Abstract
Using a bioinformatic approach, we identified a TP53INP1-related gene encoding a protein with 30% identity with tumor protein 53-induced nuclear protein 1 (TP53INP1), which was named TP53INP2. TP53INP1 and TP53INP2 sequences were found in several species ranging from Homo sapiens to Drosophila melanogaster, but orthologues were found neither in earlier eukaryotes nor in prokaryotes. To gain insight into the function of the TP53INP2 protein, we carried out a yeast two-hybrid screening that showed that TP53INP2 binds to the LC3-related proteins GABARAP and GABARAP-like2, and then we demonstrated by coimmunoprecipitation that TP53INP2 interacts with these proteins, as well as with LC3 and with the autophagosome transmembrane protein VMP1. TP53INP2 translocates from the nucleus to the autophagosome structures after activation of autophagy by rapamycin or starvation. Also, we showed that TP53INP2 expression is necessary for autophagosome development because its small interfering RNA-mediated knockdown strongly decreases sensitivity of mammalian cells to autophagy. Finally, we found that interactions between TP53INP2 and LC3 or the LC3-related proteins GABARAP and GABARAP-like2 require autophagy and are modulated by wortmannin as judged by bioluminescence resonance energy transfer assays. We suggest that TP53INP2 is a scaffold protein that recruits LC3 and/or LC3-related proteins to the autophagosome membrane by interacting with the transmembrane protein VMP1. It is concluded that TP53INP2 is a novel gene involved in the autophagy of mammalian cells.
INTRODUCTION
Tumor protein 53-induced nuclear protein 1 (TP53INP1), also known as TEAP, SIP and p53DINP1, is a recently described factor involved in cell stress response (Tomasini et al., 2001). TP53INP1 expression is induced by p53 (Tomasini et al., 2002; Okamura et al., 2001) and exerts its function mainly by inducing transcription of target genes involved in cell cycle arrest and apoptosis, as part of the cell response to genotoxic stress (Tomasini et al., 2003). Mechanistically, TP53INP1 interacts with p53 (Tomasini et al., 2003) and kinases that phosphorylate p53 (HIPK2 and PKCδ) (Tomasini et al., 2003; Yoshida et al., 2006), contributing to p53 activity regulation leading to apoptosis (Tomasini et al., 2003). TP53INP1 expression is up-regulated in different cell types upon treatment with agents inducing cell cycle arrest and/or apoptosis and overexpression of TP53INP1 induces cell cycle arrest and apoptosis, even in the absence of p53 (Tomasini et al., 2001). In that case p73 or E2F1 transcription factors, which both play a role in cell proliferation and apoptosis, seem to be implicated (Hershko et al., 2005; Tomasini et al., 2005). Altogether, currently available data point to a role of TP53INP1 in cellular homeostasis by its antiproliferative and proapoptotic activities in cell response against cellular stress. Finally, TP53INP1 could be considered as a putative tumor suppressor gene because TP53INP1-deficient mice are more susceptible to develop tumors by a mechanism involving free radicals (Gommeaux et al., 2007; Cano et al., 2008).
Using a bioinformatic approach, we identified a TP53INP1-related gene encoding a protein with 30% amino acid identity and 45% similarity with TP53INP1. This gene, located at chromosome 20q11.2 (Nowak et al., 2005), is well conserved through evolution. Interestingly, based on its chromosomal localization, TP53INP2 gene was a candidate for recessive nonprogressive infantile ataxia, although this hypothesis was eventually ruled out (Fowles et al., 2003; Bennetts et al., 2006, 2007). To gain insight into the function of the TP53INP2 protein, we carried out a yeast two-hybrid screening and found that it interacts with LC3-related proteins GABARAP and GABARAP-like2. These proteins are involved in several processes related to intracellular vesicle formation and transport, including autophagy (Tanida et al., 2004). In this article, we report that the TP53INP2 protein plays a major role in mammalian cell autophagy.
MATERIALS AND METHODS
Computer-based Identification of the TP53INP2 Gene
A nonredundant sequence database was screened with the BLASTp program (http://www.ncbi.nlm.nih.gov/BLAST/), by using as initial query the sequences of both isoforms encoded by the human TP53INP1 gene (accession no. NM_033285). This screen identified, in the Homo sapiens chromosome 20 genomic contig (accession no. NT_028392), gene 110 of the chromosome 20 open reading frame (C20orf110, NM_021202). A BLAST back-validation step confirmed its sequence homology with TP53INP1. The name of tumor protein p53-inducible nuclear protein 2 (TP53INP2) was approved by the HUGO Gene Nomenclature for C20ORF110. Exons boundaries were defined by alignment of the cDNA sequence with the sequence of the genomic region C20ORF110 (NT_028392), by using the blast2seq program available at National Center for Biotechnology Information.
Phylogenetic Reconstruction
The Ensembl database (http://www.ensembl.org/) was used to find homologues of the human TP53INP1 protein in different species. We searched with protein sequences rather than cDNA sequences to avoid sequence saturation. By using the Blast tool, we could restore the full-length sequence from fragments obtained from the Ensembl database. The Clustal X 1.83 tool (Thompson et al., 1997) was used for sequence alignment. The file containing the results was imported in the MEGA 3.1 software to establish phylogenetic trees. Phylogenetic trees were constructed with the neighbor-joining (Saitou and Nei, 1987) and the maximum of parsimony methods (Nei and Kumar, 2000) with a complete deletion and 1000 bootstraps. The sequences too short were eliminated to avoid signal loss. Groups were identified from an important genetic distance and supported by strong statistical value of the Bootstrap. Homologous sequences were used in Multiple EM for Motif Elicitation (MEME; http://meme.sdsc.edu/meme/) tools to define conserved motifs. Maximum number of motifs to find options was set at three.
Yeast Two-Hybrid Screen
Using a polymerase chain reaction (PCR)-based strategy, we subcloned the complete coding sequence of human TP53INP2 into the BamHI–SacI restriction site of the pSos vector to generate the fusion protein pSos-TP53INP2. This construct was used as bait to screen a human HeLa cDNA library (catalog no. 975212; Stratagene, La Jolla, CA) constructed into the pMyr vector according to the protocols provided by the manufacturer. pSos-TP53INP2-1-110 and pSos-TP53INP2-111-220, encoding TP53INP2 fragments from amino acids 1-110 and 111-220, respectively, were obtained by subcloning specific reverse transcription (RT)-PCR–amplified DNA fragments into the pSos vector. After cotransfection into Saccharomyces cerevisiae strain cdc25H, 2 × 106 clones were screened and several positives were identified. All clones were PCR amplified. Their sequences were identified by comparison with the GenBank repertoire. The interactions between TP53INP2 and GABARAP, and GABARAP-like2 were confirmed by transforming S. cerevisiae with both pMyr-GABARAP or pMyr-GABARAP-like2 and pSos-TP53INP2 constructs and allowing the transformants to grow on synthetic drop-out (SD) glucose and galactose agar plates lacking leucine and uracil [SD/glu(-LU) and SD/gal(-LU)] at the stringent temperature of 37°C. Clones growing on SD/gal(-LU) plates but not on SD/glu(-LU) plates at 37°C are interaction-positive clones.
