Abstract
Sister chromatid cohesion, mediated by cohesin complexes, is laid down during DNA replication and is essential for the accurate segregation of chromosomes. Previous studies indicated that, in addition to their cohesion function, cohesins are essential for completion of recombination, pairing, meiotic chromosome axis formation, and assembly of the synaptonemal complex (SC). Using mutants in the cohesin subunit Rec8, in which phosphorylated residues were mutated to alanines, we show that cohesin phosphorylation is not only important for cohesin removal, but that cohesin's meiotic prophase functions are distinct from each other. We find pairing and SC formation to be dependent on Rec8, but independent of the presence of a sister chromatid and hence sister chromatid cohesion. We identified mutations in REC8 that differentially affect Rec8's cohesion, pairing, recombination, chromosome axis and SC assembly function. These findings define Rec8 as a key determinant of meiotic chromosome morphogenesis and a central player in multiple meiotic events.
INTRODUCTION
Meiosis is the process by which diploid cells produce haploid products; these products include eggs and sperm in multicellular organisms and spores in the budding yeast Saccharomyces cerevisiae. Essentially, meiosis is a modified mitotic cell division, with the most notable modification being the presence of two chromosome segregation phases after only a single DNA replication phase. The second segregation phase (meiosis II [MII]) resembles mitosis with replicated sister chromatids segregating from each other. In contrast, the first segregation phase (meiosis I [MI]), called a reductional segregation, requires that homologous chromosomes separate. For this to occur, homologs must first be aligned and then linked through recombination (reviewed in Lee and Amon, 2001; Nasmyth, 2001; Marston and Amon, 2004).
Recombination is initiated after DNA replication by Spo11, a topoisomerase-like protein that introduces as many as 200 double-strand breaks (DSBs) into the genome (Keeney et al., 1997). In budding yeast, the initial alignment of homologs, known as pairing, also depends on the formation of DSBs (reviewed in McKee, 2004). DSBs are subsequently resected to expose 3′ single-stranded overhangs. The single-stranded DNA ends then engage in the search for homologous repair templates that is mediated by the RecA homologs Rad51 and Dmc1 (reviewed in Whitby, 2005). As DSBs are processed, a proteinaceous structure, the synaptonemal complex (SC), forms along the homologous chromosomes (reviewed in Zickler and Kleckner, 1999; Page and Hawley, 2004; Whitby, 2005). SC formation initiates by axial elements (AEs, referred to as lateral elements or LEs in the context of the SC) assembling along chromosomes where they are thought to serve as a scaffold for the progressively condensing meiotic chromosomes. Mature SC is then formed by the joining of the AEs of homologous chromosomes through transverse elements (reviewed in Zickler and Kleckner, 1998; Page and Hawley, 2004). In many organisms including budding yeast, mutants in SC formation frequently show a defect in recombination and vice versa (reviewed in Zickler and Kleckner, 1999). Components of the SC and SC initiation factors, notably budding yeast Zip1, Zip2, and Zip3 proteins, as well as the Mer3 helicase and the Msh4/Msh5 complex, are required to ensure that recombination intermediates stably invade the homologous chromosomes and mature into cross-overs (Borner et al., 2004). The process of recombination culminates in the formation of cross-over and non-cross-over products. Cross-overs result in physical links between homologous chromosomes that are manifested cytologically as chiasmata (reviewed in Zickler and Kleckner, 1999).
Central to the interaction of homologous chromosome with each other and their accurate segregation are cohesin complexes. Cohesins hold sister chromatids together from the time of their generation through DNA replication until their segregation during mitosis or meiosis. The mitotic cohesin complex consists of four core proteins: Scc3, Smc1, Smc3, and Scc1/Mcd1 (reviewed in Uhlmann, 2003). The meiotic cohesin complex contains the same proteins, with the exception that Scc1/Mcd1 is replaced by the meiosis-specific subunit Rec8 (Klein et al., 1999). At the end of meiotic prophase, homologous chromosomes are linked through chiasmata as a result of recombination, as well as cohesin linkages between sister chromatids distal to chiasmata. For homologs to segregate during MI, cohesins must be removed along chromosome arms. Cohesins are maintained at centromeres, allowing sister chromatids to continue to associate until the metaphase II–to–anaphase II transition. At this point, the remaining cohesin is removed, resulting in the formation of four balanced gametes (reviewed in Lee and Amon, 2001; Nasmyth, 2001; Marston and Amon, 2004).
The process by which cohesin is removed at the metaphase-to-anaphase transitions is well understood. A protease known as Separase is activated at the metaphase-to-anaphase transition through degradation of its inhibitor, Securin, by the APC/C (anaphase-promoting complex/cyclosome) ubiquitin ligase. Active Separase cleaves Rec8, causing removal of cohesin from chromosomes (Buonomo et al., 2000; Shonn et al., 2000; reviewed in Uhlmann, 2003). This process appears to occur through a largely identical mechanism in MI and MII. Centromeric Rec8, however, is protected from cleavage at the metaphase I-to-anaphase I transition by mechanisms that include association of centromeric Rec8 with the protector protein Shugoshin (Sgo1) and preferential phosphorylation of arm cohesins (Shonn et al., 2000; Uhlmann, 2003; Katis et al., 2004; Kitajima et al., 2004; Marston et al., 2004; Rabitsch et al., 2004; Brar et al., 2006).
Cells deleted for REC8 display defects not only in sister chromatid cohesion but also in SC formation and exit from prophase, long before cells initiate the first chromosome segregation phase (Klein et al., 1999). The prophase progression defect of cells deleted for REC8 is dependent on the creation of DSBs by Spo11, supporting a role for Rec8 in recombination. It is unclear, however, whether this requirement is simply a manifestation of Rec8's sister chromatid cohesion function or represents a specific role of Rec8 in recombination-related processes. Here we examine REC8 mutants, in which phosphorylated residues were mutated to amino acids that can no longer be phosphorylated. Their analysis confirms previous findings that phosphorylation is required for the timely onset of anaphase I (Brar et al., 2006). Furthermore, they support a role of Rec8 in the timely completion of recombination, complete pairing and SC assembly, particularly transverse element formation (Klein et al., 1999). The identification of mutations within REC8 that differentially affect Rec8's cohesion, pairing, recombination, and SC assembly functions furthermore indicates that the protein either acts in these processes via distinct mechanisms or that different levels of cohesion function are needed for the diverse prophase functions of the cohesin complex. Our discovery of a REC8 allele that is partially defective in Zip1 assembly but does not affect recombination furthermore supports the idea that in yeast SC formation is dispensable for recombination. Our results reveal the separable nature of the multiple roles for Rec8 during meiosis, and position the protein centrally in the regulation of meiotic chromosome morphogenesis and homolog interactions.
