sublines evade Bu-mediated G2-arrest and apoptosis with altered regulations of CHK2 and CDC2 proteins, constitutively up-regulated anti-apoptotic genes (BCL-XL, BCL2, BCL2L10, BAG3 and IAP2/BIRC3) and down-regulated pro-apoptotic genes (BIK, BNIP3, and LTBR). Bu-induced apoptosis is partly mediated by activation of caspases; use of the inhibitor Z-VAD-FMK completely abrogated PARP1 cleavage and reduced apoptosis by ~50%. Furthermore, Bu resistance in these cells may be attributed in part to up-regulation of HSP90 protein and activation of STAT3. Inhibition of HSP90 with geldanamycin attenuated phosphorylated STAT3 and made B5/Bu2506 and KBM3/Bu2506 more Bu-sensitive. Analysis of cells derived from patients classified as either clinically resistant or sensitive to high-dose Bu-based chemotherapy indicated alterations in gene expression that were analogous to those observed in the in-vitro model cell lines, confirming the potential clinical relevance of this model for Bu resistance.
Keywords: busulfan, myeloid leukemia, drug resistance, apoptosis, HSP90
Introduction
High-dose busulfan (1,4-butanediol dimethylsulfonate)-based conditioning therapy for hemato-poietic stem cell transplantation (HSCT) has gained increasing prominence in the last few years, mainly in the treatment of myeloid malignancies. Interest in busulfan(Bu)-based pretransplant therapy was further stimulated by a parenteral formulation that circumvented the unpredictable absorption and variable bioavailability of oral Bu that was associated with a high incidence of serious, and often lethal, venoocclusive disease [1–4]. Substitution of cyclophosphamide with fludarabine in combination with Bu further improved the clinical safety profile without appreciable loss of antileukemic activity [5, 6]. Nonetheless, relapsing leukemia remains an obstacle to patient survival. Recurrent leukemia is commonly attributed to Bu resistance, although both host and leukemic cell-derived factors may cause treatment failure. Individualized therapeutic dose guidance based on pharmacokinetics has been used to optimize systemic Bu exposure, both to increase treatment safety and to decrease the risk for recurrent leukemia. We hypothesize that some patients may suffer recurrent disease because of genetically-determined leukemic- cell drug resistance. If appropriate cellular biomarkers of Bu resistance could be identified and validated, they might be used to identify patients whose leukemia might be insensitive to high-dose Bu. A better understanding of the molecular mechanisms of cellular Bu resistance should also enable the development of strategies to circumvent such resistance, or alternatively, patients who are likely to have Bu-resistant leukemia could receive different therapy.
Cellular resistance to various DNA-alkylating agents is likely multifactorial, and some resistance mechanisms, such as altered apoptosis, may be common. Other mechanisms, however, might be specific to the inducing agent. For example, upregulation of the MGMT gene confers resistance to 1,3-bis (2-chloroethyl) nitrosourea [7], and overexpression of the ALDH1 gene results in cellular resistance to oxazaphosphorines [8, 9]. The increasing use of alkylating agent-based conditioning therapy in HSCT and the persistent problem of clinical resistance to such chemotherapy merit the establishment of human cell line models of clinically relevant degrees of resistance in order to facilitate mechanistic studies.
Using Bu as a prototype alkylating agent we established cell line models to identify signal transduction pathways and altered gene expression which might be associated with the drug-resistant phenotype. We found that Bu exposure of parental drug-sensitive cells resulted in G2-arrest and extensive apoptosis; in contrast, the resistant cells displayed markedly reduced levels of Bu-induced G2-arrest and apoptosis. Furthermore, the resistant cells were characterized by marked constitutive over-expression of CDC2, anti-apoptotic proteins, HSP90 and activated STAT3. The potential clinical relevance of these results was assessed by determining the corresponding gene-expression levels in cell samples from acute myeloid leukemia (AML) patients who were clinically considered to be either refractory or sensitive to high-dose Bu-based therapy. This communication reports the details of this in vitro Bu-resistance model with an emphasis on findings that may be of clinical relevance.
Materials and Methods
Cells and drugs
The cell lines used were KBM7/B5 (“B5”), a cloned subline of the KBM7 chronic myeloid leukemia (CML) cell line [10, 11], and KBM3, an AML cell line [12]. Bu-resistant cell lines were established as described below. Stock solution of 6 mg/ml Bu (Busulfex™) in 33.3% dimethylacetamide and 66.7% polyethylene glycol-400 solvent was obtained from PDL BioPharma, Edison, NJ. Geldanamycin (GA; Invivogen, San Diego, CA) was dissolved in dimethyl sulfoxide (DMSO). The amount of solvent was equalized in each experiment which involved exposure of cells to various concentrations of Bu or GA. In parallel, as negative controls, cells were exposed to the amount of solvent corresponding to the highest Bu or GA concentration.
Establishment of Bu-resistant cell lines
Cells were grown to logarithmic phase, washed with PBS containing 0.1% glucose, and resuspended in serum-free Iscove's Modified Dulbecco's Medium (IMDM; Sigma, St. Louis, MO). Cultures of 5 × 106 cells were exposed for 1 h to gradually increasing Bu concentrations from 25 µg/ml to 250 µg/ml. Each exposure was followed by washing in ice-cold PBS containing 0.1% glucose + 1% fetal bovine serum (FBS), then the cells were resuspended in IMDM with 20% FBS (complete IMDM) and incubated at 37°C in a humidified atmosphere of 5% CO2 in air for 2 – 3 weeks, or until a normal doubling time and viability of >98% was restored. Exposure at each drug concentration was repeated 3 times. At the final drug concentration, 250 µg/ml, the cells were exposed six times. The cell lines are referred to as B5/Bu2506 and KBM3/Bu2506.
Cytotoxicity assay
Cells (1 × 106/ml) were exposed to various concentrations of Bu at 37°C for 1 h and washed as described above. The cells were then resuspended in complete IMDM at 2 × 105/ml and aliquoted (20,000/well) in 96-well plates, incubated as above at 37°C for 4 days, and analyzed by the MTT assay [13]. Graphical analyses including calculations of IC50 were done using Prism 4 (GraphPad Software, San Diego, CA).
Fluorescence Activated Cell Sorter (FACS) Analysis
Cells were exposed to Bu at 37°C for 1 h, washed and resuspended in complete IMDM as described above. Following drug or solvent exposure and 48-h recovery the cells were centrifuged, resuspended in 70% ethanol, and fixed at −20°C overnight. Fixed cells were pelleted at 3,000 × g at room temperature, washed with PBS, and treated with 0.25 ml of 500 U/ml RNAse A in PBS containing 1.12% sodium citrate at 37°C for 30 min. After addition of 0.25 ml propidium iodide (PI, 50 µg/ml), the cells were kept in subdued light for at least 1 h prior to FACS analysis. The cellular DNA content of at least 10,000 cells was analyzed using BD FACSCalibur and CellQuest™ software (Becton Dickinson, Franklin Lakes, NJ). Histograms were analyzed using ModFit LT (Verity Software House, Topsham, ME).
FACS was also used to measure apoptosis of cells (5 × 105/ml) exposed to different concentrations of TNF-related apoptosis-inducing ligand (TRAIL; BIOMOL International, Plymouth Meeting, PA). Phosphatidyl serine externalization in apoptotic cells was determined with the Annexin V-FLUOS staining kit (Roche Diagnostics, Indianapolis, IN). Membrane integrity was simultaneously assessed by PI exclusion in the Annexin V-stained cells.
Microarray analysis
RNA was extracted using TRIzol™ Reagent (Invitrogen, Carlsbad, CA), treated with DNAse I, and purified using the RNeasy kit (Qiagen, Valencia, CA). Its purity was determined on a 1% agarose-formaldehyde gel followed by ethidium bromide staining. The TrueLabeling-AMP™ Linear RNA Amplification kit (SuperArray Bioscience, Frederick, MD) was used to synthesize biotinylated antisense RNA as a probe for hybridization. The RNA probe was purified using an ArrayGrade™ cRNA Cleanup kit (SuperArray) and the concentration was determined spectrophotometrically.
Oligo GEArray® membranes (SuperArray) were pre-hybridized, hybridized with the RNA probe and developed for chemiluminescent detection as described by the manufacturer. The signals on X-ray films were scanned using a densitometer and analyzed with ImageQuant software (both from GE Healthcare Bio-Sciences, Piscataway, NJ) and the SuperArray Analysis Suite.
Real Time PCR
The high capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) was used to synthesize cDNA. Real time PCR amplification was performed using either Taqman probe- or SYBR Green-based assay with the 7500 Real Time PCR System (Applied Biosystems). The quantification of gene expression was carried out by comparative CT methodology (i.e., threshold cycle number at which the increase in fluorescence was logarithmic); the ABL1 gene was used as an endogenous control. The genes analyzed with real time PCR were selected based on the results of the microarray analysis.
