Abstract
The molecular composition of plasma membranes is constantly remodeled by endocytosis and exocytosis. Eisosomes are large cytoplasmic protein assemblies that localize to specialized domains on the yeast plasma membrane. They are of uniform size and immobile, and their disruption leads to large aberrant plasma membrane invaginations and endocytic defects. It is unknown how eisosomes are formed or inherited and what governs their size, distribution, and location. Here we show that eisosomes are formed de novo in the bud of dividing cells. They colonize newly formed membrane at a fixed density in a polarized wave proceeding from the bud neck to the bud tip and become anchored at the site of their formation. Pil1, one of the two main eisosome subunits, emerges as the central regulator of eisosome biogenesis that determines both size and location of eisosomes. Lowering Pil1 expression leads to normal-sized eisosomes at a reduced density, suggesting that eisosomes must be of a minimal size. Conversely, raising Pil1 expression leads to larger eisosomes at a fixed density, suggesting that under these conditions eisosome nucleation sites are limiting. Pil1 expression is regulated by the cell cycle, which synchronizes eisosome formation with plasma membrane growth. Our results establish a first framework of the molecular principles that define eisosome assembly and distribution.
INTRODUCTION
All communication and exchange of metabolites, such as nutrients and catabolites, with the environment occurs across the cell's plasma membrane, which must dynamically adapt its molecular composition to changing conditions. As such, the plasma membranes of all eukaryotic cells are constantly remodeled by endocytosis and exocytosis. To accommodate these processes effectively, the plasma membrane is highly organized and laterally divided into domains of different composition and function. Plasma membrane organization has been characterized in many cell types and has been most extensively studied in polarized epithelial cells and neurons (Broadie, 2004; Rodriguez-Boulan et al., 2005). Specialized areas of the plasma membrane, such as the neuromuscular junction or the neuronal synapse, form zones of active endocytosis and exocytosis that are spatially separated, and disruption of this organization leads to impairment of membrane traffic (Koh et al., 2004; Marie et al., 2004).
Similar organizational features are found even in unicellular yeasts, such as Saccharomyces cerevisiae. In this organism, recent analysis of protein and lipid distribution revealed that the plasma membrane is laterally inhomogeneous. Examples include the polar distribution of ergosterol-rich domains to the shmoo tip during the formation of mating projections (Bagnat and Simons, 2002), the polarized distribution of plasma membrane proteins that are involved in the directed targeting of secretory vesicles to the bud tip (Valdez-Taubas and Pelham, 2003), and the nonuniform distribution of plasma membrane proteins, such as Can1 and Sur7, into punctate domains (Grossmann et al., 2007; Malinska et al., 2003, 2004). Recently, we discovered large immobile structures called eisosomes (from the Greek “eis” meaning “into” or “portal” and “some” meaning “body”), which organize these latter domains, and showed that they mark sites of endocytosis (Walther et al., 2006).
Eisosomes have fascinating features (Walther et al., 2006). They are large macromolecular assemblies of relatively uniform size, each composed of a few thousand copies of two principal subunits, Pil1 and Lsp1. They lie underneath the plasma membrane and localize in a punctate pattern that is not reminiscent of any other organelle, but colocalize with specialized membrane domains of unique protein and lipid composition (Grossmann et al., 2007). Eisosomes distribute evenly over the cell cortex, but the principles guiding this distribution are unknown. Deletion of PIL1 leads to clustering of the remaining eisosome components to one or a few spots (“eisosome remnants”) that are associated with grossly aberrant plasma membrane invaginations and causes a reduction of the endocytic rate (Walther et al., 2006). Eisosomes are immobile and, remarkably, do not exchange subunits with free cytoplasmic pools of Lsp1 or Pil1, indicating that after initial assembly, they are not subject to dynamic remodeling at the level of exchange of their major structural subunits. How eisosomes assemble, achieving both their uniform size and even distribution, is unknown. To gain first insights into eisosome biogenesis, we describe here the formation of eisosomes in dividing cells and define the rules that govern their formation.
MATERIALS AND METHODS
Yeast Strains
Genotypes of all strains used in this study are listed in Supplementary Table S1. The Pil1-GFP and Lsp1-GFP strains were described previously (TWY110 and TWY113, respectively; Walther et al., 2006). Diploid yeast strains expressing various amounts of Pil1 were generated by mating of Pil1-GFP strains with a pil1Δ strain (TWY580) or a Pil1-GFP strain of opposite mating type (to generate TWY576). In addition, an extra copy of Pil1-GFP was cloned onto a pRS306 plasmid and integrated into one (TWY581) or both URA (TWY578) loci. To generate the DAMP allele of Pil1-GFP (KEM101), a KAN-selection marker was integrated directly after the open reading frame in TWY 110 as previously described (Schuldiner et al., 2005). The Cdc28 mutant strain (KEM100) was generated by integrating the plasmid pRS406 carrying the cdc28-as2 allele in the URA locus of TWY110. The cdc28-as2 plasmid was generated by David Morgan's laboratory at the University of California, San Francisco (Ubersax et al., 2003) and was a generous gift from Gustavo Pesce (Molecular Sciences Institute, Berkeley, CA). The copper inducible strains KEM102 and KEM103 were made by replacing the endogenous promoter of PIL1 with the CUP1 promoter in TWY110 and TWY113. This was done by homologous recombination using a PCR-based modification (Janke et al., 2004).
Microscopy
Live cell microscopy was performed by mounting yeast cells on concanavalin A–covered coverslips. Images were taken at room temperature on a Zeiss LSM510 confocal microscope (Thornwood, NY) and on a Nikon TE2000U inverted microscope (Melville, NY) with a Yokogawa CSU22 Spinning Disk Confocal from Solamere Technology Group (Salt City, UT). Images were processed with ImageJ (http://rsb.info.nih.gov/ij/index.html) and MATLAB (MathWorks, Natick, MA).
Immunofluorescence was performed as described previously (Brickner and Walter, 2004). Cells were fixed in 4% formaldehyde for 45 min at room temperature, then spheroplasted with lyticase, and detergent-extracted with 0.01% Triton X-100. Cells were probed with 1:200 rabbit polyclonal anti-green fluorescent protein (GFP; Invitrogen, Carlsbad, CA; A6455 rabbit polyclonal antibody against full-length GFP) in WT buffer (1% nonfat dry milk, 0.5 mg/ml BSA, 150 mM NaCl, 50 mM HEPES, pH 7.5) for 1 h at room temperature, followed by secondary, goat anti-rabbit Alexafluor 488 (Invitrogen, A11034) antibody diluted at 1:200 in WT buffer for 1 h also at room temperature. Slides were mounted using Vectashield (Vector Laboratories, Burlingame, CA), and imaged with a Zeiss LSM510 confocal microscope.
For surface area calculation we treated yeast cells as a spheroid shape, determined the major and minor axes and calculated the surface area using the standard formula: S = 2πa2 + 2π(ab/e)sin−1 e, where a is the major axis, b is the minor axis, and e = [√]1 − (b2/a2). Mother and nocodazole-arrested cells were approximated to a sphere shape, S = 4πr2.
The measurements of eisosome distribution in the 3D reconstructed images showed that the radii varied within 10% (Supplemental Table S2), validating our strategy to approximate yeast cells as spheres for the purpose of this analysis.
Molecular Biology
For gene expression analysis, total RNA was extracted with acidic phenol and blotted for Northern analysis. Blots were probed with a radio-labeled probe representing 500 base pairs of the PIL1 mRNA amplified with primers 5′-cgatgtttccgacatcactg and 5′-gtgcgctttcagcatcaata.
