Abstract
Protein-only (prion) epigenetic elements confer unique phenotypes by adopting alternate conformations that specify new traits. Given the conformational flexibility of prion proteins, protein-only inheritance requires efficient self-replication of the underlying conformation. To explore the cellular regulation of conformational self-replication and its phenotypic effects, we analyzed genetic interactions between [PSI+], a prion form of the S. cerevisiae Sup35 protein (Sup35[PSI+]), and the three Nα-acetyltransferases, NatA, NatB, and NatC, which collectively modify ∼50% of yeast proteins. Although prion propagation proceeds normally in the absence of NatB or NatC, the [PSI+] phenotype is reversed in strains lacking NatA. Despite this change in phenotype, [PSI+] NatA mutants continue to propagate heritable Sup35[PSI+]. This uncoupling of protein state and phenotype does not arise through a decrease in the number or activity of prion templates (propagons) or through an increase in soluble Sup35. Rather, NatA null strains are specifically impaired in establishing the translation termination defect that normally accompanies Sup35 incorporation into prion complexes. The NatA effect cannot be explained by the modification of known components of the [PSI+] prion cycle including Sup35; thus, novel acetylated cellular factors must act to establish and maintain the tight link between Sup35[PSI+] complexes and their phenotypic effects.
INTRODUCTION
The transmission of phenotypes from one individual to another is a fundamental process in biology. Much of our understanding of these events arises from decades of study on nucleic acid metabolism, but new traits may also be passed between individuals without changes in nucleic acid content through a number of epigenetic mechanisms. One particularly intriguing example of such a process is the prion phenomenon, in which the activity of a protein is altered in a heritable way to transmit a new phenotype. How is such a feat accomplished? In 1967, Griffith proposed that some proteins, now known as prions (Prusiner, 1982), can adopt more than one stable form in vivo (Griffith, 1967). Since a protein's structure determines its function, two cells containing the same protein but in diffrent physical states will have distinct phenotypes. This protein-based process has been linked to a number of previously inexplicable events, including the development and spread of the transmissible spongiform encephalopathies in mammals (Prusiner, 1982) and the non-Mendelian inheritance of some traits in fungi (Wickner, 1994).
Protein-based traits can only become transmissible, however, if the inherent structural flexibility of prion proteins can be constrained by regulatory mechanisms to create an epigenetic element. For example, if each newly synthesized prion polypeptide chain independently folded to a unique form, all cells would display the same phenotype, which would reflect the average of the accessible states. The appearance of distinct protein-based phenotypes suggests that while prion proteins remain flexible enough to adopt multiple forms, their folding is somehow restricted in individual cells to yield distinct phenotypes. How cells accomplish this regulation is currently poorly understood.
One well-studied example of such a protein-based trait is the Sup35/[PSI+] prion of Saccharomyces cerevisiae. The Sup35 protein is a component of the translation termination complex whose function is modulated by a prion cycle (Zhouravleva et al., 1995; Serio and Lindquist, 1999). In the nonprion ([psi−]) or default state, Sup35 is soluble and facilitates efficient translation termination (Cox, 1965; Patino et al., 1996; Paushkin et al., 1996). However, in the prion ([PSI+]) or self-replicating form, the majority of Sup35 assembles into aggregates that establish a translation termination defect, leading to stop codon read-through (Cox, 1965; Patino et al., 1996; Paushkin et al., 1996). Prion aggregates or their component prion-state protein (Sup35[PSI+]) facilitate propagation of the [PSI+] phenotype by acting in three key roles. First, they template the conversion of newly synthesized Sup35 protein to the prion form (Patino et al., 1996; Satpute-Krishnan and Serio, 2005). Second, they are continually regenerated to create new surfaces for efficient conversion (Ness et al., 2002; Cox et al., 2003; Satpute-Krishnan et al., 2007), and third, they are partitioned to daughter cells where they continue the cycle and allow inheritance of the associated phenotype (Cox et al., 2003; Satpute-Krishnan et al., 2007). Thus, the strong link between protein state and phenotype that allows proteins to act as epigenetic elements is established and maintained through a multistep pathway of protein dynamics (Pezza and Serio, 2007).
Much of our mechanistic understanding of the Sup35/[PSI+] in vivo prion cycle has its origins in a variety of genetic screens, which identified key modulators of this process. Although a number of mutagenic (Young and Cox, 1971; Jung et al., 2000), overexpression (Chernoff et al., 1993; Chernoff et al., 1995; Kryndushkin et al., 2002), and interaction screens (Bailleul et al., 1999) have been conducted to date, additional factors impacting prion propagation continue to be identified by candidate gene approaches (Chernoff et al., 1999, 2003; Newman et al., 1999; Ganusova et al., 2006; Park et al., 2006; Fan et al., 2007; Kryndushkin and Wickner, 2007; Sadlish et al., 2008), suggesting that our current understanding of prion cycle regulation in vivo is incomplete.
In an attempt to elucidate additional factors and processes governing efficient prion propagation in vivo, we took advantage of the fact that N-terminal acetylation is critical for the normal function of many proteins (Polevoda and Sherman, 2003). Approximately 50% of the yeast proteome is cotranslationally acetylated on its N-terminus by the collective action of three enzyme complexes: NatA, NatB, and NatC (Polevoda and Sherman, 2003). Mutations in these complexes lead to a number of pleiotropic phenotypes, but in recent years, a growing number of these defects have been specifically linked to single protein substrates. For example, telomeric silencing, membrane trafficking, L-A viral assembly, and tropomyosin–actin interactions are mediated by the specific N-terminal acetylation of Orc1, Arl3, gag, and Tpm1, respectively (Tercero and Wickner, 1992; Singer and Shaw, 2003; Behnia et al., 2004; Geissenhoner et al., 2004; Setty et al., 2004). In many of these cases, the loss of acetylation reduces the affinity of these substrates for a partner protein, providing a detailed mechanistic explanation for the observed phenotype. Thus, altering N-terminal acetylation profiles is a powerful method to partially impair the activities of target proteins.
Using this approach, we have uncovered a role for the NatA complex in the [PSI+] prion cycle. In strains lacking NatA function, heritable Sup35 prion complexes are uncoupled from their phenotypic effects. In contrast to other known modulators of [PSI+] propagation that appear to act by altering prion complex dynamics (Jung et al., 2000; Wegrzyn et al., 2001; Kryndushkin et al., 2003; Song et al., 2005; Satpute-Krishnan et al., 2007), NatA null strains efficiently incorporate soluble Sup35 into existing prion complexes, but the nature of this interaction is altered, allowing faithful translation termination.
MATERIALS AND METHODS
Plasmids
SB531 (pRS304PGPDGST(UGA)DsRED-NLS) was constructed by replacing the DsRed open reading frame in pRS304GST(UGA)DsRED (Satpute-Krishnan and Serio, 2005) with DsRed-NLS as a EcoRI-XhoI fragment. The latter was generated by PCR (primers: 5′ TCGAATAAACACACATAAACAAACAA3′ and 5′TCGAGGGTCGACTTATTTAACGACAACCTTTCTCTTCTTCTTTGGCAGGAACAGGTGGTGGCG 3′) using pRS304GST(UGA)DsRed as a template. SB208 was generated by amplifying a genomic fragment of NAT1 from 74D-694 by PCR (primers: 5′GCTCTAGAGCCATTCTCGTTCGTATACC3′ and 5′GCTCTAGAGGAAGGAGTGCTAGATGTAG3′) and subcloning this region into pRS315 as an XbaI fragment. SB106 (pRS303PMFα1Sup35-GFP) contains the PMFα1 promoter amplified by PCR (primers: 5′ GGAATTCTATTGATAAGATTTAAAGGTATT3′ and 5′ CGGGATCCTCTTTTAATCGTTTATATTGTG3′) and substituted for PMFA1 as an EcoRI/BamHI fragment in pRS303PMFA1 SUP35-green fluorescent protein (GFP; Satpute-Krishnan and Serio, 2005). The sequences of all plasmids generated by PCR were confirmed. pAB2341 (Polevoda et al., 1999) and pAA2241(Whiteway and Szostak, 1985) were kind gifts from F. Sherman (University of Rochester). pUKC815, pUKC817 (UAA), pUKC818 (UAG), and pUKC819 (UGA; Stansfield et al., 1995) were kindly provided by M. Tuite (University of Kent). pXS (Sizonenko et al., 1990) was a kind gift from Y. Chernoff (Georgia Tech). pCORE (Storici et al., 2001) was a kind gift from M. Resnick (National Institutes of Health), and pRS426-Sp-PrDM(HA)EF (DePace et al., 1998) was a kind gift from J. Weissman (UCSF).
Yeast Strains
All strains used in this study are listed in Table 1 and are derivatives of 74D-694 (Chernoff et al., 1995) unless otherwise indicated. NT64 was a kind gift from C. Cullin (Université Bordeaux, France). The following disruptions were generated by PCR using pFA6aHis3MX6 (Longtine et al., 1998) as a template and the indicated primers: NAT3 (5′ATTGAGAATATTCAAGGAAAGAGACAGGAGGATTCGAGAACGGATCCCCGGGTTAATTAA3′ and 5′ATTATTATGTTCTGAGTATGAGGACGAGGTAATACATACCGAATTCGAGCTCGTTTAAAC3′), NAT5 (5′AACAAATTGGCAAAGACAATTGAAGGGAGTAGTGCGAAATCGGATCCCCGGGTTAATTAA3′ and 5′AGCAAAACCAAAAAAAAAAAAAAAATTTTTCAGCCATCTGGAATTCGAGCTCGTTTAAAC3′), and MAK3 (5′GGCCTGGGAAGAAAAAAATTCCCTCGTCGATAATATAAATCGGATCCCCGGGTTAATTAA3′ and 5′TTATTAATATATATTTTATCATCATCGAGTGTTTTTCCTTGAATTCGAGCTCGTTTAAAC3′). Disruptions of SSA2 were generated by PCR using pFA6ahphMX4 (Goldstein and McCusker, 1999) as a template (primers: 5′ CCAACAGATCAAGCAGATTTTATACAGAAATATTTATACACAGCTGAAGCTTCGTACGC3′ and 5′ AGTAAAACTTTTCGGATATTTTACAGGGCGATCGCTAAGCGCATAGGCCACTAGTGGATCTG3′).
Table 1.