Western Blot Analysis
Cells in logarithmic growth phase (1 × 106 cells/ml) were either mock-exposed or pulse-exposed to Bu at 37°C for 1 h. The cells were then washed with PBS containing 0.1% glucose + 1% FBS, and allowed to recover in complete IMDM for 48 h at 37°C as described above. They were then centrifuged, resuspended in PBS, lysed with Laemmli buffer and boiled for 5 min. Normalization of the amount of protein in the Laemmli buffer was done by immuno-blot analysis using anti-β-actin antibody. Densitometric analysis of the β-actin signals was done and protein concentrations were adjusted as appropriate. The Bradford method [14] was also used to determine protein concentrations of some cell extracts.
Western blot analysis was done by separating protein extracts on polyacrylamide-SDS gels and blotting onto Hybond ECL membranes (GE Healthcare). Immunoblot analysis by chemiluminescence was done using the ECL plus Western blotting Detection System (GE Healthcare) and the SuperSignal West Pico/Dura Substrate (Pierce, Rockford, IL). All antibodies, their sources and other relevant information are listed in Table 1.
Table 1.
List of primary antibodies, their sources and dilutions
| Antigen | Source/Cat. # | Clone type* | Dilution** |
|---|---|---|---|
| BCL-XL | Santa Cruz Biotech/8392 | mAb | 500 |
| BID | Cell Signaling/2002 | pAb | 1000 |
| BNIP3 | Abcam/Ab10433 | mAb | 1000 |
| Caspase 3 | Cell Signaling/9662 | pAb | 1500 |
| Caspase 8 | Cell Signaling/9746 | mAb | 1500 |
| CDC2 | Cell Signaling/9112 | pAb | 1500 |
| HSP90 | Axxora/ALX-804-078 | mAb | 1500 |
| p14ARF | Santa Cruz Biotech/8613 | pAb (goat) | 500 |
| p21 | Upstate Biotech/05-345 | mAb | 1000 |
| p53 | Santa Cruz Biotech/1315 | pAb | 1000 |
| PARP1 | Santa Cruz Biotech/8007 | mAb | 1000 |
| P-CDC2 (Thr14) | Cell Signaling/2543 | pAb | 1500 |
| P-CDC2 (Tyr15) | Cell Signaling/9111 | pAb | 1500 |
| P-CHK2(Thr68) | Cell Signaling/2661 | pAb | 1500 |
| P-STAT3(Tyr705) | Cell Signaling/9131 | pAb | 1000 |
| RAF1 | Santa Cruz Biotech/133 | pAb | 500 |
| STAT3 | Cell Signaling/9132 | pAb | 1000 |
| β-actin | Sigma/A5316 | mAb | 10000 |
| γ-H2AX | Upstate Biotech/05-636 | mAb | 1000 |
pAb: polyclonal antibody; used anti-rabbit IgG (or anti-goat as indicated) for secondary antibody from Bio-Rad Lab
mAb: monoclonal antibody; used anti-mouse IgG for secondary antibody from Bio-Rad Lab
Fold dilution in PBS with 0.05% Tween 20
Inhibition of caspases
Cells were pre-treated with DMSO or 40 µM Z-VAD-FMK (Axxora, LLC, San Diego, CA) for 1 h, then pulse-exposed to Bu for 1 h at 37°C and washed as described above. Cells were allowed to recover at 37°C in complete IMDM containing DMSO or 40 µM Z-VAD-FMK for 48 h and analyzed by Western blot and PI staining followed by FACS analysis.
Inhibition of HSP90
Cells were treated with DMSO or 2 µM GA for 16 h at 37°C, 5% CO2. Cells were washed with PBS/0.1% glucose, resuspended in serum-free IMDM (1 × 106 cells/ml), pulse-exposed to Bu for 1 h at 37°C and washed as described above. After 48-h recovery in complete IMDM, cells were analyzed by FACS, MTT assay and Western blot as described above.
Patient cell samples
Mononuclear cell samples from peripheral blood or bone marrow of AML patients who had intravenous Bu-based pretransplant therapy were obtained from the AML cell sample core repository of the MD Anderson Cancer Center. Samples were assigned as deriving from “clinically sensitive” or “clinically resistant” patients; patients who had achieved a complete remission (CR) after HSCT or received the HSCT in CR and remained in CR for a minimum of one year after HSCT and either were alive in ongoing CR or later died of causes unrelated to leukemia progression were considered clinically sensitive. Patients with active AML who were transplanted and either did not achieve a CR, or achieved CR but suffered a recurrence within the first 100 days, were considered clinically resistant. All analyzed pre-transplant cell samples contained at least 95% leukemic cells. CD34+ cells (normal control) were obtained from a healthy volunteer who received filgrastim for 4 days prior to peripheral blood apheresis collection of CD34+ cells. This cell product was enriched using a CD34+ selection column in an “Isolex” machine (Miltenyi Biotec, Auburn, CA). Total RNA from these cell samples was isolated using TRIzol™ as described above.
Results
Establishment and phenotypic characterization of Bu-resistant cell lines
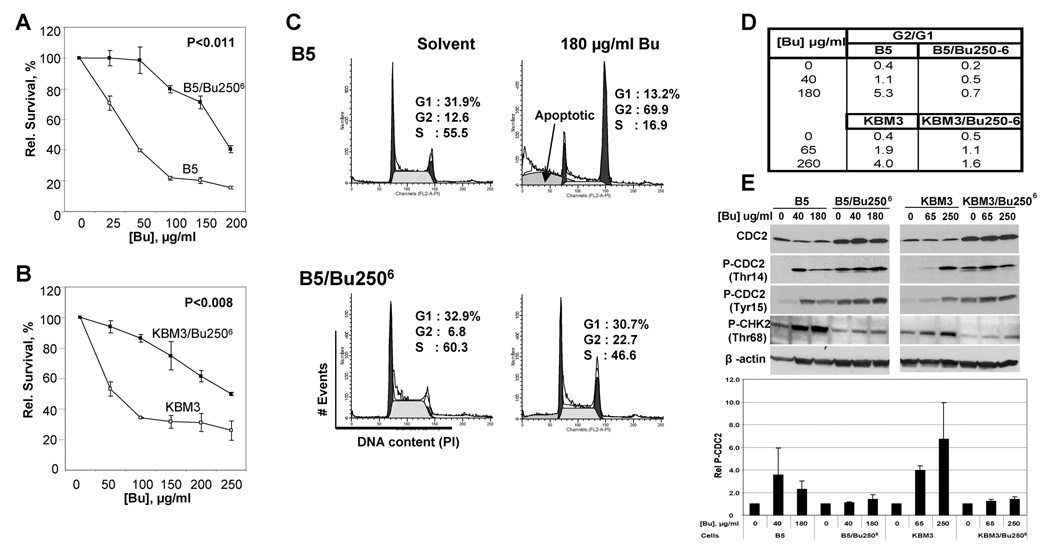
The first parental strain was KBM7/B5 (“B5”), a cloned subline of KBM7 [10]. A Bu-resistant subline was established through intermittent in-vitro exposure of B5 cells to gradually increasing drug concentrations as described above. Using the MTT assay, the Bu-resistant and -sensitive sublines have IC50 values of 180 µg/ml and 40 µg/ml Bu, respectively, giving a resistance index of ~4.5 for B5/Bu2506 cells (Figure 1A). A similar result was obtained by clonogenic assay (data not shown). This phenotypic resistance of B5/Bu2506 cells has been maintained for more than four months of in-vitro passaging without drug exposure (data not shown), suggesting genotypic stability. The resistance of B5/Bu2506 cells to Bu did not alter their growth rate; a doubling time of approximately 22 h for both B5 and B5/Bu2506 cells suggests a lack of growth advantage of B5/Bu2506 cells in the absence of drug.
Figure 1. Characterization of busulfan (Bu)-sensitive and -resistant cell lines.
(A, B) Cells were pulsed with the indicated concentration of Bu at 37°C for 1 h, washed, and plated at 20,000 cells/well. After 4 days, cell proliferation was analyzed by the MTT assay. The data represent an average of 5 replicates respective to solvent alone control. The P values were obtained using paired t test. (C) Cell cycle analysis was done by pulsing cells with solvent or 180 µg/ml Bu at 37°C for 1 h. The cells were then washed and allowed to recover for 2 days. Cells were stained with propidium iodide, sorted by flow cytometry and analyzed using the ModFit LT™ program. (D) The same procedure used in panel C was done at different concentrations of Bu using the two parental cell lines B5 and KBM3 and their respective Bu-resistant subline. Cell cycle analysis was used to compare the G2/G1 ratio of Bu-sensitive and -resistant cells. (E) Cells were pulsed-exposed to various concentrations of Bu or solvent alone for 1 h, allowed to recover for 2 days and analyzed for expression of cell cycle-related proteins by Western blot immunostaining (upper panel). Films were scanned using a densitometer and the ratio P-CDC2(Tyr15)/CDC2 was calculated for each treatment. Using the obtained ratios, the level of P-CDC2(Tyr15) was then determined relative to the solvent alone control (i.e. 0 µg/ml Bu) for each cell line (lower panel). The mean values and standard deviations were obtained from three independent Western blot analyses.