MATERIALS AND METHODS
Strains and Plasmids
All strains described are of the SK1 background of S. cerevisiae and are listed in Table 1. Deletions have all been performed by one-step gene replacement as described in Longtine et al. (1998). Meiotic depletions are achieved by one-step promoter replacement as described in Lee and Amon (2003) and Hochwagen et al. (2005). cdc6-mn is a meiotic depletion allele generated by placement of CDC6 under control of the SCC1 promoter. cdc5-mn is a meiotic depletion allele generated by placement of CDC5 under control of the CLB2 promoter. rec8-NC is described in Buonomo et al. (2000). An unstable form of Rec8 was constructed by fusing REC8 with the first ubiquitin in UBI4, leading to rapid proteasomal degradation of newly translated Rec8. The estrogen-inducible REC8 construct was generated as described for NDT80 in Carlile and Amon (2008). The Rec8 phospho mutants are described in Brar et al. (2006). rec8-6A is mutated at S197A, S386A, S387A, S245A, S521A, and T173A; rec8-psa is mutated at S197A, S386A, S387A, S136A, T173A, S199A, T249A, S410A, S179A, S215A, S465A, and S466A; rec8-29A is mutated at S197A, S386A, S387A, S136A, T173A, S199A, S245A, T249A, S521A, S522A, S314A, S410A, S179A, S215A, S465A, S466A, S285A, S494A, S421A, Y14A, S552A, T18A, T19A, S292A, S425A, S404A, S125A, T126A, and S224A. All three mutants are described in Brar et al. (2006). tetO arrays inserted at LEU2 are described in Marston et al. (2004). LYS2-located tetO arrays were generated by amplifying LYS2 from pRS317, digesting with HindIII and BglII, and cloning into pRS306tetO14. URA3-localized tetO arrays are described in Michaelis et al. (1997). CEN5-localized tetO arrays are described in Toth et al. (2000). TEL5-localized tetO arrays were described in Alexandru et al. (2001).
Table 1.
Yeast strains used in this study
| Strain number | Relevant genotype |
|---|---|
| A1556 | MATa/α his4B::LEU2 his4X::LEU2-(BamH1) |
| A1972 | MATa/α REC8-3HA::URA3 |
| A3528 | MATa/α rec8Δ::KanMX4 |
| A5111 | MATa/α leu2::LEU2::tetR-GFP-tetO::HIS3 ndt80::URA3 |
| A5844 | MATa/α cdc5::pCLB2-CDC5::KanMX6 |
| A6946 | MATa/α pURA3-tetR-GFP::LEU2 ura3::tetOx224::URA3 ndt80::URA3 |
| A6917 | MATa/α leu2::LEU2::tetR-GFP-tetO::HIS3 ndt80::URA3 dmc1::KanMX |
| A7097 | MATa/α |
| A7803 | MATa/α pURA3-tetR-GFP::LEU2 ura3::tetOx224::URA3 spo11::TRP1 ndt80::URA3 |
| A8477 | MATa/α spo11::URA3 |
| A9115 | MATa/α pURA3-tetR-GFP::LEU2 ura3::tetOx224::URA3 spo11::spo11-Y135F-HA::URA3 ndt80::LEU2 |
| A9828 | MATa/α lys2::tetOx240::URA3 leu2::LEU2-tetR-GFP ndt80::URA3 |
| A9829 | MATa/α lys2::tetOx240::URA3 (heterozygous) leu2::LEU2-tetR-GFP pURA3-tetR-GFP::LEU2 ura3::tetOx224::URA3 (heterozygous) ndt80::URA3 |
| A10404 | MATa/α pURA3-tetR-GFP::LEU2 ura3::tetOx224::URA3 ndt80::URA3 cdc6::KanMX6::pSCC1-CDC6-3HA |
| A10735 | MATa/α lys2::tetOx240::URA3 leu2::LEU2-tetR-GFP ndt80::URA3 cdc6::KanMX6::pSCC1-CDC6-3HA |
| A11469 | MATa/α lys2::tetOx240::URA3 leu2::LEU2-tetR-GFP ndt80::URA3 spo11::URA3 |
| A11268 | MATa/α lys2::tetOx240::URA3 leu2::LEU2-tetR-GFP ndt80::URA3 clb6::TRP1 clb5::KanMX6 |
| A11326 | MATa/α leu2::LEU2::tetR-GFP-tetO::HIS3 ndt80::URA3 clb6::TRP1 clb5::KanMX6 |
| A11474 | MATa/α lys2::tetOx240::URA3 (heterozygous) leu2::LEU2-tetR-GFP leu2::LEU2-tetR-GFP-tetO-HIS3 (heterozygous) ndt80::URA3 |
| A12095 | MATa/α lys2::tetOx240::URA3 leu2::LEU2-tetR-GFP ndt80::URA3 spo11::spo11-Y135F-HA::URA3 |
| A13346 | MATa/α leu2::LEU2::tetR-GFP-tetO::HIS3 ndt80::URA3 spo11-Y135F-HA::KanMX4 |
| A13539 | MATa/α pREC8::REC8-3HA-NC rec8::KanMX4 |
| A13946 | MATa/α pREC8::REC8-3HA::KanMX4::LEU2 |
| A14385 | MATa/α pREC8::rec8-29A-3HA::LEU2 rec8::KanMX4 |
| A14655 | MATa/α pREC8::REC8-3HA::KanMX4::LEU2 ubr1::KanMX4 |
| A15042 | MATa/α pREC8::rec8-6A-3HA::LEU2 rec8::KanMX4 |
| A15364 | MATa/α pREC8::rec8-psa-3HA::LEU2 rec8::KanMX4 ubr1::KanMX4 |
| A15880 | MATa/α cdc6:: pSCC1-3HA-CDC6::KanMX6 |
| A16108 | MATa/α lys2::tetOx240::URA3 leu2::LEU2-tetR-GFP ndt80::URA3 rec8::LEU2 |
| A16113 | MATa/α clb5::KanMX6 clb6::TRP1 pREC8::REC8-3HA::URA3 |
| A16126 | MATa/α lys2::tetOx240::URA3 leu2::LEU2-tetR-GFP ndt80::URA3 spo11-D290A-HA::KanMX4 |
| A16131 | MATa/α leu2::LEU2::tetR-GFP-tetO::HIS3 ndt80::URA3 rec8::LEU2 |