Yeast strains
| Strain | Genotype | Reference |
|---|---|---|
| 74D-694 | MATa (or MATα or MATa/α) [PSI+] (or [psi−]) ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 | Chernoff et al. (1995) |
| SY536 | MATα [PSI+] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 nat3::HIS3MX4 | This study |
| SY563 | MATα [psi−] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 nat3::HIS3MX4 | This study |
| SY540 | MATα [PSI+] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 mak3::HIS3MX4 | This study |
| SY567 | MATα [psi−] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 mak3::HIS3MX4 | This study |
| TRS169 | MATα [PSI+] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 ard1::kanMX4 | This study |
| TRS220 | MATα [psi−] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 ard1::kanMX4 | This study |
| SY538 | MATα [PSI+] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 nat5::HIS3MX4 | This study |
| SY565 | MATα [psi−] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 nat5::HIS3MX4 | This study |
| SY356 | MATα [PSI+] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 nat1::hphMX4 | This study |
| SY982 | MATα [psi−] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 nat1::hphMX4 | This study |
| SY319 | MATα [PSI+] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 nat1::hphMX4 ard1::kanMX4 | This study |
| SY978 | MATα [psi−] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 nat1::hphMX4 ard1::kanMX4 | This study |
| TRS47 | MATα [PSI+] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 nat1::LEU2 | This study |
| TRS51 | MATα [psi−] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 nat1::LEU2 | This study |
| W303-1A | MATa leu2-3, 112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 | Thomas and Rothstein (1989) |
| AMR1 | MATa leu2-3, 112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 nat1::LEU2 | Mullen et al. (1989) |
| TRS64 | MATa [psi−] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 sup35K556E | This study |
| TRS192 | MATa [psi−] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 sup35K556E ard1::kanMX4 | This study |
| SY80 | MATα [PSI+] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3,112 SUP35-GFP | Satpute-Krishnan and Serio (2005) |
| SY84 | MATα [psi−] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3,112 SUP35-GFP | Satpute-Krishnan and Serio (2005) |
| SY1068 | MATα [psi−] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 nat1::hphMX4 ard1::kanMX4 SUP35-GFP | This study |
| SY330 | MATα [PSI+] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 nat1::hphMX4 ard1::kanMX4 SUP35-GFP | This study |
| SY357 | MATα [psi−] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3, 112 nat1::hphMX4 | This study |
| SY1163 | MATa [psi−] ade1-14 trp1-289:: TRP1::PGPDGST(UGA)DsRED-NLS his3Δ200 ura3-52 leu2-3, 112 | This study |
| SY1181 | MATα [psi−] ade1-14 trp1-289:: TRP1::PGPDGST(UGA)DsRED-NLS his3Δ200 ura3-52 leu2-3, 112 | This study |
| SY1180 | MATα [psi−] ade1-14 trp1-289:: TRP1::PGPDGST(UGA)DsRED-NLS his3Δ200 ura3-52 leu2-3, 112 nat1::hphMX4 | This study |
| SY1308 | MATα [PSI+] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3,112 ssa1S2P | This study |
| SY1338 | MATα [PSI+] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3,112 ssa2::hphMX4 | This study |
| SY1339 | MATα [PSI+] ade1-14 trp1-289 his3Δ200 ura3-52 leu2-3,112 ssa1S2P ssa2::hphMX4 | This study |
| NT64 | MATα [URE3] trp1-1 ade2-1, his3-11,15 leu2-3,112 ura3-1 pdal5::ADE2 | Bach et al. (2003) |
| SY1444 | MATα [URE3] trp1-1 ade2-1, his3-11,15 leu2-3,112 ura3-1 pdal5::ADE2 ard1::kanMX4 | This study |
| SY360 | MATa [psi−] ade1-14 trp1-289 his3Δ200::HIS3::PMFA1SUP35-GFP ura3-52 leu2-3, 112 | Satpute-Krishnan and Serio (2005) |
| SY1370 | MATa [psi−] ade1-14 trp1-289:: trp1-289:: TRP1::PGPDGST(UGA)DsRED-NLS his3Δ200::HIS3::PMFA1SUP35-GFP ura3-52 leu2-3, 112 | This study |
| SY1371 | MATα [psi−] ade1-14 trp1-289::TRP1::PGPDGST(UGA)DsRED-NLS his3Δ200:: HIS3::PMFα1SUP35-GFP ura3-52 leu2-3, 112 nat1::hphMX4 | This study |
The PCR products were transformed into yeast, and recombinants were selected on minimal media lacking histidine (SD-his) or complete medium supplemented with hygromycin B (YPD+hyg). For ARD1 disruptions, a BamHI-XhoI fragment of pAB2341 was transformed into yeast, and recombinants were selected on YPD supplemented with G418. For NAT1, a disruption of NAT1 with LEU2 was amplified from the yeast strain AMR1 (a kind gift of R. Sternglanz, SUNY Stony Brook; Mullen et al., 1989; Thomas and Rothstein, 1989) with primers (5′CAAGGCATCGCTTCGGTAG3′ and 5′CTACCGAAGCGATGCCT3′) and transformed into yeast, selecting on media lacking leucine (SD-leu), or a PCR disruption was generated using pFA6ahphMX4 (Goldstein and McCusker, 1999) as a template (primers: 5′GACAAATACCATTGAGGAAGGCGATTGACCCTAACGAAGTCCTTGACAGTCTTGACGTGC3′ and 5′AATTAAGTAAGAGTTAATTGACACATTGAGGAGTTCGCAGGCGCACTTAACTTCGCATCTG3′). In all cases, disruptions were generated in [PSI+] diploids, which were sporulated and dissected to generate the haploid strains. [psi−] strains were obtained by guanidine HCl (GdnHCl) curing of [PSI+] variants (Tuite et al., 1981).
Disruptions were verified by PCR, by 2:2 segregation of the disruption marker, and where possible, by haploid mating defects (Whiteway and Szostak, 1985; Polevoda et al., 1999). Strains expressing GST (UGA) DsRED-NLS were generated by transforming SB531 DNA digested with Bsu36I and selecting for tryptophan prototrophy. Strains expressing Sup35-GFP from PMFα1were generated by transforming SB106 DNA digested with Eco47III and selecting for histidine prototrophy. SY1209 was generated by substituting sup35S2P for SUP35 by two-step replacement using BsrGI-digested SB548; the replacement was confirmed by PCR amplification of genomic DNA and sequencing. SY1308 was generated by targeting the core module of pCORE to the SSA1 5′ end by amplifying the cassette by PCR (primers: 5′ GTAATCAAGTATTACAAGAAACAAAAATTCAAGTAAATAACAGATAATATGGAGCTCGTTTTCGACACTGG3′ and 5′ GTGAGCAACACACGAGTATGTTGTACCTAAATCAATACCGACAGCTTTTGATCCTTACCATTAAGTTGATC3′), transforming the product into wild-type 74D-694 and selecting for uracil prototrophy. Proper integration was confirmed by resistance to G418 and by PCR analysis. The ssa1S2P allele was installed by transforming oligonucleotides (5′ GTATTACAAGAAACAAAAATTCAAGTAAATAACAGATAATATGCCAAAAGCTGTCGGTATTGATTTAGGTACAACATACTCGTGTGTTGCTC3′ and 5′ GAGCAACACACGAGTATGTTGTACCTAAATCAATACCGACAGCTTTTGGCATATTATCTGTTATTTACTTGAATTTTTGTTTCTTGTAATAC3′) into the pCORE-targeted strain and by selecting individual colonies on 5-FOA. Replacements were confirmed by PCR amplification of genomic DNA and sequencing.
Read-Through Assays
Yeast strains were transformed with pUKC815, pUKC817 (UAA), pUKC818 (UAG), or pUKC819 (UGA), grown to midlog phase in minimal media lacking uracil to select for the plasmids, and lysed as previously described (Chernoff et al., 2002). β-Galactosidase activity was determined using a chemiluminescent assay according to the manufacturer's instructions (Galacto-Light, Applied Biosystems, Foster City, CA). Percent read-through is the ratio of activity from stop codon containing constructs to that of the control construct lacking a stop codon (pUKC815). Measured activities were normalized to total protein concentrations (Bradford, Bio-Rad, Hercules, CA) and were determined in triplicate for each lysate.
Imaging, Digital Processing, and Fluorescence Recovery after Photobleaching Analysis
Cells were observed using a Zeiss Axioplan 2 (Carl Zeiss MicroImaging, Thornwood, NY) using a 100× αPlan-FLUAR objective and a Hamamatsu-ORCA camera (Hamamatsu Photonics, Bridgewater, NJ; Satpute-Krishnan and Serio, 2005). Fluorescence recovery after photobleaching (FRAP) analysis was performed and analyzed as previously described (Satpute-Krishnan et al., 2007), but in this case data were collected on a Zeiss LSM 510 confocal microscope.
Analysis of Sup35 Physical State
For fractionation, lysates were prepared from the indicated strains and separated by centrifugation as previously described (Ness et al., 2002) but with the following modifications. Lysis buffer contained 150 mM NaCl, and the lysates were spun for 30 min. After centrifugation, samples were analyzed by SDS-PAGE and anti-Sup35 immunoblotting as previously described (Satpute-Krishnan and Serio, 2005). For semidenaturing detergent agarose gel electrophoresis, lysates were prepared from the indicated strains and analyzed as previously described (Kryndushkin et al., 2003) with the following modifications. Cultures were grown to midlog phase and were lysed in buffer containing 10 mM sodium phosphate (pH 7.5). Lysates were incubated in SDS-sample buffer at 30°C for 7 min before loading on a 1.5% Tris-glycine agarose gel as previously described (Bagriantsev et al., 2006). After transfer to Immobilon-P (Millipore, Bedford, MA), proteins were analyzed by immunoblotting with anti-Sup35 serum. For analysis of prion complex stability, yeast lysates were prepared and incubated in the presence of 2% SDS at the indicated temperatures as previously described before analysis by SDS-PAGE (Satpute-Krishnan and Serio, 2005). For quantification, proteins were transferred to fluorescence optimized PVDF (Immobilon-FL, Millipore). Anti-Sup35 immunoblotting was performed using Q-dot-655 conjugated goat anti-rabbit sera (Invitrogen, Carlsbad, CA) as a secondary antibody, and fluorescence intensity was quantified on a Typhoon imager (GE Healthcare Life Sciences, Piscataway, NJ; excitation λ = 457 nm, emission λ = 670 nm). Sup35 levels were normalized to Pgk1, as assessed by anti-Pgk1 immunoblotting using Q-dot-605 conjugated goat anti-mouse sera as a secondary antibody (excitation λ = 457 nm, emission λ = 610 nm), and were expressed as a percentage of the total (100°C incubation) for each sample.
Protein Expression Analysis
For analysis of chaperone expression profiles, lysates were prepared as described above for the centrifugation assay but analyzed directly without fractionation by SDS-PAGE and immunoblotting for Hsp104 (with antiserum kindly provided by S. Lindquist, Whitehead Institute, Massachusetts Institute of Technology), Ssa1/2 (with antiserum kindly provided by E. Craig, University of Wisconsin-Madison), Ssa3/4 (with antiserum kindly provided by E. Craig, University of Wisconsin-Madison) or Pgk1 (Molecular Probes, Eugene, OR), as a loading control. For analysis of glutathione S-transferase (GST) expression, the indicated yeast strains were grown to midlog phase, and lysates were prepared by alkali-SDS lysis (Kushnirov, 2000) and analyzed by SDS-PAGE and anti-GST (Covance Laboratories, Madison, WI) immunoblotting.