Using a similar protocol of intermittent exposure to gradually increasing Bu concentrations, a Bu-resistant cell line was established from the human AML cell line KBM3 [12]. Figure 1B compares the survival of KBM3 and KBM3/Bu2506 cells after Bu exposure. The Bu-resistant subline has an IC50 of 260 µg/ml and the parental cells exhibit an IC50 of 65 µg/ml (Figure 1B), indicating a resistance index of ~4.0 similar to the B5 model (Figure 1A).
Busulfan is known to arrest cells in the G2 phase of the cell cycle [15, 16]. To determine how Bu would affect the cell cycle of B5 and B5/Bu2506 cells, PI staining and FACS analysis were conducted. Cells were exposed to 180 µg/ml Bu for 1 h and allowed to recover in complete IMDM for 48 h. A significant difference in the fraction of cells arrested in G2 was observed, as evidenced by the %G2-phase/%G1-phase (G2/G1) ratios; B5 cells exhibited a G2/G1 ratio of 5.3, while the B5/Bu2506 cells had a G2/G1 ratio of 0.7 (Figures 1C and 1D). Similar results were obtained in the KBM3 model (Figure 1D). Furthermore, ~35% of the Bu-exposed B5 cells had undergone apoptosis as indicated by the proportion of cells exhibiting a sub-G1 DNA content. Apoptosis was not observed after 24-h post-exposure (data not shown); hence, all subsequent analyses were done 48 h after Bu exposure. In contrast, the B5/Bu2506 cultures showed less sub-G1 DNA content/apoptosis 48 h after exposure to the same drug concentration (Figure 1C). Note that, in the MTT assay, this drug exposure would result in the loss of viability of more than 80% of the parental B5 cells and of about 50% of the B5/Bu2506 cells (Figure 1A). Interestingly, and in contrast to the absence of caspase and PARP1 cleavage in Bu-exposed B5/Bu2506 cells (see below), there still was a residual sub-G1 signal in these cells (Figure 1C).
At equi-cytotoxic drug concentrations and 48-h post-treatment, B5 and B5/Bu2506 cells showed a slight difference in their G2/G1 ratios, similar to that seen in the control (Figure 1D). At 40 µg/ml Bu, B5 cells show a G2/G1 ratio of 1.1 whereas B5/Bu2506 cells exhibited a G2/G1 ratio of 0.7 at 180 µg/ml Bu. Similar results were obtained in the KBM3 cell model; the G2/G1 ratios were 1.9 (KBM3) and 1.6 (KBM3/Bu2506), at 48 h after exposure to 65 µg/ml and 260 µg/ml Bu, respectively (Figure 1D). At the same drug concentration of 180 µg/ml Bu and 48-h post-exposure, B5 and B5/Bu2506 cells showed G2/G1 ratios of 5.3 and 0.7, respectively. Similar exposure at 250 µg/ml Bu of KBM3 and KBM3/Bu2506 cells resulted in G2/G1 ratios of 4.0 and 1.6, respectively (Figure 1D). The lower G2/G1 ratios for the two Bu-resistant cell lines relative to their Bu-sensitive parental cells may suggest a common mechanism of evading G2-arrest and apoptosis in the B5/Bu2506 and KBM3/Bu2506 cells.
To further understand the mechanism of Bu-mediated G2-arrest in human myeloid leukemia cells, expression of various cell cycle-related proteins was determined. CDC2, a critical protein for G2/M phase [17], was constitutively up-regulated in both Bu-resistant cell lines (Figure 1E, upper panel). Since phosphorylation of CDC2 at Thr14 and Tyr15 is a hallmark of G2-arrest [18], we determined the effects of Bu on the level of P-CDC2 relative to solvent alone control in each cell line. In general, the level of P-CDC2 is higher in the resistant cells but Bu exposure resulted in a more dramatic change in the phosphorylation of CDC2 at Thr14 and Tyr15 in Bu-sensitive B5 and KBM3 cells compared to their Bu-resistant counterparts. Exposure of B5 cells to 40 µg/ml Bu, which corresponds to its IC50 value, resulted in ~4-fold increase in P-CDC2(Tyr15) relative to solvent alone control; a similar increase in P-CDC2(Tyr15) was observed for KBM3 exposed to 65 µg/ml Bu (Figure 1E, lower panel). Exposure of the two Bu-resistant cell lines to Bu did not result in a significant change in the level of P-CDC2( Tyr15) or P-CDC2(Thr14). To identify the upstream factor that may contribute to the inhibitory phosphorylation of CDC2 kinase, we examined the level of phosphorylated CHK2 at Thr68 which is known to inactivate CDC25 phosphatase that acts on P-CDC2 [19]. Exposure of B5 and KBM3 cells to Bu increased the relative level of P-CHK2(Thr68) to a greater extent compared to B5/Bu2506 and KBM3/Bu2506 (Figure 1E, upper panel). Our data suggest that exposure of sensitive cells to Bu results in activation of the P-CHK2/CDC25/P-CDC2 pathway that leads to G2 cell cycle arrest. The same pathway is activated to a lesser extent in Bu-resistant cells and may, in addition to the up-regulation of CDC2, explain their abilities to evade G2-arrest.
Lack of caspase activation in drug-exposed B5/Bu2506 cells
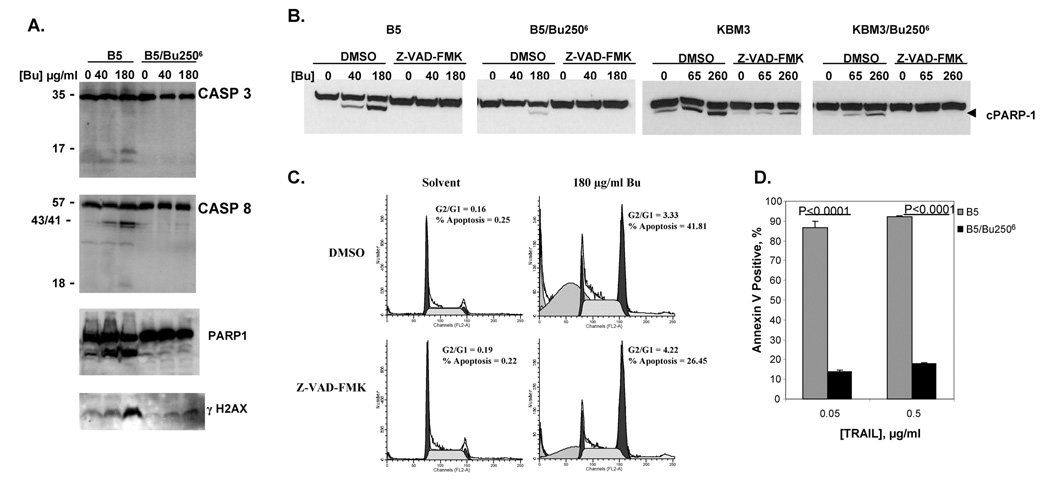
As shown in Figure 1C, the cytotoxic activity of Bu towards the parental B5 cells was to a significant extent due to activation of the apoptotic pathway, as indicated by the number of cells with sub-G1 DNA content. In contrast, the resistant B5/Bu2506 cells effectively evade this response at the same level of drug exposure. Apoptosis is intimately linked to the activation of proteolytic caspases triggered by DNA damage [20, 21]. We therefore sought confirmation and further insight into the role of mitigated apoptosis in the Bu-resistant phenotype of B5/Bu2506 cells by examining the cleavage/activation of two key apoptotic proteins, caspases 3 and 8. The data in Figure 2A indicate extensive dose-dependent activation of caspases 3 and 8 in B5 cells treated with Bu (40 and 180 µg/ml, 1 h), as indicated by their cleaved products at 48 h post-exposure. In contrast, little or no caspase cleavage was apparent in B5/Bu2506 cells following exposure to the same Bu concentrations. A similar activation of caspases 3 and 8 was observed in Bu-exposed KBM3 cells, but this effect was again less marked in KBM3/Bu2506 cells (data not shown). This caspase activation was associated with significant cleavage of the PARP1 protein in Bu-exposed parental cells (Figure 2A and 2B) and DNA fragmentation as measured by the alkaline comet assay (data not shown). DNA fragmentation following Bu exposure of B5 cells was also suggested by the observed phosphorylation of histone 2AX (H2AX, Figure 2A). Exposure of B5/Bu2506 cells to the same drug concentration caused dramatically less PARP1 cleavage and minimal phosphorylation of H2AX (Figures 2A and 2B).
Figure 2. Involvement of caspases in busulfan-mediated apoptosis.