Quantitative Western blots were done by preparation of cell extracts in 8 M urea, 50 mM HEPES, pH 7.4. Pil1-GFP was detected by a chemifluorescence method (ECF) using an antibody against GFP (Invitrogen, A6455 rabbit polyclonal antibody against full-length GFP recognizing both the native and denatured protein) at 1:10,000 dilution, a rabbit polyclonal antibody against full-length Pil1 at 1:10,000 dilution (Walther et al., 2007), or a monoclonal mouse antibody against Pgk1 (Invitrogen, A6457) at 1:5000 dilution, followed by an alkaline phosphatase-labeled secondary antibody (Amersham Biosciences, Piscataway, NJ) diluted at 1:5000. The blots were incubated with an ECF Substrate, which when dephosphorylated fluoresces and were then scanned in a Typhoon 9400 (GE Healthcare, Waukesha, WI). Individual bands were quantified using ImageQuant TL (Molecular Dynamics, Sunnyvale, CA).
RESULTS
Eisosomes Form De Novo and after an Initial Lag Phase, Colonize the Bud as a Polar Wave Initiating at the Bud Neck
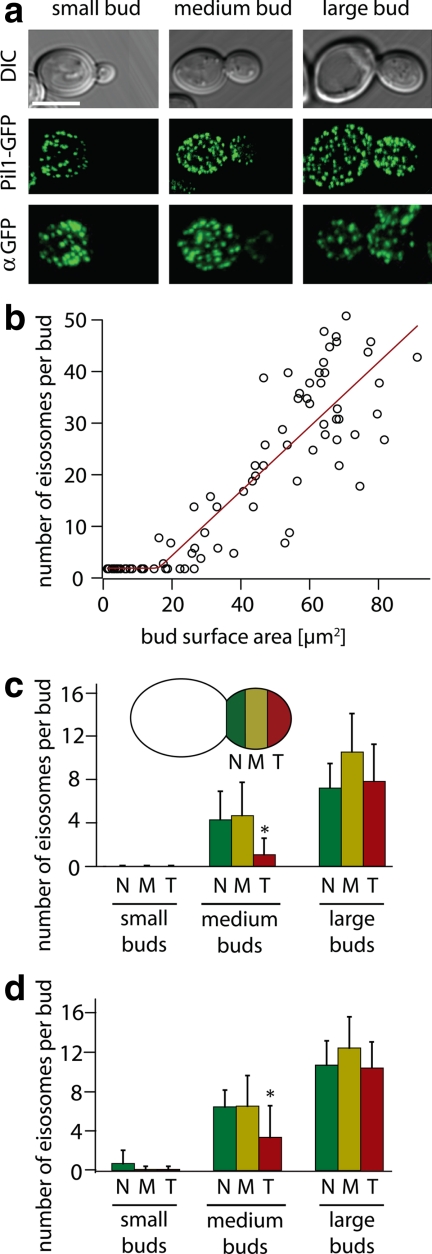
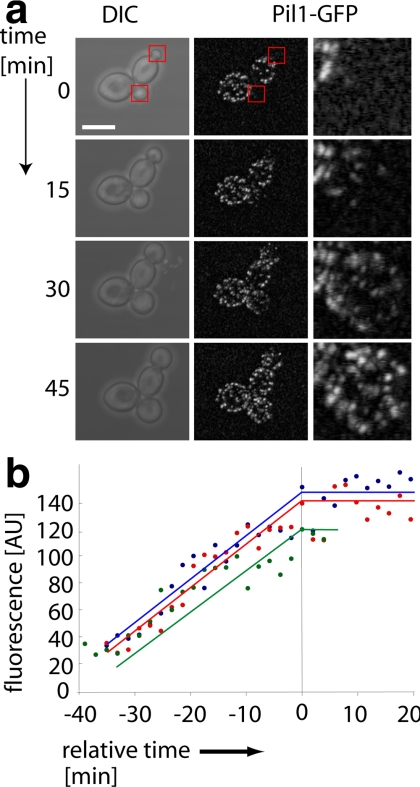
To begin to determine the rules that guide eisosome biogenesis, we first asked two simple questions: 1) are new eisosomes derived by division of existing eisosomes or are they assembled de novo, and 2) are they preassembled at some site in the cell and transported to their final location or do they form in situ? To distinguish between these possibilities, we fused GFP to PIL1 at its endogenous genomic locus and observed the formation of new eisosomes by 3D confocal time-lapse microscopy with different time intervals (Supplemental Movie S1; Figure 1). A qualitative inspection of the data shows that small buds do not contain eisosomes (Figure 1a, middle left panel). As the buds grow beyond a critical size, eisosomes form in a wave that starts close to the bud neck (Figure 1a, central panel) and continues in a polar manner toward the bud tip (Figure 1a, middle right panel). During these analyses, we never observed an eisosome forming in the mother cell, nor did we find evidence for division of preexisting eisosomes. We also never saw movement of eisosomes, excluding a model invoking transport of eisosomes to the sites of their deposition. Rather, all newly formed eisosomes were immediately immobile, indicating that they form at the sites where they are anchored.
Figure 1.
Eisosomes are assembled de novo. (a) Pil1-GFP accurately reflects the localization of eisosomes. Pil1-GFP fluorescence pictures (top row) are compared with immunofluorescence staining of Pil1-GFP with an antibody against GFP. Scale bar, 5 μm. (b) Eisosomes are formed after an initial lag phase, and their number is directly proportional to cell surface area. Eisosomes were visualized in 3D confocal stacks of cells expressing Pil1-GFP. The number of eisosomes per bud was counted and plotted against the surface area of the bud; n = 83. Eisosome number was fit to a hockey-stick progression using R (http://cran.r-project.org/) according to (Bacon and Watts, 1971). (c) Eisosomes are formed in a polar manner. Confocal stacks of cells with small- (0–17 μm2), medium- (17–60 μm2), and large- (>60 μm2) sized buds were recorded. The buds were segmented along the axis of cell polarity into three equal regions (N, bud neck; M, middle; T, bud tip), and the number of eisosome was counted for each region, n = 95. The number of eisosomes was significantly lower in the T region compared with the N and M regions (ANOVA, *p < 0.001). (d) Similar to panel c, the number of eisosomes was counted for images after immunofluorescence staining of Pil1-GFP with antibody against GFP; n = 43. Similar to panel c, the number of eisosomes was significantly lower in the T region compared with the N and M regions (ANOVA, *p < 0.001). TWY110 was used in all experiments.
Quantitative analyses validate and refine these conclusions. For example, we counted the number of eisosomes in buds of various sizes and plotted these numbers against the bud's surface area. The data in Figure 1b are well described statistically by a hockey-stick regression, which fits the data to a linear function preceded by a lag phase. This analysis shows that eisosomes are absent from buds with <17-μm2 surface area. After this lag, their number grows proportionally to the bud's surface area.
To rule out that the observed lag phase might be artifactually caused by a delay in the appearance of fluorescence due to delayed maturation of the GFP fluorophore, we visualized eisosomes by indirect immunofluorescence over the same time course, using a polyclonal antibody that recognizes GFP independent of its folding or fluorescence maturation state (Figure 1a, bottom panels). This analysis showed an indistinguishable eisosome distribution during the formation of new buds, demonstrating that the Pil1-GFP fusion is a faithful marker of eisosomes in these experiments.
Also consistent with our observations from time-lapse microscopy, quantitative analysis shows that eisosomes form in a polar wave originating at the bud neck. We counted eisosomes in small-, medium-, and large-sized buds (small, 0–17 μm2; medium, 17–60 μm2; and large >60 μm2 in surface area, respectively), for each bud assessing eisosome distribution in three zones of equal length along the polarity axis, defined as neck (N), middle (M), and tip zone (T), respectively. As shown in Figure 1c, medium-sized buds show a strongly polarized distribution of eisosomes within the bud, with fourfold more eisosomes in the neck than in the tip zone. The polar distribution becomes less pronounced as the bud grows and eventually diminishes, as seen in the approximately even distribution in large buds. We repeated this quantification on immunofluorescence images of cells stained with anti-GFP antibodies and confirmed that it parallels the deposition pattern of live cells detecting GFP fluorescence (Figure 1d). By this analysis also, small buds contained no eisosomes (Figure 1d), again confirming that GFP accurately reports on eisosome disposition in our experiments.