Isoelectric Focusing
Cultures were grown to midlog phase in YPD, harvested, and lysed with glass beads and agitation at 4°C in lysis buffer (20 mM Tris-HCl, pH 7.4, 10 mM NaCl, 1 mM PMSF, and 5 μg/ml pepstatin). Lysates were precleared at 10,000 × g for 10 min, and the supernatant was diluted 50-fold in lysis buffer with glycerol added to 5%. Samples were run on one-dimensional isoelectric focusing (IEF) gels (pH range 5–8, Bio-Rad) according to the manufacturer's instructions. After electrophoresis, the IEF gel was equilibrated in 0.7% acetic acid, transferred to Immobilon PVDF in 0.7% acetic acid, and analyzed by immunoblotting for Ssa1/2 (with antiserum kindly provided by E. Craig, University of Wisconsin-Madison).
Mass Spectrometry
The indicated yeast strains were transformed with an expression plasmid for Sup35 containing an internal hemagglutinin (HA) tag (DePace et al., 1998). Denaturing lysates were prepared as previously described (Satpute-Krishnan and Serio, 2005), and epitope-tagged Sup35 was isolated by immunoprecipitation with beads conjugated to an anti-HA rat mAb (3F10, Roche, Indianapolis, IN). After immunoprecipitation, the sample was electrophoresed onto a preparative SDS-PAGE gel that was subsequently stained with Coomassie Brilliant Blue R-250. A single dominant band, corresponding to the predicted size of Sup35-HA, was apparent, and this species was cut out and subjected to in-gel trypsin digest as previously described (Stone and Williams, 2004). After digestion, the peptide solution was dried under vacuum and resuspended in buffer A (1% acetic acid) containing 200 fmol angiotensin II as an internal standard for LC/MS/MS analysis. Tryptic peptides were analyzed by a fully automated liquid chromatography tandem mass-spectrometry (LC/MS/MS) platform-coupling peptide separation via reversed-phase chromatography with tandem mass spectrometry with static peak parking, as described previously (Ficarro et al., 2005). Briefly, the tryptic peptide preparation was loaded into an online LC autosampler (Agilent, Wilmington, DE; model 1100), which transferred the sample onto a reversed-phase precolumn (75-μm ID fused silica [Polymicro Technologies, Phoenix, AZ] with 2 cm of 5 μm Monitor C18 resin [Column Engineering, Ontario, CA] packed behind a fused silica frit). Loaded peptides were then washed with buffer A. Peptides were then eluted from the precolumn over an analytical column (75-μm ID fused silica with 12 cm of 5-μm Monitor C18 particles with an integrated 4-μm electrospray ionization (ESI) emitter tip fritted with 3 μm silica [Bangs Laboratories, Fishers, IN]) into the mass spectrometer (LTQ-FT Ultra Hybrid, Thermo Electron, Madison, WI) with an HPLC gradient (0–70% solvent B [1% acetic acid in acetonitrile]).
Static peak parking was performed via flow rate reduction from an initial 200 nl/min to 20 nl/min at the ESI tip when peptides began to elute into the mass spectrometer. A 2.0-kV electrospray voltage was applied to draw peptides into the mass spectrometer. Spectra were collected in positive ion mode in data-dependent cycles encompassing one FTICR MS scan (m/z: 400-1800, ∼1 s each), followed by 5 LTQ collisional MS/MS scans (m/z: 300-2000, 0.3 s each) of the five most abundant nonredundant ions from the MS scan. The automatic gain control was 1,000,000 for the MS scan and 10,000 for the MS/MS scans. The maximum ion time was 500 ms for the MS scans and 100 ms for the MS/MS scans. MS/MS spectra were automatically searched against the full Sup35-HA sequence using the SEQUEST algorithm provided with Bioworks 3.2SR1 (Thermo Fisher, Waltham, MA). Search parameters evaluated the following differential modifications: acetylation (+42.0106 Da) on lysine and the protein N-terminus, propionamide alkylation (+71.0371 Da) on cysteine, and oxidation (+15.9949) on methionine. SEQUEST results were filtered by Xcorr (+1 >1.5; +2 >2.0; +3 >2.5) and precursor mass error (<20 ppm window). For all Sup35 peptides exceeding these thresholds, peptide sequence assignments were manually validated, as described previously (Eng et al., 1994). Relative quantitation was performed on all validated peptides via calculation of selected ion chromatogram (SIC) peak areas with a 50-ppm mass window, with normalization to the 200 fmol angiotensin II standard that was spiked into all samples.
GdnHCl Curing
The indicated yeast strains were grown in rich media (YPD) supplemented with 3 mM GdnHCl and maintained in exponential phase for the duration of the experiment. At the indicated generation number (as determined by optical density measurements at 600 nm), aliquots of the cultures were plated on solid YPD media, and the percentage of [PSI+] colonies was determined by their color phenotype as previously described (Eaglestone et al., 2000). Sectored colonies were scored as [PSI+].
Zygote Isolation
Matings, zygote isolation, and diploid confirmation were performed as previously described (Satpute-Krishnan and Serio, 2005). Where indicated, mating cells were treated with GdnHCl for at least 2.5 h before zygote isolation.
RESULTS
NatA Null Strains Do Not Display the [PSI+] Phenotype
To explore the effects of N-terminal acetylation on prion propagation in vivo, we constructed a series of [PSI+] yeast strains individually disrupted for ARD1, NAT3, and MAK3, the catalytic subunits of the NatA, NatB, and NatC Nα-acetyltransferases, respectively (Polevoda et al., 1999), and monitored the [PSI+] phenotype using colony growth assays based on expression of the ade1-14 (UGA) allele (Chernoff et al., 1995). In [psi−] strains, the premature stop codon is utilized efficiently, creating a block in the adenine biosynthetic pathway. This adenine auxotrophy is easily scored by the production of red colonies on rich media and by the inability of the strain to grow on minimal media lacking adenine (Figure 1A). In [PSI+] cells, the premature stop codon is read-through at a low frequency, allowing the formation of white colonies on rich media and growth on minimal media lacking adenine, hallmarks of adenine prototrophy (Figure 1A).
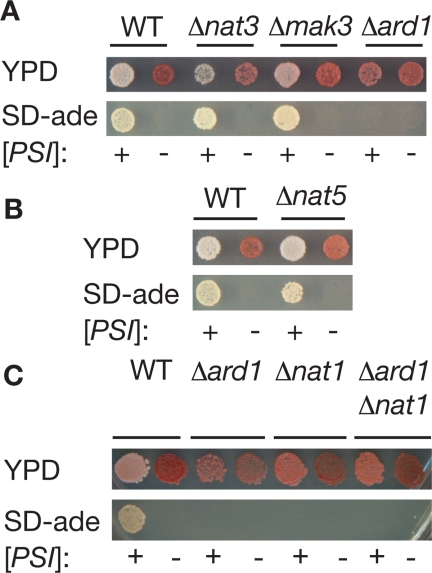
Figure 1.
Genetic interactions between Nα-acetyltransferases and the [PSI+] prion. (A) [PSI+] (+) and [psi−] (−) versions of wild-type (WT, 74D-694), Δnat3 (SY536, SY563), Δmak3 (SY540, SY567), and Δard1 (TRS169, TRS220) yeast strains were spotted onto ¼YEPD (YPD) or minimal media lacking adenine (SD-ade) to assess the [PSI+] phenotype. (B) [PSI+] (+) and [psi−] (−) versions of wild-type (WT, 74D-694) and Δnat5 (SY538, SY565) yeast strains were spotted onto ¼YEPD (YPD) or minimal media lacking adenine (SD-ade) to assess the [PSI+] phenotype. (C) [PSI+] (+) and [psi−] (−) versions of wild-type (WT, 74D-694), Δard1 (TRS169, TRS220), Δnat1 (SY356, SY982), or Δard1Δnat1 (SY319, SY978) yeast strains were spotted onto ¼YEPD (YPD) or minimal media lacking adenine (SD-ade) to assess the [PSI+] phenotype.
NAT3 and MAK3 null strains displayed the [PSI+] phenotype and were efficiently converted to the [psi−] state upon treatment with GdnHCl (Figure 1A), an inhibitor of the molecular chaperone Hsp104, which is required for prion propagation (Chernoff et al., 1995; Grimminger et al., 2004). In contrast, disruption of ARD1 in a [PSI+] strain led to a reversal of the [PSI+] phenotype; the Δard1 [PSI+] strain formed pink colonies on rich media and was unable to grow in the absence of exogenous adenine (Figure 1A). In the [psi−] state, disruption of ARD1 had no observable effects; the strain formed red colonies on rich media and failed to grow in the absence of adenine (Figure 1A). Thus, the ARD1 gene alters the expressivity of the ade1-14 allele in a [PSI+]-dependent manner.
The NatA acetyltransferase is a heterotrimer composed of Ard1, the catalytic subunit (Polevoda et al., 1999), Nat1, a factor essential for NatA activity that tethers the complex to the ribosome for cotranslational modification of substrates (Mullen et al., 1989; Park and Szostak, 1992; Gautschi et al., 2003), and Nat5, a subunit dispensable for NatA function (Gautschi et al., 2003). To further explore the importance of NatA activity on the [PSI+] phenotype, we disrupted NAT1 and NAT5 alone or in combination with ARD1 disruptions. A [PSI+] strain deleted for NAT5 formed white colonies on rich media and on minimal media lacking adenine and, after exposure to GdnHCl, formed red colonies on rich media and failed to grow in the absence of adenine, hallmarks of the [psi−] state (Figure 1B). In contrast, deletion of NAT1 in a [PSI+] strain reversed the prion phenotype, leading to the formation of pink colonies on rich media and the inability to grow in the absence of adenine (Figure 1C). Double mutant Δard1Δnat1 [PSI+] and [psi−] strains were phenotypically indistinguishable from the single disruptions (Figure 1C), consistent with the roles of Ard1 and Nat1 as subunits of a single enzyme complex (Mullen et al., 1989; Park and Szostak, 1992).
Overproduction of Nat1 has previously been linked to chromosome instability (Ouspenski et al., 1999), raising the possibility that another unknown genetic lesion leads to the observed reversal of the prion phenotype in NatA null strains. However, the NAT1 or ARD1 disruptions cosegregated with adenine auxotrophy in 100% of tetrads analyzed (11 or 16 tetrads, respectively), indicating tight linkage. Together, these observations suggest that NatA function is required for [PSI+] strains to display the prion phenotype.
