Abstract
Background
Little information is available on the immunological aspect of parasitic Gasterophilus intestinalis (Diptera, Oestridae) larvae causing horse gastric myiasis. The objectives of this research were to analyze the protein content of larval crude extracts of the migrating second and third larvae (L2 and L3) of G. intestinalis in order to characterize the immune response of horses.
Results
The proteomic profile of L2 and L3, investigated by using one and two dimensional approaches, revealed a migration pattern specific to each larval stage. Furthermore, Western blots were performed with horse sera and with sera of Balb/c mice immunised with the larval crude extracts of L2 or L3, revealing a different immune reaction in naturally infected horses vs. artificially induced immune reaction in mice. The comparisons of the immunoblot profiles demonstrate that the stage L2 is more immunogenic than the stage L3 most likely as an effect of the highest enzymatic production of L2 while migrating through the host tissues. Fifteen proteins were identified by mass spectrometry.
Conclusion
This work provides further information into the understanding of the interaction between G. intestinalis and their host and by contributing a novel scheme of the proteomic profile of the main larval stages.
Background
Nine species of Gasterophilus (Diptera, Oestridae) flies have been described causing, in the larval stage, gastrointestinal myiasis in equids. While Gasterophilus intestinalis (De Geer, 1776) and Gasterophilus nasalis (Linnaeus, 1758) are distributed worldwide and are often the only species reported in many parts of the New World, the remaining species are only reported in very limited areas of Europe, Eastern Countries [1] and Africa [2]. Adult bot flies deposit their eggs on the hosts' hair at different locations depending on the species of Gasterophilus [3]. G. pecorum is an exception as females lay their eggs on grass, leaves and stems of plants [1]. Infection occurs when eggs are introduced into horse mouth by animal licking and grooming. The first larval stage (L1) hatches, starts migrating and moulting into the second larval stage (L2) in the oral cavity [4]. Larvae of different species of Gasterophilus are specifically present in one or more regions of the gastrointestinal tract where the third larval stage (L3) remains attached to the mucosa for about 8–10 months [5]. The clinical signs associated with the migration and maturation stages of the larvae are difficult to diagnose, but it has been shown that different species of Gasterophilus can cause severe damages during their life cycle [6-9].
In the past years studies concerning the immunology and immunopathology of many oestrid myiasis causing larvae have increased because of their important implications in diagnostics and in immunisation programmes [10]. While immunological studies were mainly focused on Hypoderma cattle grub infection [11], and sheep nasal oestrosis by Oestrus ovis [12], the immunology of Gasterophilus spp. caused myiasis received little attention. This is also due to the inherent difficulties in studying immunological host-parasite interactions at the gastrointestinal mucosa interface. As a consequence, so far no major immunogens have been reported [13]. A single study discussed the development of antibodies for the diagnosis of myiasis by G. intestinalis larvae although the specificity of the immune reaction was not tested in the occurrence of concomitant horse parasitic infection [14]. More recently, many proteomics-based analyses, combined with two-dimensional gel-electrophoresis, have offered a comprehensive approach to better understand biological and immunological processes of pathogens and diseases [15-17]. The aim of this study was to characterize L2 and L3 proteins of G. intestinalis and to analyze the immune response of horses and immunized mice against larval antigens.
Results
1-D analysis of the larval crude extract (LCE) of L2 and L3
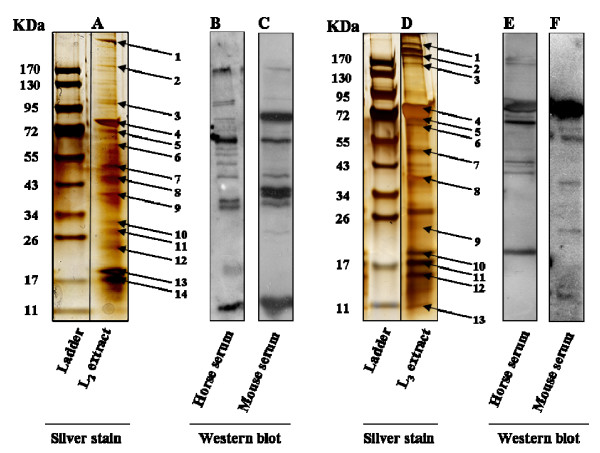
Migration of the LCE2 on the 1-D silver-stained gel showed a specific pattern (Figure 1A) with 14 bands that were isolated from the gel for further identification by mass spectrometry (MS) (Table 1). The selection of the bands was based upon the intensity of the band on the silver stained gel, as well as the immuno-reactivity observed after immunoblotting with horse serum (Figure 1B) or L2-mice serum (Figure 1C). Three proteins in 4 out of the 14 selected bands gave a significant score (p < 0.05) and were identified as actin (Figure 1A, band 8), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Figure 1A, band 9) and hemoglobin (Figure 1A, bands 13 and 14). A different migration pattern was observed for the LCE3 on the 1-D silver-stained gel (Figure 1D). Analogously, immunoblots were performed with horse sera (Figure 1E) and sera of L3-mice (Figure 1F). The selection of 13 bands, indicated with arrows (Figure 1D), was based upon the same criteria as above. They were isolated for further identification by MS (Table 2). Ten proteins out of the 13 selected bands gave a significant score (p < 0.05) and were identified as the alpha chain of larval serum protein (Figure 1D, bands 1–4), arylphorin (Figure 1D, band 6), beta chain of larval serum protein (Figure 1D, band 7), hemoglobin (Figure 1D, bands 10–12) and murein lipoprotein (Figure 1D, band 13).