Mammalian Cell Lines and Transfections
Human HeLa and 293T cell lines and the mouse NIH3T3 cell line were obtained from the American Type Culture Collection (Manassas, VA) and maintained according to American Type Culture Collection instructions. Atg5-deficient mouse embryonic fibroblasts (MEFs) (Atg5−/−) and wild-type MEFs (Atg5+/+) have been described previously (Kuma et al., 2004). Cells were transfected using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions, with pEGFP-N1 and pEYFP-C1 (Clontech, Mountain View, CA) plasmids containing the full-length human TP53INP2 cDNA (NM_021202) subcloned into SacI–BamHI restriction sites, or no insert, and pERFP-C1 containing the full-length GABARAP (NM_007278) or GABARAP-like2 (NM_007285) or LC3 (NM_032514) subcloned into the BglII–SalI restriction site. GABARAP, GABARAP-like2, and LC3 were also subcloned into the EcoRI–XhoI restriction site of the pcDNA3-FLAG vector. Full-length human cDNA encoding beclin 1 (NM_003766) was subcloned into pECFP-C1 within the EcoRI and SalI restriction sites. Construction of pcDNA4-VMP1-enhanced green fluorescent protein (EGFP) has been reported previously (Dusetti et al., 2002). Direct mutagenesis was used in GABARAP-ERFP, GABARAP-like2-ERFP, and LC3-ERFP vectors by replacing the C-terminal Gly, essential for lipidation and autophagosome formation, to Ala in positions 116, 116, and 120, respectively, as reported previously (Kabeya et al., 2004). Primers used for mutagenesis were: 5′-GACGAAAGTGTCTACGCTCTGTGAAGCTGCTGG-3′ for GABARAP, 5′-GGAGAGAACACTTTTGCCTTCTGAGGGCCATTG-3′ for GABARAP-like2, 5′-CAGGAGACGTTCGCGACAGCACTGGCT-3′ for LC3, and their reverse complementary sequence. Mutations were confirmed by DNA sequencing.
Localization of TP53INP2, GABARAP, GABARAP-like2, LC3, and Beclin1 by Fluorescence Microscopy
Cells were fixed in 4% formaldehyde and washed twice with phosphate-buffered saline (PBS). Cells were mounted in Mowiol 488 (Sigma-Aldrich, St. Louis, MO) and observed with anAxioplan 2 confocal microscope (Carl Zeiss, Jena, Germany).
LC3 Fluorescence
A rabbit anti-LC3 antibody (MBL International, Woburn, MA) was used to detect endogenous LC3 proteins, which were revealed with the goat Alexa 488-conjugated anti-rabbit secondary antibody (Invitrogen, Carlsbad, CA).
Autophagy Induction
Autophagy was induced by an amino acid/serum standard starvation protocol. Cells were washed three times with PBS and incubated with Earle's balanced salt solution (EBSS; Invitrogen) at 37°C. Alternatively, autophagy was induced by treatment with 10 μM rapamycin (LcLabs, Woburn, MA) in a nutrient-repleted medium. In some experiments, cells were pretreated with TP53INP2 small interfering RNA (siRNA) 24 h before starvation or rapamycin treatment. Wortmannin (Sigma-Aldrich) was used at 200 nM during starvation. For inhibition of autophagolysosome acidification, 200 nM bafilomycin A1 (LcLabs) was added in EBSS during starvation.
Percentage of LC3-ERFP Cells with Punctate Staining
The percentage of cells with punctate staining was determined in three independent experiments. We considered an LC3-ERFP cell to have punctate staining when all the red fluorescence was present as dots with no diffuse staining remaining in the cytoplasm. The percentage was obtained by counting cells with punctate staining in six fields chosen at random, each covering ∼100 fluorescent LC3-ERFP–transfected cells. Results were expressed as the mean ± SD of combined results.
siRNA-mediated Knockdown of TP53INP2 Expression
TP53INP2 expression was knocked-down in cultured cells with a specific siRNA by using 5′-CCGAAACCUCCCUUCUUAATT-3′ as sense, and 5′-UUAAGAAGGGAGGUUUCGGTG-3′ as antisense oligonucleotides (QIAGEN, Valencia, CA). The day before transfection cells were placed in six-well plates. After removal of the medium, cells (at 50% confluence) were washed once with serum-free medium and transfection was performed with a mixture of 10 μl of Xtremgene reagent with 1.0 μg of siRNA targeting TP53INP2 (TP53INP2-siRNA) or control scrambled siRNA (control-siRNA) in serum-free medium (total volume, 1 ml).
Immunoprecipitation and Western Blotting
The mouse monoclonal anti-FLAG (M2; Sigma-Aldrich), and mouse monoclonal anti-green fluorescent protein (GFP) (clones 7.1/13.1; Roche Diagnostics) antibodies were used for immunoprecipitation and Western blotting. Twenty-four hours after transfection, cells were lysed in lysis buffer and sonicated by 10 pulses followed by a centrifugation for 15 min at 4°C. Clear lysates were incubated with antibodies for 2 h at 4°C. Immune complexes were precipitated after 1-h incubation with protein G-Sepharose beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). After washing three times in cold lysis buffer, the complexes were resuspended in Laemmli sample buffer (Bio-Rad, Hercules, CA) and boiled for 5 min. Proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose filters. Filters were blocked overnight at 4°C in Tris-buffered saline (TBS) with 5% milk proteins and immunoblotted in the same solution with the primary antibody for 2 h. After extensive washes in 0.05% TBS Triton X-100, filters were incubated with the horseradish peroxidase-conjugated secondary antibody for 1 h in 5% TBS milk proteins. After extensive washes in 0.05% TBS Triton X-100, filters were developed using enhanced chemiluminescence (GE Healthcare) and exposure to Kodak Biomax films (Eastman Kodak, Rochester, NY). LC3 polyclonal antibody (MBL International) was used at 1/1000 on 50 μg of HeLa cell protein extracts transfected with scramble or TP53INP2 siRNA.