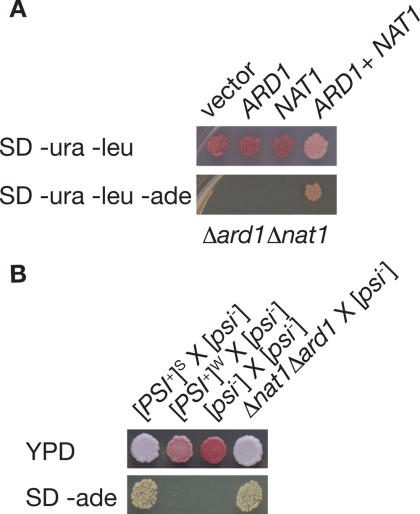
To determine the generality of this effect, we next monitored the phenotype of a second yeast prion, [URE3], using a DAL5 promoter-driven ADE2 reporter in wild-type and NatA mutant strains (Bach et al., 2003). [URE3] is a self-propagating form of the Ure2 protein, a factor that normally mediates nitrogen catabolite repression through the Gln3 transcriptional activator (Blinder et al., 1996). As is the case for [PSI+], the prion state inactivates Ure2, leading to expression of transcriptional targets including DAL5 (Brachmann et al., 2006). Under these conditions, a wild-type [URE3] strain forms white colonies on rich media and in the absence of adenine due to expression of ADE2 from the derepressed DAL5 promoter, whereas a nonprion [ure3-o] strain forms red colonies on rich media and fails to grow in the absence of adenine due to continued repression of the promoter (Supplemental Figure S1). On disruption of ARD1, [URE3] strains continue to form white colonies on rich and adenine-deficient media (Supplemental Figure S1), indicating that NatA activity is not required to detect the [URE3] phenotype. Thus, the NatA effect is specific to the Sup35/[PSI+] prion.
NatA Mutation Specifically Rescues the [PSI+]-dependent Translation Termination Defect
NatA effects on the colony growth assays used to monitor [PSI+] could arise through alteration of prion propagation or, alternately, from an assay-specific effect independent of [PSI+]. To rule out this latter possibility, we used another method to assess [PSI+]-dependent read-through of stop codons. In this case, read-through was monitored by directly assaying the production of β-galactosidase activity expressed from a Pgk1-stop-β-galactosidase reporter (Stansfield et al., 1995). In line with previous studies, each of the three stop codons (UAA, UAG, and UGA) is read through at a low frequency (<2%) in wild-type [PSI+] strains to produce β-galactosidase activity, whereas little activity is recovered in [psi−] strains expressing the same reporters (Figure 2A). The [PSI+]-dependent read-through of these stop codons is absent in Δnat1 strains, consistent with the reversal of the [PSI+] phenotype as assessed by colony growth (Figure 1, A and C). Thus, NatA activity is essential for stop codon read-through in [PSI+] strains.
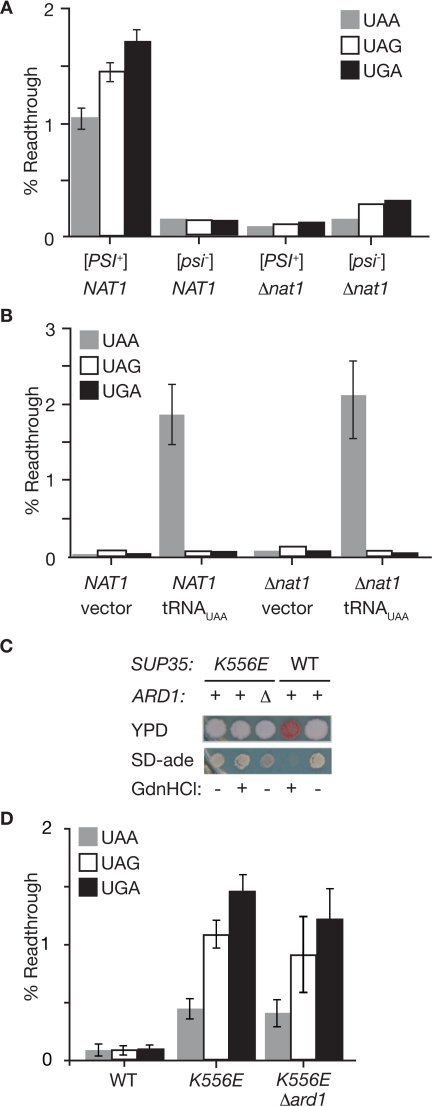
Figure 2.
Disruption of NatA restores efficient translation termination to [PSI+] strains specifically. (A) The efficiency of stop codon read-through in wild-type (NAT1, 74D-694) or Δnat1 (TRS47, TRS51) [PSI+] and [psi−] strains was determined using a series of Pgk1-stop-β-galactosidase reporters. n = 8; error bars, SEM. (B) The efficiency of stop codon read-through in wild-type (NAT1, W303) or Δnat1 (AMR1) strains that had been transformed with an empty vector (vector) or a plasmid expressing a UAA-specific tRNA suppressor (pXS) was determined by expression of β-galactosidase from Pgk1-stop-β-galactosidase reporters. n = 8; error bars, SEM. (C) Yeast strains containing a mutant version of Sup35 with a substitution of glutamic acid for lysine at position 556 (K556E) in the catalytic domain in a wild-type ARD1 (+, TRS64) or Δard1 (Δ, TRS192) background were spotted on ¼YEPD (YPD) or minimal media lacking adenine (SD-ade). For comparison, a wild-type [PSI+] strain (WT, 74D-694) is shown. Treatment with guanidine HCl (GdnHCl, +) cures the [PSI+] prion in the wild-type strain but has no effect on the phenotype of the sup35K556E strain. (D). The efficiency of stop codon read-through in wild-type (WT, 74D-694), sup35K556E (K556E, TRS64), and sup35K556E Δard1 (K556E Δard1, TRS192) [psi−] strains was determined using a series of Pgk1-stop-β-galactosidase reporters. n = 9; error bars, SEM.
Is the NatA requirement specific to nonsense suppression by [PSI+] or is it a general effect on translation termination fidelity? To distinguish between these possibilities, we monitored stop codon read-through mediated either by a tRNA suppressor (Figure 2B) or by a mutation in the Sup35 catalytic domain (Figure 2, C and D). Using the β-galactosidase reporter system, we detected efficient and stop codon-specific read-through of the UAA reporter by a UAA-specific tRNA suppressor in both wild-type and Δnat1 [psi−] strains (Figure 2B). A [psi−] strain containing an amino acid substitution in the catalytic domain of Sup35 (K556E) accumulated similar levels of Sup35 protein as that of a wild-type strain (Supplemental Figure S2), but the Sup35(K556E) protein was functionally compromised, leading to a phenocopy of the [PSI+] state in both the colony-based assays (Figure 2C) and the β-galactosidase reporter assay (Figure 2D). Consistent with its genetic rather than epigenetic basis, the sup35K556E phenotype was not reversed by treatment with GdnHCl (Figure 2C). The sup35K556E phenotype was also unaffected by disruption of ARD1 (Figure 2, C and D), in stark contrast to the effects of this disruption on [PSI+]-dependent read-through of ade1-14 (Figures 1A and 2A). Together, these observations demonstrate that NatA function is specifically required to produce [PSI+]-dependent stop codon read-through in vivo.
[PSI+] Is Propagated Cryptically in NatA Mutant Strains
Reversal of the [PSI+] phenotype in NatA-deficient strains could arise either through loss (curing) of the prion form or through masking of the prion phenotype (antisuppression) in these strains. To distinguish between these possibilities, we complemented the Δard1, Δnat1, and Δard1Δnat1 defects either by introducing plasmid-borne copies of the disrupted genes (Figure 3A and data not shown) or by mating the strains to a wild-type partner (Figure 3B and data not shown). In all cases, the [PSI+] phenotype was restored upon complementation. At the colony level, the diploid phenotype of wild-type and heterozygous mutants was indistinguishable, with both forming white colonies on rich media and in the absence of adenine (Figure 3B and data not shown), but the effects of loss of even one copy of NAT1 could still be detected at the single-cell level using a more sensitive fluorescence-based read-through reporter, with the wild-type strains exhibiting a higher level of stop codon read-through than heterozygous mutants (Supplemental Figure S3). In contrast, when a weak [PSI+] variant, which displays the same phenotype as NatA [PSI+] strains but in this case results from a change in the conformation of Sup35[PSI+] (Derkatch et al., 1996; King and Diaz-Avalos, 2004; Tanaka et al., 2004; Toyama et al., 2007), is crossed to a wild-type [psi−] strain, the resulting diploid retains the weak phenotype, forming pink colonies on rich media that fail to grow in the absence of adenine (Figure 3B). Thus, the reemergence of the [PSI+] phenotype upon complementation of the NatA defect indicates that self-replication of the parental Sup35[PSI+] conformation persists but is somehow phenotypically masked in NatA-deficient strains.
Figure 3.
[PSI+] is propagated cryptically in NatA null strains. (A) A [PSI+] strain disrupted for both ARD1 and NAT1 (Δard1Δnat1, SY319) was transformed with empty vectors (vector), an empty vector and an expression plasmids for ARD1 (pAA2241) or NAT1 (SB208), or ARD1 and NAT1 together (ARD1 + NAT1) and plated on minimal media lacking uracil and leucine (SD−ura−leu) to select for the plasmids or minimal media lacking uracil, leucine, and adenine (SD−ura−leu−ade) to select for the plasmids and for the [PSI+] phenotype. (B) Diploid strains resulting from the following crosses, strong [PSI+] × [psi−], weak [PSI+] × [psi−], [psi−] × [psi−], Δnat1Δard1 (SY319) × [psi−], were plated on ¼ YEPD (YPD) or minimal media lacking adenine (SD-ade) to select for the [PSI+] phenotype.
NatA Does Not Alter the Prion Phenotype by Modifying Known Regulators of the [PSI+] Prion Cycle
Cryptic propagation of the [PSI+] prion has been previously reported in strains with compromised Sup35, Hsp104, and Hsp70 (Ssa) activities (Young and Cox, 1971; Chernoff et al., 1995; Newman et al., 1999; Jung et al., 2000; DePace and Weissman, 2002; Jones and Masison, 2003). We next determined if NatA mediates its effects on the [PSI+] phenotype through these factors.
The prion-determining domain of Sup35 is located at the N-terminus of the protein (Ter-Avanesyan et al., 1994; Bradley and Liebman, 2004; Shkundina et al., 2006). Mutations within this region have profound effects on the efficiency of prion propagation in vivo, ranging from phenotypic masking of the prion state, as we have observed for NatA mutants, to curing (Young and Cox, 1971; DePace et al., 1998; King, 2001). The substrate specificities of the Nα-acetyltransferases are determined in large part by the second amino acid of target proteins (Polevoda et al., 1999), with proteins containing penultimate residues with a small radius of gyration such as Ser, Gly, Thr, and Ala generally targeted for modification by NatA after removal of the initiator methionine. Sup35 is a predicted substrate of NatA, and addition of an acetyl group to its N-terminus would remove a positive charge and add a new functional group to this crucial region.
