Abstract
Basal bodies organize cilia that are responsible for both mechanical beating and sensation. Nascent basal body assembly follows a series of well characterized morphological events; however, the proteins and their assembly dynamics for new basal body formation and function are not well understood. High-resolution light and electron microscopy studies were performed in Tetrahymena thermophila to determine how proteins assemble into the structure. We identify unique dynamics at basal bodies for each of the four proteins analyzed (α-tubulin, Spag6, centrin, and Sas6a). α-Tubulin incorporates only during new basal body assembly, Spag6 continuously exchanges at basal bodies, and centrin and Sas6a exhibit both of these patterns. Centrin loads and exchanges at the basal body distal end and stably incorporates during new basal body assembly at the nascent site of assembly and the microtubule cylinder. Conversely, both dynamic and stable populations of Sas6a are found only at a single site, the cartwheel. The bimodal dynamics found for centrin and Sas6a reveal unique protein assembly mechanisms at basal bodies that may reflect novel functions for these important basal body and centriolar proteins.
INTRODUCTION
Centrioles and basal bodies are responsible for organizing centrosomes and cilia, respectively. Centrosomes organize the bipolar spindle that is necessary for segregation of the duplicated genome during mitosis, and cilia generate mechanical flow and sense the surrounding cellular environment (Marshall and Nonaka, 2006). The majority of human cells contain cilia at some point during their life cycle. During cell cycle quiescence, primary cilia are organized by basal bodies that reside at the cell cortex. On cell cycle entry, cilia resorb and the basal body migrates to the nucleus where it functions as a centriole to organize centrosomes. Mutations affecting centrioles, basal bodies, and cilia are associated with several human diseases (Badano et al., 2006; Pan and Snell, 2007; Marshall, 2008). A wide range of pathologies are associated with these defects, including cystic kidneys, polydactyly, obesity, and retinal degeneration. The diversity in maladies likely results from the complex number of cellular processes in which these structures are involved (Marshall, 2008). Centrioles, basal bodies, and cilia are important for mitosis, polarity, cell division, protein trafficking, signaling, motility, and sensation. The modification of these organelles for each function involves a series of regulated events, including self-assembly of new structures. For nascent centriole and basal body assembly, the mechanism for how proteins comprising these structures are modified and regulated for their individual and collective roles is not well understood.
Nascent centriole and basal body assembly involves a series of well conserved morphological stages leading to a mature structure (Dippell, 1968; Allen, 1969; Anderson and Brenner, 1971; Cavalier-Smith, 1974; Kuriyama and Borisy, 1981; Pelletier et al., 2006). The cartwheel, a central hub with nine radial symmetric spokes, is built on the generative disk, followed by triplet microtubules assembled perpendicular to each of the nine cartwheel spokes to form the basal body cylinder. The cylinder is capped by a laminar electron density at the distal end that forms the transition zone linking the basal body triplet microtubules to the ciliary doublets. The transition zone extends a central density from which one microtubule of the ciliary central pair originates (Allen, 1969). Despite the clearly defined structural events of new basal body and centriole formation, the molecular assembly pathway is not as well understood.
Recent centriole, basal body, and cilia studies have focused on determining the many (>1000) proteins comprising these structures (Gherman et al., 2006; Inglis et al., 2006). Specifically, molecular components of centrioles and basal bodies were identified by mass spectrometry (Keller et al., 2005; Kilburn et al., 2007). We previously used immunoelectron microscopy (IEM) to determine the domains within the basal body to which newly identified components reside (Kilburn et al., 2007). The localization may reveal roles for these components within the overall assembly and organization of the structure. For example, both Sas-6 and Bld10 localize solely to cartwheels and are required for the formation of the ninefold symmetric centriole and basal body structure (Dammermann et al., 2004; Matsuura et al., 2004; Leidel et al., 2005; Pelletier et al., 2006; Kleylein-Sohn et al., 2007; Nakazawa et al., 2007; Rodrigues-Martins et al., 2007; Strnad et al., 2007; Vladar and Stearns, 2007; Yabe et al., 2007). In contrast, several components localize to more than one ultrastructural domain, suggesting multiple roles in basal body function. Characterizing protein assembly dynamics is critical to determine when such proteins function in various aspects of organelle self-assembly and organization (Misteli, 2001; Karsenti, 2008). Furthermore, analyzing the temporal and dynamic assembly of centrioles and basal bodies is important to understand their organization and function through the cell cycle.
In centrioles, specific protein components display both stable and dynamic incorporation. α-tubulin, Sas-6, and pericentrin (PACT domain) are stably incorporated, whereas Sas-4 and Sas-5 proteins exchange at centrioles (Kochanski and Borisy, 1990; Kirkham et al., 2003; Leidel and Gonczy, 2003; Martinez-Campos et al., 2004; Leidel et al., 2005; Dammermann et al., 2008). The importance of Sas-4 dynamics at centrioles is suggested by its regulation through the Caenorhabditis elegans cell cycle (Dammermann et al., 2008). Sas-4 protein at centrioles remains in dynamic equilibrium with the cytoplasmic pool until centriolar microtubules are formed, at which time Sas-4 becomes stably associated with centrioles during mitosis. Detailed studies, such as this, enable predictions for how the dynamics of centriole and basal body proteins may facilitate the structural assembly of a new organelle.
Basal body assembly has been well characterized morphologically in the ciliates, Tetrahymena and Paramecium (Dippell, 1968; Allen, 1969; Perlman, 1973; Kaczanowski, 1978; Iftode et al., 1989). The majority of basal bodies are duplicated adjacent and anterior to existing, parent basal bodies at the cell median before cell division (Perlman, 1973; Nanney, 1975; Kaczanowski, 1978; Frankel et al., 1981; Iftode et al., 1989). An advantage to studying Tetrahymena thermophila basal body assembly is the rapid timing of new basal body maturation compared with centrioles of mitotic systems. Hundreds of new basal bodies assemble during the ∼3.0-h life cycle of Tetrahymena. In addition to cortical rows of basal bodies, a new oral apparatus (OA; the cell feeding structure) containing ∼150 basal bodies is rapidly assembled immediately posterior to the cell division plane (Wolfe, 1970; Nanney et al., 1975; Frankel et al., 1984; Williams et al., 1990).
We developed tools in Tetrahymena to measure the dynamics of basal body proteins relative to their ultrastructural localization by using quantitative fluorescence imaging, fluorescence recovery after photobleaching (FRAP), and high-resolution IEM. The dynamics of four distinctly localized basal body proteins was determined. Centrin (Cen1) is found at multiple sites within and surrounding basal bodies (Stemm-Wolf et al., 2005; Kilburn et al., 2007). Sas6a localizes at the cartwheel hub and is responsible for centriole and basal body assembly and establishing the ninefold symmetry (Dammermann et al., 2004; Leidel et al., 2005; Delattre et al., 2006; Pelletier et al., 2006; Kilburn et al., 2007; Nakazawa et al., 2007; Rodrigues-Martins et al., 2007; Strnad et al., 2007; Vladar and Stearns, 2007; Yabe et al., 2007). α-Tubulin (Atu1) is found throughout the basal body cylinder comprising the triplet microtubules (Allen, 1969). Spag6 (Chlamydomonas PF16 or mammalian sperm antigen 6) is a ciliary protein associated with the microtubule central pair and is found at the basal body distal end or transition zone (Dutcher et al., 1984; Smith and Lefebvre, 1996; Kilburn et al., 2007). We identify unique dynamics at basal bodies for each of the proteins analyzed in this study.
MATERIALS AND METHODS
Strains and Cell Culture Conditions
T. thermophila strains B2086 and SB1969 (Tetrahymena Stock Center, Cornell University, Ithaca, NY) were used in this study. Cells were grown in super proteose peptone (SPP) media (2% proteose peptone, 0.1% yeast extract, 0.2% glucose, and 0.003% Fe-EDTA) at 30°C. For starvation experiments, cells were grown to mid-log phase in SPP before washing and resuspending in 10 mM Tris-HCl, pH 7.4, at 30°C for ∼14 h.
