Abstract
The present study describes a method using immunohistochemical labeling in combination with high-resolution imaging (field emission scanning electron microscopy) to investigate the spatial localization of amelogenins on apatite crystallites in developing porcine enamel. Cross-sections of developing enamel tissue from freeze-fractured pig third molar were treated with antiserum against recombinant mouse amelogenin and immunoreactivity confirmed by Western blot analysis. The samples were then treated with the goat anti-rabbit IgG conjugated with 10-nm gold particles. The control samples were treated with the secondary antibody only. The in-lens secondary electrons detector and quadrant back-scattering detector were employed to reveal the high-resolution morphology of enamel structures and gold particle distribution. The immunolabeling showed a preference of the gold particle localization along the side faces of the ribbon-like apatite crystals. The preferential localization of amelogenin in vivo on enamel crystals strongly supports its direct function in controlling crystal morphology.
Key Words: Amelogenin, Developing porcine enamel, Immunogold labeling, Field emission scanning electron microscopy, Biomineralization
Introduction
As documented by a series of in vitro and in vivo studies, the supramolecular assembly of enamel matrix proteins plays a pivotal role in the regulation of enamel biomineralization [Fincham et al., 1999; Paine et al., 2000; Gibson et al., 2001; Lyngstadaas, 2001; Kim et al., 2006; Margolis et al., 2006]. In vitro studies have indicated that amelogenins self-assemble into nanosphere structures under various conditions [Fincham et al., 1999] and that these nanospheres can further assemble into higher-order structures such as short chain-like arrays [Du et al., 2005; Margolis et al., 2006; Moradian-Oldak et al., 2006]. Observations on developing dental enamel by transmission electron microscopy (TEM) [Robinson et al., 1981; Fincham et al., 1995] have revealed linear arrays of nanometer-sized spherical particles, although the exact nature of these structures is still unclear. In in vitro crystallization systems, amelogenins increased aspect ratio (length/width) of octacalcium phosphate (OCP) crystals in a dose-dependent manner, suggesting a favorable adsorption of amelogenin nanospheres onto (010) side faces of OCP [Wen et al., 2000; Iijima and Moradian-Oldak, 2004]. Furthermore, the nanoparticles of recombinant human amelogenin aligned into short strings along the c-axis of fluoroapatite crystals and altered the growth of different crystal faces [Habelitz et al., 2004]. Wallwork et al. [2001] have shown that recombinant amelogenin nanospheres adsorbed more favorably to the side faces of the extracted enamel crystals in vitro.
Immunogold labeling together with TEM have provided high-resolution images elucidating the intra- and extracellular distribution of enamel matrix proteins [Nanci et al., 1987; Shore et al., 1995; Nanci et al., 1998]. However, the spatial relationship and localization of enamel matrix proteins on apatite crystallites at a molecular level, as well as the spatial organization of the molecular assembly in the enamel matrix, remain obscure. The present study describes a method using immunohistochemical labeling in combination with high-resolution imaging [field emission scanning electron microscopy (FESEM)] to investigate the spatial localization of amelogenins on apatite crystallites in developing porcine enamel.
Materials and Methods
Tissue Preparation
The unerupted third mandibular molars were dissected from 6-month-old pig jaws obtained through Sierra for Medical Sciences (Santa Fe Springs, Calif., USA). The cusps were washed in cold saline and freeze fractured. The cross-section of developing enamel tissue was collected on the conductive tape.
Raman Spectroscopy
Raman spectra were collected using a Renishaw Micro Raman System RM1000 (Renishaw plc, Wotton-under-Edge, UK) equipped with an Ar+ (514.5 nm) laser.