Bioluminescence Resonance Energy Transfer (BRET) Assay
Plasmid Construction. The BRET expression vectors are codon humanized pRluc C-terminus fusion protein vectors (pHRluc-C; PerkinElmer Life and Analytical Sciences, Boston, MA), defined as BRET donor, and pEYFP-C (Clontech), defined as BRET acceptor, containing the open reading frame for Rluc or for enhanced yellow fluorescent protein (EYFP), respectively. We inserted the complete coding sequence of TP53INP2 into the EcoRI/BamHI restriction sites of pHRluc-C, and GABARAP, GABARAP-like2, and LC3 were subcloned into the BglII/SalI restriction sites of pEYFP-C. HeLa cells were transfected with the TP53INP2-pHRluc-C construct alone or in combination with pEYFP-C or the GABARAP-pEYFP-C, GABARAP-like2-pEYFP-C, or LC3-pEYFP-C fusion proteins. Transfections were performed using the FuGene HD reagent (Roche Biochemicals, Mannheim, Germany) according to the manufacturer's recommendations. For each transfection, equal amounts of BRET donor and BRET acceptor vectors were used. As BRET acceptors, we used different amounts of GABARAP-pEYFP-C, GABARAP-like2-pEYFP-C, or LC3-pEYFP-C fusion proteins, and the empty vector (pEYFP-C) was used to equalize DNA amounts in each sample. Cells were transfected in 12-well culture plates with 0.8 μg of total plasmid DNA. One day later, cells were harvested and distributed in a white 96-well microplate (25,000 cells/well). On the following day, cells were treated or not, washed with PBS, and the cell-permeable Rluc substrate coelentherazin-h (Promega, Madison, WI) was added to a final concentration of 5 μM in PBS, 15 min before reading. Repeated readings were done for at least 5–10 min by using a LB 940 Mithras reader (Berthold France SA, Thoiry, France), with signal detection in the 470–490 nm (donor) and 520–540 nm (acceptor) windows. Data were analyzed as a BRET ratio, which is defined as the BRET ratio for the coexpression of the Rluc and EYFP constructs against the BRET ratio for the Rluc expression construct alone. To assess signal variation, the BRET values were determined by using the following equation, expressed in mBu (milli-BRET unit): 530 nm acceptor signal/480 nm donor signal − E0 × 1000, where E0 corresponds to the ratio 30 nm acceptor signal/480 nm donor signal obtained with the Rluc construct alone in the same experiment.
RESULTS
Cloning of the TP53INP1-related Protein TP53INP2
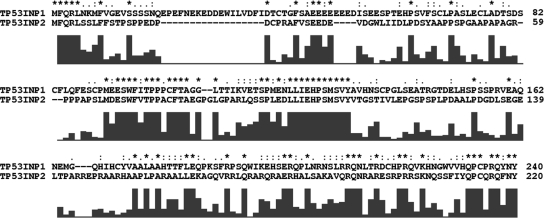
TP53INP2 is organized in five exons spanning 9 kb of genomic DNA on chromosome 20, locus q11 (Nowak et al., 2005). The predicted open reading frame of TP53INP2 codes for a 220-amino acid peptide with a predicted molecular mass of 24 kDa. Comparison of TP53INP1 and TP53INP2 amino acid sequences showed 30 and 45% identity and similarity, respectively (Figure 1).
Figure 1.
Alignment of human TP53INP1 and TP53INP2 protein sequences with the Clustal X software. A quality curve below the sequence gives a graphical representation of identical or similar residues.
Phylogenetic Tree and Conserved Regions
Homologous sequences of TP53INP1 and TP53INP2 were found in several species, ranging from H. sapiens to D. melanogaster. No homologous sequence was found in earlier eukaryotes or in prokaryotes. In Drosophila, we found only one sequence, the far ancestral sequence of the TP53INP family, named here TP53INP (cg11347). From Tetraodon and Fugu, two copies of the TP53INP gene are found. A phylogenetic tree constructed with the TP53INP1 homologues revealed that duplication of the ancestral TP53INP gene occurred at the Actinopterygian spread (420 millions years) and established the relationships between orthologues and paralogues (Figure 2). This phylogenetic analysis completed the preliminary report on genes homologous to TP53INP2 (Bennetts et al., 2007). The homologous sequences were used in the MEME tool for discovering motifs conserved during evolution (http://meme.sdsc.edu/meme/). This analysis revealed that three motifs were shared by all sequences (Figure 2).
Figure 2.
Phylogenetic tree of the TP53INP family. (a) After multiple alignments of protein sequence, phylogenetic reconstructions were made by neighbor joining and maximum parsimony. Both topographies give the history of sequences and their relationships. This display represents the topology of the tree only. Distances between sequences are not represented. We identified TP53INP2 as a human sequence homologous to TP53INP1. Genes orthologous to human TP53INP1 and TP53INP2 were all named TP53INP1 and TP53INP2, respectively. We could not find homologous sequences in early eukaryotes and in prokaryotes. (b) Schematic representation of conserved motifs of the TP53INP family in homologous sequences. The combined best matches of a sequence to a group of motifs are defined as combined p value. Consensus sequences of motifs are shown below.
TP53INP2 Interacts with GABARAP and GABARAP-like2 Proteins
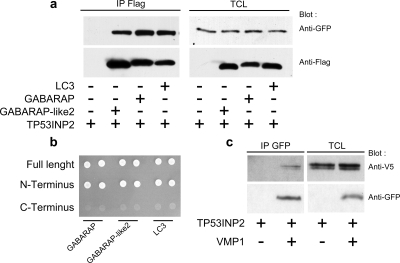
Proteins interacting with TP53INP2 were identified by yeast two-hybrid screening of a HeLa cDNA library. Results are presented in Supplemental Table 1. GABARAP and GABARAP-like2 were among putative interactors. Yeast growth in stringent conditions (see Materials and Methods) was observed when pSos-TP53INP2 was transfected with pMyr-GABARAP or pMyr-GABARAP-like2 but not when pSos-TP53INP2, pMyr-GABARAP, or pMyr-GABARAP-like2 were used separately. Negative and positive controls were grown as suggested by the manufacturer with expected results. These data show that TP53INP2 interacts with GABARAP and GABARAP-like2 and that the interaction is specific. These results were confirmed by coimmunoprecipitation assays: 293T cells were transfected with GABARAP-FLAG and GABARAP-like2-FLAG and TP53INP2-EGFP, alone or in combination. GABARAP-FLAG or GABARAP-like2-FLAG were immunoprecipitated from cell extracts with anti-FLAG antibodies and analyzed by western blot. Anti-GFP antibody was used to detect tagged TP53INP2. TP53INP2 was detected in the complex containing GABARAP or GABARAP-like2. Tags were not detected in the negative control (Figure 3).
Figure 3.
Validation of the interaction between TP53INP2 and its partners. (a) Coimmunoprecipitation assays. 293T cells were transfected with the TP53INP2-EGFP construct alone or in combination with FLAG-GABARAP, FLAG-GABARAP-like2, or FLAG-LC3 plasmids. Lysates were immunoprecipitated with a FLAG antibody and revealed by a GFP antibody. (b) Yeast two-hybrid cotransformation. pSos-TP53INP2, pSos-TP53INP2-N-term, or pSos-TP53INP2-C-term were combined with pMyr-GABARAP, pMyr-gGbarap-like2, or pMyr-LC3 to transform yeasts. Yeast cells were plated on Gal-U/L for growth selection of bait–prey interaction. (c) Interaction of TP53INP2 with VMP1. Coimmunoprecipitation assay: 293T cells were transfected with the TP53INP2-V5 construct alone or in combination with the VMP1-EGFP construct. Lysates were immunoprecipitated with a GFP antibody and revealed with a V5 antibody. IP, immunoprecipitation; TCL, total cell lysate.