Figure 1.
1-D analysis of the LCE of L2 and L3. Representative 1-D silver-stained gel of LCE of L2 (A) and L3 (D). The arrows indicate the bands that were selected for mass spectrometry. Western blot analysis of L2 incubated with horse serum (B) and mouse serum (C). Western blot analysis of L3 incubated with horse serum (E) and mouse serum (F). The protein identification by MS is presented in Table 1 and 2.
Table 1.
Mass spectrometry identification of proteins identified from the LCE of L2.
| Band ID | Protein name | Species | Accession number | MW (Da) | pI | Protein score |
| 8 | Protein similar to Actin-87E isoform 2 | Drosophila melanogaster | AAM29410 | 37816 | 5.36 | 223 |
| 9 | Glyceraldehyde-3-phosphate dehydrogenase | Drosophila hydei | S24630 | 35369 | 8.2 | 224 |
| 13 | Hemoglobin | Gasterophilus intestinalis | O96457 | 17912 | 8.44 | 440 |
| 14 | Hemoglobin | Gasterophilus intestinalis | O96457 | 17912 | 8.44 | 144 |
Spots assignments refer to Figure 1. Proteins listed have been identified with a significant probability score at p < 0.05 in MSDB.
Table 2.
Mass spectrometry identification of proteins identified from the LCE of L3.
| Band ID | Protein name | Species | Accession number | MW (Da) | pI | Protein score |
| 1 | Larval serum protein 1 alpha chain precursor | Drosophila melanogaster | LSP1A_DROME | 98802 | 5.72 | 92 |
| 2 | Larval serum protein 1 alpha chain precursor | Drosophila melanogaster | LSP1A_DROME | 98802 | 5.72 | 89 |
| 3 | Larval serum protein 1 alpha chain precursor | Drosophila melanogaster | LSP1A_DROME | 98802 | 5.72 | 98 |
| 4 | Larval serum protein 1 alpha chain precursor | Drosophila melanogaster | LSP1A_DROME | 98802 | 5.72 | 102 |
| 6 | Arylphorin subunit A4 precursor | Calliphora vicina | ARY1_CALVI | 92282 | 5.59 | 71 |
| 7 | Larval serum protein 1 beta chain precursor | Drosophila melanogaster | LSP1B_DROME | 95849 | 5.41 | 69 |
| 10 | Hemoglobin | Gasterophilus intestinalis | O96457 | 17912 | 8.44 | 247 |
| 11 | Hemoglobin | Gasterophilus intestinalis | O96457 | 17912 | 8.44 | 132 |
| 12 | Hemoglobin | Gasterophilus intestinalis | LPEBWM | 17912 | 8.44 | 112 |
| 13 | Major outer membrane lipoprotein precursor (Murein-lipoprotein) | Pectobacterium atrosepticum | LPP_ERWCT | 8396 | 9.36 | 69 |
Spots assignments refer to Figure 1. Proteins listed have been identified with a significant probability score at p < 0.05 (1, 2, 3, 4, 6, 7, 13: Expasy database; 10, 11, 12: MSDB).