Enamel Protein Extraction
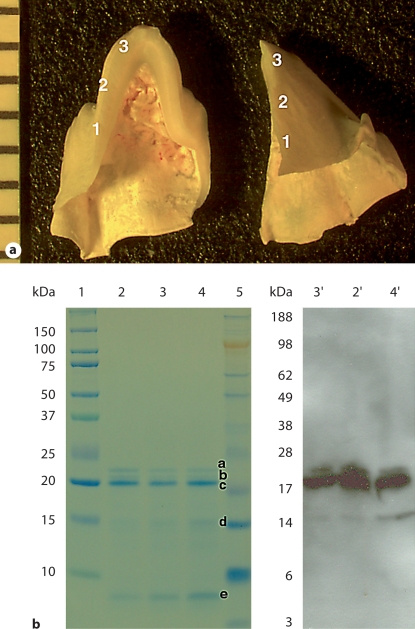
For biochemical analysis, the enamel tissue was roughly divided into 3 zones (approx. 2 mm in length each) in the direction cervical margin to cuspal edge, as shown in figure 1a. The pooled scrapings were dissolved in 10% acetic acid and protein extracts were prepared as previously reported [Fincham et al., 1994].
Fig. 1.
a Photograph of a freeze-fractured tooth cusp from a third molar crown of a 6-month-old pig. For biochemical analysis, the enamel tissue was roughly divided into 3 zones (approx. 2 mm in length each) in the direction cervical margin to cuspal edge. b SDS-PAGE pattern (left) and Western blots (right) of acetic acid extract of enamel tissue scrapings from the 3 zones. Lane 1: Bio-Rad Precision Plus standard proteins. Lane 2 and 2′: zone 1, cervical portion. Lane 3 and 3′: zone 2, middle portion. Lane 4 and 4′: zone 3, cuspal portion. Lane 5: Invitrogen SeeBlue Plus2 standard proteins. a–e = Various porcine amelogenin components, presumably 25, 23, 20, 13 kDa and tyrosine-rich amelogenin peptide (TRAP), respectively.
SDS-PAGE and Western Blots
SDS-PAGE was performed with Nupage™ 12% Bis-Tris gel (Invitrogen, Carlsbad, Calif., USA). The molecular weights of protein bands were estimated by using Precision Plus standard proteins (Bio-Rad, Hercules, Calif., USA) or SeeBlue® Plus2 standard proteins (Invitrogen). For Western blots, proteins were transferred to PVDF membranes. Blocking was performed with 2.5% nonfat milk, followed by incubation with a rabbit antiserum against recombinant mouse amelogenin (rM179) [Simmer et al., 1994] at a dilution of 1:1,000. The secondary antibody was goat anti-rabbit IgG conjugated with alkaline phosphatase at a dilution of 1:4,000, and chemiluminescent detection was performed with the Immun-Star AP Chemiluminescent Kits (Bio-Rad).
Immunogold Labeling
The samples on the conductive tape were fixed in 1% paraformaldehyde. After thorough washing, blocking was performed with 0.2% nonfat milk, followed by incubation for 1–2 h with antiserum against rM179 with a dilution of 1:100. The samples were thoroughly washed in TTBS and incubated with the secondary antibody, the goat anti-rabbit IgG conjugated with 10 nm gold particles. The control samples were treated with the secondary antibody only. The samples were thoroughly washed in TTBS, followed by a brief wash in deionized water and air dried.
FESEM Observation
The samples were carbon coated for evaluation under LEO 1550VP FESEM. The in-lens secondary electrons (SE) detector and quadrant backscattering detector were employed to reveal the high-resolution morphology of enamel structures and the gold particles distribution. Images were also recorded by mixing both backscattered electron (BSE) and SE signals.
Results
The unerupted third mandibular molar enamel of the 6-month-old pig is in the stage of matrix formation [Robinson et al., 1987]. The acetic acid extracts of the 3 enamel tissue fractions from cervical margin to cuspal edge had a similar amelogenin protein profile (fig. 1b). The predominant component was the ‘20K’ amelogenin [Fincham et al., 1994], with small amounts of full-length (‘25K’) and another proteolytic product (‘23K’) amelogenins [Yamakoshi et al., 1994], as well as low-molecular-weight protein bands around 13 and 5 kDa. On Western blots, the anti-rM179 serum strongly reacted with 20K, 23K and 25K amelogenins, and recognized the 13-kDa band (fig. 1b). In addition, there was a slight decrease in the relative amount of the higher-molecular-weight amelogenins and an increase in that of the lower-molecular-weight components from cervical margin to cuspal edge. The mineral phase was characterized as carbonated apatite from Raman spectra with the main PO43– stretch at 957 cm–1 and a complex of CO32– and PO43– at 1,068 cm–1 (data not shown).