Green fluorescent protein (GFP) strains were generated using biolistic transformation (Bruns and Cassidy-Hanley, 2000) with an N-terminal GFP fusion under the control of a metallothionein inducible promoter (pBSmttGFPgtw [GFP-Cen1, GFP-Sas6a, and GFP-Atu1] or pIGF [GFP-Spag6]). Constructs were integrated into the genome at either RPL29 or the ribosomal DNA (rDNA) chromosome, respectively. The endogenous genes were left intact in each strain. Plasmids were constructed by cloning the coding sequence of each gene into the pENTR-D Gateway Entry Vector (Invitrogen, Carlsbad, CA). The coding sequence was then Gateway cloned into pBSmttGFPgtw or pIGFgtw (Malone et al., 2008). pBSmttGFPgtw contains the inducible GFP-tagging cassette from pIGFgtw cloned upstream of a cycloheximide-resistance allele of the rpL29 gene, which allows the tagged construct to integrate into the endogenous rpL29 locus (Chalker, Washington University). Plasmid transformants in Tetrahymena were selected by growth in either cycloheximide (15 μg/ml) or paromomycin (100 μg/ml), depending on the vector. To verify that expression of each GFP fusion was not deleterious, we performed a growth curve to determine whether protein expression significantly affect the cell cycle. The cell cycle timing of GFP-Cen1 (t1/2 = 3.3 ± 0.2 h), GFP-Atu1 (t1/2 = 3.0 ± 0.4 h), GFP-Sas6a (t1/2 = 3.1 ± 0.7 h) containing cells lines was similar (p > 0.1) to the parental control strain B2086 (t1/2 = 3.1 ± 0.4 h). GFP-Spag6–containing cells exhibited slightly slower growth (t1/2 = 3.6 ± 0.4 h; p = 0.1).
GFP induction was performed by treating cell cultures with 0.125–1.0 μg/ml cadmium chloride (CdCl2), depending upon the expression construct and the experiment. Induction was carried out for 2, 4, or 8 h, depending upon the experiment. After induction, both live and fixed cells were imaged. For live cell imaging, cells were washed with 10 mM Tris-HCl, pH 7.4, and resuspended in diluted SPP (0.25×) to reduce autofluorescence associated with the media.
Immunofluorescence
For fixed cell imaging and costaining with rabbit anti-Cen1 (Stemm-Wolf et al., 2005) or mouse anti-K-antigen (10D12; Williams et al., 1990; Shang et al., 2005, provided by N. Williams and J. Frankel [University of Iowa]) antibodies, a modified ethanol fixation procedure was used (Williams et al., 1990; Shang et al., 2005). Cultured cells were pelleted and resuspended in 2.0 ml of fixative (70% ethanol and 0.2% Triton X-100) on ice for 10 min. Cells were subsequently washed and stained for immunofluorescence in suspension. Samples were then incubated with anti-Cen1 (1:1000) and anti-K-antigen (1:50) for 1 h before washing and incubating with secondary antibodies for 1 h (Texas Red-conjugated anti-rabbit or anti-mouse secondary antibodies; 1:1000). Cells were washed, incubated on poly-l-lysine–coated coverslips, and mounted in Citifluor (Citifluor, London, United Kingdom).
Image Acquisition and Fluorescence Quantification
Fluorescence imaging was performed using a Leica DMRXA upright microscope fitted with a 63× PL-APO numerical aperture (NA) 1.32 objective and a CoolSNAP hq2 charge-coupled device (CCD) camera (Photometrics, Tucson, AZ). Images were collected and analyzed using MetaMorph Imaging Software (Molecular Devices, Sunnyvale, CA). Acquisition exposure times between 100 and 500 ms were used, depending upon the GFP-fusion protein and experiment. Exposure times were constant for each individual experiment.
Basal body fluorescence intensities were determined by subtracting the background fluorescence (Fbackground) using modified methods (Hoffman et al., 2001). The 5 × 5 (inner fluorescence intensity, Fi) and 9 × 9 (outer fluorescence intensity, Fo) pixel regions were symmetrically placed around each basal body, and the total integrated fluorescence intensity was determined for each region. Fbackground was calculated based on the surrounding fluorescence that is not contained in the inner fluorescence intensity. The background fluorescence intensity was then corrected for the total region size of Fi (Fbackground = (Fo − Fi) × ). The basal body fluorescence intensity was determined by subtracting Fbackground from Fi.
Immunoelectron Microscopy
Cells were prepared for IEM by using techniques described above. Cells were either arrested by starvation in 10 mM Tris for ∼12 h (starved) before induction of GFP-fusion protein expression for 2 h or GFP-fusions were induced in cycling cells for 8 h (asynchronous). Samples were then processed for IEM.
For IEM, T. thermophila cells were centrifuged into 15% dextran (9–11 kDa; Sigma-Aldrich, St. Louis, MO) and 5% bovine serum albumin in 10 mM Tris-HCl, pH 7.4, and the resulting loose pellet was high-pressure frozen in a Bal-Tec HPM-010 (Leica Microsystems, Wetzlar, Germany). Samples were then freeze-substituted in 0.25% glutaraldehyde, 0.1% uranyl acetate in acetone at −80°C for 3 to 4 d, and embedded in Lowicryl HM20. Thin sections (60 nm) were produced and picked up on Formvar-coated nickel slot grids. For detection of the GFP-fusion proteins, the grids were incubated on a blocking solution of 1% nonfat dry milk dissolved in phosphate-buffered saline-Tween 20 (0.1%) and then incubated on anti-GFP antibodies (a gift from Dr. M. Rout, The Rockefeller University) diluted 1:500 in blocking solution. Then, 10-nm gold-conjugated anti-rabbit secondary antibody was applied to the grids (Ted Pella, Redding, CA). Grids were poststained with 2% uranyl acetate and lead citrate. Samples were imaged using a Philips CM 10 transmission electron microscope (FEI, Hillsboro, OR). Images were acquired using a Gatan BioScan digital camera and software (Gatan, Pleasanton, CA).
For each experimental condition, the domain(s) of localization was determined by imaging the basal bodies and compiling the total number of gold particles in each region on a schematic cartoon image. A minimum of 150 gold particles and 50 basal bodies were quantified for each condition.
FRAP
To determine the short-term exchange of GFP-labeled proteins at basal bodies, FRAP was used. Cells were induced for GFP-Cen1, -Sas6a, -Atu1, and -Spag6 expression for 2 h to achieve differential basal body signal (except for GFP-Spag6 where all basal bodies were equally labeled with GFP fluorescence). All experiments were performed at room temperature (∼25°C). Bright basal bodies (newly assembled) were photobleached using a targeted 13 × 13 pixel region with a 500-ms laser exposure from a Mosaic Digital Diaphragm System (Photonics Systems, St. Charles, IL). Dim basal body FRAP could not be quantified due to the low fluorescence signal causing inconsistent but almost complete recovery (data not shown). Images were collected using a TE2000U inverted microscope (Nikon, Melville, NY) fitted with a confocal system (Solamere, Salt Lake City, UT), CSU-10 confocal head (Yokogawa, Singapore), 60× Plan-Apo VC NA 1.4 objective (Nikon), and a Cascade II-512B CCD camera (Photometrics, Tucson, AZ). Acquisition exposure times ranged from 100 to 500 ms, depending on the experiment. Acquisition time intervals ranged from 1 s to 1 min and a maximum experimental length of 5 min was performed for each GFP-fusion protein. Fluorescence intensities for each time point were determined using the above-described background subtraction, and the fluorescence recovery was analyzed using Excel software (Microsoft, Redmond, WA). A single exponential recovery curve was fit to each recovery profile of the short time interval experiments and averaged for a minimum of 11 experiments for each GFP-labeled protein. The percentage of recovery, rate constant (k), and recovery half-time (t1/2) were then calculated as described previously (Salmon et al., 1984). To verify that the recovery for each protein exhibited single exponential recovery kinetics, the natural logarithm of the fluorescence recovery was plotted relative to time to determine whether a single straight line (predicting a single rate constant) was observed. The R2 value was then calculated for the regression line through each data set. R2 values for GFP-Cen1, -Sas6a, and -Spag6 are 0.95, 0.86, and 0.97, respectively. The percentage of recovery was quantified for both the short time interval experiments used to measure recovery kinetics and long interval acquisition experiments to minimize effects from photobleaching. With the short, 5-min period used in these experiments, multi-phasic recovery kinetics were not observed, however, such recovery may be found if the cells were followed through the entire cell cycle. Plots of theoretical recovery curves based on the k value and final percentage of recovery were displayed relative to the average recovery plots for comparison.