2-D analysis of the LCE of L2
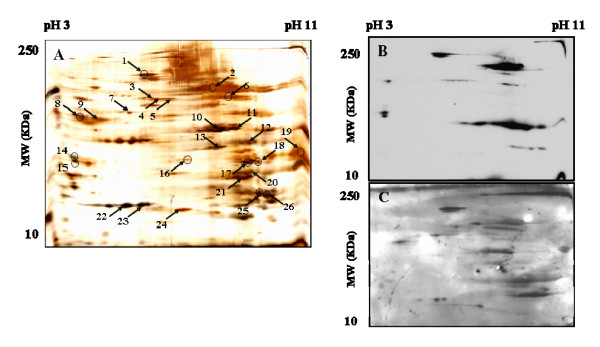
The protein migration of the LCE of L2 on a 2-D gel (Figure 2A) showed the presence of proteins all along the pH spectrum. Western blots were performed with L2-mice serum (Figure 2B) and with horse serum (Figure 2C). The silver-stained gel was used to align detected protein spots on both immunoblot profiles. A larger range of immuno-reactive proteins was observed on the horse immunoblot profile than on the L2-mice immunoblot. Circles indicate 12 spots that were considered as immuno-reactive with L2-mice serum (Figure 2A, spots: 1, 2, 6, 8, 14–20, 25) and arrows indicate 24 spots that were considered as immune reactive with horse serum (Figure 2A, spots: 1–13 and 16–26). A total of 26 spots were selected for MS analysis (Table 3). Among the 26 selected spots, 7 proteins were successfully identified (p < 0.05) by MS: paramyosin (Figure 2A, spot 1), serum albumin (Figure 2A, spot 5), tubulin (Figure 2A, spot 9), enolase (Figure 2A, spot 11), tropomyosin (Figure 2A, spot 14), GAPDH (Figure 2A, spot 19) and hemoglobin (Figure 2A, spot 20).
Figure 2.
2-D analysis of the LCE of L2. Silver-stained representative 2-D protein map of the LCE of L2 comprising pH gradient from 3 to 11 with the MWs ranking from 10 to 250 KDa (A). The silver-stained gel was used to align detected protein spots on the Western blots performed with serum of mice immunised with the LCE of L2 (B) and with horse serum (C). The circles indicate the spots that were immunolabelled with mice serum (B; spots 1,2,6,8,14–19,25,26) and the arrows indicate the spots that were immunolabelled with horse serum (C; spots 1–13 and 16–26). A total of 26 spots were isolated for further MS identification. The protein identification by MS is presented in Table 3.
Table 3.
Mass spectrometric identifications of proteins identified from the LCE of L2.
| Spot ID | Protein name | Species | Accession number | MW (Da) | pI | Protein score |
| 1 | Paramyosin | Drosophila melanogaster | S22028 | 102277 | 5.5 | 97 |
| 5 | Serum albumin precursor | Bos taurus | ABBOS | 69225 | 5.8 | 99 |
| 9 | Tubulin alpha-1 chain | Drosophila melanogaster | A26488 | 49876 | 5.0 | 260 |
| 11 | Enolase | Oryza sativa | Q7XBE4 | 47942 | 5.4 | 80 |
| 14 | Tropomyosin | Drosophila melanogaster | C25242 | 32740 | 4.7 | 78 |
| 19 | Glyceraldehyde-3-phosphate dehydrogenase | Drosophila hydei | S24630 | 35369 | 8.2 | 100 |
| 20 | Hemoglobin | Gasterophilus intestinalis | O96457 | 17912 | 8.4 | 209 |
Spots assignments refer to Figure 2. Proteins listed have been identified with a significant probability score at p < 0.05 in MSDB.
2-D analysis of the LCE of L3
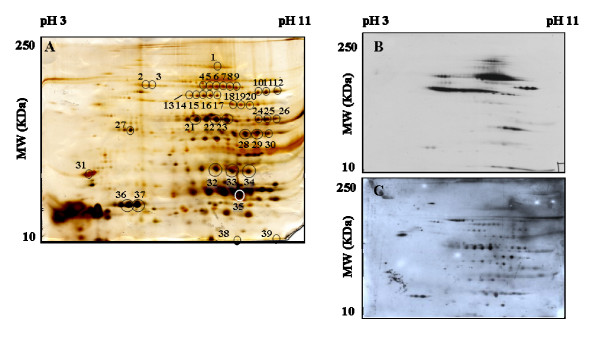
The proteomic profile of the LCE of L3 presented on figure 3A shows that most of the proteins are located in a basidic range between pH 7–11. Western blots were performed with L3-mice serum (Figure 3B) and with horse serum (Figure 3C). The intensity of the immune reaction differs when the LCE of L3 is exposed with L3-mice serum or with horse serum but the immunoblot profiles were similar. After comparison of the immunoblots with the silver-stained gel, 39 spots, indicated by circles were selected for further MS identification (Table 4). 19 out of the 39 isolated spots were successfully identified (p < 0.05), corresponding to 8 different proteins: filamin (Figure 3A, spot 1), heat shock protein (HSP-70) (Figure 3A, spot 2), serum albumin precursor (Figure 3A, spot 3,18–20), phosphoenolpyruvate carboxykinase (PEPCK) (Figure 3A, spot 8), enolase (Figure 3A, spot 22,23), fumarase (Figure 3A, spot 1), beta-actin (Figure 3A, spot 27), hemoglobin (Figure 3A, spot 32–39).