Interaction of TP53INP2 with LC3
GABARAP and GABARAP-like2 belong to a protein family involved in intracellular transport processes (Sagiv et al., 2000; Paz et al., 2000), which also include LC3, a light chain of microtubule-associated proteins MAP1A and MAP1B (Mann and Hammarback, 1994; Mann and Hammarback, 1996). GABARAP-like proteins are highly conserved among species ranging from mammals to yeast (Coyle and Nikolov, 2003). Because these proteins are members of a structurally and functionally related multigenic family we wondered whether TP53INP2 interacts with LC3. 293T cells were transfected with LC3-FLAG and TP53INP2-EGFP, alone or in combination. LC3-FLAG was immunoprecipitated from cell extracts with anti-FLAG antibody and analyzed by Western blot. Anti-GFP antibody was used to detect tagged TP53INP2. TP53INP2-EGFP was only detected in the complex containing LC3 (Figure 3). Tags were not detected in the negative control. Altogether, these results confirm that GABARAP, GABARAP-like2 and LC3 interact with TP53INP2 in 293T cells. Interestingly, the orthologues of these proteins in Drosophila also interact with each other, according to the Curagen Drosophila Interaction Database (http://portal.curagen.com/cgi-bin/interaction/flyHome.pl). In addition, Giot et al. (2003) reported a high confidence score for the probability of occurrence of an interaction between cg11347 (TP53INP), cg32672 (DmeI/Atg8a), and cg12334 (DmeI/Atg8b).
Interaction with GABARAP, GABARAP-like2, and LC3 Involves the N-terminal Part of TP53INP2
The pSos-TP53INP2-1-110 and pSos-TP53INP2-111-220 constructs that, respectively, encode the N-terminal and C-terminal parts of TP53INP2 were used separately in yeast two-hybrid experiments to identify which part of the TP53INP2 molecule is involved in GABARAP, GABARAP-like2, and LC3 bindings. The constructs were cotransfected with the pMyr-GABARAP, pMyr-GABARAP-like2, or pMyr-LC3 plasmids. Results show that these proteins interact with the N-terminal part of TP53INP2, but not with its C-terminal part (Figure 3). Because we have defined conserved regions in the N-terminal part of TP53INP2 in all homologues, and beecause this interaction is observed in Drosophila, we can speculate that restriction to the N-terminal part is conserved from Drosophila to human.
TP53INP2 Is a Nuclear Protein That Translocates to Autophagosomes after Rapamycin Treatment or Nutrient Deprivation
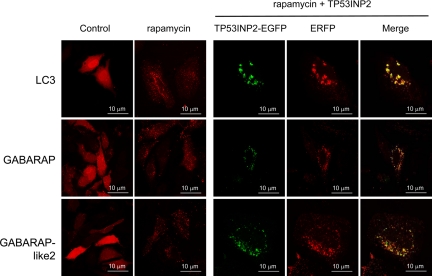
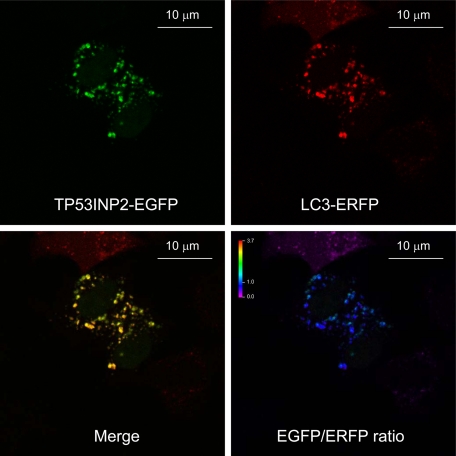
TP53INP2 localizes to the nucleus when transfected as an EGFP fusion protein in HeLa (Figure 4), NIH3T3, or 293T (data not shown) cells. This is in agreement with the computational predictions generated with the PSORT II algorithm (Horton and Nakai, 1997). To our surprise, when cells were treated with rapamycin or were nutrient-deprived, TP53INP2 localization shifted almost completely from the nucleus to the cytoplasm, where it occurred in spots, as evidenced by confocal microscope analysis (Figure 4). Because these spots resemble autophagosomes, and TP53INP2 binds LC3 and LC3-related GABARAP and GABARAP-like2 proteins that are involved in autophagy, we speculated that TP53INP2 also could be involved in that process. During autophagy, the cytosolic form of LC3 (LC3-I) undergoes C-terminal modifications (proteolysis and lipid addition) to generate the LC3-II form that translocates to the autophagosomal membrane. We investigated whether TP53INP2 was located with LC3 and LC3-related GABARAP and GABARAP-like2 proteins to the autophagosomal membrane after rapamycin treatment or nutrient-deprivation. Interestingly, TP53INP2 colocalized predominantly with LC3, GABARAP and GABARAP-like2 proteins in the cytoplasmic spot-like structures induced by rapamycin and nutrient-deprivation (Figure 5). However, TP53INP2-EGFP is not present in all LC3-ERFP–positive dots, possibly due to the higher sensitivity of EGFP (pKa = 6.0) to low pH within the lysosome (∼4.7), compared with ERFP (pKa = 4.5) (Shaner et al., 2004; Kimura et al., 2007). To validate this hypothesis, we treated the cells with Bafilomycin A1 during starvation. Bafilomycin A1 is an inhibitor of the vacuolar ATPase that blocks acidification of the lysosomes and thereby also blocks lysosomal degradation, without affecting the fusion of autophagosomes with lysosomes (Mousavi et al., 2001). As expected, we observed in these conditions a larger number of dots double positive for LC3 and TP53INP2 (Figure 6). However, in certain conditions, GFP-LC3 is reported to form puncta, independently from autophagy. To check whether colocalizations into dots were indeed associated with autophagy and did not result from nonspecific aggregation, we used, in the same experimental setup, inactive mutants of GABARAP-ERFP, GABARAP-like2-ERFP, and LC3-ERFP. The C-terminal Gly of the three proteins, essential for lipidation and autophagosome formation, was replaced to Ala as reported by Kabeya et al., 2004. As expected, TP53INP2-EGFP moved to the cytoplasm and formed dots with the endogenous LC3, but the signal was spread into the cytoplasm under rapamycin treatment when mutant LC3-deltaG-ERFP GABARAP-deltaG-ERFP or GABARAP-like2-deltaG-ERFP were used (see Supplemental Figure 1). These observations confirm that overexpression of TP53INP2-EGFP-, LC3-ERFP-, and LC3-related proteins under rapamycin treatment is followed by their colocalization in autophagic vacuoles.
Figure 4.
Subcellular localization of TP53INP2. HeLa cells were transfected with TP53INP2-EGFP and treated with rapamycin, or starved with EBSS. Quantification of TP53INP2 cytoplasmic positive spots in autophagy-induced cells, relative to control. The number of cells with punctate staining per 100 fluorescent TP53INP2-EGFP transfected cells was determined as it is described under Materials and Methods and expressed as the mean ± SD of combined results from three independent experiments. (*p < 0.05).