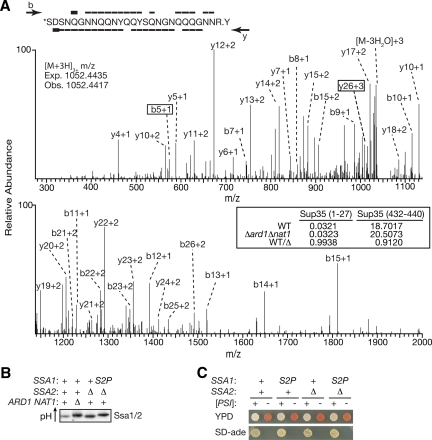
To determine if Sup35 is acetylated by NatA, we purified the protein from wild-type and NatA null strains and analyzed tryptic digests of this material by MS. The theoretical mass of the predicted tryptic peptide originating from the Sup35 N-terminus (amino acids 1-27) is 3154.3068 Da in its acetylated state, and a corresponding peptide (3154.3014 Da) was detected in the mass spectra obtained from the wild-type strain (Figure 4A), consistent with the idea that Sup35 is acetylated in this region. To unambiguously position the modification within this fragment, the b and y ions originating from this peptide were analyzed in the MS/MS spectra. The b5 ion mass is consistent with acetylation of this fragment, whereas the y26 ion mass is consistent with an unacetylated fragment (Figure 4A). Thus, the N-terminal serine residue bears the acetyl group. When Sup35 purified from a NatA null strain was similarly analyzed, the N-terminal acetylated fragment was again detected, indicating that Sup35 continues to be acetylated in the absence of NatA function (Figure 4A). To determine if the abundance of this acetylated fragment was affected by NatA function, we compared the selected ion chromatogram peak areas (SICs) corresponding to the N-terminal fragment and to an internal fragment (amino acids 432-440) between the spectra from wild-type and NatA mutants (Figure 4A, inset). To allow direct comparisons between the same peptides in the different samples, the values were first normalized to an added standard. The ratio of normalized SIC peak areas for the internal peptide approached unity and indicated that each sample contained a similar amount of Sup35 protein, consistent with Coomassie blue staining of the SDS-PAGE gels before excision of the Sup35 bands (data not shown). Similarly, the peak areas for the N-terminal acetylated fragment were nearly identical in wild-type and mutant samples, suggesting that there was minimal change in the abundance of this species. Taken together, these data indicated that the acetylation state of Sup35 is unaltered by the loss of NatA activity. Thus, the NatA effect on the [PSI+] phenotype cannot be explained by loss of Sup35 N-terminal acetylation.
Figure 4.
NatA does not mediate its effects on [PSI+] propagation through known prion regulators. (A) MS/MS spectrum, displayed on two axes to enhance detail, of the Sup35 N-terminal peptide (amino acids 1-27) after purification from a wild-type [psi−] strain (74-D694) and tryptic digestion. Sequence coverage across the peptide is represented by overlines (b-ions) or underlines (y-ions) on the peptide sequence. The boxed ions (b5 and y26) act together to define the site of N-terminal acetylation, marked with an asterisk (*) on the sequence. The expected and observed mass-to-charge ratio (m/z) for the acetylated form of this peptide is indicated on the spectra. The selected ion chromatogram peak areas for both the N-terminal peptide as well as an internal peptide (amino acids 432-440) for the spectra obtained from Sup35 purified from a wild-type (WT, 74D-694) or Δard1Δnat1 (SY978) [psi−] strain along with the ratio of these peak areas (WT/Δ) is shown in the inset. (B) Yeast lysates from a wild-type (74D-694), Δard1Δnat1 (SY319), Δssa2 (SY1338), and Δssa2 ssa1S2P (SY1339) [PSI+] strains were analyzed by IEF and immunoblotting for Sup35. The theoretical pI of Ssa1 is 5.0. (C) Wild-type (WT, 74D-694), ssa1S2P (S2P, SY1308), Δssa2 (Δ2, SY1338), or ssa1S2PΔssa2 (S2P Δ2, SY1339) [psi−] (−) or [PSI+] (+) yeast strains were spotted on ¼YEPD (YPD) or SD-ade to monitor the [PSI+] phenotype.
[PSI+] propagation is exquisitely sensitive to cellular levels of Hsp104, with either inhibition or overexpression of this chaperone leading to quantitative prion loss (Chernoff et al., 1995). Although Hsp104 is not a substrate of NatA (Boucherie et al., 1996; Perrot et al., 1999), loss of NatA activity could perturb [PSI+] propagation through Hsp104 indirectly by altering its expression level. To determine if this was the case, we monitored Hsp104 levels in yeast lysates by immunoblot. Under normal growth conditions, similar levels of Hsp104 are detected in lysates from wild-type and NatA mutant strains (Supplemental Figure S4), indicating that NatA does not modulate the [PSI+] phenotype by changing Hsp104 expression.
In contrast to Hsp104, the Hsp70 proteins Ssa1/2 are known NatA substrates (Polevoda et al., 1999). To determine if the NatA effects on the [PSI+] phenotype could be explained by modification of these factors, we first analyzed the expression pattern of Hsp70 in wild-type and NatA mutant strains. By immunoblotting, Hsp70 constitutively expressed from the SSA1 and SSA2 loci accumulated to similar levels in wild-type and NatA mutants, whereas expression of the stress-inducible forms of Hsp70 from the SSA3 and SSA4 loci were not detected in either wild-type or NatA strains under normal growth conditions (Supplemental Figure S4), indicating that loss of NatA activity did not alter Hsp70 expression profiles.
To further explore a potential role for Hsp70 N-terminal acetylation in modulating the [PSI+] phenotype, we next generated a ssa1 allele containing a Ser-to-Pro substitution at the second position, a change that has been shown to block Nα-acetylation by NatA in other proteins (Huang et al., 1987; Geissenhoner et al., 2004). To rule out the possibility that acetylated Ssa2, which is also expressed under normal growth conditions (Werner-Washburne et al., 1989), could mask any potential effects of unacetylated Ssa1(S2P) on the [PSI+] phenotype, we also disrupted SSA2 in both wild-type and ssa1S2P strains. Notably, Ssa3/4 expression was not induced under these conditions (Supplemental Figure S5); thus, the sole source of Hsp70 was the ssa1S2P allele. To confirm that this substitution did indeed alter acetylation of Ssa1, we analyzed yeast lysates on IEF gels followed by immunoblotting for Ssa1/2 (Figure 4B). Loss of a N-terminal acetyl group will shift the isoeletric point of a protein to a more basic pH because of the positive charge on the N-terminus. As expected (Polevoda et al., 1999), Ssa1/2 derived from a NatA null strain focused at a more basic pH than that derived from a wild-type strain on IEF gels (Figure 4B). Ssa1(S2P) also focused at this more basic point (Figure 4B), consistent with the prediction that the substitution of Pro for Ser at the second position would disrupt N-terminal acetylation of Ssa1.
Strains expressing the acetylation-defective Ssa1(S2P) protein were next analyzed for their ability to support the [PSI+] phenotype. In contrast to NatA mutants, the ssa1S2P and ssa1S2PΔssa2 strains both display the [PSI+] phenotype normally in the colony-based assays, forming white colonies on rich media and growing in the absence of exogenous adenine (Figure 4C). Thus, Hsp70 is not the crucial NatA substrate. Together these observations suggest that NatA mediates its effects on the [PSI+] phenotype through a novel target(s).
Sup35 Efficiently Joins Prion Complexes in the Absence of NatA Function, But Translation Termination Is Not Compromised
Our genetic analyses indicate that the [PSI+] prion persists in NatA mutants but is somehow disconnected from its normal phenotypic effects. What is the molecular basis of this uncoupling? Reversal of the prion phenotype in cells that continue to replicate the Sup35[PSI+] state could arise either from a decrease in the number or activity of prion templates (propagons), as is the case for Hsp104 inactivation (Ness et al., 2002; Satpute-Krishnan et al., 2007), or alternately from a diminished capacity of soluble Sup35 to be incorporated into prion complexes, as has been previously suggested for some point mutations within the Sup35 prion domain (DePace et al., 1998; Kochneva-Pervukhova et al., 1998).
To begin to distinguish between these possibilities, we first determined propagon levels in wild-type and NatA mutant strains. In the presence of GdnHCl, propagon replication is inhibited, and the templates existing in a given cell are redistributed to daughter cells upon division. Under these conditions, [PSI+] cultures begin to switch to the [psi−] state after a lag phase, the length of which is directly related to the number of propagons (Eaglestone et al., 2000). On treatment with GdnHCl, wild-type and NatA mutant [PSI+] strains were cured of the prion state after a similar lag time (Supplemental Figure S6), suggesting that the number of prion templates is not affected by NatA function.
Although the GdnHCl experiments suggest that the NatA effect on the prion phenotype cannot be explained by a change in the production of propagons, loss of NatA activity could impact the activity of prion templates in the conversion of soluble Sup35 to the prion state. To test this idea, we determined the converting capacities of Sup35[PSI+] in wild-type and NatA mutant strains in mating experiments. For these studies, we used a fluorescent reporter, which is expressed only upon read-through of an upstream stop codon (GST-UGA-DsRed-NLS; Satpute-Krishnan and Serio, 2005; Kawai-Noma et al., 2006). Expression of the red fluorescent protein in zygotes requires the rapid conversion of soluble, functional Sup35 to the aggregated state upon fusion of the mating partners (Satpute-Krishnan et al., 2007). Importantly, expression of this reporter was not affected by loss of NatA function independent of its effects on the [PSI+] phenotype (Supplemental Figure S7). As expected, red fluorescence was detected in the nuclei of wild-type zygotes isolated from a [psi−] × [PSI+] cross, but the proportion of zygotes expressing red fluorescence was greatly diminished when propagon replication was blocked by treatment with GdnHCl or was absent when [psi−] strains were crossed to each other (Figure 5A). However, when a Δnat1 [PSI+] strain was crossed to wild-type [psi−] strain, the number of zygotes with red fluorescent nuclei was nearly identical to that of a wild-type [psi−] × [PSI+] cross (Figure 5A), indicating that prion templates were similarly active in wild-type and NatA-deficient strains.
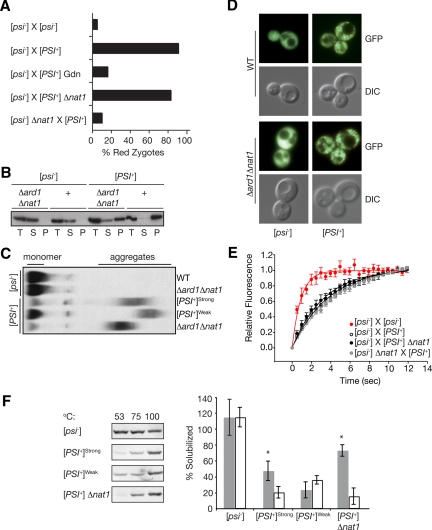
Figure 5.