Figure 3.
2-D analysis of the LCE of L3. Silver-stained representative 2-D protein map of the LCE of L3 comprising pH gradient from 3 to 11 with the MWs ranking from 10 to 250 KDa (A). The silver-stained gel was used to align detected protein spots on the Western blots performed with serum of mice immunised with the LCE of L3 (B) and with horse serum (C). 39 spots immunolabelled with both sera were isolated for further MS identification. The protein identification by MS is presented in Table 4.
Table 4.
Mass spectrometric identifications of proteins identified from the LCE of L3.
| Spot ID | Protein name | Species | Accession number | MW (Da) | pI | Protein score |
| 1 | Filamin 1 | Drosophila melanogaster | Q8T3K7 | 151931 | 5.72 | 76 |
| 2 | Heat shock 70 kDa protein 70C | Drosophila melanogaster | HSP7A_DROME | 70871 | 5.34 | 70 |
| 3 | serum albumin | Bos taurus | AAN17824 | 71274 | 5.82 | 171 |
| 8 | Phosphoenolpyruvate carboxykinase | Drosophila melanogaster | PPCK_DROME | 71882 | 6.07 | 79 |
| 18 | Serum albumin precursor | Bos taurus | ALBU_BOVIN | 71244 | 5.82 | 108 |
| 19 | Serum albumin precursor | Bos taurus | ALBU_BOVIN | 71244 | 5.82 | 117 |
| 20 | Serum albumin precursor | Bos taurus | ALBU_BOVIN | 71244 | 5.82 | 165 |
| 22 | Enolase (Fragment) | Drosophila subobscura | O44101 | 44548 | 5.92 | 142 |
| 23 | Enolase | Schistosoma mansoni | ENO_SCHMA | 47421 | 6.18 | 163 |
| 24 | Fumarase | Drosophila melanogaster | Q9VTI5 | 51239 | 8.47 | 111 |
| 27 | Beta-actin | Danio rerio | ACTB1_BRARE | 42082 | 5.3 | 184 |
| 32 | Hemoglobin | Gasterophilus intestinalis | O96457 | 18026 | 8.44 | 95 |
| 33 | Hemoglobin | Gasterophilus intestinalis | O96457 | 18026 | 8.44 | 113 |
| 34 | Hemoglobin | Gasterophilus intestinalis | O96457 | 18026 | 8.44 | 131 |
| 38 | Hemoglobin | Gasterophilus intestinalis | O96457 | 18026 | 8.44 | 415 |
| 39 | Hemoglobin | Gasterophilus intestinalis | O96457 | 18026 | 8.44 | 410 |
Spots assignments refer to Figure 3. Proteins listed have been identified with a significant probability score at p < 0.05 (2, 8, 18, 19, 20, 23, 27: Expasy database; 1, 3, 22, 24, 32, 33, 34, 38, 39: MSDB).
Discussion
The comparison of the migration patterns of the LCE2 and LCE3 in both dimensional analysis (1-D and 2-D), indicates that the larval proteinic profile is stage specific, thus suggesting a different composition in larval metabolism and antigenic properties. The 1-D silver-stained gels of the larval crude extracts indicated a very dense concentration of proteins; consequently the alignment of the immuno-reactive bands detected by horse and mice might present some differences. The proteomic approach confirmed the specific protein profile of both larval stages and allowed better identification of the different spots.
Mice were artificially immunised against a crude protein extract from whole larvae. A large number of proteins that are normally not directly in contact with horses were presented to the immune system of the mice. Furthermore, unlike natural infection eliciting a horse immune reaction, subcutaneous injection in mice induces a systemic immune reaction shifted to a Th1 response by the adjuvant. This difference in immune reaction explains the fact that most of the L2 and L3 proteins that reacted with mice sera showed a more intense signal when compared with the reaction with horse sera. Since the lifecycle of G. intestinalis occurs in the gastro-intestinal tract of horses, the mucosal immune system is in contact with the larvae and thus exposed to excreted or secreted substances during larval migration and development [18]. Although L2 migration patterns from mouth to stomach remains unclear, the immune reaction detected in horses and in experimentally immunized mice might suggest that the larval stage L2 possesses more antigenic proteins than the larval stage L3, probably useful for larval enzymatic migration [19]. Accordingly, enolase was identified by MS and has previously been reported as an important enzyme localized on the surface of several pathogens when invading tissue [20]. L2 is a stage inducing a strong host immune response so that its development into L3 in the stomach might be a defence mechanism of the larvae to bypass the horse immune defences. Conversely, the fact that L3 remains attached for 8–10 months to the stomach wall suggests a hypometabolic status, or reduced immunogenic properties. This can explain the weak immune reaction observed in the presence of horse serum against the L3.