| A16132 | MATa/α pREC8::pREC8-SCC1-3HA::LEU2 rec8::KanMX4 his4B::LEU2 his4X::LEU2-(BamH1) |
| A16133 | MATa/α lys2::tetOx240::URA3 leu2::LEU2-tetR-GFP ndt80::URA3 spo11-Y135F-HA::KanMX4 |
| A16147 | MATa/α lys2::tetOx240::URA3 leu2::LEU2-tetR-GFP ndt80::URA3 SPO11-HA::KanMX4 (heterozygous) spo11-Y135F::KanMX4 (heterozygous) |
| A16148 | MATa/α lys2::tetOx240::URA3 leu2::LEU2-tetR-GFP ndt80::URA3 SPO11-HA::KanMX4 (heterozygous) spo11-Y135F-HA::KanMX4 (heterozygous) |
| A16149 | MATa/α lys2::tetOx240::URA3 leu2::LEU2-tetR-GFP ndt80::URA3 mer2::mer2-S30A::URA3 |
| A16290 | MATa/α lys2::tetOx240::URA3 leu2::LEU2-tetR-GFP ndt80::URA3 msh5::HIS3 |
| A16292 | MATa/α lys2::tetOx240::URA3 leu2::LEU2-tetR-GFP ndt80::URA3 SPO11-HA::KanMX4 |
| A16360 | MATa/α leu2::LEU2::tetR-GFP-tetO::HIS3 (heterozygous) CENV::tetOx224::HIS3 (heterozygous) leu2::pURA3-tetR-GFP ndt80::URA3 |
| A16362 | MATa/α CENV::tetOx224::HIS3 leu2::pURA3-tetR-GFP ndt80::URA3 |
| A16366 | MATa/α pURA3-tetR-GFP::LEU2 TELV::tetOx224::URA3 ndt80::URA3 |
| A16376 | MATa/α leu2::LEU2::tetR-GFP-tetO::HIS3 ndt80::URA3 SPO11-HA::KanMX4 |
| A16386 | MATa/α leu2::LEU2::tetR-GFP-tetO::HIS3 ndt80::URA3 SPO11-HA::KanMX4 (heterozygous) spo11-Y135F-HA::KanMX4 (heterozygous) |
| A16391 | MATa/α leu2::LEU2::tetR-GFP-tetO::HIS3 ndt80::URA3 SPO11-HA::KanMX4 (heterozygous) spo11-Y135F::KanMX4 (heterozygous) |
| A16399 | MATa/α leu2::LEU2::tetR-GFP-tetO::HIS3 ndt80::URA3 spo11-D290A-HA::KanMX4 |
| A16412 | MATa/α lys2::tetOx240::URA3 leu2::LEU2-tetR-GFP ndt80::URA3 pREC8::rec8-29A-3HA::LEU2 rec8::KanMX4 |
| A16446 | MATa/α lys2::tetOx240::URA3 leu2::LEU2-tetR-GFP ndt80::URA3 cdc6::KanMX6::pSCC1-CDC6-3HA rec8::LEU2 |
| A16460 | MATa/α leu2::LEU2::tetR-GFP-tetO::HIS3 ndt80::URA3 cdc6::KanMX6::pSCC1-CDC6-3HA rec8::LEU2 |
| A16533 | MATa/α leu2::LEU2::tetR-GFP-tetO::HIS3 ndt80::URA3 cdc6::KanMX6::pSCC1-CDC6-3HA |
| A16535 | MATa/α pURA3-tetR-GFP::LEU2 TELV::tetOx224::URA3 ndt80::URA3 rec8::LEU2 |
| A16537 | MATa/α CENV::tetOx224::HIS3 leu2::pURA3-tetR-GFP ndt80::URA3 rec8::LEU2 |
| A16538 | MATa/α pURA3-tetR-GFP::LEU2 ura3::tetOx224::URA3 ndt80::URA3 rec8::LEU2 |
| A16664 | MATa/α rec8::KanMX4 |
| A17021 | MATa/α cdc6:: pSCC1-3HA-CDC6::KanMX6 rec8::KanMX4 |
| A18933 | MATa/α his4B::LEU2 his4X::LEU2-(BamH1) rec8::KanMX4 |
| A18936 | MATa/α his4B::LEU2 his4X::LEU2-(BamH1) pREC8::rec8-6A-3HA::LEU2 rec8::KanMX4 |
| A19798 | MATa/α spo11Δ::URA3 ndt80Δ::URA3 ura3::pGPD1-GAL4(848)-ER::URA3 rec8::pGAL-REC8-UP-3HA::KanMX6 lys2::tetOx240::URA3 leu2::tetR-GFP::LEU2 |
| A19800 | MATa/α spo11Δ::URA3 ndt80Δ::URA3 ura3::pGPD1-GAL4(848)-ER::URA3 rec8::pGAL-REC8-3HA::KanMX6 lys2::tetOx240::URA3 leu2::tetR-GFP::LEU2 |
| A20066 | MATa/α pREC8::REC8-3HA::KanMX4::LEU2 |
| A20072 | MATa/α pREC8::REC8-T173D S197D S245D S386D S387D S521D-3HA::KanMX4::LEU2 |
| A20075 | MATa/α MATa/α pREC8::REC8-T173E S197E S245E S386E S387E S521E-3HA::KanMX4::LEU2 |
| A20151 | MATa/α pREC8::rec8-6A-3HA::LEU2 rec8::KanMX4 pch2Δ::KanMX4 |
| A20153 | MATa/α pREC8::REC8-3HA::KanMX4::LEU2 pch2::KanMX4 |
| A20154 | MATa/α pREC8::rec8-6A-3HA::LEU2 rec8Δ::KanMX4 mek1::KanMX |
| A20156 | MATa/α pREC8::REC8-3HA::KanMX4::LEU2 mek1::KanMX |
| A20157 | MATa/α pREC8::rec8-29A-3HA::LEU2 rec8::KanMX4 mek1::KanMX |
| A20163 | MATa/α pSCC3::pCLB2-SCC3 scc3::KanMX6 |
| A20164 | MATa/α pREC8::rec8-29A-3HA::LEU2 rec8::KanMX4 pch2::KanMX4 |
| A21230 | MATa/α ura3::pGPD1-GAL4(848)-ER::URA3 rec8::KanMX4::REC8-3HA::LEU2 pGAL-NDT80::TRP1 |
| A21232 | MATa/α ura3::pGPD1-GAL4(848)-ER::URA3 rec8::KanMX4::rec8-29A-3HA::LEU2 pGAL-NDT80::TRP1 |
| A21234 | MATa/α ura3::pGPD1-GAL4(848)-ER::URA3 rec8::KanMX4::rec8-17A-3HA::LEU2 pGAL-NDT80::TRP1 |
| A21463 | MATa/α spo11::TRP1 pREC8::rec8-6A-3HA::LEU2 rec8::KanMX4 leu2::tetR-GFP-tetO::LEU2::HIS3 spo13::KanMX |
| A21464 | MATa/α spo11::TRP1 pREC8::rec8-6A spo13::KanMX -3HA::LEU2 rec8::KanMX4 leu2::tetR-GFP-tetO::LEU2::HIS3 |
| A21465 | MATa/α spo11::TRP1 pREC8::rec8-6A-3HA::LEU2 rec8::KanMX4 leu2::tetR-GFP-tetO::LEU2::HIS3 spo13::KanMX |
| A21466 | MATa/α spo11::TRP1 pREC8::REC8-3HA::LEU2 leu2::tetR-GFP-tetO::LEU2::HIS3 spo13::KanMX |
| A21467 | MATa/α spo11::TRP1 pREC8::REC8-3HA::LEU2 leu2::tetR-GFP-tetO::LEU2::HIS3 spo13::KanMX |
| A21468 | MATa/α spo11::TRP1 pREC8::REC8-3HA::LEU2 leu2::tetR-GFP-tetO::LEU2::HIS3 spo13::KanMX |