(A and B) Total cell extracts from cells previously pulsed with busulfan (Bu) with or without Z-VAD-FMK exposure were analyzed by Western blot. (C) B5 cells were pre-treated with 40 µM Z-VAD-FMK for 1 h, pulse-exposed to 180 µg/ml busulfan or its solvent for 1 h, allowed to recover for 48 h in the presence or absence of 40 µM Z-VAD-FMK, and analyzed by propidium iodide staining followed by flow cytometry. The proportion of cells in different phases of the cell cycle (including apoptotic cells) was determined using a ModFit LT™ software. (D) Cells were exposed to different concentrations of TRAIL for 18 h and Annexin V-positive cells were measured by flow cytometry. The graph represents an average of three independent experiments and P values were calculated using unpaired t test.
Thus, Bu exposure triggers activation of caspases 3 and 8, PARP1 cleavage, DNA strand breakage and H2AX phosphorylation in Bu-sensitive cells. Perhaps the most intriguing finding, however, is the virtual absence of apoptosis activation in the Bu-resistant cells after exposure to an IC50 drug concentration (180 or 260 µg/ml Bu). This finding suggests that the Bu-induced cytotoxicity observed in the resistant cells is mediated by mechanisms other than apoptosis.
It was reported that Bu selectively induces cellular senescence but not apoptosis in WI38 fibroblasts [22]. We sought to determine if Bu-resistant cells exposed to Bu might undergo accelerated senescence. However, analysis of expression of senescence-associated p14ARF and staining for β-galactosidase did not show any difference between untreated and treated (180 µg/ml Bu) Bu-resistant cells.
Abrogation of caspase activation by the inhibitor Z-VAD-FMK
The role of caspases in Bu-mediated apoptosis in the two cell-line models was confirmed using the inhibitor Z-VAD-FMK. Figure 2B shows significant inhibition of caspase-mediated PARP1 cleavage in cells pre-treated with 40 µM Z-VAD-FMK for 1 h, followed by a 1-h exposure to Bu, and a 48-h recovery in complete IMDM containing 40 µM Z-VAD-FMK. FACS analysis showed an approximately 50% reduction in Bu-induced apoptosis in the Z-VAD-FMK-exposed B5 cells (Figure 2C). The partial inhibition of apoptosis (Figure 2C) despite the almost complete abrogation of PARP1 cleavage (Figure 2B) suggests that caspase-independent pathways contribute to cell death.
The lack of Bu-mediated caspase activation in B5/Bu2506 cells suggests possible cross-resistance to TRAIL or Apo2L. Indeed, treatment of B5 and B5/Bu2506 cells with 0.05 µg/ml TRAIL for 18 h resulted in 87% and 14% Annexin V-positive cells, respectively. A 10-fold increase in TRAIL concentration did not cause significant apoptosis of B5/Bu2506 cells (Figure 2D). Similar results were obtained for the KBM3 model (data not shown). Although the TRAIL signaling pathway might be initially different from Bu-mediated apoptosis, the results suggest that insufficient activation of caspases might be a common feature and thus may explain the resistance of B5/Bu2506 and KBM3/Bu2506 cells to apoptosis in the presence of TRAIL or Bu.
Gene expression profiling
To further understand the mechanism(s) of Bu resistance, we identified differentially expressed genes in B5 versus B5/Bu2506 cells, as well as KBM3 versus KBM3/Bu2506 cells, at steady state in the absence of drug exposure, using the apoptosis-specific gene expression profiling system from SuperArray. Real-time PCR and Western blot analyses were used to confirm the microarray results. Table 2 shows the major differences in expression of apoptosis-related genes in Bu-resistant cells relative to their Bu-sensitive counterparts. At the mRNA level, the biggest differences were observed for the pro-apoptotic genes BIK, TNFSF7, BNIP3, LTBR, BID, and BAK1, which were all down-regulated in Buresistant B5/Bu2506 cells. BIK, BNIP3 and LTBR were also down-regulated whereas TNFSF7, BID and BAK1 were either unchanged or slightly up-regulated in KBM3/Bu2506 cells. Anti-apoptotic genes that were up-regulated in both Bu-resistant lines include IAP2/BIRC3, BAG3, BCL2L10, BCL-XL, and BCL2 (Table 2).
Table 2.
Differentially expressed apoptosis genes in Bu-resistant cells relative to their Bu-sensitive parental lines by real time RT-PCR.
| Gene Symbol | Description | GenBank # | Fold difference | |
|---|---|---|---|---|
| B5/Bu250-6 | KBM3/Bu250-6 | |||
| BIK | BCL2-interacting killer | NM_001197 | 0* | 0* |
| TNFSF7 | Tumor necrosis factor (ligand) superfamily,mem 7CD27L/CD70 | NM_001252 | 0.1 | 1.0 |
| BNIP3 | BCL2/adenovirus E1B 19kDa interacting protein 3 | NM_004052 | 0.2 | 0.3 |
| LTBR | Lymphotoxin beta receptor (TNFR superfam, 3) | NM_002342 | 0.2 | 0.5 |
| BID | BH3 interacting domain death agonist | NM_001196 | 0.3 | 2.0 |
| BAK1 | BCL2 antagonist/killer 1 | NM_001188 | 0.4 | 1.0 |
| BCL2 | B-cell CLL/Lymphoma 2 | NM_000633 | 3.4 | 2.4 |
| BCL-XL | BCL-XL/BCL2-like 1 | NM_138578 | 3.7 | 1.5 |
| BCL2L10 | BCL2-like 10 | NM_020396 | 5.2 | 7.0 |
| BAG3 | BCL2-associated athanogene 3 | NM_004281 | 10.2 | 16.0 |
| IAP2/BIRC3 | Inhibitor of apoptosis protein 2/baculoviral IAP repeat-containing 3 | NM_001165 | 86.0 | 2.5 |
Undetectable in the Bu-resistant cells
Levels of pro- and anti-apoptotic proteins with and without Bu exposure
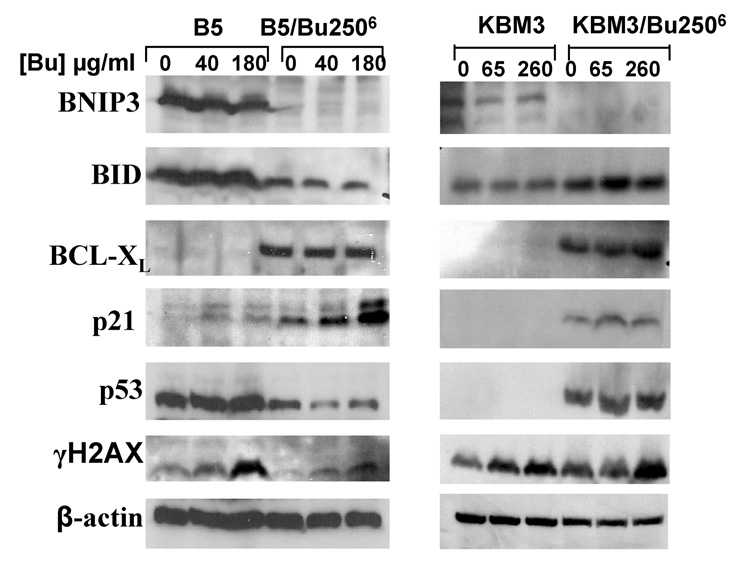
Having established major differences in the constitutive expression of key apoptosis-regulating genes at the mRNA level, we sought to determine the corresponding protein levels and how they are affected by Bu exposure. We analyzed mock- or Bu-exposed cells using concentrations close to their IC50 values: 40 µg/ml for B5, 180 µg/ml for B5/Bu2506, 65 µg/ml for KBM3, and 260 µg/ml for KBM3/Bu2506. Figure 3 shows constitutive down-regulation of the pro-apoptotic proteins BNIP3 and BID and up-regulation of the anti-apoptotic protein BCL-XL in Bu-resistant B5/Bu2506 cells. The protein levels of BNIP3 and BCL-XL, but not BID, show a similar pattern in the KBM3/Bu2506 cells. This pattern is similar to that observed for the mRNA levels (Table 2).
Figure 3. Levels of expression of apoptosis-related proteins.
Cells were pulse-exposed to the indicated concentrations of busulfan (Bu) for 1 h and allowed to recover for 48 h prior to extraction of total protein and Western blot analysis.
As shown in Figure 3, BNIP3 protein was strongly constitutively expressed in both parental cell lines but was not induced following Bu exposure, and was essentially absent in the resistant cells. BCL-XL protein showed the opposite pattern, being strongly constitutively expressed in the resistant cells while not being induced following Bu exposure and being essentially absent in the parental cell lines. The expression of another pro-apoptotic protein BID was not induced by Bu but was constitutively higher in the B5 compared to the B5/Bu2506 cells. It is surprising that BID was up-regulated at both the mRNA and protein levels (Table 2 and Figure 3) in KBM3/Bu2506 cells and the phenotypic implication of this finding is not clear. The CDK inhibitor p21/WAF1/Cip1/CDKN1A was up-regulated in B5/Bu2506 cells and exposure to Bu further induced its expression. This induction of p21 is p53-independent as suggested by the lower expression of p53 in B5/Bu2506 cells and absence of p53 induction in the presence of Bu (Figure 3). The inducibility of p21 expression in B5/Bu2506 cells might be responsible for the inhibition of cell growth seen at high Bu concentrations (Figure 1A), which correlates with less apoptosis than in the B5 parental cell line (Figure 1C and Figure 2A). Expression of p21 in the KBM3 cell-line model suggests a different mechanism. Whereas the p21 and p53 proteins were both constitutively up-regulated in KBM3/Bu2506 cells, expression of the two proteins was not induced by Bu exposure.