As a further control, we tagged the second principal eisosome core component, Lsp1, with GFP and analyzed its distribution. We found that the number of Lsp1-GFP–marked eisosomes per bud when plotted against the bud surface area is very similar to eisosomes marked by Pil1-GFP, first showing a lag phase and then a steady increase of eisosome number proportional to surface area (Pearson correlation = 0.925, Supplemental Figure S1a). In addition, the distribution of Lsp1 eisosomes in the three different regions of growing buds, as analyzed in Figure 1, c and d, for Pil1, is indistinguishable from the distribution of Pil1 (Supplemental Figure S1b). In time-lapse microscopy experiments following the kinetics of Lsp1-GFP localization compared with Pil1 tagged with mCherry, we did not observe a significant difference in localization or kinetics of deposition between Pil1 and Lsp1, indicating that, within the precision of our measurements, both subunits assemble synchronously.
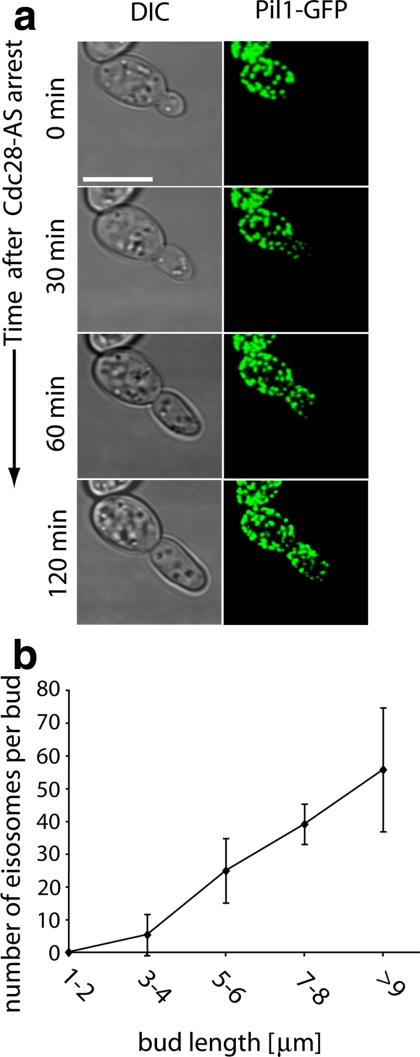
The apparent lack of eisosomes in small buds (<17 μm2 surface area) could be due to a cell cycle–regulated switch in expression or assembly, or it could be due to a time lag between the formation of new plasma membrane and new eisosomes. To distinguish between these possibilities, we arrested cells at a specific stage in the cell cycle, G2, by inactivating a drug-sensitized mutant of Cdc28 kinase (cdc28-AS2) with low levels of a cognate inhibitor, 1NM-PP1 (Bishop et al., 2000). Under these conditions, bud growth continues in a polar manner for extended periods of time because the switch from polar to isotropic growth that normally occurs after the transition to M phase does not take place (Lew and Reed, 1993). This defect results in formation of a cylindrical rather than the normal spherical shape of the bud. As expected, the G2-arrested cdc28 mutant cells extended elongated buds (Figure 2a, DIC). The cells formed new eisosomes in the bud (Figure 2a, Pil1-GFP). Indeed, eisosome formation still proceeded in a wave-like pattern starting at the bud neck, which because of the elongated shape of the buds resulted in an even more striking appearance than that observed in wild-type cells. Eisosome formation never caught up with the bud tip during the time course of the experiment. Eisosome counting revealed that their number was roughly proportional to the cell length (Figure 2b), which for a cylindrical shape in approximation is proportional to the surface area. Together these observations suggest that eisosome formation proceeds at the same rate as plasma membrane growth and that the transition past G2/M phase per se is not required for their formation.
Figure 2.
Eisosome assembly occurs in a polar manner. (a) Cells expressing Pil1-GFP and harboring a drug-sensitized cdc28 allele (KEM100) were incubated with 10 μM of the inhibitor 1NM-PP1 (Bishop et al., 2000) and were followed over time by confocal live-cell microscopy at room temperature. Representative 3D projection images are shown. Scale bar, 5 μm. (b) Quantitation of the eisosome number from experiments described in panel a plotted against bud length along the axis of polarization; n = 75.
Thus the results presented so far suggest that 1) eisosomes are assembled de novo, 2) they are formed in situ, 3) their formation proceeds in a wave-like pattern, with new eisosomes being assembled next to existing ones, leading to the polar colonization of the newly formed plasma membrane, and 4) there must be intricate regulation of the process to achieve a constant eisosome number per membrane surface area.
Eisosome Number Is Proportional to the Cell Surface Area
The data in Figure 1b show that in the colonized regions of the cell surface the number of eisosomes linearly correlates (p = 0.0002) to the surface area. By contrast, a fit of the eisosome count to cell volume shows poor correlation (p = 0.43). These observations suggest that eisosome density is tightly controlled and linked to plasma membrane expansion.
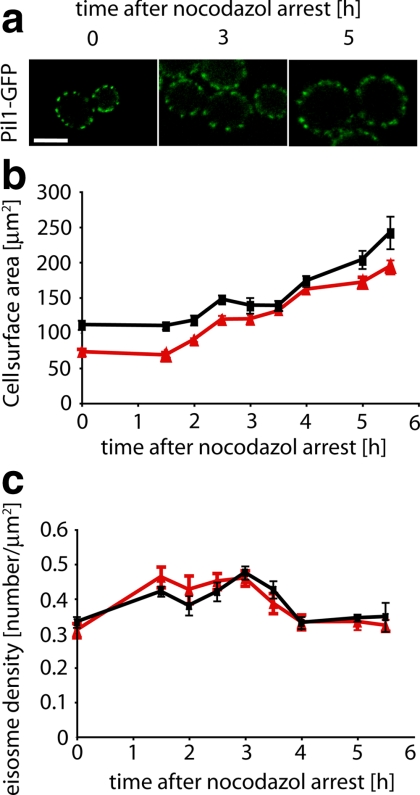
To seek further evidence for eisosome density control, we sought experimental conditions under which the cell surface area would change. To this end, we arrested yeast cells with the microtubule-depolymerizing drug nocodazole, which blocks budding and causes isotropic growth (nonpolar growth enlarging the plasma membrane in all directions) yielding large, round cells. Incubation with nocodazole leads to an increase of cell surface area that is first apparent after ∼3 h and reaches 2.5-fold after 5.5 h (Figure 3b). Remarkably, we found that the eisosome density of these giant yeast cells remains constant when compared with the density observed in wild-type mother cells (Figure 3c, t = 0; 0.33 ± 0.06 eisosomes/μm2 surface area) between 0.32 and 0.34 eisosomes/μm2 surface area as the cells grow (Figure 3c).
Figure 3.
Eisosome density is constant and regulated to a set point. (a) Cells expressing Pil1-GFP (TWY110) were arrested in M-phase by the addition of 30 μM nocodazole at 30°C and followed over time by confocal life cell microscopy. Representative images are shown. Scale bar, 5 μm. (b) Increase of yeast cell surface area over time after nocodazole arrest in daughter cells (red lines) and mother cells (black lines). (c) Development of eisosome density over time after nocodazole arrest in daughter cells (red lines) and mother cells (black lines). Error bars, SDs from the mean; n = 112.
In these experiments, we reproducibly observed a ∼20% “overshoot” of increased eisosome density at the onset of cell growth (Figure 3b, 3 h), yet 1 h later (Figures 3b, 4 h) this trend was reversed, returning the density back to the starting value. A similar situation was observed during the cell cycle of normally growing cells, where the density briefly reached 0.5 eisosomes/μm2 in large buds before returning to the set value in mother cells (Figure 1b, eisosome density for small buds = 0.01 ± 0.04 eisosomes/μm2 surface area; for medium buds = 0.32 ± 0.2 eisosomes/μm2 surface area and for large buds = 0.5 ± 0.13 eisosomes/μm2 surface area). A possible interpretation of these results is that there is a set point of eisosome density that is maintained by feedback regulation. When eisosome density becomes smaller than the set point after an initial increase in cell size, the regulatory loop overshoots leading to a transient increase in eisosome density.