RESULTS
Centrin Incorporates Bimodally at Basal Bodies
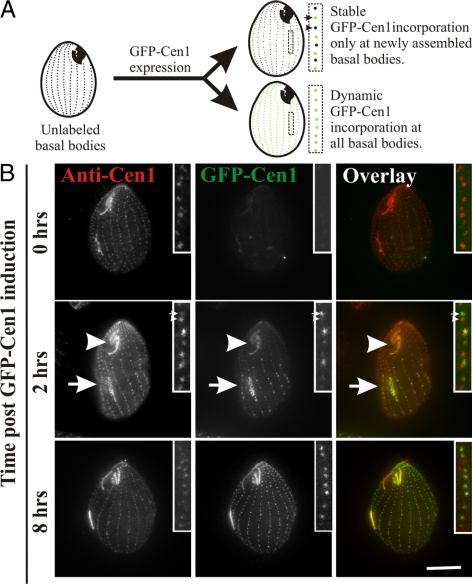
Centrin (Cen1) localizes at equal levels to all Tetrahymena basal bodies based on antibody staining and GFP-Cen1 expression (Guerra et al., 2003; Stemm-Wolf et al., 2005; Kilburn et al., 2007). Cen1 localizes to multiple sites within the basal body structure, namely, the distal end, the cylinder midpoint, and the nascent site of basal body assembly as determined by IEM (Stemm-Wolf et al., 2005; Kilburn et al., 2007). GFP-tagged Cen1 (GFP-Cen1) and endogenous Cen1 localize to the same basal body domains, indicating that the GFP tag does not interfere with localization (Stemm-Wolf et al., 2005; Kilburn et al., 2007). However, the mechanism by which each of these subpopulations is assembled into basal bodies is not clear. To determine the dynamics and timing of new Cen1 incorporation at basal bodies, a conditional GFP-Cen1 fusion was expressed and localization was monitored in cells also containing endogenous CEN1 (Figure 1A). If GFP-Cen1 is only incorporated during new basal body assembly, then only the fraction of basal bodies that are assembled after GFP-Cen1 expression is initiated will be visible (green) (Figure 1A; Stable, top cell inset, arrow). Alternatively, if Cen1 protein exchanges at basal bodies regardless of basal body assembly status, then all basal bodies will be labeled equally (Figure 1A; Dynamic, bottom cell).
Figure 1.
Differential centrin assembly at basal bodies. (A) Two possible outcomes for GFP-Cen1 expression: 1) stable incorporation only during new basal body assembly (top cell) or 2) dynamic incorporation throughout the cell cycle (bottom cell). Differentially labeled (arrow) and unlabeled (arrowhead) basal bodies indicate GFP-Cen1 assembly occurs conservatively during new basal body assembly (top cell). Basal body duplication occurs in the medial region of dividing cells with new, daughter basal bodies (inset, arrow) assembling anterior to old, parent basal bodies (inset, arrowhead). Alternatively, uniformly labeled basal bodies (lower cell) indicate proteins are incorporated independent of basal body assembly. (B) GFP-Cen1 expression for 2 h caused all basal bodies to be labeled. A fraction of the basal bodies within the cell median exhibit increased fluorescence compared with the majority of the basal bodies. GFP-Cen1 (green) was followed relative to basal bodies labeled with anti-Cen1 (red) antibodies showing uniform protein levels. The OAs (arrows), composed of basal bodies, are differentially labeled so that the old OA (arrowhead) at the cell anterior has decreased fluorescence relative to the new OA (arrow) at the cell median. After 8 h of GFP-Cen1 expression (∼3 to 4 cell cycles), the majority of basal bodies were equally labeled, indicating that most basal bodies have duplicated, incorporated, and retained high levels of GFP-Cen1. Bar, 10 μm.
Before GFP-Cen1 expression, almost no signal (green) was observed at the basal bodies, whereas anti-Cen1 antibody staining (red) uniformly labeled all basal bodies (Figure 1B, 0 h). After 2 h of GFP-Cen1 expression, a fraction of the basal bodies near the cell median exhibited bright GFP fluorescence, whereas the remaining basal bodies contained dim fluorescence signal that was significantly brighter than that observed before expression (Figure 1B, 2 h). Two hours is less than a single cell cycle (∼3.0 h). Thus, only a limited number of new basal bodies undergo assembly during the period when GFP-Cen1 is present. To quantify the difference in the two basal body populations, a ratio of fluorescence intensity for dim relative to bright basal bodies was calculated. For induced GFP-Cen1, the average ratio was 0.47 (dim/bright), whereas anti-Cen1 exhibited equivalent levels with a ratio of 0.95 (Table 1). In addition, the newly developing OA (arrow) near the division furrow, a site of new basal body assembly was more brightly labeled with GFP-Cen1 compared with the old, pre-existing OA (arrowhead) at the cell anterior (Figure 1B, 2 h). These results are consistent with the observation that bulk radiolabeled proteins assemble at the developing OA in Tetrahymena (Williams et al., 1969) and Oxytricha (Grimes and Gavin, 1987). After GFP-Cen1 expression for several cell cycles, the GFP-Cen1 fluorescence was generally uniform between basal bodies (Figure 1B, 8 h) and the new and pre-existing OAs (Supplemental Figure S1). At this late time, the fluorescence ratios of basal bodies in proximity to each other and at the cell median (likely newly duplicated) were ∼1.0 for both GFP-Cen1– and anti-Cen1–stained basal bodies (Table 1). The uniform GFP-Cen1 at this late time point indicates that most of the basal bodies have undergone a cell cycle event in the presence of GFP-Cen1 to facilitate bright basal body fluorescence. We predict this event is basal body assembly. When GFP-Cen1 expression is turned off for several cell cycles, fluorescence is lost from newly assembled basal bodies and OAs but retained at old basal bodies (Supplemental Figure S1).
Table 1.
Ratio of fluorescence intensity for dim relative to bright basal bodies
| Time following GFP–Cen1 induction |
||||
|---|---|---|---|---|
| 2.0 h |
8.0 h |
|||
| α-Cen1 | GFP-Cen1 | α-Cen1 | GFP-Cen1 | |
| Fluorescence ratio (posterior/anterior)a | 0.95 ± 0.30 | 0.47 ± 0.21 (43) | 1.07 ± 0.35 | 1.11 ± 0.44 (31) |
a The fluorescence ratio is the corrected fluorescence for the posterior (dim) basal body divided by the anterior (bright) basal body fluorescence. Parentheses describe the number (n) of basal body pairs for each experiment.
The bimodal basal body fluorescence at the 2-h time-point suggests that GFP-Cen1 dynamics correlate with the basal body duplication cycle. Based on these data, we hypothesize that Cen1 populations localize to distinct basal body domains exhibit unique incorporation dynamics. One fraction is stable through the cell cycle, whereas the second exchanges continuously with the cytoplasmic pool of Cen1. The bright signal at the cell median and newly assembled OAs suggests that the majority of Cen1 protein is deposited only during nascent basal body assembly. Results described below suggest that the second population exhibits dynamic exchange.
New Basal Body Assembly Is Required for Differential Protein Loading of GFP-Cen1
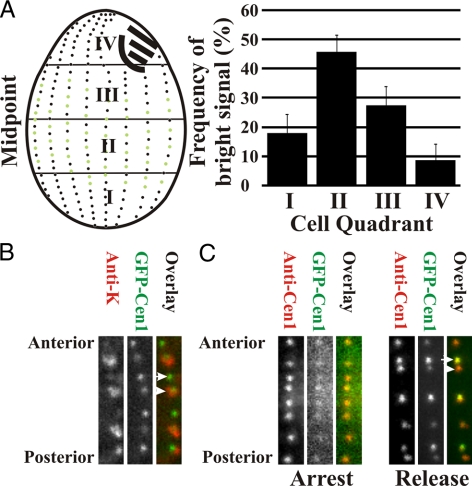
The majority of new basal bodies are assembled within the medial quadrants of Tetrahymena cells, whereas less duplication occurs within the anterior and posterior of the cell (Kaczanowski, 1978; Thazhath et al., 2004). The relative cellular location of bright GFP-Cen1–labeled basal bodies was quantified to verify that the bright GFP-Cen1 label coincides with newly assembled basal bodies. Most bright GFP-Cen1–labeled basal bodies localized to quadrants II (46%) and III (27%), equating to the medial region of the cell (Figure 2A).