NatA modulates [PSI+] propagation by ensuring efficient conversion of soluble Sup35 to the prion form. (A) The percentage of zygotes displaying red nuclear fluorescence from a PGPD GST-UGA-DsRed-NLS reporter were scored following the indicated crosses: [psi−] (SY1163 or SY1181) × [psi−], [psi−] (SY1163 or SY1181) × [PSI+] untreated or in the presence of GdnHCl, [psi−] (SY1163) × [PSI+]Δnat1 (SY356), [psi−]Δnat1 (SY1180) × [PSI+]; n ≥ 37. (B) Lysates from Δard1Δnat1 (SY319, SY978) or wild-type (WT) [PSI+] and [psi−] strains were fractionated by centrifugation and analyzed by SDS-PAGE and immunoblotting for Sup35. (C) Lysates from wild-type (WT, [PSI+]Strong, [PSI+]Weak) or Δard1Δnat1 (SY319, SY978) strains were analyzed by semidenaturing detergent agarose gel electrophoresis (SDD-AGE) and immunoblotting for Sup35. (D). Shown are fluorescent (GFP) and bright-field (DIC) images of wild-type (WT, SY80, and SY84) and Δard1Δnat1 (SY1068 and SY330) strains expressing a Sup35-GFP fusion as the sole copy of Sup35. (E). Wild-type (SY360 or SY1370) or Δnat1 (SY1371) [psi−] strains expressing Sup35-GFP were mated to unmarked wild-type (74D-695) or Δnat1 (SY356) [PSI+] strains, and the resulting zygotes were analyzed by FRAP. The recovery half-times (t1/2) were determined by curve-fitting as previously described (Satpute-Krishnan et al., 2007): [psi−] × [psi−] (t1/2 = 0.7 s, R2 = 0.98, n = 10); [psi−] × [PSI+] (t1/2 = 3.1 s, R2 = 0.96, n = 60); [psi−] × [PSI+] Δnat1 (t1/2 = 2.7 s, R2 = 0.95, n = 60); and [psi−] Δnat1 × [psi−] (t1/2 = 3.3 s, R2 = 0.98, n = 71). Error bars, SEM. (F). Lysates from wild-type (74D-695) [psi−], [PSI+]strong, or [PSI+]weak variants or from a [PSI+] Δnat1 mutant were incubated in the presence of SDS at 53, 75, or 100°C before SDS-PAGE and immunoblotting for Sup35 (left). Quantification of the immunoblots, expressed as the percentage of Sup35 at 53°C (white) or 75°C (gray) relative the 100°C sample is indicated in the graph on the right. n ≥ 6; error bars, SD; *p < 0.001.
Because NatA does not appear to alter the number or activity of prion complexes, we next determined if loss of NatA function altered the establishment of a translation termination defect upon conversion of soluble Sup35 to the prion state using a similar single-cell mating experiment. In this case, the reporter was expressed in a Δnat1 [psi−] strain that was crossed to a wild-type [PSI+] strain. Under these conditions, red fluorescent zygotes were observed much less frequently than those isolated from the reciprocal Δnat1 [PSI+] × [psi−] cross (Figure 5A). Thus, the asymmetric effect of NatA on [PSI+] and [psi−] cells in this assay strongly suggests that NatA function modulates the prion phenotype by specifically influencing the pool of Sup35[psi−].
To determine if the faithful termination of translation in [PSI+] strains lacking NatA activity (Figures 1, A and C, and 2A) arose from an alteration in the distribution of Sup35 between the soluble and aggregated pools, we analyzed lysates from wild-type and NatA mutant strains by differential centrifugation and semidenaturing agarose gel electrophoresis (SDD-AGE). As expected (Patino et al., 1996; Paushkin et al., 1996; Kryndushkin et al., 2003), in lysates from wild-type and NatA mutant [psi−] strains, Sup35 was found almost exclusively in the supernatant fraction after differential centrifugation (Figure 5B) and migrated as a monomer near the bottom of the gel by SDD-AGE (Figure 5C). By contrast, in lysates from wild-type [PSI+] strains, Sup35 partitioned almost exclusively to the pellet after centrifugation (Figure 5B), and the majority of the protein migrated as an oligomer in an extended smear toward the top of the gel by SDD-AGE (Figure 5C). In Δard1, Δnat1, and Δard1Δnat1 [PSI+] lysates, however, the physical state of Sup35 was significantly different from the wild-type form (Figure 5, B and C, and data not shown). By differential centrifugation two forms of Sup35 were detected (Figure 5B). The majority of Sup35 was found in the pellet, consistent with the persistence of heritable [PSI+] in these strains (Figure 3, A and B). However, an appreciable portion of the total Sup35 also fractionated to the supernatant in NatA null [PSI+] lysates (Figure 5B). By SDD-AGE, Sup35 oligomeric complexes migrated farther on the gel when lysates were prepared from Δard1, Δnat1, or Δard1Δnat1 strains than from wild-type strains (Figure 5C and data not shown), indicating a NatA-dependent change in this species. Surprisingly, an increase in soluble Sup35 was not detected in Δard1, Δnat1, or Δard1Δnat1 [PSI+] lysates by SDD-AGE (Figure 5C and data not shown), although this technique could readily detect accumulation of soluble Sup35 in a weak [PSI+] lysate (Figure 5C).
In an attempt to reconcile the disparate results of the two biochemical assays with regard to the soluble pool of Sup35, we analyzed the physical state of a Sup35-GFP fusion protein in wild-type and NatA [PSI+] and [psi−] cells by microscopy (Figure 5D). As we have previously shown (Satpute-Krishnan and Serio, 2005), fluorescence from this fusion is diffusely distributed throughout the cytoplasm in [psi−] strains but coalesces into foci in [PSI+] strains (Figure 5D). In NatA-deficient strains, Sup35-GFP fluorescence patterns are distinct in [psi−] and [PSI+] strains (i.e., diffuse vs. punctate, respectively), and these patterns are indistinguishable from those of wild-type strains (Figure 5D). Despite the appearance of fluorescent Sup35-GFP foci in the NatA mutant strain, a pool of soluble Sup35 could coexist with these complexes, as suggested by differential centrifugation (Figure 5B), but be difficult to detect given the density of the prion complexes in the cytoplasm. To rule out this possibility, we monitored the mobility of Sup35-GFP in wild-type and NatA mutant strains using FRAP experiments, which are capable of quantitatively detecting soluble Sup35-GFP in individual cells containing a mixture of protein forms (Satpute-Krishnan et al., 2007). When a [psi−] cell expressing Sup35-GFP is mated to an unmarked [psi−] cell and the resulting zygote is analyzed by FRAP, fluorescence recovery in the bleached area is rapid, with a half-time (t1/2) ∼0.7 s. However, when the same Sup35-GFP expressing [psi−] strain is mated to an unmarked wild-type [PSI+] strain, fluorescence recovery is slowed (t1/2 ∼3.1 s) because of the incorporation of soluble Sup35-GFP into prion complexes (Song et al., 2005; Satpute-Krishnan et al., 2007). When a [psi−] strain expressing Sup35-GFP was mated to an unmarked NatA mutant [PSI+] strain, the rate of fluorescence recovery by FRAP was indistinguishable from the wild-type [psi−] × [PSI+] cross (t1/2 ∼2.7 s, Figure 5E), consistent with the rapid incorporation of soluble Sup35 in this cross (Figure 5A). In the reciprocal [psi−] Δnat1 × [PSI+] cross, Sup35-GFP recovered with a half-time consistent with the presence of prion complexes (t1/2 ∼3.3 s, Figure 5E), despite the accurate translation termination observed in these zygotes (Figure 5A). Thus, an increase in soluble Sup35 cannot explain reversion of the [PSI+] phenotype in NatA mutant strains.
How can the efficient self-assembly of Sup35 (Figure 5, C and E) be reconciled with the accurate termination of translation in NatA-deficient [PSI+] strains (Figures 1, A and C, 2A, and 5A)? We reasoned that perhaps this uncoupling arose from a change in the nature of the interaction between Sup35 and prion complexes in NatA mutant strains. To directly test this idea, we assessed the stability of wild-type and NatA mutant prion complexes by their sensitivity to detergent solubilization at different temperatures (Tanaka et al., 2004). For these studies, we prepared lysates and incubated them in the presence of 2% SDS at 53, 75, or 100°C before SDS-PAGE and anti-Sup35 immunoblotting (Figure 5F). When [psi−] lysates are treated in this manner, equal amounts of Sup35 entered the gel regardless of the incubation temperature, consistent with their monomeric state. When [PSI+] lysates were subjected to the same conditions, <20% of the total Sup35 entered the gel at 53°C (Figure 5F), and this proportion rose to ∼50% when the lysates were incubated at 75°C before electrophoresis (Figure 5F), presumably because of the partial disruption of prion complexes at this temperature. For prion complexes derived from a NatA mutant strain, the proportion of Sup35 that entered the gel was comparable to that for a wild-type strain after incubation at 53°C (Figure 5F), consistent with the similar levels of soluble Sup35 in these strains (Figure 5, D and E). However, after incubation at 75°C, a greater proportion of Sup35 entered the gel from a NatA mutant strain than from a wild-type strain (∼75 vs. ∼50%, Figure 5F). This increase in recovered Sup35 indicates that [PSI+] NatA complexes are less stable than wild-type complexes under these conditions and provides an explanation for the discrepancy between the centrifugation and SDD-AGE assays, because the lysates are incubated in the presence of detergent for a longer period of time with the former approach. Thus, despite the efficient recruitment of soluble Sup35 to prion complexes in the absence of NatA function (Figure 5, C and E), the strength of this association is diminished, creating a situation that allows efficient translation termination (Figures 1, A and C, 2A, and 5A).
DISCUSSION
The accepted model for the [PSI+] phenotype posits that the incorporation of soluble and functional Sup35 into oligomeric complexes upon conversion to the Sup35[PSI+] prion form compromises the normal activity of this protein, leading to a defect in translation termination (Patino et al., 1996; Paushkin et al., 1996). However, our studies on the genetic interaction between NatA and [PSI+] have, for the first time, uncoupled the process of Sup35 aggregation from the prion-associated phenotype. A similar proportion of Sup35 protein is incorporated into prion complexes in wild-type and NatA mutant strains at steady state (Figure 5, C, E, and F), but the stability of these aggregates is diminished in the absence of NatA function (Figure 5F). This change in stability does not alter the activity of these complexes as prion templates, as the number of propagons in and the converting capacity of Sup35[PSI+] in wild-type and mutant strains are nearly identical (Supplemental Figure S6 and Figure 5A). Rather, this change in stability reflects a failure to establish a translation termination defect upon the incorporation of soluble Sup35 into prion complexes (Figures 1, A and C, and 5A).