Figure 5.
Localization of TP53INP2 with GABARAP, GABARAP-like2 and LC3 after autophagy induction. Left, HeLa cells were transfected with LC3-ERFP, GABARAP-ERFP, or Gabarap-like2-ERFP plasmids with or without rapamycin treatment (10 μM for 4 h). Right, TP53INP2-EGFP was used in combination with LC3-ERFP, GABARAP-ERFP, or GabArap-like2-ERFP with or without rapamycin treatment (10 μM for 4 h).
Figure 6.
Increase in the number of double-positive dots after treatment with bafilomycin A1 during starvation, in cell cotransfected with TP53INP2-EGFP and LC3-ERFP. The EGFP/ERFP ratio of intensities is shown (bottom right). The value of the ratio is given by color scale, as indicated on the top left.
TP53INP2 Binds to the Transmembrane VMP1 Protein
VMP1 is a transmembrane protein whose expression is induced by standard inducers of autophagy. VMP1, through its binding to beclin1, is absolutely required for autophagosome formation (Ropolo et al., 2007). However, VMP1 does not bind to LC3. We speculated that TP53INP2, which binds to LC3 and LC3-related proteins, might also interact with VMP1. It would therefore act as a scaffold protein, allowing the recruitment of LC3 or LC3-related proteins to the autophagosome. To test this hypothesis, 293T cells were transfected with EGFP-tagged VMP1 and V5-tagged TP53INP2, alone or in combination. VMP1-EGFP was immunoprecipitated from cell extracts with anti-EGFP antibody and analyzed by Western blot. Anti-V5 antibody was used to detect tagged TP53INP2. TP53INP2-V5 was only detected in the complex containing VMP1 (Figure 3). Tags were not detected in the negative control. Together, these results show that VMP1 interacts with TP53INP2 in 293T cells.
TP53INP2 Is Required for Autophagosome Formation and Processing
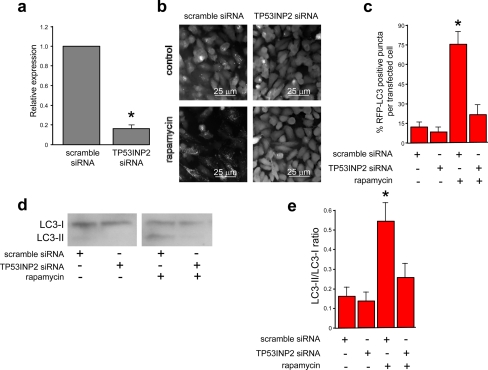
To establish whether TP53INP2 is required for autophagy, we reduced the expression of TP53INP2 by using an siRNA strategy. HeLa cells were transfected with TP53INP2-siRNA and subjected to rapamycin treatment or to nutrient deprivation to induce autophagy. TP53INP2 expression was efficiently knocked down (Figure 7). We found that autophagosome formation was almost completely inhibited in TP53INP2-siRNA–transfected cells under both treatments, as evidenced by the distribution of the LC3-ERFP fluorescent fusion protein (Figure 7). In three independent experiments for each treatment, the percentage of ERFP-positive punctuated cells in TP53INP2-siRNA–transfected cells was drastically reduced, compared with cells transfected with scrambled siRNA (Figure 7). Similar results were obtained with endogenous LC3 (see Supplemental Figure 2). Inhibition of TP53INP2 expression by siRNA during starvation leads to decreased activation of LC3 (LC3-I to LC3-II) (Figure 7). These findings show that TP53INP2 is required for the translocation of LC3 and LC3-related proteins to the autophagosome membrane.
Figure 7.
TP53INP2 silencing blocks the formation of LC3-positive vacuoles. (a) The effect of siRNA-mediated TP53INP2 silencing was quantified in HeLa cells by real-time PCR. (b) HeLa cells were plated and, the next day, they were transfected with TP53INP2 siRNA or scrambled siRNA as a control, and grown for 24 additional hours before LC3-ERFP transfection. Twenty-four hours later, cells were treated with rapamycin (10 μM) for an additional 4-h period. (c) Relative quantification of LC3-ERFP–positive vesicles, in rapamycin-treated or untreated cells, transfected with siRNA TP53INP2 or scrambled siRNA as a control. The number of cells with punctate staining per 100 fluorescent LC3-ERFP–transfected cells was determined as described under Materials and Methods and expressed as the mean ± SD of combined results from three independent experiments (*p < 0.05). (d) Western blotting of endogenous LC3 from HeLa cells transfected with scrambled or TP53INP2 siRNA and in rapamycin-treated or untreated cells. (e) LC3-II/LC3-I ratio was calculated from three independent Western blotting performed as described above and expressed as the mean ± SD of combined results (*p < 0.05).
TP53INP2 and Beclin 1
Autophagy is mediated by a set of evolutionarily conserved gene products (termed the Atg proteins) originally discovered in yeast (reviewed in Khalfan and Klionsky, 2002). The mammalian autophagy protein beclin 1 is an orthologue of yeast Atg6, and it is part of a complex including the class III phosphoinositide-3-kinase Vps34, responsible for autophagosome formation. We tried to establish whether expression of TP53INP2 is necessary for preautophagosome formation, by monitoring the translocation of a beclin 1 fusion protein to the preautophagosome. As expected, we found that beclin 1 colocalized with TP53INP2 in the spot-like structures induced by rapamycin treatment and nutrition deprivation, as shown in Figure 8. Nevertheless, coimmunoprecipitation experiments showed that TP53INP2 and beclin1 did not bind to each other (data not shown). Then, we studied whether TP53INP2 is necessary for the recruitment of beclin 1 to the preautophagosome (phagophore) membrane, as it is for the recruitment of LC3. To this end, expression of TP53INP2 was knocked down in HeLa cells with a TP53INP2 siRNA; then, cells were transfected with cyan fluorescent protein (CFP)-tagged beclin 1 and treated with rapamycin to induce the formation of autophagic vacuoles. In TP53INP2-knocked down cells, beclin 1 was unable to translocate to the vacuoles that formed upon rapamycin treatment, as shown in Figure 8. Hence, TP53INP2 is required for autophagosome formation, suggesting that TP53INP2 is a key gene in the autophagic process.
Figure 8.
TP53INP2 silencing blocks beclin-1 recruitment to rapamycin-induced autophagic vacuoles. (a) HeLa cells were cotransfected with beclin 1-CFP, TP53INP2-EYFP, and LC3-ERFP plasmids and treated with rapamycin (10 μM for 4 h). TP53INP2 is in green, LC3 is in red, and beclin 1 is in blue. (b) HeLa cells were plated then, the next day, they were transfected with TP53INP2 siRNA or scrambled siRNA as a control and grown for an additional 24 h before beclin 1-CFP transfection. Twenty-four hours later, cells were treated with rapamycin (10 μM) for an additional 4-h period. (c) Relative quantification of beclin 1-CFP–positive vesicles in rapamycin-treated or untreated cells transfected with TP53INP2 siRNA or with scrambled siRNA as a control.