Figure 2.
GFP-Cen1 incorporates coincident with new basal body assembly. (A) The brightly labeled basal bodies were quantified relative to quadrants (I, II, III, and IV) within dividing cells. The greatest frequencies of bright basal bodies were found in quandrants II and III, consistent with the cellular locations of most new basal bodies (Kaczanowski, 1978). The frequency (percentage) of bright GFP-Cen1–labeled basal bodies in each quadrant is described for 15 cells (1204 basal bodies). (B) Antibodies to K-antigens (red) were used to define old basal bodies compared with GFP-Cen1 label (green). Each panel contains a segment of a representative ciliary row. The bright GFP-Cen1 signal is found at new basal bodies (arrow; low or no K-antigen), whereas dim GFP-Cen1 is coincident with K-antigen signal marking older basal bodies (arrowhead). (C) Cells were arrested by starvation before induction of GFP-Cen1 for 2 h. GFP-Cen1 exhibited low fluorescence signal at each basal body. On release into the cell cycle by refeeding with media for 2 h (release) differential labeling by GFP-Cen1 was observed. Panel width, 2.1 μm.
To show directly that the bright GFP-Cen1 marks newly duplicated basal bodies, we took advantage of a marker for mature basal bodies. K-antigens are proteins that associate only with mature basal bodies (Williams et al., 1990; Shang et al., 2005). Colocalization experiments 2 h after GFP-Cen1 induction show that the bright GFP-Cen1 signal (green) associates with immature basal bodies lacking the K-antigen label (red) (Figure 2B). Thus, the bright GFP-Cen1 label is associated only with new basal bodies. New basal body assembly occurs immediately adjacent and anterior to a parent basal body and is followed by separation of the new basal body from the old along the cortical ciliary row toward the cell anterior (Allen, 1969). Consistent with this, the bright GFP-Cen1 basal bodies found in proximity to those that are dim were oriented in this anterior position (Figures 1 and 2B).
To determine whether basal body assembly and differential GFP-Cen1 labeling requires cell cycle progression, GFP-Cen1 was expressed in cells arrested by starvation (Figure 2C, arrest). Equally low GFP-Cen1 fluorescence was observed at all basal bodies, indicating that the dynamic population of Cen1 does not require cell cycle progression, whereas differential labeling does. Cells released from the arrest exhibited differential labeling, showing the bright signal at new basal bodies (Figure 2C, release).
Therefore, new basal body assembly is required to generate the bright GFP-Cen1 signal. Although the majority of Cen1 is stably incorporated at basal bodies during assembly, the dim signal incorporated at mature basal bodies represents a smaller, more dynamic population of Cen1. Once the GFP-Cen1 is incorporated at new basal bodies, it remains there through additional cell cycles as indicated by constant GFP-Cen1 levels.
Cen1, Sas6a, Atu1, and Spag6 Basal Body Incorporation Dynamics
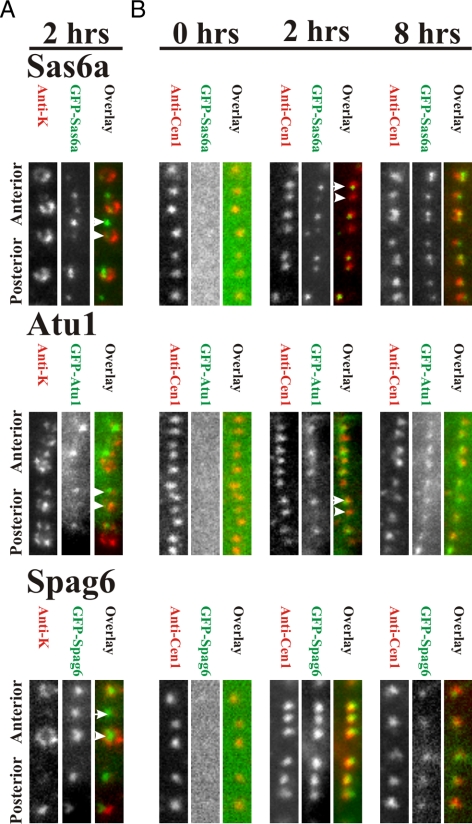
To determine whether other basal body proteins exhibit bimodal incorporation dynamics similar to centrin, GFP-Sas6a, -Atu1, and -Spag6, each were examined relative to anti-K-antigen signal (Figure 3A). Like Cen1, Sas6a and Atu1 exhibited differential basal body signals that are dependent upon new basal body assembly (Figure 3A). Bright GFP signal (green) was found at basal bodies lacking K-antigens (red) (Figure 3A). In contrast, the distal end localizing protein, Spag6, labeled both old and new basal bodies at similar intensities, indicating Spag6 incorporates at basal bodies independent of new assembly. Similar results were observed for incorporation at the OA, in which the new OA had brighter GFP-Cen1, -Sas6a, and -Atu1 signal compared with the old OA, whereas GFP-Spag6 decorated both new and old OAs with similar fluorescence intensities (Supplemental Figure S2).
Figure 3.
Distinct classes of basal body protein dynamics. (A) GFP-Sas6a and -Atu1 are incorporated into new basal bodies (arrow) that are not labeled with anti-K-antigen (arrowhead; old basal body). GFP-Spag6 exhibits equal intensity at new and old basal bodies. (B) GFP-Sas6a, -Atu1, and -Spag6 fluorescence intensity (green) relative to anti-Cen1 (red) was determined at 0, 2, and 8 h after GFP fusion protein expression. After 2 h, anti-Cen1 labeling (red) remained constant, whereas GFP-tagged proteins exhibited three distinct classes of assembly at basal bodies. As with Cen1, Sas6a associated with basal bodies to a low level (arrowhead), whereas the majority of incorporation occurred during new basal body assembly (arrow). Atu1 associated with basal bodies only during new basal body assembly (arrow). Spag6 incorporation at basal bodies is independent of new basal body assembly. Panel width, 2.1 μm.
To measure the incorporation dynamics of GFP-Sas6a, -Atu1, and -Spag6, each protein was observed at 0, 2, and 8 h after induction (Figure 3B). As for GFP-Cen1, no GFP signal was detected at 0 h for GFP-Sas6a, whereas differential signal was found at 2 h (Figure 3B). After expression for 8 h, GFP-Sas6a levels were high at all basal bodies (Figure 3B). Thus, GFP-Sas6a is predominantly a stable basal body protein with a fraction of the protein population exhibiting dynamic exchange at each basal body. This suggests that Sas6a, like Cen1, is responsible for multiple functions at basal bodies. GFP-Sas6a (green) localized as a focused spot surrounded by a larger cylinder of anti-Cen1 (red). This localization is consistent with Sas6a as a cartwheel protein, whereas Cen1 resides at multiple sites in and surrounding the basal body (Stemm-Wolf et al., 2005; Kilburn et al., 2007; Nakazawa et al., 2007).
GFP-Atu1 localized brightly at newly assembled basal bodies, whereas existing basal bodies remained almost undetectable (Figure 3B). The low level of GFP-Atu1 at existing or unduplicated basal bodies is consistent with previous reports of a conservative mechanism for α-tubulin assembly at mammalian centrioles (Kochanski and Borisy, 1990). The continued bright signal after 8 h of induction indicates that Atu1 is stably retained with little turnover through the rest of the basal body life cycle (Figure 3B). GFP-Atu1 was also found to selectively assemble at a subpopulation of cilia, even with cilia attached to basal bodies that were not labeled with GFP-Atu1 (old basal bodies; data not shown) suggesting α-tubulin assembly at cilia is distinct from basal body incorporation. Transverse and longitudinal microtubules and contractile vacuole pores were labeled in cells where GFP-Atu1 was expressed for 2 h, indicating a moderate level of tubulin exchange within these cortical structures.
In contrast to Cen1, Sas6a, and Atu1, GFP-Spag6 localized at equal levels to all basal bodies after expression for 2 or 8 h (Figure 3). Thus, Spag6 incorporation at basal bodies is independent of new basal body assembly. After induction, GFP-Spag6 assembles at all basal bodies before localizing additionally to cilia (Supplemental Figure S3). Thus, the basal body, specifically the distal end, is a dynamic loading site for Spag6 before localizing to cilia.