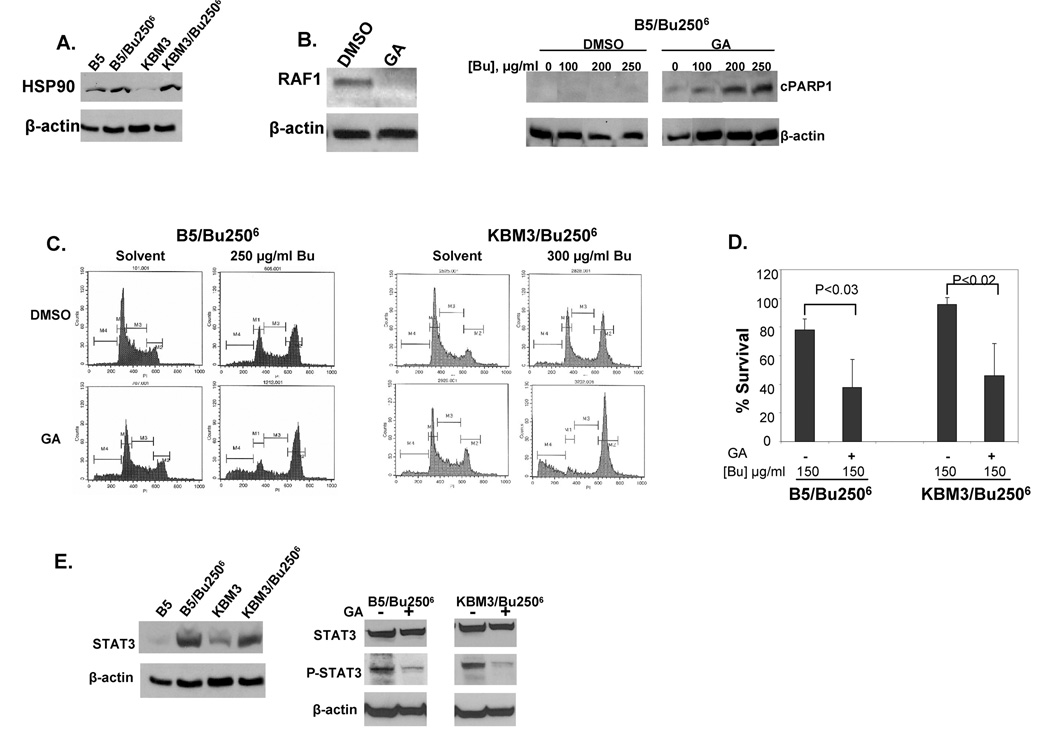
Overexpression of HSP90 in Bu-resistant cells contributes to Bu resistance
Further analysis of the expression of survival-related proteins in these cell lines shows constitutive up-regulation of HSP90 protein in B5/Bu2506 and KBM3/Bu2506 cells (Figure 4A) relative to the parental cell lines. Analysis of HSP90 mRNA by real-time RT-PCR did not show any significant difference between Bu-sensitive and Bu-resistant cells, suggesting post-transcriptional control of HSP90 expression. HSP90 was not inducible by Bu exposure in any of the cell lines (data not shown). To determine whether this constitutive up-regulation may contribute to Bu resistance, cells were pre-exposed at 37°C for 16 h to 2 µM GA, an inhibitor of HSP90 [23]. The level of RAF-1 protein was analyzed to determine if GA disrupts the function of HSP90 under our experimental conditions. RAF-1 is a client protein of HSP90 and inhibition of HSP90 causes misfolding and degradation of RAF-1 [24]. Figure 4B shows that exposure of B5/Bu2506 cells to GA resulted in the disappearance of RAF-1, suggesting efficient inhibition of HSP90 activity. Pre-exposure to GA partially restored Bu sensitivity in both B5/Bu2506 (Figure 4B) and KBM3/Bu2506 (data not shown) cells as suggested by PARP1 cleavage (cPARP1 in Figure 4B). Cell cycle analysis showed a marked G2-arrest of GA-exposed B5/Bu2506 and KBM3/Bu2506 cells following exposure to 250 and 300 µg/ml Bu, respectively (Figure 4C). Pre-exposure to GA increased the G2/G1 ratio up to 14-fold and the proportion of cells in sub-G1 up to 3-fold (Figure 4C). These results strongly suggest that disruption of cellular HSP90 activity partially restores Bu sensitivity in the resistant cells. This interpretation is further supported by a 2-fold decrease in the survival of GA-exposed Bu-resistant cells in the presence of 150 µg/ml Bu (Figure 4D). Overall, these findings suggest that Bu resistance in B5/Bu2506 and KBM3/Bu2506 cells may be partly due to their constitutive overexpression of HSP90.
Figure 4. HSP90 and STAT3 contribute to the resistance of B5/Bu2506 and KBM3/Bu2506 cells to busulfan (Bu).
(A) Western blot analysis showing constitutive overexpression of HSP90 in Bu-resistant cells compared to their parental cell lines. (B) Cells were exposed overnight to DMSO or 2 µM geldanamycin (GA) to inhibit HSP90 activity and a fraction of the cells was used to determine the level of RAF1 protein by Western blot analysis. The remaining fraction of pre-treated cells was pulse-exposed to the indicated concentrations of Bu for 1 h and allowed to recover for 2 days prior to (B) extraction of total protein and Western blot analysis for cleaved PARP1 (cPARP1), (C) cell cycle analysis, and (D) proliferation assay. Results shown for % survival (D) are means of three independent experiments and P values were determined using unpaired t test. (E) Constitutively up-regulated expression of STAT3 protein in Bu-resistant B5/Bu2506 and KBM3/Bu2506 cells relative to their parental cell lines. Overnight exposure to 2 µM GA inhibits the phosphorylation of STAT3 at Tyr-705 (P-STAT3) in Bu-resistant cells.
To determine a possible mechanism of HSP90-mediated Bu resistance and myeloid leukemia cell survival, we compared the levels of expression of STAT3 in the Bu-sensitive and -resistant cells. A recent report links HSP90 and STAT3 to drug resistance and survival of myeloma cells [25]. Figure 4E shows greatly up-regulated constitutive expression of STAT3 protein in both B5/Bu2506 and KBM3/Bu2506 cells compared to their parental cells. We then examined the activation of STAT3 protein in Bu-resistant cells exposed to GA. Figure 4E shows that inhibition of HSP90 activity by GA is associated with attenuated levels of phosphorylated STAT3, whereas the levels of total STAT3 were minimally influenced under these conditions. The attenuated STAT3 and HSP90 activities might contribute to the observed restoration of Bu sensitivity of B5/Bu2506 and KBM3/Bu2506 cells.