Translocation of TP53INP2 to the Autophagosome
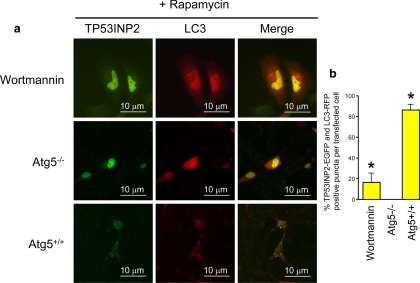
We investigated the mechanism by which TP53INP2 translocates to the autophagosomes after induction of autophagy. Autophagosome formation is highly dependent on phosphatidylinositol 3 (PI3)-kinase activity and wortmannin, an inhibitor of that kinase, is actually a potent inhibitor of autophagy (Blommaart et al., 1997). HeLa cells were transfected with TP53INP2-EGFP and LC3-ERFP. The next day, autophagy was induced by treating cells with rapamycin or starving them for 4 h. Treatment with wortmannin inhibited almost completely the translocation of TP53INP2 from the nucleus to the autophagosome (Figure 9), demonstrating that it is PI3-kinase dependent. A similar observation was made upon treatment with another PI3-kinase inhibitor, 3-methyladenine (data not shown). These data indicate that translocation of TP53INP2 from the nucleus to autophagosomes is a PI3-kinase activity-dependent process.
Figure 9.
TP53INP2 autophagy-dependent translocation. (a) HeLa cells were transfected with LC3-ERFP in combination with TP53INP2-EGFP and treated with wortmannin. Autophagy was induced by rapamicyn. (b) Atg5+/+ and Atg5−/− MEFs cells were transfected with LC3-ERFP in combination with TP53INP2-EGFP. Autophagy was induced by rapamicyn.
Autophagy is regulated by Atgs, which are mostly involved in the process of autophagosome formation. Among the Atg proteins, the role of Atg5 is well established (Mizushima et al., 1998). In fact, Atg12 covalently links Atg5. The mode of conjugation of Atg12 to Atg5 is similar to that of ubiquitination, because Atg12 is first activated by Atg7 (= ubiquitin-activating enzyme E1) and then transferred to Atg10 (= ubiquitin-activating enzyme E2) before it binds to Atg5. This process is essential for autophagy. We monitored TP53INP2 translocation to the autophagosome in Atg5+/+ and Atg5−/− cells. In Atg5+/+ cells, TP53INP2 translocation to the autophagosome was similar to that observed in HeLa cells. However, in Atg5−/− cells TP53INP2 remained located into the nucleus, whereas LC3 spread into the cytoplasm, like after wortmannin treatment (Figure 9). These results indicate that TP53INP2 translocation is dependent on autophagy, the involved mechanism occurring downstream from the Atg12–Atg5 step.
Analysis by BRET of the Interaction between TP53INP2 and LC3, GABARAP, or GABARAP-like2 in Cells
For BRET assays, HeLa cells were transfected with TP53INP2-pHRluc-C and GABARAP-pEYFP-C, GABARAP-like2-pEYFP-C or LC3-pEYFP-C constructs respectively as described in Materials and Methods. Energy transfer was quantified using the BRET ratio, defined as the ratio between the emissions at 530 nm (Rluc + EYFP fusion proteins) and at 490 nm (Rluc protein alone). The TP53INP2-pHRluc-C construct, overexpressed alone, gave a control value (without energy transfer) for the BRET ratio. Results were very reproducible. The BRET signal was higher than control with GABARAP-like2-pEYFP-C and LC3-pEYFP-C, albeit a little weaker for LC3-pEYFP-C (Figure 10). Because the observed energy transfer between TP53INP2-pHrluc-C and the pEYFP-C fusion proteins was not the result of a strong overexpression of EYFP in the cells, the detected BRET signals reflected their interactions in living cells. We wondered whether the BRET signal obtained between TP53INP2-Phrluc-C and GABARAP-pEYFP-C, GABARAP-like2-pEYFP-C, or LC3-pEYFP-C could be affected by autophagy modulation. As expected, induction of autophagy (starvation) increased the BRET signal. Conversely, blocking starvation-induced autophagy with the PI3-kinase inhibitor wortmannin decreased the signal. These data show that interactions between TP53INP2 and the Atg8-like proteins are associated with autophagy.
Figure 10.
TP53INP2 interaction with GABARAP, GABARAP-like2, and LC3 proteins is dependent on autophagy. (a–c) Relative quantification of cytoplasmic TP53INP2-positive dots in autophagy-induced cells, in the presence or absence of wortmannin. HeLa cells were cotransfected with TP53INP2-EGFP and GABARAP-ERFP (a), GABARAP-like2-ERFP (b), or LC3-ERFP (c) and treated with EBSS (starvation) in combination with wortmannin (d–f). TP53INP2-pHRluc-C construct was used as BRET donor, and different amounts of GABARAP-pEYFP-C, GABARAP-like2-pEYFP-C or LC3-pEYFP-C fusion proteins were used as BRET acceptors. Cells were transfected in 12-well culture plates, and 1 day later, they were harvested and distributed in 96-well microplates. On the following day, the Rluc substrate coelentherazin-h was added 15 min before reading. Data are expressed as BRET ratio as described in Materials and Methods. (d) To study the interaction between TP53INP2 and GABARAP, GABARAP-like2, or LC3, cells were transfected with the BRET donor (TP53INP2-pHRluc-C) and BRET acceptors (GABARAP-EYFP, GABARAP-like2-EYFP, or LC3-EYFP) constructs, and autophagy was induced by starvation (e) or inhibited by wortmannin treatment (f). BRET values are expressed in milli-BRET units. Data are expressed as the mean ± SD of results from three independent experiments (*p < 0.05).
DISCUSSION
Autophagy is an evolutionarily preserved degradation process of cytoplasmic constituents that serves as a survival mechanism in starved cells (Yorimitsu and Klionsky, 2005). This catabolic process probably plays a protective role in aging, cell death, defense against intracellular pathogens, neurodegenerative diseases and tumorigenesis, which highlights its biological and medical importance (Shintani and Klionsky, 2004). Most of the detailed molecular mechanistic work on autophagy has been carried out in yeast to show that a series of factors are involved in the sequential steps of this process (Levine and Klionsky, 2004). Recent studies have reported new factors involved in mammalian autophagy regulation (Liang et al., 2006). Yet, autophagosome formation in higher eukaryotes is a complex process and neither the mechanism of vesicle formation nor all the implicated genes are known. In this study, we show that TP53INP2 is a new autophagic factor that binds to and allows inclusion of LC3 and LC3-related GABARAP and GABARAP-like2 proteins into autophagic vacuoles. LC3 is an autophagosomal orthologue of yeast Atg8. A lipidated form of LC3, named LC3-II, is essential for autophagy and is used as an autophagosomal marker in mammals (reviewed in Tanida et al., 2004). The other LC3-related homologues, GABARAP and GABARAP-like2 proteins are also modified by the same mechanism, their role in autophagy being assumed but not completely understood (reviewed in Tanida et al., 2004).