| A21469 | MATa/α spo11::TRP1 pREC8::rec8-29A-3HA::LEU2 rec8::KanMX4 leu2::tetR-GFP-tetO::LEU2::HIS3 spo13::KanMX |
| A21470 | MATa/α spo11::TRP1 pREC8::rec8-29A-3HA::LEU2 rec8::KanMX4 leu2::tetR-GFP-tetO::LEU2::HIS3 spo13::KanMX |
| A21471 | MATa/α spo11::TRP1 pREC8::rec8-29A-3HA::LEU2 rec8::KanMX4 leu2::tetR-GFP-tetO::LEU2::HIS3 spo13::KanMX |
| A21472 | MATa/α spo11::TRP1 rec8::KanMX4 leu2::tetR-GFP-tetO::LEU2::HIS3 spo13::KanMX |
| A21473 | MATa/α spo11::TRP1 rec8::KanMX4 leu2::tetR-GFP-tetO::LEU2::HIS3 spo13::KanMX |
| A21474 | MATa/α spo11::TRP1 rec8::KanMX4 leu2::tetR-GFP-tetO::LEU2::HIS3 spo13::KanMX |
| A21618 | MATa/α his4B::LEU2 his4X::LEU2-(BamH1) pREC8::rec8-29A-3HA::LEU2 rec8::KanMX4 |
| A21668 | MATa/α lys2::tetOx240::URA3 leu2::LEU2-tetR-GFP ndt80::URA3 pREC8::rec8-6A-3HA::LEU2 rec8::KanMX4 |
| A21669 | MATa/α leu2::LEU2::tetR-GFP-tetO::HIS3 ndt80::URA3 pREC8::rec8-29A-3HA::LEU2 rec8::KanMX4 |
| A21670 | MATa/α leu2::LEU2::tetR-GFP-tetO::HIS3 ndt80::URA3 pREC8::rec8-6A-3HA::LEU2 rec8::KanMX4 |
Synchronous Meiosis
Cells were grown to saturation in YPD (YEP + 2% glucose) for 24 h, diluted into YPA (YEP + 2% KAc) at OD600 = 0.3, and grown overnight. Cells were then washed with water and resuspended in SPO medium (0.3% KAc, pH = 7.0) at OD600 = 1.9 at 30°C to induce sporulation. The NTD80 block–release experiments were performed as described in Carlile and Amon (2008).
Irradiation
Irradiation was performed using 1-min exposures on a Gammacell 220E Cesium irradiator to yield 20 Krad. Cells were exposed to irradiation in 500-μl uncovered sporulation cultures in 5-ml Erlenmeyer flasks.
Southern Blot Analysis
Southern blot analysis was conducted as described by Hunter and Kleckner (2001). Blots were quantified using ImageQuant software (Amersham Biosciences, Piscataway, NJ).
Meiotic Spreads and Immunofluorescence
Chromosome spreads and immunofluorescence were performed as described in Marston et al. (2003) Rad51 was visualized with the (y-180) rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA) at a 1:200 dilution. Zip1 was visualized with a rabbit antibody that was a generous gift of S. Roeder (Yale University, New Haven, CT) and F. Klein (University of Vienna, Vienna, Austria) at a 1:200 dilution. Hop1 was visualized with a rabbit antibody that was a generous gift of S. Roeder at a 1:200 dilution. Rec8-HA was visualized with an HA.11 (16B12) mouse antibody (Covance Laboratories, Madison, WI) at a 1:200 dilution. The “percentage of mononucleates with Zip1” in graphs describes the sum of cells with partially and fully assembled Zip1 as defined in Figure 4A. One hundred mononucleate cells were counted per strain per time point unless otherwise noted.
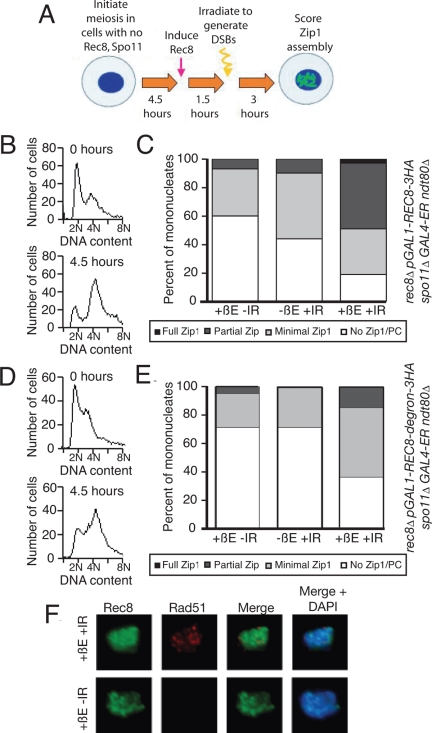
Figure 4.
Postreplicative Rec8 is sufficient for Zip1 assembly in the presence of DSBs. (A) The experimental scheme used in B–E. rec8::pGAL1-REC8-3HA spo11Δ rec8Δ GAL4-ER cells were induced to sporulate, allowed 4.5 h to progress through meiosis, and then treated with 1 μM β-estradiol (+βE). After another 1.5 h incubation in sporulation medium, cells were γ-irradiated with 20KRad (+IR) to induce DSBs. Cells were then kept in sporulation medium for another 3 h, when they were harvested and assayed for α-Zip1 and α-HA staining. A sample was also taken at 6.5 h to assay for DSBs by α-Rad51 staining. (B and C) rec8::pGAL1-REC8-3HA spo11Δ rec8Δ GAL4-ER ndt80Δ (A19800) cells were induced to sporulate and treated as described in A. At 9 h, cells with the indicated treatments were harvested, and chromosome spreads were assayed for Zip1 staining (C). DNA content was determined by flow cytometry analysis of cells harvested at 0 and 4.5 h of sporulation (B). (D and E) rec8::pGAL1-REC8-3HA-degron spo11Δ GAL4-ER ndt80Δ (A19798) cells were induced to sporulate and treated as described in A. At 9 h, cells with the indicated treatments were harvested, and chromosome spreads were assayed for Zip1 staining (E). DNA content was determined by flow cytometry analysis of cells harvested at 0 and 4.5 h of sporulation (D). (F) Examples of Rec8 and Rad51 staining in cells treated as described in A. At 6.5 h, cells were harvested and chromosome spreads were stained for Rec8, Rad51, and DNA. α-HA is shown in green, α-Rad51 in red, and DNA in blue.