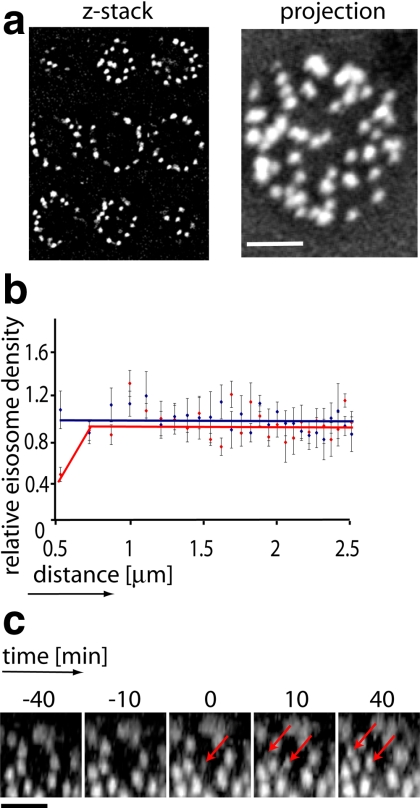
Figure 4.
Eisosomes are distributed randomly. High-resolution confocal stacks were acquired for strain TWY110, and representative images are shown in panel a. The coordinates of the centers of all eisosomes of a cell were determined (Supplemental Table S2), and the distribution of the density of eisosomes related to the distance from the center of an eisosome was calculated. Five independent measurements were taken, each with an average of 39 eisosomes. The resulting values were averaged and are shown as a red line in panel b. The blue line shows the result for the same analysis performed on simulated random coordinates. (c) De novo eisosome formation in nocodazole-arrested cells occurs in apparent gaps. Cells (TWY110) were treated as described in Figure 3, and the formation of new eisosomes in the mother cells was followed by confocal live-cell microscopy at room temperature. A representative optical surface section is shown. Arrows indicate the formation of new eisosomes between already existing ones on the mother cell plasma membrane. Scale bars, (a and c) 2 μm.
Eisosomes Are Arranged Randomly
The wave-like appearance of newly made eisosomes suggests that eisosomes form next to existing ones, perhaps at specific distances and thus resulting in a spatial pattern on the plasma membrane. Because eisosomes are immediately immobilized at the location where they are formed, their distribution pattern in cells reflects possible constraints imposed during their biogenesis, such as the distribution of putative nucleation sites. On first principle, eisosomes could be 1) randomly distributed with respect to each other, 2) fixed on a regular grid, or 3) distributed nonrandomly with less obvious constraints, such as preferred distance or orientation. To distinguish among these scenarios, we acquired complete 3D stacks of images of individual yeast cells and assigned polar coordinates to individual eisosomes (Supplemental Table S2). Figure 4a shows a representative stack. We then calculated the distances between all eisosomes on the surface of the cells and calculated the pairwise correlation function g(r) to determine whether there are any constraints on the distribution of eisosomes. In brief, this function gives the ratio of the density of eisosomes at any distance from any given eisosome to the average density of eisosomes over the entire surface. If eisosomes were randomly distributed, the local density would be the same everywhere and g(r) would be constant; if eisosomes were positioned at the nodes of a regular grid, the local density would have preferred distances and g(r) would have an oscillating form.
Figure 4b shows a comparison between the experimentally determined g(r) (red line) and a function derived for the same number of simulated random distributions of points on a sphere (blue line). With the important exception of the single point representing the smallest distance bin (0–0.5 μm), the two functions do not differ from each other significantly, indicated by the overlap of the SEs, providing positive evidence that eisosomes are randomly distributed over the surface of the cell. By contrast, the closest distance measured between eisosome centers, 0.5 μm, shows a clear underrepresentation of eisosomes: it is fourfold less likely to find an eisosome at that distance than is predicted from a random distribution. Thus eisosomes appear to be distributed randomly but are restricted to a minimal distance from one another.
To understand better the nature of distance constraints on eisosome distribution, we experimentally forced new eisosome assembly on an already colonized cell membrane. As demonstrated above, this scenario does not occur under normal physiological conditions, where eisosomes are exclusively formed at the newly assembled plasma membrane in the bud. However, the experiments discussed above, arresting cells in M phase by nocodazole arrest, showed that the cell surface grew 2.5-fold, yet maintained the same density of eisosomes (Figure 3, b and c). Therefore, new eisosome assembly must occur under these conditions at the already eisosome-covered plasma membrane. To observe these events, we imaged cells after 3 h of nocodazole treatment, a time when the effect of the arrest on cell size became first apparent. As shown in Figure 4c, we observed that, as the plasma membrane expanded, gaps in the eisosome pattern formed. The gaps expanded as the cells grew further and after a variable amount of time new eisosomes formed in the gaps. Similar to eisosome insertion during budding, the formation of these new eisosomes in arrested cells occurred de novo and not by a division or preassembly transport mechanism.
Pil1 Controls Eisosome Number and Size
Regulation of eisosome number must be coordinated with control of eisosome size, which, given the large size of these macromolecular assemblies, is maintained within a remarkably narrow window, only varying within a factor of two (Walther et al., 2006). To begin addressing how such tight control is achieved, we analyzed confocal 3D time-lapse movies of yeast cells expressing GFP-tagged Pil1 (Figure 5a). We then selected individual eisosomes in the recorded movies and ascertained that they had reached a steady level of fluorescence, indicating that they had reached a final state of Pil1 assembly. We then went back to preceding frames and quantitated the fluorescence intensities to derive the kinetics of assembly of individual eisosomes. The graph in Figure 5b shows four representative examples of eisosome growth. The data show that the formation of individual eisosomes is characterized by a steady linear increase of fluorescence that takes on average 40 ± 4 min to reach steady state. Pil1 assembly into eisosomes therefore appears to occur as a continuous process of subunit addition.
Figure 5.
Eisosomes are formed in a continuous process. (a) Formation of eisosomes over a complete cell cycle was followed by confocal time-lapse microscopy of cells expressing Pil1-GFP (TWY110) at room temperature. Representative images are shown. Scale bar, 5 μm. (b) The rate of eisosome formation is constant. The increase of fluorescence of individual eisosomes was quantified over time. The vertical line indicates the time when each eisosome has reached its final fluorescence intensity.