In this work, we demonstrate that 1) TP53INP1 and TP53INP2 are homologous sequences found in several species, ranging from H. sapiens to D. melanogaster, but no orthologue was found in earlier eukaryotes or prokaryotes; 2) TP53INP2 binds to LC3 as well as to LC3-related proteins GABARAP and GABARAP-like2; 3) this interaction is autophagy-dependent because it is stimulated by autophagy enhancers and reduced by the PI3-kinase inhibitor wortmannin; 4) TP53INP2 binds to the autophagosome transmembrane protein VMP1 but not to beclin 1; 5) the protein translocates from the nucleus to the autophagosome after activation of autophagy by rapamycin or starvation. The mechanism requires a PI3-kinase activity and is dependent on Atg5 expression; and 6) TP53INP2 expression is necessary for autophagosome development. Together, these data strongly suggest that TP53INP2 is a novel gene involved in autophagy, with a function already established in Drosophila with the ancestral TP53INP.
We investigated the role of TP53INP2 in autophagy. We found that when TP53INP2 was knocked down in vitro, LC3 and LC3-related proteins were not recruited to the autophagosomes induced by rapamycin or starvation. This has two possible explanations: 1) TP53INP2, by its binding to LC3 and LC3-related proteins, is necessary for their recruitment to the autophagosome through its interaction with the autophagosome transmembrane protein VMP1; and 2) TP53INP2 is necessary for autophagosome formation or development by a way independent from its interaction with LC3 or LC3-related proteins. Another interesting point is that knocking down TP53INP2 prevented the recruitment of beclin 1 to the rapamycin- or starvation-induced autophagosome, although that protein does not bind directly to TP53INP2. Beclin 1 binding to PI3-kinase (Liang et al., 2006) is required for autophagosome development. An integrative hypothesis could be that beclin 1, through its PI3-kinase–associated activity (Kihara et al., 2001), allows autophagosome development after binding to VMP1 (Ropolo et al., 2007) provided LC3 (or LC3-related proteins) are located near the autophagosome membrane (Tanida et al., 2004). A proautophagic stimulus induces the formation of a complex between TP53INP2 and LC3 (or LC3-related proteins). TP53INP2, acting as a scaffold protein, is integrated at the preautophagosome surface through its binding to the transmembrane protein VMP1. In the absence of TP53INP2 (after TP53INP2 siRNA treatment), LC3 is not recruited to the proautophagosome membrane, which prevents further expansion. Thus, proautophagosomes remaining undetectable, which accounts for the absence of visible beclin 1 immunostaining.
We analyzed the regulation in the living cell of the interactions between TP53INP2 and LC3 or LC3-related proteins. We performed coimmunoprecipitations between TP53INP2 and LC3 and LC3-related proteins in control cells or after a treatment with EBSS that induces autophagy, and we found no significant differences (data not shown). This was not a complete surprise because coimmunoprecipitation tests require cell lysis followed by long incubation times with antibodies. During that process, proteins previously stored in different cellular compartments are allowed to interact. To circumvent this problem we used another technique, named BRET, which allows real time quantitation of protein interactions in living cells under experimental conditions. BRET can occur when two compatible optical probes are brought into proximity (50–80 Å). To probe for an interaction between two given partner proteins, each protein is genetically fused to the blue light emitting humanized Renilla luciferase (donor), and to a blue light absorbing yellow fluorescent protein (acceptor), respectively. If the two hybrid proteins interact, the emission energy of the luciferase can be transferred to the fluorescent protein, resulting in an easily detected yellow-shift in the luminescence spectrum. Using this approach, we found that the studied interactions were enhanced after induction of autophagy but inhibited by treatment with the PI3-kinase inhibitor agent wortmannin (an autophagy inhibitor), indicating that these interactions are essentially controlled by the status of autophagy.
We recently proposed that TP53INP1 is a tumor suppressor gene because it is involved in cell cycle arrest and apoptosis (Tomasini et al., 2001). It is a target of p53 but can also enhance p53 activity (Okamura et al., 2001; Tomasini et al., 2002, 2003), and its deficiency is associated with increases tumor development in mice (Gironella et al., 2007; Gommeaux et al., 2007). In spite of its homology with TP53INP1, TP53INP2 is not a tumor suppressor per se, because it is apparently not induced by p53 (data not shown) and its forced overexpression did not alter the cell cycle or apoptosis (data not shown). However, our data suggest that TP53INP2 could actually be involved in the control of tumor development through its capacity at modulating autophagy, because cellular autophagic activity is inversely correlated with malignancy and autophagy is even suppressed in many cancer cells (Ogier-Denis and Codogno, 2003, Levine, 2007). We found in a preliminary set of experiments that TP53INP1, like TP52INP2, could bind LC3 and LC3-related proteins and that its knocking down by specific siRNAs inhibited the formation of autophagosomes that occurs after treatment of the cells by starvation or rapamycin. Inhibition was not as strong as when TP53INP2 was targeted (data not shown), but yet it was significant, suggesting that TP53INP1 is also involved in the development of autophagy. Another interesting point is that TP53INP2, which is expressed during development in the nervous system, could be involved in neuronal development. During mouse development, its spatiotemporal expression correlates with that of activating molecule in beclin1-regulated autophagy 1 protein (Ambra1). Ambra1 is a vertebrate gene that regulates autophagy, suggesting a novel role for autophagy in neurodevelopment (Fimia et al., 2007).
In conclusion, we report that a novel nuclear factor, named TP53INP2, is absolutely required for autophagy development. Our results strongly suggest that TP53INP2 is a scaffold protein that recruits LC3 and the LC3-related proteins GABARAP and GABARAP-like2 and brings them to the autophagosome membrane by interacting with the VMP1 transmembrane protein where, in cooperation with the beclin 1-PI3-kinase class III complex, they trigger autophagosome development.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Patricia Spoto and Patrice Berthezene for technical help. This work was supported by Institut National de la Santé et de la Recherche Médicale, Canceropole PACA, and Ligue Contre le Cancer (to J.L.I.) and Conicet and Anpcyt (to M.I.V.) and Fondation de la Recherche Médicale.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-07-0671) on December 3, 2008.