Whole Cell Immunofluorescence
Indirect in situ immunofluorescence was carried out as described in Visintin et al. (1998). Rat anti-tubulin antibodies (Oxford Biotechnology, Kidlington, United Kingdom) and anti-rat FITC antibodies (Jackson ImmunoResearch, West Grove, PA) were used at a 1:100 dilution. Two hundred cells were counted per strain per time point. “Unassembled spindles” refer to cells that have not yet assembled a visible meiotic spindle (showing separated spindle poles). This category encompasses cells that are premeiotic, are undergoing DNA replication and are in meiotic prophase. Cells with “separated spindle poles” have assembled a meiotic spindle. This category includes cells that have reached metaphase I and beyond.
Flow Cytometry
Flow cytometric analysis of DNA content was performed as described in Visintin et al. (1998).
Live In Vivo Pairing Assay
Diploid cells were induced to sporulate and assayed as described in Figure 1A. Samples were taken regularly to monitor proximity of green fluorescent protein (GFP) dots live. Cells were scanned through on the z-axis to visualize entire cell volume. No fixation or staining was utilized. One hundred cells were counted per time point per strain. All pairing strains discussed within were deleted for NDT80 to arrest cells in pachytene.
Figure 1.
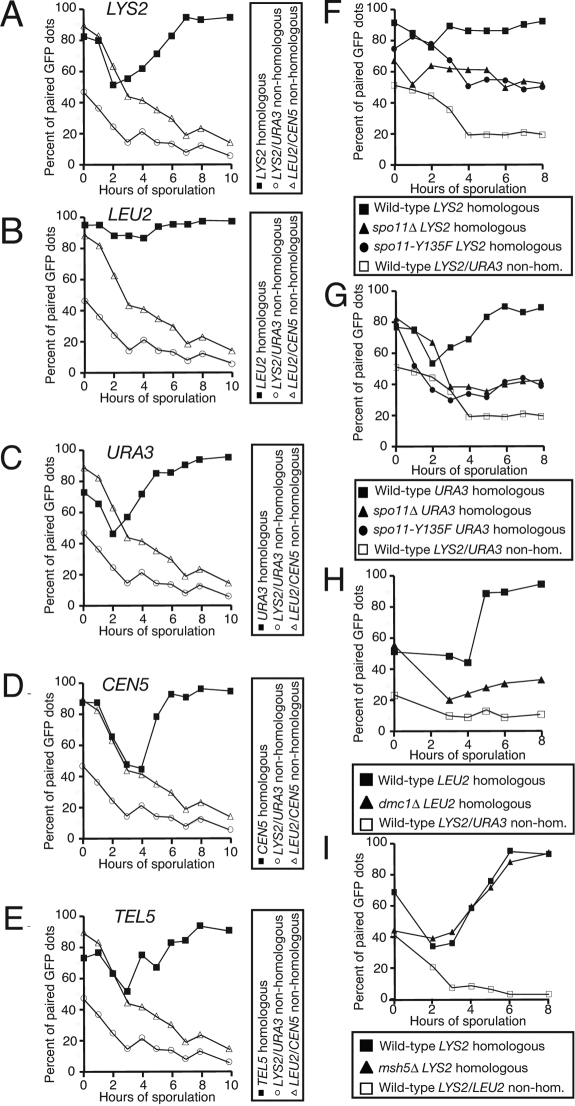
An assay to monitor pairing in live cells. (A) Wild-type cells with homologous LYS2 dots (A9828, ■), wild-type cells with nonhomologous LYS2/URA3 dots (A9829, ○), and wild-type cells with nonhomologous LEU2/CEN5 dots (A16360, ▵), all deleted for NDT80, were introduced into sporulation medium. At the indicated times, samples were taken and assayed for pairing live on aliquots that are taken at the indicated times as cells progress through prophase. Paired GFP signals are not distinguishable, and only a single GFP dot is visible. Nonhomologous arrays are included in each experiment as a control for clustering of the tet operators. Note that the same nonhomologous dot controls are used in A–E as the data from these panels were generated from the same experiment. (B) Wild-type cells with homologous LEU2 dots (A5111, ■), wild-type cells with nonhomologous LYS2/URA3 dots (A9829, ○), and wild-type cells with nonhomologous LEU2/CEN5 dots (A16360, ▵), all deleted for NDT80, were assayed for pairing as described in A. (C) Wild-type cells with homologous URA3 dots (A6946, ■), wild-type cells with nonhomologous LYS2/URA3 dots (A9829, ○), and wild-type cells with nonhomologous LEU2/CEN5 dots (A16360, ▵), all deleted for NDT80, were assayed for pairing as described in A. (D) Wild-type cells with homologous CEN5 dots (A16362, ■), wild-type cells with nonhomologous LYS2/URA3 dots (A9829, ○), and wild-type cells with nonhomologous LEU2/CEN5 dots (A16360, ▵), all deleted for NDT80, were assayed for pairing as described in A. (E) Wild-type cells with homologous TEL5 dots (A16366, ■), wild-type cells with nonhomologous LYS2/URA3 dots (A9829, ○) and wild-type cells with nonhomologous LEU2/CEN5 dots (A16360, ▵), all deleted for NDT80, were assayed for pairing as described in A. (F) Wild-type cells with homologous LYS2 dots (A9828, ■), spo11Δ cells with homologous LYS2 dots (A11469, ▴), spo11-Y135F cells with homologous LYS2 dots (A12095, ·), and wild-type cells with nonhomologous LYS2/URA3 dots (A9829, □), all deleted for NDT80, were assayed for pairing as described in Figure (A). Note that the same nonhomologous dot control strain is used in F and G as the data from these panels were generated from the same experiment. (G) Wild-type cells with homologous URA3 dots (A6946, ■), spo11Δ cells with homologous URA3 dots (A7803, ▴), spo11-Y135F cells with homologous URA3 dots (A9115, ·), and wild-type cells with nonhomologous LYS2/URA3 dots (A9829, □), all deleted for NDT80, were assayed for pairing as described in A. (H) Wild-type cells with homologous LEU2 dots (A5111, ■), dmc1Δ cells with homologous LEU2 dots (A6917, ▴), and wild-type cells with nonhomologous LEU2/LYS2 dots (A11474, □), all deleted for NDT80, were assayed for pairing as described in A. (I) Wild-type cells with homologous LYS2 dots (A9828, ■), msh5Δ cells with homologous LYS2 dots (A16290, ▴) and wild-type cells with nonhomologous URA3/LYS2 dots (A9829, □), all deleted for NDT80, were assayed for pairing as described in A.
Western Blot Analysis
Cells were harvested, incubated in 5% trichloroacetic acid (TCA) and lysed as described in Moll et al. (1991). Immunoblots were performed as described in Cohen-Fix et al. (1996). Pgk1 was detected using a mouse anti-PGK1 antibody (Molecular Probes) at a 1:5000 dilution. Rec8-HA was detected using a mouse anti-HA antibody (HA.11, Covance) at a 1:1000 dilution.