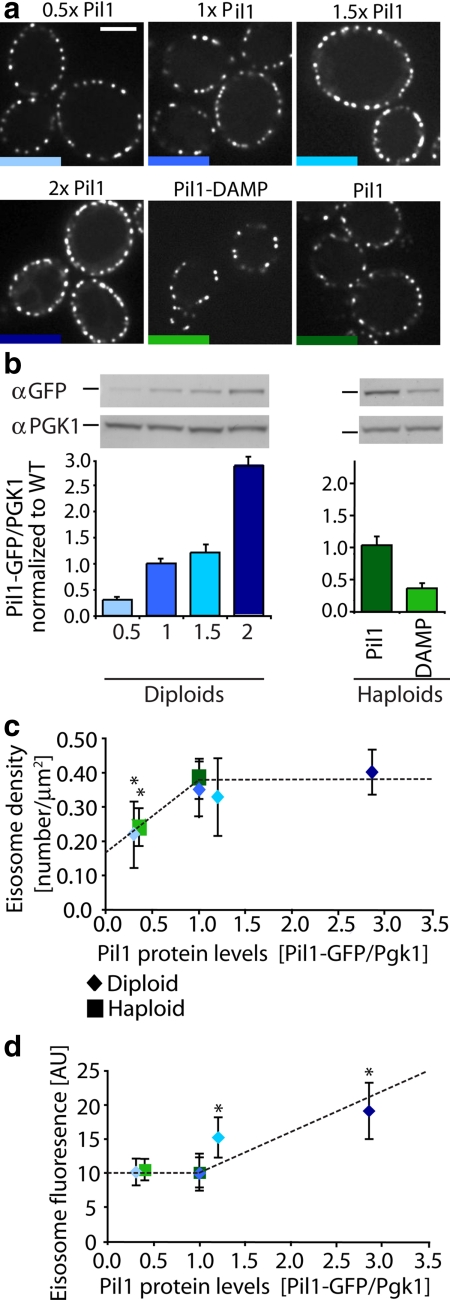
To gain mechanistic insights into the process, we focused on PIL1, because deletion of genes encoding other known eisosome components, such as the highly homologous LSP1 or the combined deletion of the SUR7 gene family members, has no dramatic effect on eisosome size or number (T. C. Walther and P. Walter, unpublished data). By contrast, deletion of PIL1 disrupts eisosome formation, size, and density, causing clustering of the remaining eisosome components to one or a few loci at the cell periphery (Walther et al., 2006; Grossmann et al., 2007). To control the amount of Pil1 expressed in cells, we generated diploid yeast cells bearing one, two, three, or four copies of PIL1-GFP (Figure 6a) that replace the wild-type gene. To assess the relative expression level of Pil1-GFP in these strains, we performed quantitative Western blotting against GFP, measured Pil1 protein levels relative to Pgk1 levels, the constitutively expressed 3-phosphoglycerate kinase, and normalized the signal to wild-type diploid cells. This analysis showed that the Pil1 expression level parallels the gene dose in the diploid strains (Figure 6b, left panel).
Figure 6.
Eisosome number and size is controlled by the levels of Pil1. (a) Diploid cells were engineered to express either one (TWY580), two (TWY576), three (TWY581), or four copies (TWY578) of Pil1-GFP, and images were acquired by spinning disk confocal microscopy. Pil1-GFP expression was decreased in haploid cells by expressing a destabilized form of PIL1 mRNA by deletion of the Pil1-GFP mRNA 3′-UTR (Pil1-DAMP, KEM101). Scale bar, 5 μm. (b) Western blot against GFP of diploid cells with increasing copies of Pil1-GFP, and wild-type haploid cells expressing one copy of Pil1-GFP, and against Pgk1, the constitutively expressed 3-phosphoglycerate kinase are shown (top). The graph shows the quantification of Pil1-GFP protein levels relative to Pgk1 and normalized to the wild-type diploid protein level levels (bottom left, blue bars) or to the wild-type haploid protein levels (bottom right, green bars), n = 3. Error bars, SD from the mean. (c) Eisosome density was calculated for cells such as shown in panel a and plotted as a function of Pil1 protein levels, measured by Pil1-GFP/Pgk1 ratios, which are normalized to the wild-type diploid protein levels (blue diamonds) or wild-type haploid protein levels (green squares). ANOVA,* p < 0.0001 for samples with densities lower than wild type. (c) The relative fluorescence intensity was measured and plotted as a function of protein levels as described in panel c. Error bars, the SDs from the mean. Diploid cells (blue diamonds) 0.5, n = 9; 1, n = 16; 1.5, n = 17; 2, n = 22; and haploid cells (green squares) Pil1, n = 17; DAMP, n = 28. Dashed lines represent trend lines.
Consistent with the notion of a “set-point” of eisosome density, the bigger diploid yeast cells bearing two or more copies of PIL1-GFP displayed an almost identical density of eisosomes on the cell surface as did haploid cells (0.39 ± 0.07 eisosomes per μm2 for diploids vs. 0.33 ± 0. 06 for haploids). By contrast, decreasing the gene dose to one-half of normal (i.e., one PIL1-GFP copy per diploid cell) resulted in a significant decrease in eisosome density, yet each eisosome was assembled to a normal size as judged by their Pil1-GFP intensity (Figure 6, c and d; Pil1-GFP/Pgk1 levels <1, blue diamonds). This observation suggests a minimal eisosome size is maintained independent of the amount of available Pil1; under conditions of limiting Pil1 availability, fewer eisosomes assemble at this minimal size. To obtain further evidence for this notion, we generated a haploid strain bearing a “dampened” allele of PIL1-GFP. In this construct, the 3′-untranslated region (UTR) was deleted, resulting in a destabilized PIL1-GFP mRNA and reduced Pil1 expression to roughly 45% of the wild type (Figure 6b, right panel). Consistent with the observations made in PIL1/pil1Δ diploid cells, we found that reduction of Pil1 expression levels also leads to a decrease of eisosome density and the size of individual eisosomes remains unchanged (Figure 6, b and c, Pil1-GFP/Pgk1 levels <1, green squares).
By contrast, overexpression of Pil1-GFP by increasing the gene dose by 1.5- and 2-fold maintained eisosome density at the set-point (0.33 ± 0.11 and 0.40 ± 0.06 eisosomes/μm2, respectively), yet their sizes (as assessed by Pil1-GFP fluorescence per eisosome) increased proportionately to the gene dosage (Figure 6, c and d, Pil1-GFP/Pgk1 levels >1, blue diamonds; ANOVA, p < 0.0001). These results suggest that if Pil1 is available in excess, nucleation sites become limiting and eisosomes grow larger to use up the available Pil1 building blocks. Together, these data establish Pil1 as a regulator of eisosome formation, with regard to both number and size.
Cell Cycle Regulation of Pil1 Determines the Polar Formation of Eisosomes
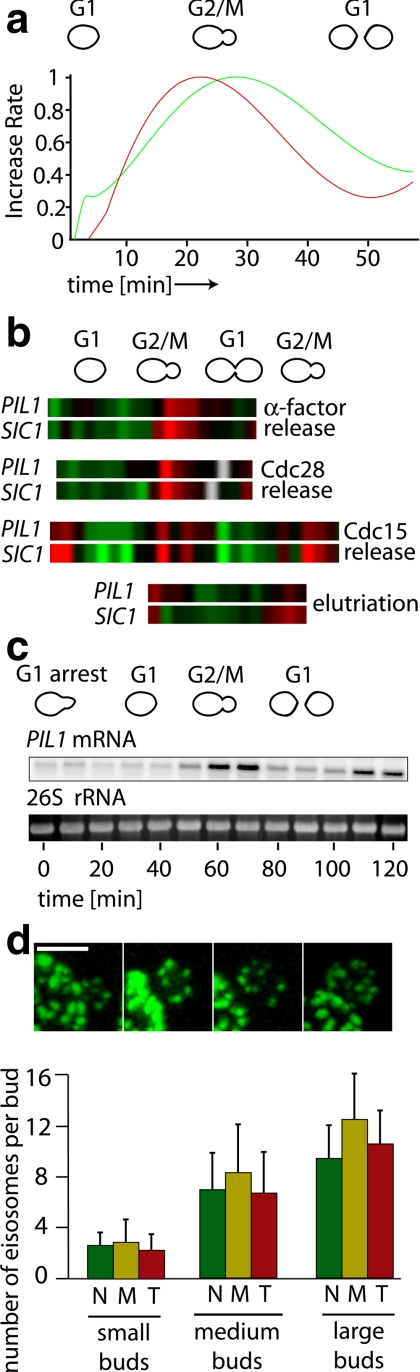
As yeast cells grow, their plasma membrane expands nonlinearly. We have estimated the size of the plasma membrane by measuring cell size microscopically throughout the cell cycle and calculated the rate of membrane addition from the derivative (d[membrane increase]/d[time]) of these data (Figure 7a, plotted as the green curve). This analysis shows that most membrane addition occurs at the G2/M transition, when the rate of bud expansion peaks. We then determined the rate of total fluorescence change in cells expressing Pil1-GFP (Figure 7a, red curve) and found that Pil1 synthesis also occurs in cell cycle–regulated bursts. As is apparent from the similarity of the curves, plasma membrane expansion and Pil1-GFP expression correlate well through the cell cycle. Consistent with this observation, inspection of published microarray gene expression data (Spellman et al., 1998) reveal that PIL1 mRNA abundance is cell cycle regulated. Clustering analysis of genes showing expression patterns similar to PIL1 identifies SIC1 as the gene with the most similar expression pattern. SIC1 is an inhibitor of the Cdc28-cyclin kinase complex, and its expression serves as a marker for the G2/M transition (Figure 7b; Oehlen et al., 1998). The strong similarity between PIL1 and SIC1 expression patterns was observed in experiments utilizing a variety of cell cycle synchronization methods, such as release from α-factor arrest, release from cdc28ts and cdc15ts arrests, as well as by elutriation (Figure 7b). To confirm the conclusions derived from the published high-throughput data sets, we arrested yeast cells in G1 by incubation with α-factor for 3 h, washed the cells in fresh YPD to release the block, and took samples at 10-min intervals as cells progressed through one and a half subsequent cell cycles. Northern blot analysis of these samples showed that PIL1 mRNA levels oscillate with a threefold increase in amplitude through the cell cycle (Figure 7c).