Gene-expression studies in clinical material
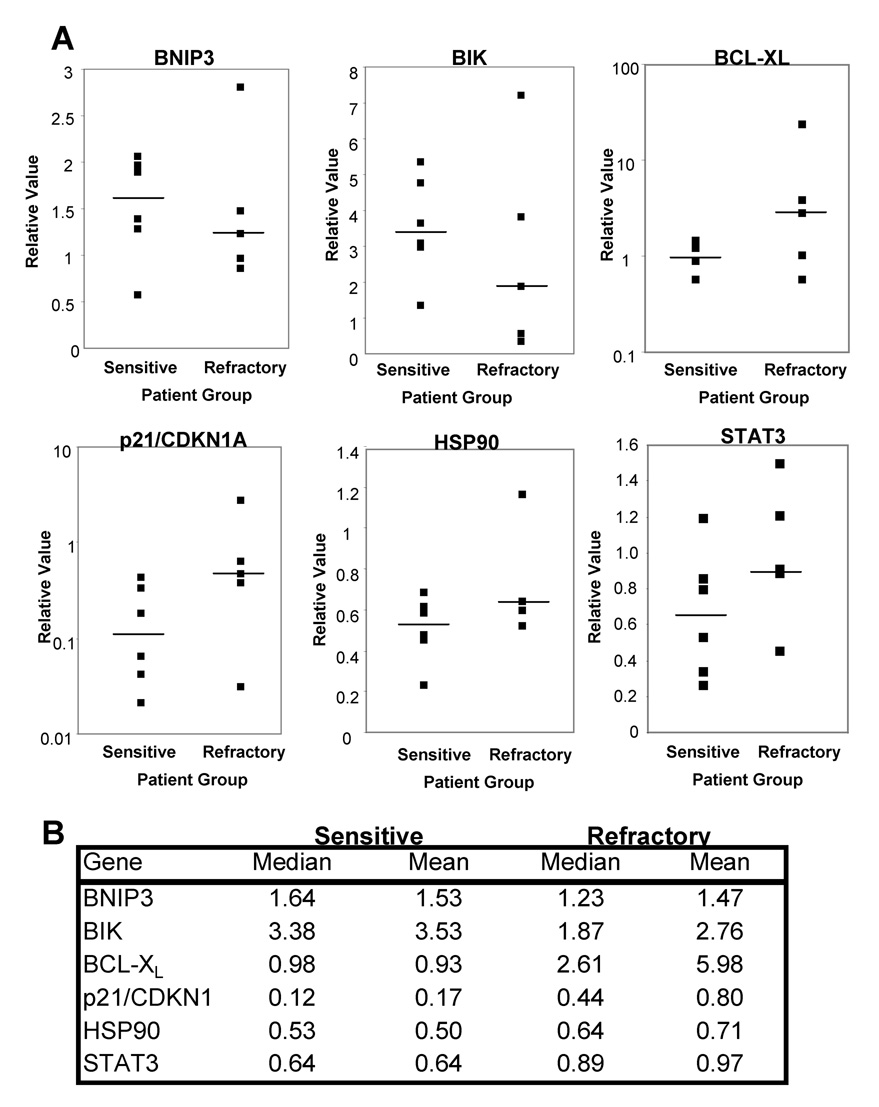
To determine the possible clinical relevance of these results, we used real-time PCR to determine the levels of expression of BNIP3, BIK, BCL-XL, p21/CDKNIA, HSP90 and STAT3 in clinically sensitive (n = 6) and resistant (n = 5) patient-derived cell samples (see Materials and Methods for classification of patients from whom these samples were obtained). Comparison of their median relative values shows down-regulation of pro-apoptotic BNIP3 and BIK in resistant cell samples and up-regulation of anti-apoptotic BCL-XL and p21/CDKN1A (Figure 5), findings which are consistent with the results obtained using the cell-line models (Figure 3). HSP90 and STAT3, which are up-regulated in Bu-resistant B5/Bu2506 and KBM3/Bu2506 cells (Figure 4), may have slightly higher levels of expression in refractory patient leukemia cells than the sensitive patient samples.
Figure 5. Gene expression analysis using samples from clinically sensitive or resistant AML patients.
cDNAs were prepared using total RNA extracted from patient cell samples and expressions of specific genes were analyzed by real time RT-PCR. The bar shows the median value for each group of patient samples (A). The median and mean values are summarized in panel B.
Discussion
Clinical “drug resistance” is a major obstacle to long-term control of myeloid leukemia after HSCT. A large fraction of high-risk AML and CML patients who achieve CR after transplant eventually relapse, suggesting that suboptimal/incomplete eradication of leukemic cells contributes to leukemic recurrence. It has been demonstrated that higher intensity pretransplant conditioning programs translate into better disease-free survival [26]. Bu treatment is one of the most widely used conditioning therapies and Bu resistance is a problem in HSCT. As a bifunctional alkylating agent, Bu is known to cause DNA intra- and inter-strand crosslinks [27, 28]. The molecular mechanisms of its cytotoxic action, and more importantly, potential mechanisms underlying the subsequent emergence of tumor-cell resistance to this agent, are not well understood. We hypothesize that a better understanding of genetically determined cellular resistance mechanisms to alkylating agents will forward our ability to control the malignancy/leukemia. While we recognize that resistance to DNA-alkylating agents might be multifactorial, and cross-resistance to various cytotoxic agents is possible, it is important to understand the cellular mechanisms involved in conferring phenotypic resistance. It might be possible to manipulate such mechanisms to increase therapeutic response. Alternatively, relevant surrogate resistance markers can be used to follow the development of resistant cell populations over time and alternative treatment modalities can also be offered to patients carrying genetic markers of drug resistance.
To dissect the genetic mechanism(s) responsible for cellular Bu resistance we established cell-line models through intermittent exposures to incrementally increasing drug concentrations of the myeloid leukemia cell lines B5 and KBM3 [10, 12]). This approach has been extensively used to induce cellular drug resistance to DNA-alkylating agents including melphalan [29, 30], cyclophosphamide [31], and 2-chloroethyl-N-nitrosourea [32]. This procedure resulted in cell lines with clinically relevant degrees of Bu resistance compared with the parent lines (Figure 1). Based on our results and previously published findings for other DNA-alkylating agents [9, 33], it appears likely that cellular resistance to Bu is multifactorial. In this report we focused on the role of altered apoptosis and the up-regulated expression of STAT3 and HSP90 as causative factors in the emergence of Bu resistance.
Our data suggest that Bu causes a G2 cell cycle arrest and apoptosis in these myeloid leukemia cells (Figure 1C), consistent with previous reports [16, 34]. However, the magnitude of the G2 arrest and the apoptotic response was much reduced in both of the Bu-resistant cell lines compared with their parental counterparts. Thus, a more dramatic increase in the G2/G1 ratio was observed in the parental cells compared to their Bu-resistant sublines when cells were exposed to 180–260 µg/ml Bu (Figure 1D). We reasoned that the ability of Bu-resistant cells to evade Bu-mediated G2-arrest and cell death might be due to up-regulated expression of CDC2, abrogation of its phosphorylation at Thr14 and Tyr15, and/or differential expression of genes involved in mediating apoptosis. Exposure of sensitive cells to Bu resulted in a more dramatic increase in P-CHK2(Thr68) and P-CDC2(Tyr15) relative to mocktreated control. These two proteins are critical in controlling the G2 phase of cell cycle. Phosphorylation at Thr68 activates CHK2 kinase activity which inhibits CDC25 phosphatase activity resulting in a higher level of inactive P-CDC2(Tyr15) and ultimately in G2-arrest. In contrast, exposure of resistant cells to Bu resulted in a less significant change in the phosphorylation of these two proteins (Figure 1E). These results suggest that low expression and increased phosphorylation of CDC2 are associated with Buinduced G2 cell cycle arrest of Bu-sensitive cells which might be followed by apoptosis, whereas the upregulated expression of CDC2 and its lower phosphorylation may allow Bu-resistant cells to exit from G2-arrest and enter mitosis, thereby evading apoptosis. Indeed, this phenotype is most likely mediated by the observed constitutive down-regulation of pro-apoptotic genes including BNIP3, BIK, LTBR and TNFSF7 and up-regulation of anti-apoptotic genes including BCL2, BCL-XL (or BCL2L1), BAG3, BCL2L10 and IAP2/BIRC3 in the Bu-resistant cells (Table 2 and Figure 3).
The Bu-mediated signal transduction events that lead to apoptosis of B5 and KBM3 cells include the activation of caspases 3 and 8, eventually leading to the cleavage of PARP1 and DNA fragmentation. Such activation is less marked in resistant B5/Bu2506 and KBM3/Bu2506 cells (Figure 2), substantiating the observation that these cells acquired the ability to evade apoptosis. The inability of the caspase inhibitor Z-VAD-FMK to completely prevent apoptosis in B5 and KBM3 cells (Figures 2B and 2C) suggests that Bu induces apoptosis through additional mechanism(s) in these cells. In fact, the phosphorylation of H2AX is not completely inhibited by Z-VAD-FMK (data not shown) suggesting the presence of DNA nicks or cleavages probably mediated by mechanisms other than caspase activation.
A possible contributing mechanism to explain how Bu-resistant cells evade Bu-mediated toxicity is through up-regulation of stress response genes. The ability of cells to tolerate stressful conditions contributes to cellular drug resistance [35, 36]. One such stress factor is the HSP90 protein, which catalyzes proper folding of some pro-survival proteins, and was substantially up-regulated in the Bu-resistant cells (Figure 4A). Furthermore, inhibition of HSP90 activity by GA rendered B5/Bu2506 and KBM3/Bu2506 cells more sensitive to Bu as demonstrated by an increase in the G2/G1 ratio, increase of cells in sub-G1, increased cleavage of PARP1, and a decreased survival (Figure 4). HSP90 client proteins include AKT, RAF1, BCR-ABL, RIP, JAKs and cyclin dependent kinases, all of which are involved in cell survival and apoptosis [37, 38]. We have identified STAT3 signaling as a possible HSP90-dependent pathway (Figure 4E) which contributes to Bu resistance and myeloid leukemia cell survival. These results suggest that Janus kinases (JAKs), which phosphorylate STAT3, are dependent on HSP90 activity, and this observation is consistent with a previous report on the interaction of JAK and HSP90 [39]. Other specific HSP90 client proteins that might be of importance for the development of Bu resistance remain to be identified.