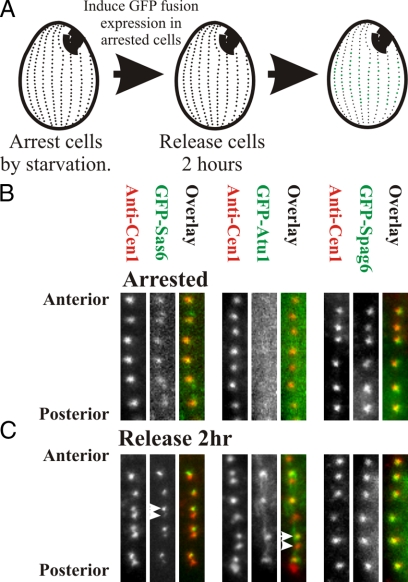
As a second test that the bimodal incorporation of GFP-fusion proteins reflects new basal body assembly, GFP-fusions were expressed in cells arrested by starvation (Figure 4A). As found for GFP-Cen1, GFP-Sas6a showed a low and uniform level of fluorescence at all basal bodies (Figures 2C and 4B). GFP-Atu1 was not detectable, whereas GFP-Spag6 exhibited high levels of loading at all basal bodies (Figure 4B). After the 2 h of GFP-fusion protein expression in starved cells, cells were released from arrest into the cell cycle for 2 h. As found for unperturbed cells after 2 h of induction, GFP-Sas6a, and -Atu1 differentially assembled at basal bodies, whereas GFP-Spag6 equally labeled all basal bodies (Figure 4B). Thus, the low levels of incorporation can occur in the absence of cell cycle progression (for GFP-Cen1 and -Sas6a), whereas the bright signal found for GFP-Cen1, -Sas6a, and -Atu1 requires cell cycle progression presumably because it represents incorporation by dynamics and new basal body assembly.
Figure 4.
Duplication dependent basal body protein incorporation requires cell cycle progression. (A) To determine whether basal body assembly and the differential labeling of basal bodies by each GFP-labeled protein is dependent upon cycle progression, cells were starved for ∼14 h before GFP-labeled proteins were expressed for 2 h. Cells were then refed media and grown for 2 h to determine whether the expressed protein was incorporated at basal bodies. (B) Low levels of homogenous signal (i.e., no differentially bright and dim labeled OAs) were detected at GFP-Sas6a– and -Atu1–labeled basal bodies. GFP-Spag6 brightly labeled basal bodies uniformly. (C) On release for 2 h, GFP-Sas6a and -Atu1 incorporated at basal bodies with dim and bright fluorescence. GFP-Spag6 levels of protein incorporation were constant at all basal bodies. Panel width, 2.1 μm.
Cen1 and Sas6a Bimodal Populations Localize to Distinct Basal Body Domains
The differential fluorescence intensities observed with GFP-Cen1 and GFP-Sas6a with mature, dim basal bodies and newly assembled, bright basal bodies suggest there may be protein subpopulations within the basal body that exhibit discrete protein dynamics. To determine whether the dynamic population localizes to a distinct domain within the basal body, we used arrest and release experiments in conjunction with IEM localization of the GFP-tagged fusion proteins (Figures 4 and 5). The GFP-fusion proteins were expressed first, during starvation, so that only low level incorporation occurred without new basal body assembly and second, during asynchronous growth, so that by 8 h, the majority of basal bodies were brightly labeled by incorporation of GFP-fusion as a result of new basal body assembly. GFP-fusion protein localization was then measured by IEM for each condition.
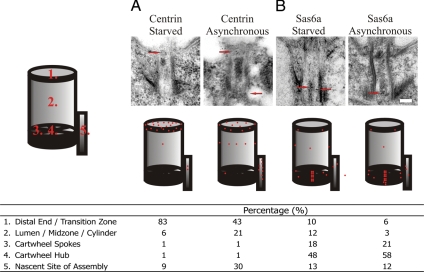
Figure 5.
Domain localization of bimodal Centrin and Sas6a basal body incorporation. (A) Centrin IEM localization in starved (no basal body duplication) or asynchronous (incorporation at duplicated basal bodies) cells expressing GFP-Cen1. The majority of Cen1 loaded at the basal body distal end, independent of new basal body assembly. GFP-Cen1 incorporated at all three major Cen1-localizing domains after stable and dynamics incorporation during new basal body assembly. (B) Both dynamic (starved) and dynamic and stable (asynchronous) populations of GFP-Sas6a predominantly localize to the cartwheel hub and spokes. On new basal body assembly, the GFP-Sas6a is more focused at the cartwheel. Each condition represents a total count of at least 150 gold particles for at least 30 basal bodies. Cartoons display the relative fraction of gold localization to each domain with a total of 25 red spots. Bar, 100 nm.
Cen1 localizes to three discrete domains within the basal body (the distal end, the cylinder midpoint, and the nascent site of basal body assembly) (Stemm-Wolf et al., 2005; Kilburn et al., 2007). To determine whether there is a discrete site to which the assembly-independent population incorporates, cells were arrested by media starvation and GFP-Cen1 was expressed for 2 h. We found that most GFP-Cen1 incorporated only at the distal end, whereas only a small fraction incorporated at the midpoint and nascent site of assembly (Figure 5A; Centrin Starved). In contrast, when GFP-Cen1 was expressed through multiple rounds of cell division, such that most basal bodies were uniformly labeled in a basal body assembly-dependent manner, all three domains were strongly labeled with GFP-Cen1 (Figure 5A, Centrin Asynchronous). Thus, the basal body distal end is the site of dynamic Cen1 exchange, whereas stable populations are assembled at the midpoint and the nascent site of assembly. However, we cannot rule out the possibility that dynamic exchange of GFP-Cen1 occurs at these later domains during normal cell growth. Our kinetic data (see below), however, suggests that there is a single population of GFP-Cen1 dynamics during normal cell growth. We believe this to be the population that resides at the transition zone.
Unlike Cen1, we found that the majority of GFP-Sas6a localizes to the cartwheel (Figure 5B; Kilburn et al., 2007). The dynamic GFP-Sas6a population, as assessed in starvation-arrested cells is also primarily incorporated at the cartwheel hub and spokes with a low level of signal through the rest of the structure (Figure 5B). Thus, Sas6a and the cartwheel are at least partly dynamic basal body components in which incorporation can occur independent of basal body assembly. When stable and dynamic populations that are dependent on new basal body assembly and exchange, respectively, were observed in asynchronously growing cultures, localization was also found predominantly at the cartwheel hub and spokes. Cartwheel localization was more specific to these structures than that observed in arrested cells (Figure 5B). Our results indicate that there are two classes of Sas6a at basal body cartwheels, that which is stable and that which is dynamic.
Quantification of Stable and Dynamic Protein Components during Basal Body Assembly
To determine the fraction of GFP labeled proteins that are dynamic, the relative fluorescence of new and old basal bodies adjacent to each other were quantified in live cells after 2 h of GFP-fusion protein expression. The signal at the old basal body represents protein incorporated during the time that GFP-labeled proteins are expressed but not as a result of new basal body assembly. The GFP-Cen1 ratio (old/new fluorescence) was 0.32, indicating ∼32% of the GFP-Cen1 exchanges with basal bodies independent of nascent basal body assembly, whereas the remaining 68% is incorporated during new basal body formation (Table 2). The discrepancy between this ratio and that observed for GFP-Cen1 in Table 1 is likely due to the difference between fixed and live cells. Similar to GFP-Cen1, the GFP-Sas6a ratio was 0.38 (Table 2). The GFP-Atu1 ratio was 0.11, indicating that the majority (89%) of Atu1 is assembled only during new basal body assembly (Table 2). We predict the low level of fluorescence at old basal bodies is due to turnover at other microtubule-rich structures surrounding basal bodies (Frankel, 2000; Thazhath et al., 2004). Finally, the GFP-Spag6 ratio was 0.98, indicating that complete protein exchange of Spag6 at the basal body does not require new basal body assembly (Table 2).
Table 2.