RESULTS
An Assay to Examine the Role of Rec8 in Homolog Pairing
Cells lacking the meiosis-specific cohesin subunit Rec8 show significant meiotic defects and low spore viability (Klein et al., 1999). In addition to a defect in sister chromatid cohesion, these cells exhibit a substantial delay in prophase progression, SC assembly, and recombination (Klein et al., 1999). However, although cohesin is required for recombination, a sister chromatid is not. Cells depleted for the DNA replication initiation factor Cdc6 during meiosis (cdc6-mn) do not undergo meiotic DNA replication, but are able to create mature recombination products, at least at the LEU2-HIS4 hotspot, with only a modest delay (Hochwagen et al., 2005). The observation that REC8 but not sister chromatid cohesion was required for recombination raised the possibility that cohesin's cohesion function was separate from its role in recombination. To see if Rec8's role in providing sister chromatid cohesion can similarly be separated from its functions in pairing and SC formation, these processes were compared in strains lacking either REC8 or sister chromatids.
To examine the consequences of loss of sister chromatid cohesion or cohesin on pairing, we developed an assay to monitor pairing in live cells. We utilized strains with an array of tet operator (tetO) sequences inserted near homologous sites in diploid cells that carry a tet repressor (tetR)-GFP fusion construct, making tagged loci visible as GFP dots using fluorescent microcopy (Straight et al., 1996; Michaelis et al., 1997). If the tagged chromosomes are closely juxtaposed, only one dot is discernable because of the proximity of the two GFP signals. In contrast, if the homologues are not closely juxtaposed, two distinct GFP dots are distinguishable. By assessing the ratio of one versus two dots visible per cell in a population at a particular time, we can determine the level of pairing at that time point.
We used strains with tetO arrays inserted at five different loci: LYS2, LEU2, URA3, CEN5, and TEL5. LYS2 is located midarm on chromosome 2, LEU2 is situated ∼22 kb from the centromere of chromosome 3, URA3 is ∼36 kb from the centromere of chromosome 5, CEN5 is adjacent to the centromere of chromosome 5, and TEL5 is 30 kb from the telomere of chromosome 5. As a control for clustering of arrays, a strain with tetO arrays at nonhomologous chromosomal sites was examined in all experiments. Finally, we used strains that were deleted for NDT80, the gene encoding the transcription factor that promotes exit from the pachytene stage in late prophase and entry into the meiotic divisions (Xu et al., 1995; Chu et al., 1998). This feature allowed comparison of the maximal pairing levels between strains that may progress through prophase at different rates (Weiner and Kleckner, 1994; Peoples et al., 2002).
By FISH, it has been observed that early meiotic cells display residual somatic pairing which decreases shortly after cells enter meiosis. This is followed by an increase in pairing as cells progress through prophase, reaching a maximum as cells reach pachytene (Weiner and Kleckner, 1994). Using our live in vivo pairing assay, this pattern is observed at all five sites examined, with chromosomes showing a high level of somatic clustering at the beginning of each experiment (0-h time point), then dispersal in early stages of the experiment, and reassociation of homologous sites during later stages of the time course (Figure 1, A–E). Nonhomologous GFP dot controls showed clustering as cells enter meiosis, but GFP dots dissociated as cells progress through meiotic S phase and prophase (Figure 1, A–E). The timing of pairing of the three centromere-adjacent loci, CEN5, URA3, and LEU2, appears roughly similar, but the LEU2 locus undergoes significantly less dispersal of homologous sites early in meiosis (Figure 1, B–D, Supplemental Figure S1). It is not clear why this is the case. It is possible that the position of the LEU2 locus near an active DSB hotspot somehow alters the pairing dynamics at this site (Symington et al., 1991; Storlazzi et al., 1995; Gerton et al., 2000; Blitzblau et al., 2007). All five loci examined, however, show similar patterns of pairing, indicating no gross differences in pairing behavior between centromeres, telomeres, and arm loci (Figure 1, A–E). This live pairing assay reveals variability in meiotic timing and synchrony that is frequently observed in studies of meiosis in budding yeast. Nonetheless, representative patterns emerge that allow robust analysis (Supplemental Figure S1).
To determine whether this pairing assay reliably detected pairing defects, we analyzed cells deleted for factors previously implicated in pairing. spo11Δ cells, which do not initiate DSBs, exhibit severe pairing defects by multiple assays (Weiner and Kleckner, 1994; Peoples et al., 2002). Our pairing assay recapitulated these results, with both the URA3 and LYS2 loci showing dramatically reduced levels of pairing compared with wild-type controls (Figure 1, F and G). Interestingly, cells that were carrying a catalytically dead version of Spo11, Spo11-Y135F-HA, showed a pairing defect at both URA3 and LYS2 that was as severe as that observed in cells deleted for SPO11 (Figure 1, F and G). These data are consistent with studies using an exogenous Cre/loxP-based pairing assay, but differ from results obtained using FISH to assay pairing, where the authors observed normal levels of pairing in spo11-Y135F cells (Cha et al., 2000; Peoples et al., 2002). Furthermore, we find that pairing levels are roughly dependent on the levels of DSBs created in cells. Cells carrying SPO11 alleles or combinations of alleles that allowed at least 4–12% of normal DSB levels exhibited significant levels of pairing. In contrast, in cells with 0.1% of normal levels of DSBs, homologues did not pair (Supplemental Figure S2, A and B). In cells that made between 0.6 and 4% of wild-type DSB levels locus-specific effects existed. At these levels of DSB formation, pairing was wild-type at LEU2 but was greatly reduced at LYS2 (Supplemental Figure S2, A and B). This difference may be due to a recombination hotspot being located close to LEU2. We conclude that pairing depends on DSBs in a manner that is roughly dependent on the number of DSBs initiated in the genome.
Cells deleted for S-phase cyclins, CLB5 and CLB6, fail to replicate DNA and are unable to form DSBs because Clb5/CDKs phosphorylate Mer2, a factor that promotes DSB formation through recruitment of Spo11 to the sites of DSBs (Henderson et al., 2006; Dirick et al., 1998; Stuart and Wittenberg, 1998; Smith et al., 2001). Cells deleted for CLB5 and CLB6 showed severe pairing defects at LEU2 and LYS2, consistent with the importance of DSBs in pairing even in the absence of a sister chromatid (Supplemental Figure S2, C–H). mer2-S30A cells, which produce only a nonphosphorylatable version of Mer2, were also unable to pair homologous chromosomes at LYS2 (Supplemental Figure S2I). Finally, as described previously, pairing was also impaired in dmc1Δ cells that are able to form DSBs and resect them, but fail to invade the repair template (Figure 1H; Bishop et al., 1992; Weiner and Kleckner, 1994; Peoples et al., 2002). In contrast, pairing was unaffected in cells lacking the late recombination factor Msh5 (Figure 1I; Peoples et al., 2002; Peoples-Holst and Burgess, 2005). We conclude that the GFP dot system can be used to examine homolog pairing in real time. Furthermore, our data confirm in live cells that DSB formation and early stages of recombination are essential for homolog pairing.