Two important larval proteins, arylphorin and LSP-2 (larval serum protein), respectively homologous to Calliphora vicina and Drosophila melanogaster, were identified in the L3. In holometabolous insects the construction of adult tissues during metamorphosis requires a large amount of energy. It is known that before formation of the puparium, the fat body cells reabsorb proteins and other macromolecules that have accumulated in the haemolymph during the larval feeding period [21]. The major fraction of incorporated proteins consists of arylphorins and LSP-2 [22]. In addition, hemoglobin was identified in both larval stages of G. intestinalis. This abundant and circulating molecule is present in highly tracheated cells forming the posterior spiracular plate and allows the larvae to make better use of intermittent contact when air is swallowed with food [23].
Most of the other identified proteins in L2 (paramyosin, tubulin, tropomyosin, GAPDH and a protein similar to actin) or in L3 (filamin, fumarase, PEPCK, HSP-70, enolase) are shared with those of Drosophila spp. suggesting that structural or metabolic homologies do exist between these species.
During this research, different intestinal parasites (Anoplocephala perfoliata, Parascaris equorum, Cyathostominae) were simultaneously present with G. intestinalis in the gastro-intestinal tract of the slaughtered horses. The risk of cross-reactivity has still to be evaluated. But unlike some immunological studies about intestinal helminths in equids [24-26], the cross-reactivity between G. intestinalis and gastro-intestinal parasites has not yet been studied.
This work provides further information into the understanding of the interaction between G. intestinalis and their host and by contributing a novel scheme of the proteomic profile of the main larval stages. Thus our results further demonstrate the complexity of this host-parasite interaction. Indeed, this study reveals the necessity to develop a reliable serological tool to detect infested horses, particularly because the only means to detect a G. intestinalis infestation is by necropsy.
The identification of most of the proteins will be the next step to define their role, their cellular or tissue localisation and their potential antigenic properties.
Methods
Larval collection and antigen preparation
Gasterophilus spp. L2 and L3 were collected from the pyloric portion of the stomach of horses originating from two farms located in the District of the Swiss Jura; Delémont: N 47°21'; E 7°20', Switzerland. Simultaneously, the gastro-intestinal tract of each animal was examined and the presence of P. equorum, A. perfoliata and Cyathostominae was observed in any case.
All the larvae collected were washed in a sterile phosphate saline buffer (PBS 0.1 M, pH 7.2), identified as G. intestinalis on the basis of morphological keys [1] and frozen at -20°C.
For preparation of the larval crude extracts, 10 L2 larvae harvested on two different horses of a same herd, and seven L3 larvae harvested on one horse originating from the second herd, were sonicated and homogenised on ice under sterile conditions. The homogenate was extracted overnight in a 0.1 M pH 9.6 carbonate buffer containing 1 mM phenylmethylsulfonyl fluorid (PMSF) and 5 mM ethylenediamin tetraacetic acid (EDTA) by further adding 1 ml/gr of a protease inhibitor (Sigma-Aldrich, Buchs, Switzerland). The extracts were centrifuged at 20'000 × g for 30 minutes (4°C). The supernatant containing the antigens was collected and the final protein content determined by spectrometry (Bradford method, BioRad). LCE was lyophilized and stored at -20°C.
Horse serum samples
Blood samples were taken on a group of twenty horses originating from the two farms in the above described area. Although blood samples and larval collection could not be made on the same animals our observations made during a simultaneously performed epidemiological survey (Roelfstra et al, in prep.), including field observations on live animals and in slaughterhouses, confirmed the high level of endemicity of G. intestinalis previously described by Brocard [19] in the same area and allow us to presume that all horses used in this study are infested by G. intestinalis. Foetal horse serum was used as a negative control for the Western blots. The blood was centrifuged at 3500 × g for 15 minutes at room temperature and the sera were stored at -20°C.
Immunisation of mice and serum samples
Balb/c mice were immunised with LCE of L2 (L2 mice) or with LCE of L3 (L3 mice) from G. intestinalis. Two grams of L3 and two grams of L2 were sonicated, homogenised in a sterile PBS buffer pH 7.2 and centrifuged at 20'000 × g. The supernatant was then emulsified in equal volume of Freund's Incomplete Adjuvant (Sigma-Aldrich, Buchs, Switzerland).