An interesting observation here is the up-regulated expression of the cyclin-dependent kinase inhibitor 1A (p21/CDKN1A) gene, which encodes the p21 or CIP1 protein, in B5/Bu2506 and KBM3/Bu2506 cells (Figure 3). The lower level of expression of p53 in B5/Bu2506 cells compared with B5 cells (Figure 3) suggests that the expression of the p21/CDKN1A gene is p53-independent in this cell line, although a different scenario was apparent in the KBM3 cell lines where both p21 and p53 were constitutively up-regulated in the Bu-resistant cells. P21 protein is an important effector of cell cycle arrest in response to DNA damage [40, 41]. Moreover, the anti-apoptotic activity of p21 is attributed to its binding with apoptosis signal-regulating kinase 1 (ASK-1) in the cytoplasm, which consequently down-regulates the stress-induced MAPK cascade and mediates cellular resistance to genotoxic agentinduced apoptosis [42, 43]. Up-regulated p21 also has been observed in other drug-resistant leukemia and breast cancer cell lines [44, 45], but the exact mechanism of this effect is unclear.
In conclusion, we have established human myeloid leukemia cell line models of clinically relevant (i.e., “low-degree”) and stable Bu resistance that appears multifactorial. We have determined expression levels of possible genetic biomarkers for Bu resistance related to alteration of the apoptotic pathways. Moreover, the similarities in altered gene expression between the experimental model and the clinically-derived samples are intriguing and support the clinical relevance of our model which will be used to investigate the molecular mechanism(s) of Bu resistance. Although the small number of patient samples in the present study warrants the need for a prospective study of cells from a larger patient population, our results suggest a possible relevance for gene expression profiling as part of HSCT-pretreatment planning, ultimately leading to individualized pre-transplant conditioning programs in AML. For example, up-regulation of the HSP90 and STAT3 genes in clinically resistant AML, similar to their high levels of expression in B5/Bu2506 and KBM3/Bu2506 cells, would suggest that GA or a small molecule analog thereof and an inhibitor of STAT3 activation could be combined with Bu to obtain synergistic cytotoxicity in myeloablative treatment. Similarly, manipulation of the apoptotic signaling mechanisms using antisense oligonucleotides, BH3-peptides and small molecular weight chemicals combined with a bifunctional alkylating agent such as Bu might provide a more effective treatment modality for high-risk leukemia patients. If Bu resistance gene markers are established in cells from high-risk AML patients, alternative treatment strategies could be assigned based on putative collateral sensitivity patterns.
Acknowledgements
This study was supported by NIH grants P01 CA055164 and CCSG Core CA16672, and The Stephen L. and Lavinia Boyd Fund for Leukemia Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributions. B. Valdez contributed to the concept and design, and drafted the manuscript. D. Murray helped in experimental design, data interpretation and writing the manuscript. L. Ramdas did the initial genetic screening and analysis of results. M. de Lima, R. Jones and R. Champlin provided expertise on the interpretation and analysis of results and selection of patient samples. S. Kornblau provided the patient cell samples. D. Betancourt and Y. Li provided technical support. B. Andersson, the corresponding author, was responsible for the research approach, funding, analysis of data, and final version of the article.
References
- 1.Jones RJ, Lee KS, Beschorner WE, Vogel VG, Grochow LB, Braine HG, Vogelsang GB, Sensenbrenner LL, Santos GW, Saral R. Venoocclusive disease of the liver following bone marrow transplantation. Transplantation. 1987;44:778–783. doi: 10.1097/00007890-198712000-00011. [DOI] [PubMed] [Google Scholar]
- 2.Bhagwatwar HP, Phadungpojna S, Chow DS, Andersson BS. Formulation and stability of busulfan for intravenous administration in high-dose chemotherapy. Cancer Chemother Pharmacol. 1996;37:401–408. doi: 10.1007/s002800050404. [DOI] [PubMed] [Google Scholar]
- 3.Andersson BS, Kashyap A, Gian V, Wingard JR, Fernandez H, Cagnoni PJ, Jones RB, Tarantolo S, Hu WW, Blume KG, Forman SJ, Champlin RE. Conditioning therapy with intravenous busulfan and cyclophosphamide (IV BuCy2) for hematologic malignancies prior to allogeneic stem cell transplantation: a phase II Study. Biol Blood Marrow Transplant. 2002;8:145–154. doi: 10.1053/bbmt.2002.v8.pm11939604. [DOI] [PubMed] [Google Scholar]
- 4.Kashyap A, Wingard J, Cagnoni P, Roy J, Tarantolo S, Hu W, Blume K, Niland J, Palmer JM, Vaughan W, Fernandez H, Champlin R, Forman S, Andersson BS. Intravenous versus oral busulfan as part of a busulfan/cyclophosphamide preparative regimen for allogeneic hematopoietic stem cell transplantation: decreased incidence of hepatic venoocclusive disease (HVOD), HVOD-related mortality, and overall 100-d mortality. Biol Blood Marrow Transplant. 2002;8:493–500. doi: 10.1053/bbmt.2002.v8.pm12374454. [DOI] [PubMed] [Google Scholar]
- 5.de Lima M, Couriel D, Thall PF, Wang X, Madden T, Jones R, Shpall EJ, Shahjahan M, Pierre B, Giralt S, Korbling M, Russell JA, Champlin RE, Andersson BS. Once-daily intravenous busulfan and fludarabine: clinical pharmacokinetic results of a myeloablative, reduced-toxicity conditioning regiment for allogeneic stem cell transplantation in AML and MDS. Blood. 2004;104:857–864. doi: 10.1182/blood-2004-02-0414. [DOI] [PubMed] [Google Scholar]
- 6.de Lima M, Wang X, Thall PF, Couriel D, Jones RB, Shpall EJ, Popat U, Giralt S, Korbling M, Champlin RE, Andersson BS. Long-term follow-up of IV busulfan (Bu) with fludarabine (Flu) vs IV Bu with cyclophosphamide (Cy) as pre (allogeneic) transplant conditioning therapy for AML/MDS. Blood. 2006;108(11):322. [Google Scholar]
- 7.Allay JA, Dumenco LL, Koc ON, Liu L, Gerson SL. Retroviral transduction and expression of the human alkyltransferase cDNA provides nitrosourea resistance to hematopoietic cells. Blood. 1995;85:3342–3351. [PubMed] [Google Scholar]
- 8.Sládek NE, Kollander R, Sreerama L, Kiang DT. Cellular levels of aldehyde dehydrogenases (ALDH1A1 and ALDH3A1) as predictors of therapeutic responses to cyclophosphamide-based chemotherapy of breast cancer: a retrospective study. Rational individualization of oxazaphos-phorine-based cancer chemotherapeutic regimens. Cancer Chemother Pharmacol. 2002;49:309–321. doi: 10.1007/s00280-001-0412-4. [DOI] [PubMed] [Google Scholar]
- 9.Andersson BS, Murray D. Model studies of cyclophosphamide resistance in human myeloid leukemia. Cancer Treat Res. 2002;112:211–235. doi: 10.1007/978-1-4615-1173-1_11. [DOI] [PubMed] [Google Scholar]
- 10.Andersson BS, Collins VP, Kurzrock R, Larkin DW, Childs C, Ost A, Cork A, Trujillo JM, Freireich EJ, Siciliano MJ, et al. KBM-7, a human myeloid leukemia cell line with double Philadelphia chromosomes lacking normal c-ABL and BCR transcripts. Leukemia. 1995;9:2100–2108. [PubMed] [Google Scholar]
- 11.Munker R, Zhao S, Jiang S, Snell V, Andreeff M, Andersson BS. Further characterization of cyclophosphamide resistance: expression of CD95 and of bcl-2 in a CML cell line. Leuk Res. 1998;22:1073–1077. doi: 10.1016/s0145-2126(98)00093-9. [DOI] [PubMed] [Google Scholar]
- 12.Andersson BS, Bergerheim US, Collins VP, Childs C, Beran M, Sen S, Arden K, Pathak S, Siciliano MJ, Ost A, et al. KBM-3, an in vitro model of human acute myelomonocytic leukemia. Exp Hematol. 1992;20:361–367. [PubMed] [Google Scholar]
- 13.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 14.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 15.Millar BC, Tilby MJ, Ormerod MG, Payne AWR, Jinks S, Loverock PS. Comparative studies of total cross-linking, cell survival and cell cycle perturbations in Chinese hamster cells treated with alkylating agents in vitro. Biochem Pharmacol. 1986;35:1163–1169. doi: 10.1016/0006-2952(86)90155-3. [DOI] [PubMed] [Google Scholar]