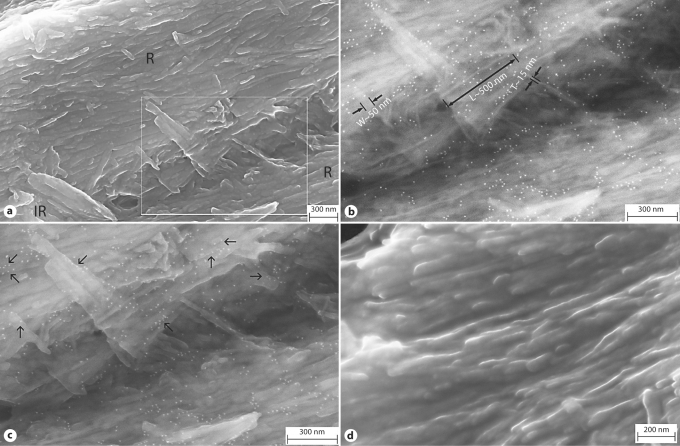
The high-resolution FESEM micrographs of the enamel tissue after immunogold labeling are shown in figure 2a–c. Figure 2d shows the control sample that omitted the incubation with anti-rM179 serum. The images were taken from areas within the middle portion (zone 2 in fig. 1a). The images using SE revealed the high-resolution features of enamel structures (fig. 2a and d). Figure 2a shows the rods, the boundary between the rods and pieces of interrod structures. The crystals appeared as highly oriented parallel ribbons within either rod or interrod structures. The gold particles were discernable on the surface of the crystal ribbons. Using the BSE signals, the strong elementary contrast highlighted the gold particles as spherical white spots of 10 nm diameter and the calcium phosphate minerals as thin long ribbons (fig. 2b). Systematic measurements of crystal dimensions from the BSE images (fig. 2b) gave a thickness of 14 ± 3 nm (n = 13) and a width of 44 ± 10 nm (n = 10). The length of the ribbons had a large variety. The measurement of several relatively long ribbons gave the length of 425 ± 197 nm (n = 7). It should be noted, however, that the maximum length of the crystals could not be established with certainty because the long crystal could be fractured during the sample preparation or deeply embedded in the tissue. Images recorded by mixing both SE and BSE signals overlay the gold nanoparticle distribution on the enamel features (fig. 2c). As indicated in figure 2c, the gold nanoparticles showed a strong preference to localize close to the edge or side faces of the mineral ribbons rather than the plate side (arrows in fig. 2c). It appeared that a group of gold nanoparticles gave a short chain-like appearance in the local vicinity. Gold particles were rarely found on the control sample (fig. 2d).
Fig. 2.
FESEM micrographs of the enamel tissue after immunogold labeling. a SE image. b BSE image of the enlargement of the square area in a. c Image recorded by mixing both SE and BSE signals. d SE image of a control sample without the anti-rM179 serum incubation. R = Rod; IR = interrod. Arrows highlight the localization of gold nanoparticles close to the edge or side face of the mineral ribbons.
Discussion
We have successfully developed a technique to reveal the spatial localization of amelogenins on apatite crystals in the developing enamel tissue. Based on the chemical composition, the enamel tissue of the third mandibular molar of porcine was in the stage of matrix formation from 3 up to 50 weeks of the animal's age [Robinson et al., 1987]. Our results of SDS-PAGE and Western blot analysis on samples from the 6-month-old animal further showed a subtle progressive difference in the relative amount of protein components along the length of the cusp. The subsequent studies were therefore performed on the middle portion of the cusp to focus on a defined stage of enamel matrix formation.