A dose of 100 μg protein/mouse, in a volume of 200 μl, was injected by intramuscular route every 2 weeks through 6 weeks. Animals were bled seven weeks after the first immunisation. Control mice were inoculated with a combination of PBS and Freund's Incomplete Adjuvant every time above. Sampled blood was centrifuged at 3500 × g for 15 minutes at room temperature and sera stored at -20°C. All procedures were approved by the responsible local committee on animal experimentations.
One dimensional electrophoresis (1-D)
The proteins of L2 (5 μg/well) and L3 (5 μg/well) were separated on gradient SDS-PAGE gels (4–20%) under reducing conditions (2-β-mercaptoethanol, 95°C for 5 min). Electrophoresis was performed at 80/100 V for 30 min/2 hrs. One set of the gels was stained with silver for mass spectrometry, and the second was transferred onto nitrocellulose membranes (GE Healthcare, Uppsala, Sweden) for Western blot analysis.
Two dimensional electrophoresis (2-D)
Lyophilized LCE samples were solubilised in 2-DE lysis buffer (9 M urea, 2 M thiourea, 1% dithioerythriol, 4% CHAPS, 2.5 μM EGTA, and 2.5 μM EDTA). Immobiline dry strips pH 3–11 non linear, 11 cm (GE Healthcare, Uppsala, Sweden) were immersed overnight in lysis buffer containing 75 μg protein sample, additional 1% Pharmalyte pH 3–10 (GE Healthcare, Uppsala, Sweden), and 0.5% bromphenol blue. IEF on a Multiphor (GE Healthcare) for 15 kV·h at 20°C was followed by separation on gradient SDS-PAGE gels (9–15%) at constant 45 V per gel. One set of gels was stained with silver for MS and the second was transferred onto nitrocellulose membranes (GE Healthcare, Uppsala, Sweden) for Western blot analysis.
Western blot analysis (WB)
Non-specific binding was blocked with 1% polyvinylpyrrolidone in PBS-Tween for 1 hour. Blots were subsequently incubated with primary antibody in PBS-Tween (overnight at 4°C; horse sera or mice sera 1:1000) and washed. The immunoreactive spots were detected using a goat anti-mouse IgG (H+L) (1:20000, Nordic Immunology Laboratories, Tilburg, The Netherlands) or an anti-horse IgG (1:10000, Sigma-Aldrich, Buchs, Switzerland) antibody conjugated with horseradish peroxidase. Signals were detected with ECL (enhanced chemiluminescence) on Hyperfilm ECL (GE Healthcare, Uppsala, Sweden) [27]. After ECL detection, the blots were subsequently stained with colloidal gold in order to match the visible spots to overall pattern on silver-stained gels.
Mass spectrometry (MS)
Selected spots were excised from 2-D gels, destained, processed by proteolysis with trypsin [28] and analyzed by MALDI-TOF and MS/MS on a MALDI-TOF/TOF tandem mass spectrometer (ABI 4700 Proteomics Analyzer, Applied Biosystems). Combined PMF (peptide mass fingerprint) and MS/MS queries were done with MASCOT© Database search engine v1.9 [29] (Matrix Science) embedded into GPS-Explorer Software (version 3.6, Applied Biosystem) on the Swiss-Prot database (version 20051206; 201594 sequences; 73123101 residues) or MSDB (version 20040703; 1501893 sequences; 480537664 residues). Protein identification was considered positive (Tables 1, 2, 3 and 4) if (i) the probability-based MOWSE score [30] obtained from both MS and MS/MS analysis was significant (i.e. scores > 66 were significant at p < 0.05 for Expasy database, and scores > 74 significant at p < 0.05 for MSDB database; confidence interval > 99% as given by GPS explorer, version 3.6); (ii) the matched peptide masses were abundant in the spectrum; and (iii) the theoretical molecular weights (MW) of the significant hits fit the experimental observed values.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
LR and KP conceived and designed the study. LR carried out the acquisition of the material and the morphological data as well as the immunoassays and drafted the manuscript. KP supervised the work and helped to write the manuscript. CD outlined and supervised all the proteomic features and helped to draft the manuscript. SMH carried out the mass spectrometry and analysed the data. CB has supported LR in all the laboratory analysis. MM conceived and carried out the immunological study with mice. BB participated in the design and coordination of the study and helped to analyse the results. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This work was supported by SFB 571 grant A5 Deeg. The authors are grateful to Prof. D. Otranto and Prof. D.D. Colwell for their critical comments.
Contributor Information
Liselore Roelfstra, Email: liselore.roelfstra@unine.ch.
Cornelia A Deeg, Email: deeg@tiph.vetmed.uni-muenchen.de.
Stefanie M Hauck, Email: hauck@helmholtz-muenchen.de.