REC8 But Not Sister Chromatid Cohesion Is Required for Homolog Pairing
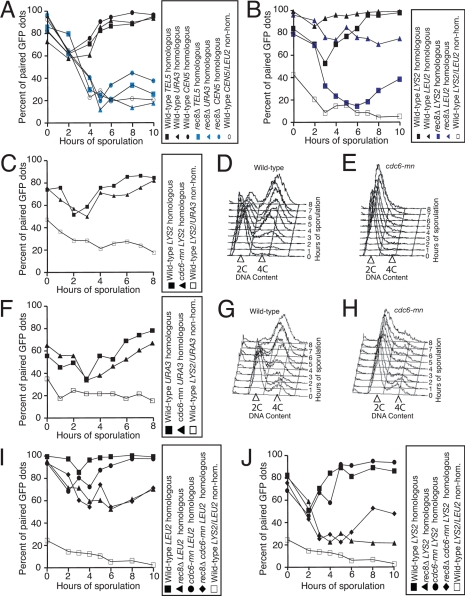
Having established a reliable live-cell assay for homolog pairing, we examined the role of cohesins and sister chromatid cohesion in this process. Work in Schizosaccharomyces pombe, Arabidopsis thaliana, Caenorhabditis elegans, and mouse indicates that Rec8 is required for homolog pairing (Molnar et al., 1995; Klein et al., 1999; Pasierbek et al., 2001; Cai et al., 2003; Xu et al., 2005; Golubovskaya et al., 2006). To examine whether REC8 was required for pairing in yeast, we examined pairing in rec8Δ cells using the GFP dot system. The LYS2, TEL5, CEN5, and URA3 loci all show a dramatic pairing defect in cells deleted for REC8 (Figure 2, A and B). Interestingly, pairing was not as affected at the LEU2 locus (Figure 2B) raising the possibility that the lack of meiotic cohesins does not affect pairing equally across the genome. In contrast to the severe pairing defect of rec8Δ cells, the absence of a sister chromatid did not dramatically affect pairing. Cells depleted for Cdc6 (cdc6-mn; Hochwagen et al., 2005) were able to pair well at URA3 and LYS2, despite undergoing little DNA replication (Figure 2, C–H).
Figure 2.
REC8 but not sister chromatid cohesion is required for pairing. (A) Wild-type cells with homologous TEL5 dots (A16366, ■), or with homologous URA3 dots (A6946, ▴), or with homologous CEN5 dots (A16362, ·), rec8Δ cells with homologous TEL5 dots (A16535, blue rectangle), or with homologous URA3 dots, (A16538, blue triangles), or with homologous CEN5 dots (A16537, blue circles), and wild-type cells with nonhomologous CEN5/LEU2 dots (A16360, ○), all deleted for NDT80, were assayed for pairing as described in Figure 1A. (B) Wild-type cells with homologous LYS2 dots (A9828, ■), or with homologous LEU2 dots (A5111, ▴), rec8Δ cells with homologous LYS2 dots (A16108, blue rectangles), or with homologous LEU2 dots, (A16131, blue triangles), and wild-type cells with nonhomologous LYS2/LEU2 dots (A11474, □), all deleted for NDT80, were assayed for pairing as described in Figure 1A. (C–E) Wild-type (A9828, ■) or cdc6-mn cells with homologous LYS2 dots (A10735, ▴) and wild-type cells with nonhomologous URA3/LYS2 dots (A9829, □), all deleted for NDT80, were induced to sporulate to assayed pairing as described in Figure 1A (C) or DNA content by flow cytometry analysis (D and E). (F–H) Wild-type cells with homologous URA3 dots (A6946, ■), cdc6-mn cells with homologous URA3 dots (A10404, ▴), wild-type cells with nonhomologous URA3/LYS2 dots (A9829, □), all deleted for NDT80, were induced to sporulate to assayed pairing as described in Figure 1A (F) or DNA content by flow cytometry analysis (G and H). (I) Wild-type cells with homologous LEU2 dots (A5111, ■), rec8Δ cells with homologous LEU2 dots (A16131, ▴), cdc6-mn cells with homologous LEU2 dots (A16533, ·), rec8Δ cdc6-mn cells with homologous LEU2 dots (A16460, ♦), and wild-type cells with nonhomologous LEU2/LYS2 dots (A11474, □), all deleted for NDT80, were assayed for pairing as described in Figure 1A. Note that the time courses shown in I and J were performed at the same time, and the nonhomologous dot controls are shown in both experiments. (J) Wild-type cells with homologous LYS2 dots (A9828, ■), rec8Δ cells with homologous LYS2 dots (A16108, ▴), cdc6-mn cells with homologous LYS2 dots (A10735, ·), rec8Δ cdc6-mn cells with homologous LYS2 dots (A16446, ♦), and wild-type cells with nonhomologous LEU2/LYS2 dots (A11474, □), all deleted for NDT80, were assayed for pairing as described in Figure 1A.
Because rec8Δ cells display some premature sister chromatid separation in prophase (Klein et al., 1999), we were concerned that our pairing assay might be detecting GFP dots from separated sister chromatids rather than unpaired homologs in the rec8Δ cells. To address this possibility, we examined pairing in rec8Δ cdc6-mn cells. Depletion of Cdc6 did not rescue the pairing defect of rec8Δ cells at LEU2, indicating that sister chromatids were not separating to an appreciable level in these cells (Figure 2I). This is consistent with the finding that factors other than Rec8 can hold centromere-adjacent regions together (Monje-Casas et al., 2007). At the arm site, LYS2, depleting Cdc6 partially suppressed the pairing defect of rec8Δ cells (Figure 2J), indicating that at sites away from centromeres, a fraction of nonpaired GFP dots is due to premature sister chromatid separation. It is, however, important to note that pairing was still substantially below that of wild-type or Cdc6-depleted cells. We conclude that, as is the case for recombination, Rec8 is required for pairing, but its cohesive function is not.
Sister Chromatid Cohesion Is Not Required for SC Formation
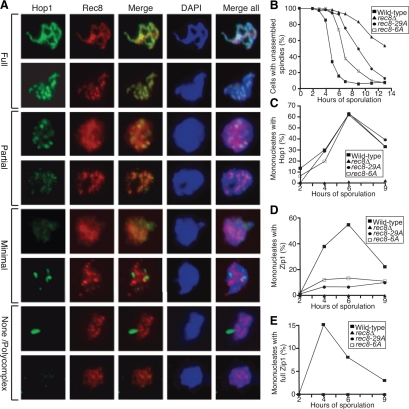
Having established that cohesin but not cohesion was important for homolog pairing, we next examined the effects of cohesins and sister chromatid cohesion on synapsis. Zip1 is the major component of the transverse element of the SC and undergoes stereotypical cytological changes in chromatin association that can be used to assess SC assembly (Sym and Roeder, 1995). During early prophase Zip1 is initially undetectable. Zip1 then forms foci on chromosomes and eventually forms visible ribbons as it zips the axial elements together. After recombination, in late prophase, Zip1 ribbons disappear from chromosomes. When SC formation is impaired, Zip1 clusters known as polycomplexes (PCs) are detected (reviewed in Zickler and Kleckner, 1998).