The preliminary immunogold labeling using the anti-rM179 serum showed gold particle localization along the side faces of the ribbon-like apatite crystals. It is noteworthy that the anti-amelogenin antibody used in the present study does not cross-react with nonamelogenin proteins [Simmer et al., 1994]. It has been shown that the lower- molecular-weight amelogenin, such as the 13-kDa fragment, mainly resided in enamel fluid as soluble moiety [Aoba et al., 1987]. The observed immunogold labeling on the crystal ribbons thus mostly represented the distribution of the higher-molecular-weight amelogenins (20–25 kDa). Our in vivo finding corroborates with previous in vitro atomic force microscopy observation, suggesting a more favorable binding of a full-length recombinant mouse amelogenin to the side faces of the extracted enamel crystals in the form of nanosphere assembly [Wallwork et al., 2001]. Those authors suggested an electrostatic interaction mechanism between the protein and the charge domains on the crystal surface [Kirkham et al., 2000]. In other studies, the morphology changes of OCP crystals grown in amelogenin gel-like matrix from ribbon-like to rod-like, with a significant reduction in the width, was attributed to the hydrophobic interaction between the amelogenin and the (010) face [Iijima and Moradian-Oldak, 2004]. Although the current observation cannot resolve the exact mechanism, it provides direct evidence of a preferential localization of amelogenins on the developing enamel crystals in vivo.
In summary, FESEM together with immunogold labeling is a powerful tool to provide high-resolution images of enamel structures with the information of molecular localization of enamel proteins. Our observation regarding a preferential localization of amelogenin in vivo with respect to enamel crystals strongly supports its direct function in controlling crystal morphology.
Acknowledgements
We are grateful to Professor George Rossman from the Division of Geological and Planetary Sciences, California Institute of Technology, Pasadena, Calif., USA, for access to Raman spectroscopy. This study was supported by NIH-NIDCR grants (DE-13414, E-15644).
Abbreviations used in this paper
- BSE
backscattered electron
- FESEM
field emission scanning electron microscopy
- OCP
octacalcium phosphate
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SE
secondary electron
- TEM
transmission electron microscopy
References
- Aoba T., Tanabe T., Moreno E.C. Proteins in the enamel fluid of immature porcine teeth. J Dent Res. 1987;66:1721–1726. doi: 10.1177/00220345870660120501. [DOI] [PubMed] [Google Scholar]
- Boyde A. The development of enamel structure. Proc R Soc Med. 1967;60:923–928. doi: 10.1177/003591576706000965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C., Falini G., Fermani S., Abbott C., Moradian-Oldak J. Supramolecular assembly of amelogenin nanospheres into birefringent microribbons. Science. 2005;307:1450–1454. doi: 10.1126/science.1105675. erratum in Science 309: 2166. [DOI] [PubMed] [Google Scholar]
- Fincham A.G., Moradian-Oldak J., Sarte P.E. Mass-spectrographic analysis of a porcine amelogenin identifies a single phosphorylated locus. Calcif Tissue Int. 1994;55:398–400. doi: 10.1007/BF00299322. [DOI] [PubMed] [Google Scholar]
- Fincham A.G., Moradian-Oldak J., Diekwisch T.G., Lyaruu D.M., Wright J.T., P. Bringas, Jr., H.C. Slavkin. Evidence for amelogenin ‘nanospheres’ as functional components of secretory-stage enamel matrix. J Struct Biol. 1995;115:50–59. doi: 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- Fincham A.G., Moradian-Oldak J., Simmer J.P. The structural biology of the developing dental enamel matrix. J Struct Biol. 1999;126:270–299. doi: 10.1006/jsbi.1999.4130. [DOI] [PubMed] [Google Scholar]
- Gibson C., Yuan Z.A., Hall B., Longenecker G., Chen E., Thyagarajan T., Sreenath T., Wright J.T., Decker S., Piddington R., Harrison G., Kulkarni A.B. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- Habelitz S., Kullar A., Marshall S.J., DenBesten P.K., Balooch M., Marshall G.W., Li W. Amelogenin-guided crystal growth on fluoroapatite glass-ceramics. J Dent Res. 2004;83:698–702. doi: 10.1177/154405910408300908. [DOI] [PubMed] [Google Scholar]