Christina Buse, Email: chrissibuse@yahoo.com.
Mathieu Membrez, Email: mathieu.membrez@gmail.com.
Bruno Betschart, Email: bruno.betschart@unine.ch.
Kurt Pfister, Email: Kurt.Pfister@tropa.vetmed.uni-muenchen.de.
References
- Zumpt F. Myasis in man and animals in the old world. Butterworths, London, United Kingdom; 1965. [Google Scholar]
- Horak IG, De Vos V, De Klerk BD. Parasites of domestic and wild animals in South Africa. XVII. Arthropod parasites of Burchell's zebra, Equus burchelli, in the eastern Transvaal Lowveld. Onderstepoort J Vet Res. 1984;51:145–154. [PubMed] [Google Scholar]
- Soulsby EJL. Helminths, arthropods and protozoa of domesticated animals. 7. Eastbourne, United Kingdom: Baillière, Tindall & Cassell; 1982. [Google Scholar]
- Cogley TP, Cogley MC. Inter-relationship between Gasterophilus larvae and the horse's gastric and duodenal wall with special reference to penetration. Vet Parasitol. 1999;86:127–142. doi: 10.1016/S0304-4017(99)00119-3. [DOI] [PubMed] [Google Scholar]
- Otranto D, Milillo P, Capelli G, Colwell DD. Species composition of Gasterophilus spp. (Diptera, Oestridae) causing equine gastric myiasis in southern Italy: parasite biodiversity and risks for extinction. Vet Parasitol. 2005;133:111–118. doi: 10.1016/j.vetpar.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Cogley TP. Effects of migrating Gasterophilus intestinalis larvae (Diptera: Gasterophilidae) on the mouth of the horse. Vet Parasitol. 1989;31:317–331. doi: 10.1016/0304-4017(89)90081-2. [DOI] [PubMed] [Google Scholar]
- Cogley TP, Anderson JR, Cogley LJ. Migration of Gasterophilus intestinalis larvae (Diptera:Gasterophilidae) in the equine oral cavity. Int J Parasitol. 1982;12:473–480. doi: 10.1016/0020-7519(82)90079-0. [DOI] [PubMed] [Google Scholar]
- Principato M, Tosti M. Scanning electron microscope observations on the anterior thoracic and post-abdominal spiracles of Gasterophilus larvae (Diptera: Gasterophilidae) Int J Parasitol. 1988;18:191–196. doi: 10.1016/0020-7519(88)90060-4. [DOI] [PubMed] [Google Scholar]
- Shefstad DK. Scanning electron microscopy of Gasterophilus intestinalis lesions of the equine stomach. J Am Vet Med Assoc. 1978;172:310–313. [PubMed] [Google Scholar]
- Otranto D. The immunology of myiasis: parasite survival and host defense strategies. Trends Parasitol. 2001;17:176–182. doi: 10.1016/S1471-4922(00)01943-7. [DOI] [PubMed] [Google Scholar]
- Boldbaatar D, Xuan X, Kimbita E, Huang X, Igarashi I, Byambaa B, Battsetseg B, Battur B, Battsetseg G, Batsukh Z, et al. Detection of antibodies to Hypoderma lineatum in cattle by Western blotting with recombinant hypodermin C antigen. Vet Parasitol. 2001;99:147–154. doi: 10.1016/S0304-4017(01)00457-5. [DOI] [PubMed] [Google Scholar]
- Alcaide M, Reina D, Frontera E, Navarrete I. Analysis of larval antigens of Oestrus ovis for the diagnosis of oestrosis by enzyme-linked immunosorbent assay. Med Vet Entomol. 2005;19:151–157. doi: 10.1111/j.0269-283X.2005.00555.x. [DOI] [PubMed] [Google Scholar]
- Colwell DD. Part C: Gasterophilinae Host-Parasite Interactions. In: Colwell DD, Hall MJR, Scholl PJ, editor. The Oestrid Flies Biology, Host-parasite relationships, impact and management. Oxfordshire, United Kingdom: CABI Publishing; 2006. pp. 189–191. [Google Scholar]
- Escartin-Pena M, Bautista-Garfias CR. Comparison of five tests for the serologic diagnosis of myiasis by Gasterophilus spp. larvae (Diptera: Gasterophilidae) in horses and donkeys: a preliminary study. Med Vet Entomol. 1993;7:233–237. doi: 10.1111/j.1365-2915.1993.tb00682.x. [DOI] [PubMed] [Google Scholar]