Relative fluorescence of new and old basal bodies adjacent to each other in live cells after 2 h of GFP-fusion protein expression
| Fluorescence ratio (old/new)a |
|
|---|---|
| GFP-Cen1 | 0.32 ± 0.13 (230) |
| GFP-Sas6a | 0.38 ± 0.18 (211) |
| GFP-Atu1 | 0.11 ± 0.20 (323) |
| GFP-Spag6b | 0.98 ± 0.32 (213) |
a The fluorescence ratio is the corrected fluorescence for the old (dim) basal body divided by the new (bright) basal body fluorescence. This ratio defines the fraction of basal body protein that is assembled independent of nascent basal body assembly. Basal body pairs in close but resolvable proximity were quantified if they were present in either quadrant II or quadrant III in cells with a developing oral apparatus. Parentheses describe the number (n) of basal body pairs for each experiment.
b It was not possible to discern new and old basal bodies with GFP-Spag6 due to dynamic protein exchange throughout the cell cycle causing equal fluorescence intensities.
Live Cell Analysis of Basal Body Protein Exchange
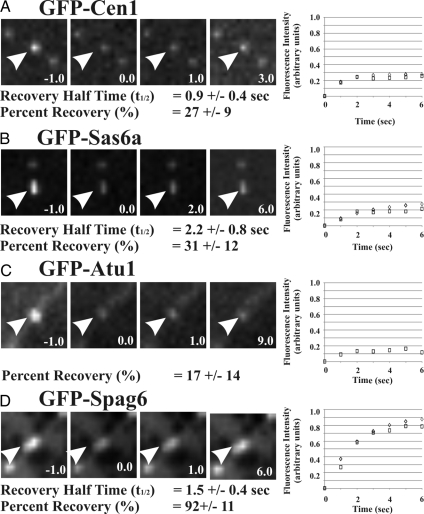
The ratios shown in Table 2 reflect the average level of protein dynamics over the course of the basal body life cycle. Our studies predict that the basal body duplication independent assembly is a dynamic class of protein incorporation due to exchange. To determine whether this is the case, the rate of protein exchange over short time intervals was measured using FRAP. GFP-labeled proteins were induced for 2 h, and brightly labeled basal bodies were exposed to a short laser exposure to photobleach the GFP conjugated to Cen1, Sas6a, Atu1, and Spag6. Fluorescence recovery was then measured over time to determine the dynamic exchange of bleached, basal body localized protein for unbleached, GFP-labeled proteins residing in the cytoplasmic pool. GFP-Cen1 recovered to an average of 27% of its fluorescence with a rapid average half-time to recovery (t1/2 = 0.9 s) (Figure 6A). The percentage of recovery is similar to the ratio of dim (old)/bright (new) GFP-Cen1–labeled basal bodies described in Table 2 (0.32), indicating that most of the dynamic fraction of Cen1 is exchanging rapidly with a t1/2 of less than one sec. Similarly, GFP-Sas6a recovered to an average of 31%, with a t1/2 of 2.2 s (Figure 6A), also consistent with the predicted exchange of 38% based on the above-mentioned ratio studies (Table 2). The slower recovery time indicates that Sas6a is exchanging at basal bodies in a manner distinct from Cen1 (p < 0.01), despite the similar total levels of dynamic protein incorporation found for both Sas6a and Cen1. GFP-Atu1 exhibited a lower percentage of recovery (17%) for which we were unable to calculate a recovery rate (Figure 6C). The Atu1 % recovery was highly variable (Figure 6C), with the majority of basal bodies (20 of 31) recovering <15%. The variability may be due to the large number of microtubule structures surrounding the basal body (Frankel, 2000), making fluorescence quantification of basal body localized α-tubulin difficult. In particular, longitudinal microtubules, although present through the cell cycle, are dynamic relative to other microtubule-containing, cortical structures (Thazhath et al., 2004).
Figure 6.
Rates of dynamic basal body protein exchange. Protein exchange at duplicated basal bodies was measured using FRAP to determine the GFP-fusion protein dynamics over a short time period (5 min). GFP-labeled Cen1 (A), -Sas6a (B), -Atu1 (C), and -Spag6 (D) (prebleach; t = −0.1 s) were photobleached with a short laser exposure (postbleach; t = 0.0 s) before the recovery was quantified in subsequent time points. The average fluorescence recovery is shown for each protein (squares) relative to the best fit exponential recovery (diamonds). GFP-Cen1 and -Sas6a exhibit similar percent recoveries but different recovery rates. Alternatively, GFP-Atu1 shows low levels of recovery, whereas GFP-Spag6 has a high level of rapid recovery. Arrows indicate the photobleached basal body. The average t1/2 recovery rate and percentage of recovery are indicated for each protein. Time, seconds. Panel width, 4.0 μm.
Finally, GFP-Spag6 exhibited the greatest level of exchange, as predicted from the ratio studies (Table 2). An average of 92% of the photobleached fluorescence signal recovered, with a half-time (t1/2) recovery rate of 1.5 s (Figure 6D). The FRAP studies described here provide a quantitative means for studying the dynamics of basal body proteins. In addition, they confirm the results of our ratio studies (Table 2) and show that the recovery of the dim populations of GFP-Cen1, -Sas6a, -Atu1, and -Spag6 occurs within a short time interval (within seconds) and not over the course of the cell cycle. The rapid recovery indicates that the dynamic incorporation found for Cen1, Sas6, and Spag6 is the result of exchange and not additive protein incorporation. The consistent results between the FRAP and the ratio studies suggest that the same population of dynamic GFP-labeled protein is observed using both experimental strategies.
DISCUSSION
Using high-resolution light and electron microscopy approaches, we have defined the incorporation dynamics of several conserved basal body and centriole proteins. We observed different mechanisms of association because several proteins require new basal body assembly for complete incorporation of new protein into basal bodies (Cen1, Sas6a, and Atu1), whereas one protein (Spag6) does not. Specifically, there are three modes of protein incorporation: 1) stable incorporation during new basal body assembly (α-tubulin), 2) dynamic exchange regardless of assembly status (Spag6), and 3) bimodal incorporation comprising both stable and dynamic mechanisms of assembly (Cen1 and Sas6a). The molecular dynamics of components residing at several key basal body and centriole domains was identified (the cartwheel, the microtubule cylinder, the distal end, and the nascent site of assembly). Our studies suggest that protein localization at distinct basal body structural domains correlates with specific dynamic properties.
Representative proteins from different structural subdomains of the basal body exhibit distinct assembly dynamics that may reflect their roles in the execution of basal body assembly, maintenance, and function. The basal body distal end or transition zone is a key domain for assembling the ciliary axoneme and to recruit proteins for cilia formation and function. We find that transition zone localizing, Spag6 and centrin exhibit rapid protein turnover. In contrast, protein populations of α-tubulin and centrin that localize to the basal body microtubule cylinder incorporate stably at basal bodies in a manner that is dependent upon new basal body assembly. The nascent site of basal body assembly seems to exhibit both classes of protein dynamics. A low level of dynamic Cen1 is found at the nascent site of assembly, whereas a larger fraction seems to stably incorporate during new basal body assembly. Thus, the Cen1 dynamic populations are dependent upon where they reside within the basal body ultrastructure. Unlike Cen1, the Sas6a protein dynamics are limited to the cartwheel in which it exhibits both dynamic and stable protein incorporation. Here, we map discrete domains within the basal body structure that exhibit both dynamic and stable regions of the basal body. Furthermore, the cartwheel, the first evident assembly structure containing the ninefold symmetry that is the hallmark of basal bodies and axonemes has both stable and dynamic elements. Thus, even basal body core domains, such as the cartwheel, exhibit dynamic exchange.