Figure 7.
Pil1 expression is cell cycle regulated. TWY110 cells were followed over an entire cell cycle by confocal live-cell imaging. The increase in cell surface over time was calculated from the images, and the total GFP fluorescence per cell was measured. The derivatives of the resulting curve fits indicating rate of change in surface area (red) and rate of change of GFP fluorescence (green) are displayed. The curves show that Pil1-GFP expression correlates with membrane growth. (b) Data mining revealed the strong cell cycle regulation of PIL1 in several different synchronization methods (Spellman et al., 1998). Expression higher than the mean is shown in red and lower than the mean is shown in green. (c) PIL1 mRNA expression is cell cycle controlled. MATa cells (CRY2) were arrested in G1 phase of the cell cycle by addition of α-factor for 1 h and then released. Total RNA from the indicated time points was extracted and analyzed for PIL1 mRNA levels by Northern blotting. The respective cell cycle stage is indicated graphically above the time points. (d) Delay in eisosome deposition is explained by PIL1 cell cycle control. Confocal 3D reconstructions of small buds expressing PIL1 under the CUP1 promoter, KEM102 (top). Eisosomes are present in small buds. Confocal stacks of cells grown in the presence of 25 μM CuSO4 concentration (bottom). Scale bar, 2 μm. Small-, medium-, and large-sized buds were recorded as in Figure 1. The buds were segmented along the axis of cell polarity into three equal regions, and the number of eisosomes was counted for each region; n = 159.
These data show that the expression of Pil1 occurs in 20-min bursts that correlate with the phase of the cell cycle when eisosome formation initiates, e.g., when the small bud starts first acquiring eisosomes. Interestingly, LSP1 shows no cell cycle regulation, whereas SUR7 does (Spellman et al., 1998); however, neither LSP1 nor SUR7 gene deletions displays a dramatic effect on eisosome organization. These observations underline the proposed role of Pil1 in controlling eisosome biogenesis and suggest that cell cycle regulation of PIL1 mRNA contributes to the regulation of eisosome formation.
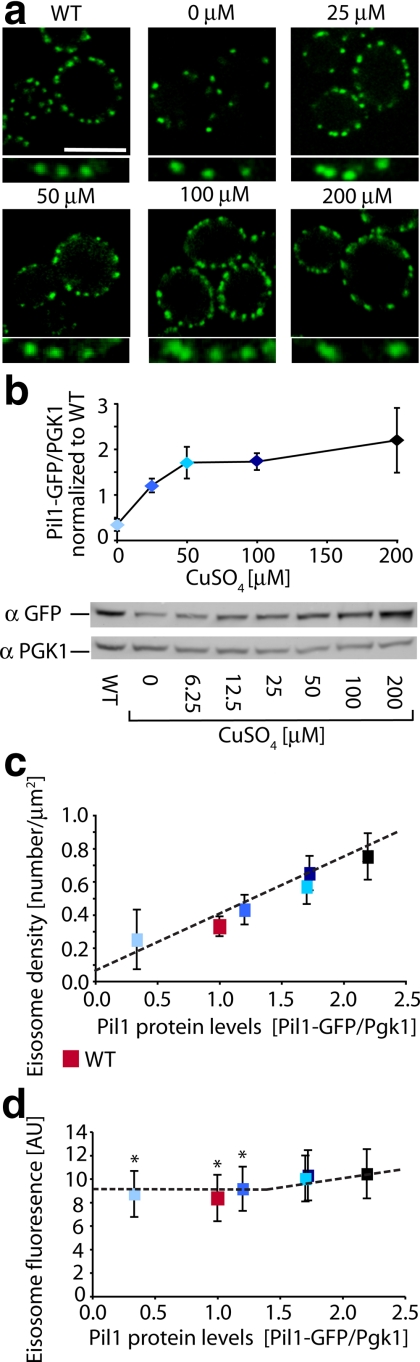
To test the importance of cell cycle regulation of Pil1 expression and eisosome biogenesis, we placed PIL1-GFP under control of the CUP1 promoter, which can be regulated by the addition of copper to the growth medium. Importantly, CUP1 is not cell cycle regulated (Spellman et al., 1998). We found that at 25 μM CuSO4, Pil1 was expressed in the cell population at levels similar to that in wild-type cells, as assessed by quantitative Western blotting (Figure 8b). Moreover, eisosome morphology and deposition looked normal, and eisosome density was within error at the set point of wild-type cells (0.43 ± 0.09 eisosomes per μm2 at 25 μM CuSO4).
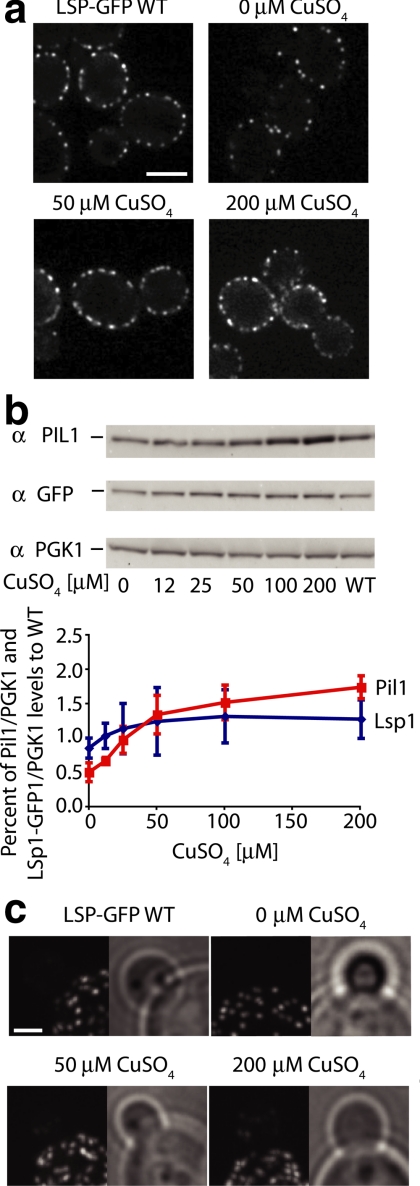
Figure 8.
Pil1-GFP expression from the CUP1 promoter. (a) Confocal cross sections of cells grown at increasing concentrations of CuSO4 at 30°C for 4 h and KEM102 cells expressing Pil1-GFP driven from the CUP1 promoter are shown. Scale bar, 5 μm. (b) Titration of CuSO4 of cells expressing Pil1-GFP under control of the CUP1 promoter. The original Western blots and the quantification of relative abundance of Pil1-GFP as measured by Pil1-GFP/Pkg1 ratios normalized to wild-type protein levels are shown; n = 3. (c) Eisosome density was measured and plotted as a function of Pil1-GFP protein levels as measured in panel b. (d) Fluorescence intensities of Pil1-GFP in individual eisosomes was measured and plotted against Pil1 protein levels as measured in panel b. Blue squares and diamonds, copper-inducible strain KEM102; red square, wt strain TWY110. Dashed lines represent trend line. ANOVA, *p = 0.147. Error bars, the SD from the mean; n = 56 at each CuSO4 concentration.