REFERENCES
- Bennetts J. S., Fowles L. F., Butterfield N. C., Berkman J. L., Teasdale R. D., Simpson F., Wicking C. Identification and analysis of novel genes expressed in the mouse embryonic facial primordia. Front. Biosci. 2006;11:2631–2646. doi: 10.2741/1997. [DOI] [PubMed] [Google Scholar]
- Bennetts J. S., Rendtorff N. D., Simpson F., Tranebjaerg L., Wicking C. The coding region of TP53INP2, a gene expressed in the developing nervous system, is not altered in a family with autosomal recessive non-progressive infantile ataxia on chromosome 20q11–q13. Dev. Dyn. 2007;236:843–852. doi: 10.1002/dvdy.21064. [DOI] [PubMed] [Google Scholar]
- Blommaart E. F., Krause U., Schellens J. P., Vreeling-Sindelárová H., Meijer A. J. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- Cano C., et al. Tumor protein 53-induced nuclear protein 1 is a major mediator of p53 antioxidant function. Cancer Res. 2008 doi: 10.1158/0008-5472.CAN-08-2320. (in press) [DOI] [PubMed] [Google Scholar]
- Coyle J. E., Nikolov D. B. GABARAP: lessons for synaptogenesis. Neuroscientist. 2003;9:205–216. doi: 10.1177/1073858403009003013. [DOI] [PubMed] [Google Scholar]
- Dusetti N. J., et al. Cloning and expression of the rat vacuole membrane protein 1 (VMP1), a new gene activated in pancreas with acute pancreatitis, which promotes vacuole formation. Biochem. Biophys. Res. Commun. 2002;290:641–649. doi: 10.1006/bbrc.2001.6244. [DOI] [PubMed] [Google Scholar]
- Fimia G. M., et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- Fowles L. F., Bennetts J. S., Berkman J. L., Williams E., Koopman P., Teasdale R. D., Wicking C. Genomic screen for genes involved in mammalian craniofacial development. Genesis. 2003;35:73–87. doi: 10.1002/gene.10165. [DOI] [PubMed] [Google Scholar]
- Giot L., et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Gironella M., et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc. Natl. Acad. Sci. USA. 2007;104:16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommeaux J., et al. Colitis and colitis-associated cancer are exacerbated in mice deficient for tumor protein 53-induced nuclear protein 1. Mol. Cell Biol. 2007;27:2215–2228. doi: 10.1128/MCB.01454-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko T., Chaussepied M., Oren M., Ginsberg D. Novel link between E2F and p 53, proapoptotic cofactors of p53 are transcriptionally upregulated by E2F. Cell Death Differ. 2005;12:377–383. doi: 10.1038/sj.cdd.4401575. [DOI] [PubMed] [Google Scholar]
- Horton P., Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Khalfan W. A., Klionsky D. J. Molecular machinery required for autophagy and the cytoplasm to vacuole targeting (Cvt) pathway in S. cerevisiae. Curr. Opin. Cell Biol. 2002;14:468–475. doi: 10.1016/s0955-0674(02)00343-5. [DOI] [PubMed] [Google Scholar]
- Kihara A., Kabeya Y., Ohsumi Y., Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–747. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Liang C., Feng P., Ku B., Dotan I., Canaani D., Oh B. H., Jung J. U. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat. Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Mann S. S., Hammarback J. A. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J. Biol. Chem. 1994;269:11492–11497. [PubMed] [Google Scholar]
- Mann S. S., Hammarback J. A. Gene localization and developmental expression of light chain 3, a common subunit of microtubule-associated protein 1A(MAP1A) and MAP1B. J. Neurosci. Res. 1996;43:535–544. doi: 10.1002/(SICI)1097-4547(19960301)43:5<535::AID-JNR3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M. D., Klionsky D. J., Ohsumi M., Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mousavi S. A., Kjeken R., Berg T. O., Seglen P. O., Berg T., Brech A. Effects of inhibitors of the vacuolar proton pump on hepatic heterophagy and autophagy. Biochim. Biophys. Acta. 2001;1510:243–257. doi: 10.1016/s0005-2736(00)00354-0. [DOI] [PubMed] [Google Scholar]
- Nei M., Kumar S. Molecular Evolution and Phylogenetics. Oxford, United Kingdom: Oxford University Press; 2000. [Google Scholar]
- Nowak J., Depetris D., Iovanna J. L., Mattei M. G., Pebusque M. J. Assignment of the tumor protein p53 induced nuclear protein 2 (TP53INP2) gene to human chromosome band 20q11.2 by in situ hybridization. Cytogenet. Genome Res. 2005;108:362. doi: 10.1159/000081534. [DOI] [PubMed] [Google Scholar]
- Ogier-Denis E., Codogno P. Autophagy: a barrier or an adaptive response to cancer. Biochim. Biophys. Acta. 2003;1603:113–128. doi: 10.1016/s0304-419x(03)00004-0. [DOI] [PubMed] [Google Scholar]
- Okamura S., Arakawa H., Tanaka T., Nakanishi H., Ng C. C., Taya Y., Monden M., Nakamura Y. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol. Cell. 2001;8:85–94. doi: 10.1016/s1097-2765(01)00284-2. [DOI] [PubMed] [Google Scholar]
- Paz Y., Elazar Z., Fass D. Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J. Biol. Chem. 2000;275:25445–25450. doi: 10.1074/jbc.C000307200. [DOI] [PubMed] [Google Scholar]
- Ropolo A., et al. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J. Biol. Chem. 2007;282:37124–37133. doi: 10.1074/jbc.M706956200. [DOI] [PubMed] [Google Scholar]
- Sagiv Y., Legesse-Miller A., Porat A., Elazar Z. GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 2000;19:1494–1504. doi: 10.1093/emboj/19.7.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shintani T., Klionsky D. J. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I., Ueno T., Kominami E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R., Samir A. A., Carrier A., Isnardon D., Cecchinelli B., Soddu S., Malissen B., Dagorn J. C., Iovanna J. L., Dusetti N. J. TP53INP1s and homeodomain-interacting protein kinase-2 (HIPK2) are partners in regulating p53 activity. J. Biol. Chem. 2003;278:37722–37729. doi: 10.1074/jbc.M301979200. [DOI] [PubMed] [Google Scholar]
- Tomasini R., Samir A. A., Pebusque M. J., Calvo E. L., Totaro S., Dagorn J. C., Dusetti N. J., Iovanna J. L. P53-dependent expression of the stress-induced protein (SIP) Eur. J. Cell Biol. 2002;81:294–301. doi: 10.1078/0171-9335-00248. [DOI] [PubMed] [Google Scholar]
- Tomasini R., Samir A. A., Vaccaro M. I., Pebusque M. J., Dagorn J. C., Iovanna J. L., Dusetti N. J. Molecular and functional characterization of the stress-induced protein (SIP) gene and its two transcripts generated by alternative splicing. SIP induced by stress and promotes cell death. J. Biol. Chem. 2001;276:44185–44192. doi: 10.1074/jbc.M105647200. [DOI] [PubMed] [Google Scholar]
- Tomasini R., Seux M., Nowak J., Bontemps C., Carrier A., Dagorn J. C., Pebusque M. J., Iovanna J. L., Dusetti N. J. TP53INP1 is a novel p73 target gene that induces cell cycle arrest and cell death by modulating p73 transcriptional activity. Oncogene. 2005;24:8093–8104. doi: 10.1038/sj.onc.1208951. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D. J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Liu H., Miki Y. Protein kinase C delta regulates Ser46 phosphorylation of p53 tumor suppressor in the apoptotic response to DNA damage. J. Biol. Chem. 2006;281:5734–5740. doi: 10.1074/jbc.M512074200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.