We predict that the characteristics of basal body and centriole protein dynamics will prove to be important for their roles in basal body assembly and/or function. The stable assembly of crucial structural proteins, such as tubulin, is essential to form and maintain the general organization of the basal body cylinder. This is consistent the stable assembly of tubulin at centrioles of mammalian cells (Kochanski and Borisy, 1990). However, the microtubule cylinder and basal body stability is lost when γ-tubulin expression is turned off in Tetrahymena, suggesting that microtubules may not be stable at basal bodies (Shang et al., 2002). Our studies confirm the stable assembly of basal body microtubules. It will be interesting to determine what factors, in addition to γ-tubulin, are responsible for this stability. Alternatively, the dynamic exchange of proteins at basal bodies may be required for remodeling and repair through the cell cycle, particularly because the basal body life cycle extends for multiple cell cycles. Such dynamics have been predicted for budding yeast spindle poles (Yoder et al., 2003). Two modes of Sas6a incorporation into basal bodies exist in Tetrahymena, suggesting more than one function exists for this protein. However, because GFP-Sas6a resides only at the cartwheel central hub and spokes in Tetrahymena and Chlamydomonas basal bodies (Kilburn et al., 2007; Nakazawa et al., 2007), any additional functions are likely limited to this domain. Tetrahymena Sas6a antibodies localize to kinetodesmal fibers that extend from basal bodies, in addition to cartwheels (Culver and Winey, personal communication), however, we do not observe such localization in cells expressing GFP-Sas6a. Although this may be the result of deleterious effects of the GFP tag, our studies indicate that there are multiple populations of Sas6a dynamics at the basal body cartwheel.
In contrast to the bimodal dynamics found for Tetrahymena Sas6a, C. elegans Sas-6 is a stable centriole protein incorporated during new assembly in C. elegans (Leidel et al., 2005) and Sas-6 levels increase as the centriole matures, reaching peak levels during S-phase before decreasing by ∼60% during mitotic prophase (Dammermann et al., 2008). Consistent with this decrease, HsSas-6 is lost from mature centrioles and is only found at the daughter procentriole (Strnad et al., 2007). Sas-6 is critical for the formation of the first detectable structural intermediate in C. elegans (Pelletier et al., 2006), the central tube, whereas it localizes to and is necessary for normal cartwheel and centriole assembly (Kilburn et al., 2007; Nakazawa et al., 2007; Rodrigues-Martins et al., 2007). These structures may be functionally analogous in guiding the early stages of assembly. As levels of Sas-6 decrease through the cell cycle, it becomes harder to detect the C. elegans cylinder and human cartwheel structure (Alvey, 1986; Strnad and Gonczy, 2008). However, in Tetrahymena, we do not observe the decreased levels of TtSas6a, presumably because the cartwheel structure remains stable in mature basal bodies (Allen, 1969). Furthermore, the dynamic population of Sas6a that is not observed in C. elegans centrioles may execute secondary functions within existing basal bodies.
A recent study has identified dynamic exchange of the C. elegans centriole component, Sas-4, through the cell cycle (Dammermann et al., 2008). During centriole maturation, Sas-4 is dynamic until the formation of the centriole microtubules are formed, at which time, Sas-4 dynamics cease and the protein becomes stably incorporated at the centriole. One exciting aspect of this assembly process is that binding and dissociation must be regulated and is responsive to the state of the structural assembly (Pelletier et al., 2006; Dammermann et al., 2008). In this case, temporal protein dynamics identify a key stage in centriole assembly. How basal body protein binding sites become accessible and/or inaccessible in the course of the addition of proteins during structural assembly may reveal critical mechanisms for the organization of the assembly process. Although the protein dynamics studied here reflect that of developed basal bodies, future studies will be directed to define the maturation of developing Tetrahymena basal bodies.
Using GFP-fusion protein pulse experiments with the endogenous protein present, we measure protein incorporation at basal bodies as it is expressed during the Tetrahymena cell cycle. This allowed us to measure when proteins are able to bind to their basal body binding sites by competing with the endogenous protein in the cytoplasmic pool. GFP-fusion protein incorporation independent of new basal body assembly is indicative of dynamic interactions with basal body binding sites. The majority of the proteins studied here exhibit at least some level of dynamic incorporation. Furthermore, the protein dynamics are rapid, with a half-life of several seconds. All of Spag6 and a portion of the total protein populations of Cen1 and Sas6a are capable of binding and dissociating from their respective binding sites with rapid kinetics. Atu1, Cen1, and Sas6a also exhibit stable protein incorporation during new basal body assembly. We predict that the stable populations are important to generate the conserved, stable basal body and centriole structure. Each protein is likely assembled and organized into the overall structure by a series of protein additions that reflect the well characterized structural intermediates observed by transmission electron microscopy.
We expect to expand our work by measuring the dynamics of additional basal body components to build a molecular map of basal body protein dynamics. In addition to analyzing protein incorporation and basal body domain dynamics, these strategies can identify basal body duplication and/or assembly defects during nascent basal body assembly in live Tetrahymena cells. Mutations affecting basal body assembly and maintenance can be quantifiably detected by measuring the frequency of new (bright) and pre-existing (dim) basal bodies for bimodally incorporated proteins. Furthermore, to measure the discrete stages perturbed by mutations affecting basal bodies, early (Sas6a) and late (Spag6) proteins can be tested for proper incorporation. The inability to incorporate early markers of basal body assembly may reveal upstream and important components for new basal body and centriole assembly. Mutations that inhibit loading of only late assembling proteins may identify discrete structural assembly stages and their molecular requirements. As the dynamics of additional proteins are determined, the defects associated with various mutations can be described in greater molecular detail. We believe this will prove to be a powerful system for centriole and basal body study.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. D. Chalker, M. Rout, J. Frankel, and N. Williams for generous gifts of reagents and helpful discussion; Drs. J. Gaertig and D. Wloga for helpful discussion; Dr. G. Voeltz for equipment; J. Meehl for electron microscopy expertise; and A. Stemm-Wolf and Drs. J. Deluca and S. Jones for helpful discussions and critical comments on the manuscript. C.G.P. is supported by Damon Runyon Cancer Research Foundation grant 1879-05 and M. Winey is supported by National Institutes of Health grant GM-074746.
Abbreviations used:
- GFP
green fluorescent protein
- IEM
immunoelectron microscopy
- OA
oral apparatus, t1/2, half-time.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0835) on December 3, 2008.
REFERENCES
- Allen R. D. The morphogenesis of basal bodies and accessory structures of the cortex of the ciliated protozoan Tetrahymena pyriformis. J. Cell Biol. 1969;40:716–733. doi: 10.1083/jcb.40.3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvey P. L. Do adult centrioles contain cartwheels and lie at right angles to each other? Cell Biol. Int. Rep. 1986;10:589–598. doi: 10.1016/0309-1651(86)90136-0. [DOI] [PubMed] [Google Scholar]
- Anderson R. G., Brenner R. M. The formation of basal bodies (centrioles) in the Rhesus monkey oviduct. J. Cell Biol. 1971;50:10–34. doi: 10.1083/jcb.50.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano J. L., Mitsuma N., Beales P. L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Bruns P. J., Cassidy-Hanley D. Biolistic transformation of macro- and micronuclei. Methods Cell Biol. 2000;62:501–512. doi: 10.1016/s0091-679x(08)61553-8. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardii. J. Cell Sci. 1974;16:529–556. doi: 10.1242/jcs.16.3.529. [DOI] [PubMed] [Google Scholar]
- Dammermann A., Maddox P. S., Desai A., Oegema K. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the gamma-tubulin-mediated addition of centriolar microtubules. J. Cell Biol. 2008;180:771–785. doi: 10.1083/jcb.200709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A., Muller-Reichert T., Pelletier L., Habermann B., Desai A., Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Delattre M., Canard C., Gonczy P. Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 2006;16:1844–1849. doi: 10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- Dippell R. V. The development of basal bodies in paramecium. Proc. Natl. Acad. Sci. USA. 1968;61:461–468. doi: 10.1073/pnas.61.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S. K., Huang B., Luck D. J. Genetic dissection of the central pair microtubules of the flagella of Chlamydomonas reinhardtii. J. Cell Biol. 1984;98:229–236. doi: 10.1083/jcb.98.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel J. Cell biology of Tetrahymena thermophila. Methods Cell Biol. 2000;62:27–125. doi: 10.1016/s0091-679x(08)61528-9. [DOI] [PubMed] [Google Scholar]