Which target(s) of NatA are crucial to link the [PSI+] prion phenotype to Sup35 complexes in vivo? Although our mass spectrometry studies indicate that Sup35 is N-terminally acetylated, it is not a substrate for NatA (Figure 4A); thus, reversion of the prion phenotype in NatA strains (Figure 1, A and C) cannot be attributed to a change in Sup35 modification. Other possible targets include Hsp104, Hsp70, and cochaperones that modulate the ATPase activity of Hsp70. Mutations in all of these factors produce a similar reversion of the [PSI+] phenotype (Chernoff et al., 1995; Jones and Masison, 2003; Fan et al., 2007; Kryndushkin and Wickner, 2007; Sadlish et al., 2008), but several lines of evidence suggest that these proteins are not the crucial target(s) of NatA. First, our expression profiling, genetic, and biochemical analyses indicate that Hsp104 and Hsp70 are neither the direct nor indirect targets of this pathway (Supplemental Figure S4 and Figure 4, B and C). Second, strains with impaired Hsp104 or Hsp70 activity are defective in the generation of new propagons (Ness et al., 2002; Song et al., 2005), whereas NatA mutants sustain wild-type propagon levels (Supplemental Figure S6). Third, the steady-state size of SDS-resistant Sup35 aggregates increases in [PSI+] strains deficient in Hsp104, Hsp70, or Hsp70 cochaperones (Eaglestone et al., 2000; Wegrzyn et al., 2001; Cox et al., 2003; Kryndushkin et al., 2003; Jones et al., 2004; Song et al., 2005; Fan et al., 2007; Kryndushkin and Wickner, 2007; Satpute-Krishnan et al., 2007; Sadlish et al., 2008), an observation that is completely opposed to our findings for NatA mutants (Figure 5C). In addition to these factors, the ubiquitin-conjugating enzyme Ubc4 and the Hsp70 family members Ssb1 and Ssb2 are predicted or proven substrates for NatA (Huang et al., 1987; Polevoda et al., 1999), are modifiers of the [PSI+] prion cycle (Chernoff et al., 1999; Allen et al., 2007), and have demonstrated synthetic interactions with NatA (Gautschi et al., 2003; Pan et al., 2006). Despite these intriguing connections, Δubc4 and Δssb1Δssb2 strains, unlike NatA null strains, display the [PSI+] nonsense suppression phenotype, indicating that they are unlikely to be the crucial NatA targets. Taken together, these observations point to a previously uncharacterized activity that is crucial for establishing the translation termination defect associated with Sup35 prion complexes in vivo.
By what mechanism may such a trans-regulator function? The asymmetric complementation of the NatA defect at the single-cell level likely provides significant insight. At the point of fusion, zygotes derived from either Δnat1 [PSI+] × [psi−] or [PSI+] × Δnat1 [psi−] crosses are predicted to have the same mix of acetylated and nonacetylated proteins, yet soluble Sup35 only directs the establishment of a translation termination defect if it is derived from a wild-type strain (Figure 5A). These observations indicate that the soluble pools of Sup35 in wild-type and NatA strains are fundamentally different. As our mass spectrometry studies indicate that the NatA null phenotype is not a manifestation of the loss of a N-terminal acetyl group from Sup35 (Figure 4A), we suggest that a trans-factor(s) binds to soluble Sup35 and facilitates the establishment of a translation termination defect upon the introduction of prion complexes. This effect could represent a change in the physical state of Sup35 induced by the trans-factor or alternately the recruitment of additional proteins into prion complexes via their association with soluble Sup35. In either scenario, loss of the N-terminal acetyl group from the factor would impair either its activity on or association with soluble Sup35, impacting both the biochemical properties of prion complexes (Figure 5, C and F) as well as the link between Sup35 aggregation and translation termination efficiency (Figure 5A).
How does a change in aggregate stability restore accurate termination to [PSI+] strains without increasing the soluble pool of Sup35? There are several possible scenarios. First, reducing the stability of prion complexes could allow enhanced exchange of Sup35 between the inactive, aggregated form and the functional, soluble form, thereby rescuing the translation termination defect in these strains by continually creating an active but transient pool. Consistent with this idea, dynamic exchange of subunits has been detected for self-replicating complexes of various proteins in vitro (Carulla et al., 2005; Maji et al., 2008). Second, the reduced stability of prion complexes in NatA mutant strains could reflect a partial or complete loss of factors other than Sup35 from these aggregates. If these factors normally inhibit interaction of Sup35 with other components of the translation termination machinery, their loss could allow Sup35 to function within the context of the aggregate. In support of this idea, the functional domain of Sup35 is located at the C-terminus of the protein and remains accessible in the absence of other factors upon self-assembly in vitro (Glover et al., 1997; Serio et al., 2000), and other proteins, such as GFP (Figure 5E) (Patino et al., 1996), retain their normal fold and activity despite incorporation into prion complexes when expressed as C-terminal fusions to the Sup35 prion domain. Alternately, if these factors are themselves components of the translation termination machinery (Paushkin et al., 1997; Czaplinski et al., 1998) even their partial release from prion complexes in NatA mutant strains could increase translation termination fidelity. Future studies to identify the crucial NatA substrate(s) will shed additional light on this previously uncharacterized, N-terminal acetylation-dependent event and thereby the processes that allow protein-only genetic determinants to establish and maintain novel phenotypes.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Sternglanz (SUNY Stony Brook), Y. Chernoff (Georgia Institute of Technology), S. Lindquist (Whitehead Institute, Massachusetts Institute of Technology), M. Resnick (National Institutes of Health), E. Craig (University of Wisconsin–Madison), and C. Cullin (Université Bordeaux, France) for reagents and for helpful discussions, G. Funk for techical assistance, and J. Laney and members of the Laney and Serio labs for helpful discussions and comments on the manuscript. This work was supported by grants from the Pew Scholars Program in the Biomedical Sciences (3274sc), the National Cancer Institute (CA096402), and the National Institute of General Medical Sciences (NIGMS) Grant GM069802 to T.R.S. J.A.P. is supported by a National Research Service Award Grant F32 GM080907 from the NIGMS.
Abbreviations used:
- GdnHCl
guanidine HCl
- FRAP
fluorescence recovery after photobleaching.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-04-0436) on December 10, 2008.
REFERENCES
- Allen K. D., Chernova T. A., Tennant E. P., Wilkinson K. D., Chernoff Y. O. Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J. Biol. Chem. 2007;282:3004–3013. doi: 10.1074/jbc.M609597200. [DOI] [PubMed] [Google Scholar]
- Bach S., Talarek N., Andrieu T., Vierfond J. M., Mettey Y., Galons H., Dormont D., Meijer L., Cullin C., Blondel M. Isolation of drugs active against mammalian prions using a yeast-based screening assay. Nat. Biotechnol. 2003;21:1075–1081. doi: 10.1038/nbt855. [DOI] [PubMed] [Google Scholar]
- Bagriantsev S. N., Kushnirov V. V., Liebman S. W. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
- Bailleul P. A., Newnam G. P., Steenbergen J. N., Chernoff Y. O. Genetic study of interactions between the cytoskeletal assembly protein sla1 and prion-forming domain of the release factor sup35 (eRF3) in Saccharomyces cerevisiae. Genetics. 1999;153:81–94. doi: 10.1093/genetics/153.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R., Panic B., Whyte J. R., Munro S. Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat. Cell Biol. 2004;6:405–413. doi: 10.1038/ncb1120. [DOI] [PubMed] [Google Scholar]
- Blinder D., Coschigano P. W., Magasanik B. Interaction of the GATA factor Gln3p with the nitrogen regulator Ure2p in Saccharomyces cerevisiae. J. Bacteriol. 1996;178:4734–4736. doi: 10.1128/jb.178.15.4734-4736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucherie H., Sagliocco F., Joubert R., Maillet I., Labarre J., Perrot M. Two-dimensional gel protein database of Saccharomyces cerevisiae. Electrophoresis. 1996;17:1683–1699. doi: 10.1002/elps.1150171106. [DOI] [PubMed] [Google Scholar]
- Brachmann A., Toombs J. A., Ross E. D. Reporter assay systems for [URE3] detection and analysis. Methods. 2006;39:35–42. doi: 10.1016/j.ymeth.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Bradley M. E., Liebman S. W. The Sup35 domains required for maintenance of weak, strong or undifferentiated yeast [PSI+] prions. Mol. Microbiol. 2004;51:1649–1659. doi: 10.1111/j.1365-2958.2003.03955.x. [DOI] [PubMed] [Google Scholar]
- Carulla N., Caddy G. L., Hall D. R., Zurdo J., Gairi M., Feliz M., Giralt E., Robinson C. V., Dobson C. M. Molecular recycling within amyloid fibrils. Nature. 2005;436:554–558. doi: 10.1038/nature03986. [DOI] [PubMed] [Google Scholar]
- Chernoff Y. O., Derkach I. L., Inge-Vechtomov S. G. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr. Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- Chernoff Y. O., Lindquist S. L., Ono B., Inge-Vechtomov S. G., Liebman S. W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [PSI+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Chernoff Y. O., Newnam G. P., Kumar J., Allen K., Zink A. D. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone Ssb in formation, stability, and toxicity of the [PSI] prion. Mol. Cell. Biol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff Y. O., Uptain S. M., Lindquist S. L. Analysis of prion factors in yeast. Methods Enzymol. 2002;351:499–538. doi: 10.1016/s0076-6879(02)51867-x. [DOI] [PubMed] [Google Scholar]
- Chernova T. A., Allen K. D., Wesoloski L. M., Shanks J. R., Chernoff Y. O., Wilkinson K. D. Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due to depletion of the free ubiquitin pool. J. Biol. Chem. 2003;278:52102–52115. doi: 10.1074/jbc.M310283200. [DOI] [PubMed] [Google Scholar]
- Cox B. [PSI], a cytoplasmic suppressor of super-suppression in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- Cox B., Ness F., Tuite M. Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria M. J., Paushkin S. V., Han X., Weng Y., Perlick H. A., Dietz H. C., Ter-Avanesyan M. D., Peltz S. W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePace A. H., Santoso A., Hillner P., Weissman J. S. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- DePace A. H., Weissman J. S. Origins and kinetic consequences of diversity in Sup35 yeast prion fibers. Nat. Struct. Biol. 2002;9:389–396. doi: 10.1038/nsb786. [DOI] [PubMed] [Google Scholar]
- Derkatch I. L., Chernoff Y. O., Kushnirov V. V., Inge-Vechtomov S. G., Liebman S. W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doel S. M., McCready S. J., Nierras C. R., Cox B. S. The dominant PNM2- mutation which eliminates the psi factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics. 1994;137:659–670. doi: 10.1093/genetics/137.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone S. S., Ruddock L. W., Cox B. S., Tuite M. F. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI(+)] of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2000;97:240–244. doi: 10.1073/pnas.97.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J., McCormack A., Yates J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrosc. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Fan Q., Park K. W., Du Z., Morano K. A., Li L. The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics. 2007;177:1583–1593. doi: 10.1534/genetics.107.077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro S. B., Salomon A. R., Brill L. M., Mason D. E., Stettler-Gill M., Brock A., Peters E. C. Automated immobilized metal affinity chromatography/nan-liquid chromatography/electrospray ionization mass spectrometry platform for profiling protein phosphorylation sites. Rapid Commun. Mass Spectrom. 2005;19:57–71. doi: 10.1002/rcm.1746. [DOI] [PubMed] [Google Scholar]