- Iijima M., Moradian-Oldak J. Interactions of amelogenins with octacalcium phosphate crystal faces are dose dependent. Calcif Tissue Int. 2004;74:522–531. doi: 10.1007/s00223-002-0011-3. [DOI] [PubMed] [Google Scholar]
- Kim J.W., Simmer J.P., Lin B.P.L., Seymen F., Bartlett J.D., Hu J.C.C. Mutational analysis of candidate genes in 24 amelogenesis imperfecta families. Eur J Oral Sci. 2006;114(suppl 1):3–12. doi: 10.1111/j.1600-0722.2006.00278.x. [DOI] [PubMed] [Google Scholar]
- Kirkham J., Zhang J., Brookes S.J., Shore R.C., Ryu O.H., Wood S.R., Smith D.A., Wallwork M.L., Robinson C. Evidence for charge domains on developing enamel crystal surfaces. J Dent Res. 2000;79:1943–1947. doi: 10.1177/00220345000790120401. [DOI] [PubMed] [Google Scholar]
- Lyngstadaas S.P. Synthetic hammerhead ribozymes as tools in gene expression. Crit Rev Oral Biol Med. 2001;12:469–478. doi: 10.1177/10454411010120060201. [DOI] [PubMed] [Google Scholar]
- Margolis H.C., Beniash E., Fowler C.E. Role of macromolecular assembly of enamel matrix proteins in enamel formation. J Dent Res. 2006;85:775–793. doi: 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J., Du C., Falini G. On the formation of amelogenin microribbons. Eur J Oral Sci. 2006;114(suppl 1):289–296. doi: 10.1111/j.1600-0722.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Nanci A., Slavkin H.C., Smith C.E. Application of high-resolution immunocytochemistry to the study of the secretory, resorptive, and degradative functions of ameloblasts. Adv Dent Res. 1987;1:148–161. doi: 10.1177/08959374870010020301. [DOI] [PubMed] [Google Scholar]
- Nanci A., Zalzal S., Lavoie P., Kunikata M., Chen W.Y., Krebsbach P.H., Yamada Y., Hammarström L., Simmer J.P., Fincham A.G., Snead M.L., Smith C.E. Comparative immunochemical analyses of the developmental expression and distribution of ameloblastin and amelogenin in rat incisors. J Histochem Cytochem. 1998;46:911–934. doi: 10.1177/002215549804600806. [DOI] [PubMed] [Google Scholar]
- Paine M.L., Zhu D.H., Luo W., P. Bringas, Jr., M. Goldberg, S.N. White, Y.P. Lei, M. Sarikaya, H.K. Fong, M.L. Snead. Enamel biomineralization defects result from alterations to amelogenin self-assembly. J Struct Biol. 2000;132:191–200. doi: 10.1006/jsbi.2000.4324. [DOI] [PubMed] [Google Scholar]
- Robinson C., Fuchs P., Weatherell J.A. The appearance of developing rat incisor enamel using a freeze fracturing technique. J Cryst Growth. 1981;53:160–165. [Google Scholar]
- Robinson C., Kirkham J., Weatherell J.A., Richards A., Josephsen K., Fejerskov O. Developmental stages in permanent porcine enamel. Acta Anat. 1987;128:1–10. doi: 10.1159/000146306. [DOI] [PubMed] [Google Scholar]
- Shore R.C., Robinson C., Kirkham J., Brookes S.J. Structure of developing enamel. In: Robinson C., Kirkham J., Shore R.C., editors. Dental Enamel: Formation to Destruction. Boca Raton: CRC Press; 1995. pp. 135–150. [Google Scholar]
- Simmer J.P., Lau E.C., Hu C.C., Aoba T., Lacey M., Nelson D., Zeichner-David M., Snead M.L., Slavkin H.C., Fincham A.G. Isolation and characterization of a mouse amelogenin expressed in Escherichia coli. Calcif Tissue Int. 1994;54:312–319. doi: 10.1007/BF00295956. [DOI] [PubMed] [Google Scholar]
- Wallwork M.L., Kirkham J., Zhang J., Smith D.A., Clarkson B.H., Brookes S.J., Shore R.C., Wood S.R., Ryu O., Robinson C. Binding of matrix proteins to developing enamel crystals: an atomic force microscopy study. Langmuir. 2001;17:2508–2513. [Google Scholar]
- Wen H.B., Moradian-Oldak J., Fincham A.G. Dose-dependent modulation of octacalcium phosphate crystal habit by amelogenins. J Dent Res. 2000;79:1902–1906. doi: 10.1177/00220345000790111501. [DOI] [PubMed] [Google Scholar]
- Yamakoshi Y., Tanabe T., Fukae M., Shimizu M. Porcine amelogenins. Calcif Tissue Int. 1994;54:69–75. doi: 10.1007/BF00316293. [DOI] [PubMed] [Google Scholar]