On the basis of the results in Figure 8b, we chose 25 μM CuSO4, as a concentration that resulted in a cellular Pil1 level comparable to that of wild-type cells. In marked contrast to wild-type cells, we found that under these conditions eisosomes formed in small buds and showed no apparent polarization of deposition (Figure 7d). Thus taken together, the data suggest that the cell cycle regulation of PIL1 expression is required for the wave-like colonization of newly made plasma membrane seen in wild-type cells.
In light of the results presented above, we were surprised to find that eisosome density increased as more Pil1 was produced under the higher CuSO4 concentration conditions (Figure 8c), whereas the size of individual eisosomes (as measured by integrating Pil1-GFP fluorescence intensity) increased only slightly (ANOVA, p < 0.0001, Figure 8d). Overexpression of Pil1 therefore can have qualitatively different consequences: increasing the gene dosage, which retains cell cycle control, leads to larger eisosomes at wild-type density (Figure 6), whereas increasing Pil1 expression from a heterologous promoter, which eliminates cell cycle control, leads to wild-type size eisosomes at an increased density. Formally however, we cannot rule out that the observed difference results more indirectly from the different ploidy of the cells.
To test whether Pil1 expression under the CUP1 promoter directed assembly of other eisosome components in small buds, we followed the expression of Lsp1-GFP, in a strain in which endogenous (untagged) PIL1 was placed under the control of the CUP1 promoter (Figure 9). Consistent with the parallel experiments following Pil1-GFP in Figure 8, we observed a reduction in Lsp1 containing eisosome density (0.24 ± 0.06 μm2) as Pil1 abundance was titrated down. Moreover, eisosome size was similar to that found in wild-type cells (data not shown). Lsp1-labeled eisosome density increased to wild-type levels as Pil1 abundance was raised to wild-type levels (Figure 9a). We did not observe a significant increase in size or density of Lsp1-labeled eisosomes when Pil1 levels were increased; however, despite the increase in Pil1-labeled eisosomes observed in Figure 8c (data not shown).
Figure 9.
Pil1 expression from the CUP1 promoter in LSP1-GFP cells. (a) Spinning disk confocal cross sections of cells grown at increasing concentrations of CuSO4 at 30°C for 4 h, expressing Pil1 driven from the CUP1 promoter in LSP1-GFP cells (KEM103) are shown and Lsp1-GFP with Pil1 under its endogenous promoter (Lsp1-GFP wild type, TWY113). Scale bar, 5 μm. (b) Titration of CuSO4 of cells expressing Pil1 under control of the CUP1 promoter. Shown are the original Western blots and the quantitation of relative abundance of Pil1 to Pgk1 and Lsp1-GFP to Pkg1 normalized to wild-type levels of Pil1 and Lsp1-GFP, respectively; n = 3. Error bars, the SD from the mean. WT lane contains extracts from Lsp1-GFP (TWY113) cells with Pil1 under its endogenous promoter. (c) Lsp1 eisosome deposition in small buds does not follow Pil1 deposition. Shown are 3D reconstructions of small buds in cells expressing Lsp1-GFP and Pil1 under its endogenous promoter, TWY113 (left panel, Lsp1-GFP WT) and Lsp1-GFP with Pil1 under the CUP1 promoter, KEM103. Scale bar, 2 μm.
We measured the levels of Pil1 and Lsp1-GFP by quantitative Western blotting and found that the expression of Lsp1-GFP decreased when Pil1 levels decreased, indicating that Lsp1 is unstable if it cannot assemble with Pil1. By contrast, Lsp1-GFP levels plateau as Pil1 levels increased (Figure 9b), indicating that Lsp1 becomes limiting under these conditions. In parallel experiments, we did not observe Lsp1-GFP in small buds when Pil1 was expressed from the CUP1 promoter (Figure 9c). We conclude that Pil1 provides the upper limit for the density of Lsp1-containing eisosomes, yet its early deposition in small buds is not sufficient to recruit the early deposition of Lsp1. This observation implies that Lsp1 deposition is governed by an unknown process coupled to the cell cycle, perhaps requiring a certain cell cycle–dependent phosphorylation state.
DISCUSSION
Eisosomes are large macromolecular assemblies underlying the plasma membrane that help sequester a subgroup of plasma membrane proteins into discrete membrane domains, which colocalize with sites of endocytosis (Grossmann et al., 2007; Walther et al., 2006). They are present in cells in relatively constant numbers and built of 2000–5000 copies each of Pil1 and Lsp1, their two major subunits. Here we describe a series of experiments and observations that establish a basic framework of eisosome biogenesis. In particular, we found that eisosomes are formed de novo, colonizing the plasma membrane of the growing bud in a wave-like manner. We currently do not know the molecular mechanism that leads to the polar eisosome deposition: either active nucleation sites form after a lag time as newly deposited membrane matures, or deposition is excluded from the very bud tip, perhaps because it is too crowded by other molecular machinery. Their deposition results in a remarkably constant density that is maintained in both haploid and diploid cells, as well as under conditions of aberrant cell growth during G1 cell cycle arrest. From this and previous studies, Pil1 emerges as the central player of eisosome organization: 1) among the known eisosome components, PIL1 is regulated in a cell cycle dependent manner that synchronizes eisosome assembly with membrane expansion, 2) lowering or increasing the gene dose of PIL1 leads to a reduction in eisosome number or increase in eisosome size, respectively, and 3) deletion of PIL1 (but not of LSP1) leads to a collapse of eisosome remnants (Walther et al., 2006; Grossmann et al., 2007).
Modulation of Pil1 levels suggests that eisosomes have a minimal size. When the relative expression level of PIL1 was reduced, both Pil1 and Lsp1 labeled eisosome size leveled off at the expense of eisosome density. In addition Lsp1-GFP levels decreased instead of assembling larger Lsp1 containing eisosomes. One possible explanation of this finding is that eisosomes have a regular structure—perhaps a toroid or an icosahedron—that is not stable when it cannot be formed completely, and thus it provides a maximum limit for the assembly of other eisosome components, resulting in the destabilization of eisosome components in excess of Pil1. In addition, we did not find a difference in the kinetics of deposition between Pil1 and Lsp1 under wild-type conditions, suggesting that these two components are assembled into eisosomes at the same time. As such, eisosomes provide an interesting paradigm for molecular self-assembly, and it will be intriguing to determine the molecular design principles that govern the assembly of Pil1 and other components into eisosomes. Equally surprising, we found that when Pil1 levels were increased moderately, any excess of Pil1 was incorporated into eisosomes leading to gradual size increases, whereas their density on the plasma membrane remained constant. Thus the capping mechanism that restricts incorporation of excess Pil1 subunits into eisosomes can be overcome—maybe forming additional “layers” onto their minimal structure. Pil1 assembly in small buds was not sufficient to recruit Lsp1 to the small buds, suggesting additional regulatory mechanisms can affect the assembly of other eisosome components. Although remarkably tight under conditions where Pil1 expression remained cell cycle controlled, eisosome density was not constant when Pil1 was expressed at elevated levels from the copper-regulated promoter. These observations suggest that the mechanism that limits eisosome nucleation is normally restricted to the timing of new bud formation to prevent additional nucleation in the mother cell. Continuously elevated expression of Pil1 in the copper regulated strains overcomes this regulation, resulting in more nucleation sites. We do not know if eisosome components other than Pil1 are also recruited to these new nucleation sites. Similarly, the tight regulation of PIL1 expression normally leads to a relatively uniform eisosome size, despite the fact that eisosomes can, in principle, grow larger as shown in the strains with increased PIL1 copy number. Thus, our results suggest that the cell cycle regulation of Pil1 contributes to the fidelity of eisosome size, eisosome density, and orderly eisosome deposition in the wave-like pattern observed in growing buds.