- 16.Ritter CA, Sperker B, Grube M, Dressel D, Kunert-Keil C, Kroemer HK. Overexpression of glutathione S-transferase A1-1 in ECV 304 cells protects against busulfan mediated G2-arrest and induces tissue factor expression. Br J Pharmacol. 2002;137:1100–1106. doi: 10.1038/sj.bjp.0704972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norbury C, Nurse P. Animal cell cycles and their control. Ann Rev Biochem. 1992;61:441–470. doi: 10.1146/annurev.bi.61.070192.002301. [DOI] [PubMed] [Google Scholar]
- 18.Krek W, Nigg EA. Mutations of p34cdc2 phosphorylation sites induce premature mitotic events in HeLa cells: Evidence for a double block to p34cdc2 kinase activation in vertebrates. EMBO J. 1991;10:3331–3341. doi: 10.1002/j.1460-2075.1991.tb04897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlsson-Rosenthal C, Millar JB. Cdc25: mechanisms of checkpoint inhibition and recovery. Trends Cell Biol. 2006;16:285–292. doi: 10.1016/j.tcb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Reed JC. Apoptosis mechanisms: implications for cancer drug discovery. Oncology (Williston Park) 2004;18:11–20. [PubMed] [Google Scholar]
- 21.Hail N, Jr, Carter BZ, Konopleva M, Andreeff M. Apoptosis effector mechanisms: A requiem performed in different keys. Apoptosis. 2006;11:889–904. doi: 10.1007/s10495-006-6712-8. [DOI] [PubMed] [Google Scholar]
- 22.Probin V, Wang Y, Bai A, Zhou D. Busulfan selectively induces cellular senescence but not apoptosis in WI38 fibroblasts via a p53-independent but extracellular signal-regulated kinase-p38 mitogen-activated protein kinase-dependent mechanism. J Pharm Expt Ther. 2006;310:551–560. doi: 10.1124/jpet.106.107771. [DOI] [PubMed] [Google Scholar]
- 23.Neckers L. Chaperoning oncogenes: Hsp90 as a target of geldanamycin. Handbk Exp Pharmacol. 2006;172:259–277. doi: 10.1007/3-540-29717-0_11. [DOI] [PubMed] [Google Scholar]
- 24.Stancato LF, Silverstein AM, Owens-Grillo JK, Chow YH, Jove R, Pratt WB. The hsp90-binding antibiotic geldanamycin decreases Raf levels and epidermal growth factor signaling without disrupting formation of signaling complexes or reducing the specific enzymatic activity of Raf kinase. J Biol Chem. 1997;272:4013–4020. doi: 10.1074/jbc.272.7.4013. [DOI] [PubMed] [Google Scholar]
- 25.Chatterjee M, Jain S, Stuhmer T, Andrulis M, Ungethum U, Kuban RJ, Lorentz H, Bommert K, Topp M, Kramer D, Muller-Hermelink HK, Einsele H, Greiner A, Bargou RC. STAT3 and MAPK signaling maintain overexpression of heat shock proteins 90alpha and beta in multiple myeloma cells, which critically contribute to tumor-cell survival. Blood. 2007;109:720–728. doi: 10.1182/blood-2006-05-024372. [DOI] [PubMed] [Google Scholar]
- 26.Shimoni A, Hardan I, Shem-Toy N, Yeshurun M, Yerushalmi R, Avigdor A, Ben-Bassat I, Nagler A. Allogeneic hematopoietic stem-cell transplantation in AML and MDS using myeloablative versus reduced-intensity conditioning: the role of dose intensity. Leukemia. 2006;20:322–328. doi: 10.1038/sj.leu.2404037. [DOI] [PubMed] [Google Scholar]
- 27.Tong WP, Ludlum DB. Crosslinking of DNA by busulfan. Formation of diguanyl derivatives. Biochim Biophys Acta. 1980;608:174–181. doi: 10.1016/0005-2787(80)90145-8. [DOI] [PubMed] [Google Scholar]
- 28.Iwamoto T, Hiraku Y, Oikawa S, Mizutani H, Kojima M, Kawanishi S. DNA intrastrand cross-link at the 5’-GA-3’ sequence formed by busulfan and its role in the cytotoxic effect. Cancer Sci. 2004;95:454–458. doi: 10.1111/j.1349-7006.2004.tb03231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellamy WT, Dalton WS, Gleason MC, Grogan TM, Trent JM. Development and characterization of a melphalan-resistant human multiple myeloma cell line. Cancer Res. 1991;51:995–1002. [PubMed] [Google Scholar]
- 30.Harada N, Nagasaki A, Hata H, Matsuzaki H, Matsuno F, Mitsuya H. Down-regulation of CD98 in melphalan-resistant myeloma cells with reduced drug uptake. Acta Haematol. 2000;103:144–151. doi: 10.1159/000041037. [DOI] [PubMed] [Google Scholar]
- 31.Andersson B, Mroue M, Britten RA, Murray D. The role of DNA damage in the resistance of human chronic myeloid leukemia cells to cyclophosphamide analogues. Cancer Res. 1994;54:5394–5400. [PubMed] [Google Scholar]
- 32.Tano K, Dunn WC, Darroudi F, Shiota S, Preston RJ, Natarajan AT, Mitra S. Amplification of the DNA repair gene O6-methylguanine-DNA methyltransferase associated with resistance to alkylating drugs in a mammalian cell line. J Biol Chem. 1997;272:13250–13254. doi: 10.1074/jbc.272.20.13250. [DOI] [PubMed] [Google Scholar]
- 33.Jones RJ. Clinical pharmacology of melphalan and its implications for clinical resistance to anti-cancer agents. Cander Treat Res. 2002;112:305–322. doi: 10.1007/978-1-4615-1173-1_15. [DOI] [PubMed] [Google Scholar]
- 34.Hassan Z, Hassan M, Hellstrom-Lindberg E. The pharmacodynamic effect of busulfan in the p39 myeloid cell line in vitro. Leukemia. 2001;15:1240–1247. doi: 10.1038/sj.leu.2402193. [DOI] [PubMed] [Google Scholar]
- 35.Dent P, Yacoub A, Contessa J, Caron R, Amorino G, Valerie K, Hagan MP, Grant S, Schmidt-Ullrich R. Stress and radiation-induced activation of multiple intracellular signaling pathways. Radiat Res. 2003;159:283–300. doi: 10.1667/0033-7587(2003)159[0283:sariao]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 36.Grant S, Fisher PB, Dent P. The role of signal transduction pathways in drug and radiation resistance. Cancer Treat Res. 2002;112:89–108. doi: 10.1007/978-1-4615-1173-1_5. [DOI] [PubMed] [Google Scholar]
- 37.Miyata Y. Hsp90 inhibitor geldanamycin and its derivatives as novel cancer chemotherapeutic agents. Curr Pharm Des. 2005;11:1131–1138. doi: 10.2174/1381612053507585. [DOI] [PubMed] [Google Scholar]
- 38.Georgakis GV, Li Y, Rassidakis GZ, Martinez-Valdez H, Medeiros LF, Younes A. Inhibition of heat shock protein 90 function by 17-allylamino-17-demethoxy-geldanamycin in Hodgkin’s lymphoma cells down-regulates Akt kinase, dephosphorylates extracellular signal-regulated kinase, and induces cell cycle arrest and cell death. Clin Cancer Res. 2006;12:584–590. doi: 10.1158/1078-0432.CCR-05-1194. [DOI] [PubMed] [Google Scholar]
- 39.Shang L, Tomasi TB. The heat shock protein 90-CDC37 chaperone complex is required for signaling by types I and II interferons. J Biol Chem. 2006;281:1876–1884. doi: 10.1074/jbc.M509901200. [DOI] [PubMed] [Google Scholar]
- 40.Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 41.Stivala LA, Riva F, Cazzalini O, Savio M, Prosperi E. p21waf1/cip1-null human fibroblasts are deficient in nucleotide excision repair downstream the recruitment of PCNA to DNA repair sites. Oncogene. 2001;20:563–570. doi: 10.1038/sj.onc.1204132. [DOI] [PubMed] [Google Scholar]
- 42.Asada MY, Yamada T, Ichijo H, Delia D, Miyazono K, Fukumuro K, Mizutani S. Apoptosis inhibitory activity of cytoplasmic p21 (Cip1/WAF1) in monocytic differentiation. EMBO J. 1999;18:1223–1234. doi: 10.1093/emboj/18.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou BP, Li Y, Hung MC. HER-2/Neu signaling and therapeutic approaches in breast cancer. Breast Dis. 2002;15:13–24. doi: 10.3233/bd-2002-15103. [DOI] [PubMed] [Google Scholar]
- 44.Cory AH, Somerville L, Cory JG. Blockage of cyclin cdk’s, PKC and phosphoinositol 3-kinase pathways leads to augmentation of apoptosis in drug-resistant leukemia cells: evidence for interactive effects of flavopiridol, LY294002, roscovitine, wortmannin and UCN-01. Anticancer Res. 2005;25:101–106. [PubMed] [Google Scholar]
- 45.Menendez JA, Mehmi I, Lupu R. Heregulin-triggered Her-2/neu signaling enhances nuclear accumulation of p21WAF1/CIP1 and protects breast cancer cells from cisplatin-induced genotoxic damage. Int J Oncol. 2005;26:649–659. [PubMed] [Google Scholar]