- Dea-Ayuela MA, Bolas-Fernandez F. Two-dimensional electrophoresis and mass spectrometry for the identification of species-specific Trichinella antigens. Vet Parasitol. 2005;132:43–49. doi: 10.1016/j.vetpar.2005.05.018. [DOI] [PubMed] [Google Scholar]
- Jungblut PR, Zimny-Arndt U, Zeindl-Eberhart E, Stulik J, Koupilova K, Pleissner KP, Otto A, Muller EC, Sokolowska-Kohler W, Grabher G, Stoffler G. Proteomics in human disease: cancer, heart and infectious diseases. Electrophoresis. 1999;20:2100–2110. doi: 10.1002/(SICI)1522-2683(19990701)20:10<2100::AID-ELPS2100>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Shin YS, Shin GW, Kim YR, Lee EY, Yang HH, Palaksha KJ, Youn HJ, Kim JH, Kim DY, Marsh AE, et al. Comparison of proteome and antigenic proteome between two Neospora caninum isolates. Vet Parasitol. 2005;134:41–52. doi: 10.1016/j.vetpar.2005.06.021. [DOI] [PubMed] [Google Scholar]
- Baron RW, Colwell DD. Mammalian immune responses to myiasis. Parasitol Today. 1991;7:353–355. doi: 10.1016/0169-4758(91)90219-E. [DOI] [PubMed] [Google Scholar]
- Brocard P, Pfister K. [The epidemiology of gasterophilosis of horses in Switzerland] Schweiz Arch Tierheilkd. 1991;133:409–416. [PubMed] [Google Scholar]
- Bernal D, de la Rubia JE, Carrasco-Abad AM, Toledo R, Mas-Coma S, Marcilla A. Identification of enolase as a plasminogen-binding protein in excretory-secretory products of Fasciola hepatica. FEBS Lett. 2004;563:203–206. doi: 10.1016/S0014-5793(04)00306-0. [DOI] [PubMed] [Google Scholar]
- Hansen IA, Meyer SR, Schafer I, Scheller K. Interaction of the anterior fat body protein with the hexamerin receptor in the blowfly Calliphora vicina. Eur J Biochem. 2002;269:954–960. doi: 10.1046/j.0014-2956.2001.02736.x. [DOI] [PubMed] [Google Scholar]
- Telfer WH, Kunkel JG. The function and evolution of insect storage hexamers. Annu Rev Entomol. 1991;36:205–228. doi: 10.1146/annurev.en.36.010191.001225. [DOI] [PubMed] [Google Scholar]
- Dewilde S, Blaxter M, Van Hauwaert ML, Van Houte K, Pesce A, Griffon N, Kiger L, Marden MC, Vermeire S, Vanfleteren J, et al. Structural, functional, and genetic characterization of Gasterophilus hemoglobin. J Biol Chem. 1998;273:32467–32474. doi: 10.1074/jbc.273.49.32467. [DOI] [PubMed] [Google Scholar]
- Dowdall SM, Proudman CJ, Klei TR, Mair T, Matthews JB. Characterisation of IgG(T) serum antibody responses to two larval antigen complexes in horses naturally- or experimentally-infected with cyathostomins. Int J Parasitol. 2004;34:101–108. doi: 10.1016/j.ijpara.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Hoglund J, Ljungstrom BL, Nilsson O, Uggla A. Enzyme-linked immunosorbent assay (ELISA) for the detection of antibodies to Anoplocephala perfoliata in horse sera. Vet Parasitol. 1995;59:97–106. doi: 10.1016/0304-4017(94)00755-2. [DOI] [PubMed] [Google Scholar]
- Proudman CJ, Trees AJ. Use of excretory/secretory antigens for the serodiagnosis of Anoplocephala perfoliata cestodosis. Vet Parasitol. 1996;61:239–247. doi: 10.1016/0304-4017(95)00837-3. [DOI] [PubMed] [Google Scholar]
- Buse C, Altmann F, Amann B, Hauck SM, Poulsen Nautrup C, Ueffing M, Stangassinger M, Deeg CA. Discovering novel targets for autoantibodies in dilated cardiomyopathy. Electrophoresis. 2008;29:1325–1332. doi: 10.1002/elps.200700686. [DOI] [PubMed] [Google Scholar]
- Deeg CA, Pompetzki D, Raith AJ, Hauck SM, Amann B, Suppmann S, Goebel TW, Olazabal U, Gerhards H, Reese S, et al. Identification and functional validation of novel autoantigens in equine uveitis. Mol Cell Proteomics. 2006;5:1462–1470. doi: 10.1074/mcp.M500352-MCP200. [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Pappin DJ. Peptide mass fingerprinting using MALDI-TOF mass spectrometry. Methods Mol Biol. 1997;64:165–173. doi: 10.1385/0-89603-353-8:165. [DOI] [PubMed] [Google Scholar]