The cell cycle regulation of PIL1 is likely to be direct. A cursory inspection of the PIL1 promoter revealed the presence of a binding site for the cell cycle regulated transcription factor Swi5 located 410 base pairs upstream of the PIL1 ORF. Swi5 is known to activate the transcription of genes in G2 phase and the G2/M boundary after phosphorylation by Cdc28 kinase. This argues that at least some of the regulation of Pil1 is directly mediated by the cell cycle via binding of Cdc28 activated Swi5 to a binding site in the PIL1 promoter. In addition, we conclude from the effects on eisosome assembly when Pil1 is expressed from a heterologous promoter that posttranscriptional and cell cycle-regulated control may also have a role in eisosome assembly. Either way, a checkpoint-like feedback overrules the cell cycle control to ensure that the eisosome density remains constant (disrupted here experimentally in the copper-regulated strains). Evidence for this notion comes from the observation that cells arrested at different points in the cell cycle maintain a constant eisosome density. Under these conditions the newly formed eisosomes are generated in apparent gaps in the plasma membrane that open up as the cells expand.
Because eisosome density does not increase under conditions of excess Pil1, when expressed under its endogenous promoter, it is likely that nucleation sites on the plasma membrane are limiting. The observed wave of eisosome colonization in which all new eisosomes form adjacent to preexisting ones would be consistent with short filamentous structures that may link eisosomes, binding to existing ones with one end and providing nucleation sites for new eisosomes at the other end. Thus eisosomes may form at vertices of a meshwork of yet-to-be-identified filamentous proteins, perhaps resembling in its characteristics the spectrin networks that underlie membrane systems in metazoan cells. A mechanism utilizing filamentous spacers could explain the constant eisosome density on the cell surface and account for the minimal distance observed between eisosomes. In G1 arrest cells, which grow isotropically, the network may rupture, allowing assembly of new filaments in the gaps that then form new nucleation sites. Alternatively, eisosome formation may be inhibited in a zone surrounding preexisting eisosomes, thus determining eisosome density by exclusion. These two mechanisms are not mutually exclusive, and currently available data do not allow us to distinguish between them.
The plasma membrane domains associated with eisosome are specialized both in protein and lipid composition (Malinska et al., 2003; Malinska et al., 2004; Grossmann et al., 2007). Moreover, Pil1 and Lsp1 are differentially phosphorylated in response to an increase of long-chain bases, which are the precursors of sphingolipids, by Pkh1/2, two kinases that localize to eisosomes (Zhang et al., 2004; Walther et al., 2007; Luo et al., 2008). Perturbation of the Pil1 phosphorylation status affects Pil1 assembly into eisosomes: specifically, eisosomes disassemble if Pil1 is hyperphosphorylated, and more Pil1 assembles into eisosomes if Pil1 is hypophosphorylated. The phosphorylation status of Pil1 therefore may be an additional factor that modulates eisosome assembly, possibly using the membrane's lipid composition as a controlling feature. Alternatively, however, Pil1 phosphorylation may primarily serve under physiological conditions to effect local structural rearrangements within an eisosome to regulate, for example, its activity to recruit endocytic effectors. As such, phosphorylation events may not globally affect assembly/disassembly properties as observed upon extreme hypo- and hyperphosphorylation observed under experimental conditions (Walther et al., 2007).
By providing on the plasma membrane a constant density of specialized domains, eisosomes may ensure an even distribution of potential endocytic sites. This organization principle is important because mutations in many parts of the system, such as in Pkh1/2 and eisosome components, show endocytic defects, impairing the rate and the localization of endocytic events. pil1Δ strains, for example, show large, aberrant plasma membrane invaginations. A massing of many endocytic sites in close proximity may result in too much actin pulling force at the same place, thereby deforming the membrane in a disorganized and uncontrolled manner. The organization provided by constant density and minimal distance between eisosomes might serve to avoid such chaotic events by guaranteeing sufficient spatial separation between individual endocytic events.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mark Segal for help with the statistical analysis of eisosome density. We also thank Ken Dill, David Gliden, Arne Gennerich, and the Walt(h)er labs for valuable discussions, and Gustavo Pesce for providing reagents. Data for this study were acquired at the Nikon Imaging Center at UCSF. T.C.W. was supported by the International Human Frontiers Science Program (HFSPO) and the Deutsche Forschungsgemeinschaft Grant WA 1669/2-1. K.E.M. is a recipient of an aging training grant from National Institutes of Health (NIH), National Institute on Aging. In addition, this work was supported by grants from the NIH to P.W. P.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-03-0313) on November 26, 2008.
REFERENCES
- Bacon D. W., Watts D. G. Estimating the transition between two intersecting straight lines. Biometrika. 1971;58:525–534. [Google Scholar]
- Bagnat M., Simons K. Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. USA. 2002;99:14183–14188. doi: 10.1073/pnas.172517799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A. C., et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Brickner J. H., Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2(11):e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadie K. Synapse scaffolding: intersection of endocytosis and growth. Curr. Biol. 2004;14:R853–R855. doi: 10.1016/j.cub.2004.09.042. [DOI] [PubMed] [Google Scholar]
- Grossmann G., Opekarova M., Malinsky J., Weig-Meckl I., Tanner W. Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J. 2007;26:1–8. doi: 10.1038/sj.emboj.7601466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;12:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Koh T. W., Verstreken P., Bellen H. J. Dap160/intersectin acts as a stabilizing scaffold required for synaptic development and vesicle endocytosis. Neuron. 2004;43:193–205. doi: 10.1016/j.neuron.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 1993;120(6):1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Gruhler A., Liu Y., Jensen O. N., Dickson R. C. The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover. J. Biol. Chem. 2008;283((16)):10433–10444. doi: 10.1074/jbc.M709972200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinska K., Malinsky J., Opekarova M., Tanner W. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell. 2003;14:4427–4436. doi: 10.1091/mbc.E03-04-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinska K., Malinsky J., Opekarova M., Tanner W. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J. Cell Sci. 2004;117:6031–6041. doi: 10.1242/jcs.01493. [DOI] [PubMed] [Google Scholar]
- Marie B., Sweeney S. T., Poskanzer K. E., Roos J., Kelly R. B., Davis G. W. Dap160/intersectin scaffolds the periactive zone to achieve high-fidelity endocytosis and normal synaptic growth. Neuron. 2004;43:207–219. doi: 10.1016/j.neuron.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Oehlen L. J., Jeoung D. I., Cross F. R. Cyclin-specific START events and the G1-phase specificity of arrest by mating factor in budding yeast. Mol. Gen. Genet. 1998;258:183–198. doi: 10.1007/s004380050722. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., Kreitzer G., Musch A. Organization of vesicular trafficking in epithelia. Nat. Rev. Mol. Cell Biol. 2005;6:233–247. doi: 10.1038/nrm1593. [DOI] [PubMed] [Google Scholar]
- Schuldiner M., et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K., Eisen M. B., Brown P. O., Botstein D., Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., Shah K., Shokat K. M., Morgan D. O. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- Valdez-Taubas J., Pelham H. R. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr. Biol. 2003;13:1636–1640. doi: 10.1016/j.cub.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Walther T. C., Brickner J. H., Aguilar P. S., Bernales S., Pantoja C., Walter P. Eisosomes mark static sites of endocytosis. Nature. 2006;439:998–1003. doi: 10.1038/nature04472. [DOI] [PubMed] [Google Scholar]
- Walther T. C., Aguilar P. S., Froehlich F., Chu F., Moreira K., Burlingame A. L., Walter P. Pkh-kinases control eisosome assembly and organization. EMBO J. 2007;26((24)):4946–4955. doi: 10.1038/sj.emboj.7601933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lester R. L., Dickson R. C. Pil1p and Lsp1p negatively regulate the 3-phosphoinositide-dependent protein kinase-like kinase Pkh1p and downstream signaling pathways Pkc1p and Ypk1p. J. Biol. Chem. 2004;279:22030–22038. doi: 10.1074/jbc.M400299200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.