- Ganusova E. E., Ozolins L. N., Bhagat S., Newnam G. P., Wegrzyn R. D., Sherman M. Y., Chernoff Y. O. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol. Cell. Biol. 2006;26:617–629. doi: 10.1128/MCB.26.2.617-629.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M., Just S., Mun A., Ross S., Rucknagel P., Dubaquie Y., Ehrenhofer-Murray A., Rospert S. The yeast N(alpha)-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol. Cell. Biol. 2003;23:7403–7414. doi: 10.1128/MCB.23.20.7403-7414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissenhoner A., Weise C., Ehrenhofer-Murray A. E. Dependence of ORC silencing function on NatA-mediated Nalpha acetylation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:10300–10312. doi: 10.1128/MCB.24.23.10300-10312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J. R., Kowal A. S., Schirmer E. C., Patino M. M., Liu J. J., Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Griffith J. S. Self-replication and scrapie. Nature. 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- Grimminger V., Richter K., Imhof A., Buchner J., Walter S. The prion curing agent guanidinium chloride specifically inhibits ATP hydrolysis by Hsp104. J. Biol. Chem. 2004;279:7378–7383. doi: 10.1074/jbc.M312403200. [DOI] [PubMed] [Google Scholar]
- Huang S., et al. Specificity of cotranslational amino-terminal processing of proteins in yeast. Biochemistry. 1987;26:8242–8246. doi: 10.1021/bi00399a033. [DOI] [PubMed] [Google Scholar]
- Jones G., Song Y., Chung S., Masison D. C. Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol. Cell. Biol. 2004;24:3928–3937. doi: 10.1128/MCB.24.9.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G. W., Masison D. C. Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+] Genetics. 2003;163:495–506. doi: 10.1093/genetics/163.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G., Jones G., Wegrzyn R. D., Masison D. C. A role for cytosolic hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai-Noma S., Ayano S., Pack C. G., Kinjo M., Yoshida M., Yasuda K., Taguchi H. Dynamics of yeast prion aggregates in single living cells. Genes Cells. 2006;11:1085–1096. doi: 10.1111/j.1365-2443.2006.01004.x. [DOI] [PubMed] [Google Scholar]
- King C. Y. Supporting the structural basis of prion strains: induction and identification of [PSI] variants. J. Mol. Biol. 2001;307:1247–1260. doi: 10.1006/jmbi.2001.4542. [DOI] [PubMed] [Google Scholar]
- King C. Y., Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- Kochneva-Pervukhova N. V., Paushkin S. V., Kushnirov V. V., Cox B. S., Tuite M. F., Ter-Avanesyan M. D. Mechanism of inhibition of PSI+ prion determinant propagation by a mutation of the N-terminus of the yeast Sup35 protein. EMBO J. 1998;17:5805–5810. doi: 10.1093/emboj/17.19.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D., Wickner R. B. Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:2149–2154. doi: 10.1091/mbc.E07-02-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D. S., Alexandrov I. M., Ter-Avanesyan M. D., Kushnirov V. V. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- Kryndushkin D. S., Smirnov V. N., Ter-Avanesyan M. D., Kushnirov V. V. Increased expression of Hsp40 chaperones, transcriptional factors, and ribosomal protein Rpp0 can cure yeast prions. J. Biol. Chem. 2002;277:23702–23708. doi: 10.1074/jbc.M111547200. [DOI] [PubMed] [Google Scholar]
- Kushnirov V. V. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maji S. K., Schubert D., Rivier C., Lee S., Rivier J. E., Riek R. Amyloid as a depot for the formulation of long-acting drugs. PLoS Biol. 2008;6:e17. doi: 10.1371/journal.pbio.0060017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J. R., Kayne P. S., Moerschell R. P., Tsunasawa S., Gribskov M., Colavito-Shepanski M., Grunstein M., Sherman F., Sternglanz R. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 1989;8:2067–2075. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness F., Ferreira P., Cox B. S., Tuite M. F. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol. Cell. Biol. 2002;22:5593–5605. doi: 10.1128/MCB.22.15.5593-5605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman G. P., Wegrzyn R. D., Lindquist S. L., Chernoff Y. O. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouspenski I. I., Elledge S. J., Brinkley B. R. New yeast genes important for chromosome integrity and segregation identified by dosage effects on genome stability. Nucleic Acids Res. 1999;27:3001–3008. doi: 10.1093/nar/27.15.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Ye P., Yuan D. S., Wang X., Bader J. S., Boeke J. D. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Park E. C., Szostak J. W. ARD1 and NAT1 proteins form a complex that has N-terminal acetyltransferase activity. EMBO J. 1992;11:2087–2093. doi: 10.1002/j.1460-2075.1992.tb05267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K. W., Hahn J. S., Fan Q., Thiele D. J., Li L. De novo appearance and “strain” formation of yeast prion [PSI+] are regulated by the heat-shock transcription factor. Genetics. 2006;173:35–47. doi: 10.1534/genetics.105.054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino M. M., Liu J. J., Glover J. R., Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- Paushkin S. V., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. Propagation of the yeast prion-like [PSI+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Paushkin S. V., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. Interaction between yeast Sup45p (eRF1) and Sup35p (eRF3) polypeptide chain release factors: implications for prion-dependent regulation. Mol. Cell. Biol. 1997;17:2798–2805. doi: 10.1128/mcb.17.5.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot M., Sagliocco F., Mini T., Monribot C., Schneider U., Shevchenko A., Mann M., Jeno P., Boucherie H. Two-dimensional gel protein database of Saccharomyces cerevisiae (update 1999) Electrophoresis. 1999;20:2280–2298. doi: 10.1002/(SICI)1522-2683(19990801)20:11<2280::AID-ELPS2280>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Pezza J. A., Serio T. R. Prion propagation: the role of protein dynamics. Prion. 2007;1:36–43. doi: 10.4161/pri.1.1.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B., Norbeck J., Takakura H., Blomberg A., Sherman F. Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. EMBO J. 1999;18:6155–6168. doi: 10.1093/emboj/18.21.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B., Sherman F. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol. 2003;325:595–622. doi: 10.1016/s0022-2836(02)01269-x. [DOI] [PubMed] [Google Scholar]
- Prusiner S. B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- Sadlish H., Rampelt H., Shorter J., Wegrzyn R. D., Andreasson C., Lindquist S., Bukau B. Hsp110 chaperones regulate prion formation and propagation in S. cerevisiae by two discrete activities. PLoS ONE. 2008;3:e1763. doi: 10.1371/journal.pone.0001763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpute-Krishnan P., Langseth S. X., Serio T. R. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol. 2007;5:e24. doi: 10.1371/journal.pbio.0050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpute-Krishnan P., Serio T. R. Prion protein remodelling confers an immediate phenotypic switch. Nature. 2005;437:262–265. doi: 10.1038/nature03981. [DOI] [PubMed] [Google Scholar]
- Serio T. R., Cashikar A. G., Kowal A. S., Sawicki G. J., Moslehi J. J., Serpell L., Arnsdorf M. F., Lindquist S. L. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- Serio T. R., Lindquist S. L. [PSI+]: an epigenetic modulator of translation termination efficiency. Annu. Rev. Cell Dev. Biol. 1999;15:661–703. doi: 10.1146/annurev.cellbio.15.1.661. [DOI] [PubMed] [Google Scholar]
- Setty S. R., Strochlic T. I., Tong A. H., Boone C., Burd C. G. Golgi targeting of ARF-like GTPase Arl3p requires its Nalpha-acetylation and the integral membrane protein Sys1p. Nat. Cell Biol. 2004;6:414–419. doi: 10.1038/ncb1121. [DOI] [PubMed] [Google Scholar]
- Shkundina I. S., Kushnirov V. V., Tuite M. F., Ter-Avanesyan M. D. The role of the N-terminal oligopeptide repeats of the yeast Sup35 prion protein in propagation and transmission of prion variants. Genetics. 2006;172:827–835. doi: 10.1534/genetics.105.048660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J. M., Shaw J. M. Mdm20 protein functions with Nat3 protein to acetylate Tpm1 protein and regulate tropomyosin-actin interactions in budding yeast. Proc. Natl. Acad. Sci. USA. 2003;100:7644–7649. doi: 10.1073/pnas.1232343100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizonenko G. I., Chernov Iu O., Kulikov V. N., Karpova T. S., Kashkin P. K., Pavlov I., Bekhtereva T. A., Sakharova E. V., Tikhodeev O. N., Inge-Vechtomov S. G. A new class of ochre-suppressors in Saccharomyces: mutations in the tRNA-gln gene. Doklady Akademii nauk SSSR. 1990;310:1480–1484. [PubMed] [Google Scholar]
- Song Y., Wu Y. X., Jung G., Tutar Y., Eisenberg E., Greene L. E., Masison D. C. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot. Cell. 2005;4:289–297. doi: 10.1128/EC.4.2.289-297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I., Akhmaloka , Tuite M. F. A mutant allele of the SUP45 (SAL4) gene of Saccharomyces cerevisiae shows temperature-dependent allosuppressor and omnipotent suppressor phenotypes. Curr. Genet. 1995;27:417–426. doi: 10.1007/BF00311210. [DOI] [PubMed] [Google Scholar]
- Stone K. L., Williams K. R. Enzymatic digestion of proteins in gels for mass spectrometric identification and structural analysis. In: Coligan J. E., editor. Current Protocols in Protein Science. Hoboken, NJ: John Wiley and Sons, Chapter 11, Unit 11-13; 2004. [DOI] [PubMed] [Google Scholar]
- Storici F., Lewis L. K., Resnick M. A. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Chien P., Naber N., Cooke R., Weissman J. S. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan M. D., Dagkesamanskaya A. R., Kushnirov V. V., Smirnov V. N. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [PSI+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero J. C., Wickner R. B. MAK3 encodes an N-acetyltransferase whose modification of the L-A gag NH2 terminus is necessary for virus particle assembly. J. Biol. Chem. 1992;267:20277–20281. [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Toyama B. H., Kelly M. J., Gross J. D., Weissman J. S. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- Tuite M. F., Mundy C. R., Cox B. S. Agents that cause a high frequency of genetic change from [PSI+] to [psi−] in Saccharomyces cerevisiae. Genetics. 1981;98:691–711. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn R. D., Bapat K., Newnam G. P., Zink A. D., Chernoff Y. O. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol. Cell. Biol. 2001;21:4656–4669. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M., Becker J., Kosic-Smithers J., Craig E. A. Yeast Hsp70 RNA levels vary in response to the physiological status of the cell. J. Bacteriol. 1989;171:2680–2688. doi: 10.1128/jb.171.5.2680-2688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M., Szostak J. W. The ARD1 gene of yeast functions in the switch between the mitotic cell cycle and alternative developmental pathways. Cell. 1985;43:483–492. doi: 10.1016/0092-8674(85)90178-3. [DOI] [PubMed] [Google Scholar]
- Wickner R. B. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Young C., Cox B. Extrachromosomal elements in a super-suppression system of yeast. I. A nuclear gene controlling the inheritance of the extrachromosomal elements. Heredity. 1971;26:413–422. [Google Scholar]
- Zhouravleva G., Frolova L., Le Goff X., Le Guellec R., Inge-Vechtomov S., Kisselev L., Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.