- Frankel J., Nelsen E. M., Bakowska J., Jenkins L. M. Mutational analysis of patterning of oral structures in Tetrahymena. II. A graded basis for the individuality of intracellular structural arrays. J. Embryol. Exp. Morphol. 1984;82:67–95. [PubMed] [Google Scholar]
- Frankel J., Nelsen E. M., Martel E. Development of the ciliature of Tetrahymena thermophila. II. Spatial subdivision prior to cytokinesis. Dev. Biol. 1981;88:39–54. doi: 10.1016/0012-1606(81)90217-7. [DOI] [PubMed] [Google Scholar]
- Gherman A., Davis E. E., Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat. Genet. 2006;38:961–962. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- Grimes G. W., Gavin R. H. Ciliary protein conservation during development in the ciliated protozoan, Oxytricha. J. Cell Biol. 1987;105:2855–2859. doi: 10.1083/jcb.105.6.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C., Wada Y., Leick V., Bell A., Satir P. Cloning, localization, and axonemal function of Tetrahymena centrin. Mol. Biol. Cell. 2003;14:251–261. doi: 10.1091/mbc.E02-05-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D. B., Pearson C. G., Yen T. J., Howell B. J., Salmon E. D. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iftode F., Cohen J., Ruiz F., Rueda A. T., Chen-Shan L., Adoutte A., Beisson J. Development of surface pattern during division in Paramecium. I. Mapping of duplication and reorganization of cortical cytoskeletal structures in the wild type. 1989;105:191–211. [Google Scholar]
- Inglis P. N., Boroevich K. A., Leroux M. R. Piecing together a ciliome. Trends Genet. 2006;22:491–500. doi: 10.1016/j.tig.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Kaczanowski A. Gradients of proliferation of ciliary basal bodies and the determination of the position of the oral primordium in Tetrahymena. J. Exp. Zool. 1978;204:417–430. doi: 10.1002/jez.1402040313. [DOI] [PubMed] [Google Scholar]
- Karsenti E. Self-organization in cell biology: a brief history. Nat. Rev. Mol. Cell Biol. 2008;9:255–262. doi: 10.1038/nrm2357. [DOI] [PubMed] [Google Scholar]
- Keller L. C., Romijn E. P., Zamora I., Yates J. R., 3rd, Marshall W. F. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Kilburn C. L., Pearson C. G., Romijn E. P., Meehl J. B., Giddings T. H., Jr, Culver B. P., Yates J. R., 3rd, Winey M. New Tetrahymena basal body protein components identify basal body domain structure. J. Cell Biol. 2007;178:905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M., Muller-Reichert T., Oegema K., Grill S., Hyman A. A. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 2003;112:575–587. doi: 10.1016/s0092-8674(03)00117-x. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.-D., Nigg E. A. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Kochanski R. S., Borisy G. G. Mode of centriole duplication and distribution. J. Cell Biol. 1990;110:1599–1605. doi: 10.1083/jcb.110.5.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R., Borisy G. G. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J. Cell Biol. 1981;91:814–821. doi: 10.1083/jcb.91.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., Gonczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Leidel S., Gonczy P. SAS-4 is essential for centrosome duplication in C elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell. 2003;4:431–439. doi: 10.1016/s1534-5807(03)00062-5. [DOI] [PubMed] [Google Scholar]
- Malone C. D., et al. Nucleus-specific importin alpha proteins and nucleoporins regulate protein import and nuclear division in the binucleate Tetrahymena thermophila. Eukaryot. Cell. 2008;7:1487–1499. doi: 10.1128/EC.00193-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W. F. The cell biological basis of ciliary disease. J. Cell Biol. 2008;180:17–21. doi: 10.1083/jcb.200710085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W. F., Nonaka S. Cilia: tuning in to the cell's antenna. Curr. Biol. 2006;16:R604–R614. doi: 10.1016/j.cub.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Martinez-Campos M., Basto R., Baker J., Kernan M., Raff J. W. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 2004;165:673–683. doi: 10.1083/jcb.200402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura K., Lefebvre P. A., Kamiya R., Hirono M. Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J. Cell Biol. 2004;165:663–671. doi: 10.1083/jcb.200402022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. The concept of self-organization in cellular architecture. J. Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y., Hiraki M., Kamiya R., Hirono M. SAS-6 is a cartwheel protein that establishes the 9-Fold symmetry of the centriole. Curr. Biol. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Nanney D. L. Patterns of basal body addition in ciliary rows in Tetrahymena. J. Cell Biol. 1975;65:503–512. doi: 10.1083/jcb.65.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanney D. L., Chow M., Wozencraft B. Considerations of symmetry in the cortical integration of tetrahymena doublets. J. Exp. Zool. 1975;193:1–14. doi: 10.1002/jez.1401930102. [DOI] [PubMed] [Google Scholar]
- Pan J., Snell W. The primary cilium: keeper of the key to cell division. Cell. 2007;129:1255–1257. doi: 10.1016/j.cell.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Pelletier L., O'Toole E., Schwager A., Hyman A. A., Muller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- Perlman B. S. Basal body addition in ciliary rows of Tetrahymena pyriformis. J. Exp. Zool. 1973;184:365–368. doi: 10.1002/jez.1401840310. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Bettencourt-Dias M., Riparbelli M., Ferreira C., Ferreira I., Callaini G., Glover D. M. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 2007;17:1465–1472. doi: 10.1016/j.cub.2007.07.034. [DOI] [PubMed] [Google Scholar]
- Salmon E. D., Leslie R. J., Saxton W. M., Karow M. L., McIntosh J. R. Spindle microtubule dynamics in sea urchin embryos: analysis using a fluorescein-labeled tubulin and measurements of fluorescence redistribution after laser photobleaching. J. Cell Biol. 1984;99:2165–2174. doi: 10.1083/jcb.99.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Li B., Gorovsky M. A. Tetrahymena thermophila contains a conventional gamma-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J. Cell Biol. 2002;158:1195–1206. doi: 10.1083/jcb.200205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Tsao C. C., Gorovsky M. A. Mutational analyses reveal a novel function of the nucleotide-binding domain of gamma-tubulin in the regulation of basal body biogenesis. J. Cell Biol. 2005;171:1035–1044. doi: 10.1083/jcb.200508184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. F., Lefebvre P. A. PF16 encodes a protein with armadillo repeats and localizes to a single microtubule of the central apparatus in Chlamydomonas flagella. J. Cell Biol. 1996;132:359–370. doi: 10.1083/jcb.132.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemm-Wolf A. J., Morgan G., Giddings T. H., Jr, White E. A., Marchione R., McDonald H. B., Winey M. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol. Biol. Cell. 2005;16:3606–3619. doi: 10.1091/mbc.E04-10-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Gonczy P. Mechanisms of procentriole formation. Trends Cell Biol. 2008;18:389–396. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Strnad P., Leidel S., Vinogradova T., Euteneuer U., Khodjakov A., Gonczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thazhath R., Jerka-Dziadosz M., Duan J., Wloga D., Gorovsky M. A., Frankel J., Gaertig J. Cell context-specific effects of the beta-tubulin glycylation domain on assembly and size of microtubular organelles. Mol. Biol. Cell. 2004;15:4136–4147. doi: 10.1091/mbc.E04-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar E. K., Stearns T. Molecular characterization of centriole assembly in ciliated epithelial cells. J. Cell Biol. 2007;178:31–42. doi: 10.1083/jcb.200703064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams N. E., Honts J. E., Kaczanowska J. The formation of basal body domains in the membrane skeleton of Tetrahymena. Development. 1990;109:935–942. doi: 10.1242/dev.109.4.935. [DOI] [PubMed] [Google Scholar]
- Williams N. E., Michelsen O., Zeuthen E. Synthesis of cortical proteins in Tetrahymena. J. Cell Sci. 1969;5:143–162. doi: 10.1242/jcs.5.1.143. [DOI] [PubMed] [Google Scholar]
- Wolfe J. Structural analysis of basal bodies of the isolated oral apparatus of Tetrahymena pyriformis. J. Cell Sci. 1970;6:679–700. doi: 10.1242/jcs.6.3.679. [DOI] [PubMed] [Google Scholar]
- Yabe T., Ge X., Pelegri F. The zebrafish maternal-effect gene cellular atoll encodes the centriolar component sas-6 and defects in its paternal function promote whole genome duplication. Dev. Biol. 2007;312:44–60. doi: 10.1016/j.ydbio.2007.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder T. J., Pearson C. G., Bloom K., Davis T. N. The Saccharomyces cerevisiae spindle pole body is a dynamic structure. Mol. Biol. Cell. 2003;14:3494–3505. doi: 10.1091/mbc.E02-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.