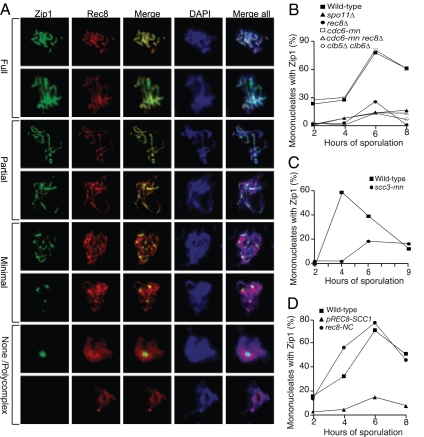
We scored Zip1 staining on meiotic chromosome spreads by employing four categories of staining pattern: none/PC, minimal, partial, and full (Figure 3A). Partial SC formation was observed as early as 2 h after transfer of cells into meiosis-inducing conditions, and the number of cells with fully assembled SCs peaked 4 h thereafter (Figure 3B, Supplemental Figures S3A and S4A). Consistent with previous results, very little Zip1 assembly was observed in spo11Δ cells or cells lacking REC8 (Figure 3B, Supplemental Figures S3A and S4, B and C; Klein et al., 1999; Henderson and Keeney, 2004). Cells lacking the S-phase cyclins, CLB5 and CLB6, which neither undergo meiotic DNA replication nor DSB formation (Smith et al., 2001), also showed poor Zip1 assembly (Figure 3B; Supplemental Figures S3A and S4D). On the basis of these data, we conclude that recombination and either Rec8 or the presence of a nearby sister chromatid is essential for Zip1 assembly.
Figure 3.
Meiotic cohesin complexes are required for Zip1 assembly. (A) Examples of meiotic cells that were harvested and assayed for Zip1 staining on chromosome spreads. Cells carry a Rec8-3HA construct. α-Zip1 staining is shown in green, α-HA in red, and DNA in blue. (B) Wild-type (A7097, ■), spo11Δ (A8477, ▴), rec8Δ (A16664, ·), cdc6-mn (A15880, □), cdc6-mn rec8Δ (A17021, ▵), and clb5Δ clb6Δ (A16113, ○) were induced to sporulate. At the indicated times, cells were harvested and chromosome spreads were assayed for Zip1 staining. In this and subsequent experiments, the “percentage of mononucleates with Zip1” encompasses cells with partially and fully assembled Zip1 as defined in A. The percentage of cells in the individual Zip1 categories is shown in Supplemental Figure S4. The meiotic progression of these strains is shown in Supplemental Figure S3A. In this and subsequent experiments 100 mononucleate cells were counted per strain per time point. (C) Wild-type (A1972, ■) and scc3-mn (A20163, ·) were induced to sporulate. At the indicated times, cells were harvested and chromosome spreads were assayed for Zip1 staining. The percentage of cells in the individual Zip1 categories is shown in Supplemental Figure S5. The meiotic progression of these strains is shown in Supplemental Figure S3B. (D) REC8-3HA (A13946, ■), pREC8-SCC1-3HA (A16132, ▴), and REC8-NC (A13539, ·) were induced to sporulate. At the indicated times, cells were harvested and chromosome spreads were assayed for Zip1 staining. The percentage of cells in the individual Zip1 categories is shown in Supplemental Figure S6. The meiotic progression of these strains is shown in Supplemental Figure S3C.
To determine whether Rec8's cohesion function was important for SC formation, we examined Zip1 assembly in cdc6-mn cells. We found that cdc6-mn cells assemble Zip1 in a pattern nearly identical to that of wild-type cells (Figure 3B; Supplemental Figures S3A and S4E). These data indicate that as in Coprinus cinereus (Pukkila and Skrzynia, 1995) the presence of a sister chromatid is dispensable for SC formation and that, though cohesin-functional Rec8 depends on DNA replication, prophase-functional Rec8 does not (reviewed in Forsburg, 2002; Uhlmann, 2003).
To exclude the possibility that the differential Zip1 assembly in rec8Δ cells and cdc6-mn cells was due to the interference of Zip1 assembly by free sister chromatids present in rec8Δ cells but not those cells lacking Cdc6, we examined Zip1 assembly in cells lacking both Cdc6 and Rec8 (cdc6-mn rec8Δ). We found that in these cells SC was assembled as poorly as in cells deleted for REC8 (Figure 3B; Supplemental Figures S3A and S4F), indicating that the severe Zip1 assembly defect in rec8Δ cells reflects a role for Rec8 protein in recombination, which is a prerequisite for SC assembly and/or Zip1 assembly itself, rather than simply a need for properly tethered sister chromatids as the SC is formed.
The Meiotic Cohesin Complex Is Required for Zip1 Assembly
Our results indicate that the cohesin component Rec8 but not cohesion between sister chromatids or even the presence of a sister chromatid is required for SC formation. We therefore wanted to determine whether Rec8's contribution to Zip1 assembly required other cohesin complex components. The cohesin complex component Smc3 associates along with Rec8 on chromosome axes and is required for SC formation (Klein et al., 1999), indicating that this is likely to be the case. We found that cells depleted for the cohesin complex component Scc3 also showed severe defects in Zip1 assembly (scc3-mn; Figure 3C; Supplemental Figures S3B and S5). Furthermore, Rec8's mitotic counterpart Scc1/Mcd1 cannot fulfill Rec8's role in SC formation. Cells expressing Scc1/Mcd1 instead of Rec8 (pREC8-SCC1) during meiosis, fail to assemble SCs (Figure 3D; Supplemental Figures S3C and S6B), despite the ability of Scc1/Mcd1 to substitute for Rec8 in its meiosis I cohesion role (Lee and Amon, 2003).
Having confirmed that Rec8 likely acts in the context of the cohesin complex to bring about Zip1 assembly, we next determined whether cohesin cleavage contributes to this process. To this end, we examined Zip1 assembly in cells expressing a version of Rec8 that is resistant to cleavage by Separase (rec8-NC; Buonomo et al., 2000) and found that these cells assemble wild-type patterns of Zip1 (Figure 3D; Supplemental Figures S3C and S6C). Our results show that the meiotic but not the mitotic cohesin complex is required for Zip1 assembly. This Zip1 assembly function is not mediated through cohesin's cohesive function and does not require cohesin cleavage.
Rec8 Can Support Zip1 Assembly after DNA Replication
For cohesins to generate sister chromatid cohesion they need to be assembled onto chromosomes during DNA replication. However, Rec8's role in recombination and Zip1 assembly is independent of DNA replication. We therefore wanted to determine whether Rec8 can support Zip1 assembly if supplied to cells after DNA replication. Mitotic cohesin can be loaded onto chromosomes in a DSB-dependent, but replication-independent manner, but Rec8 containing cohesin complexes, at least during mitosis, cannot (Strom et al., 2004; Unal et al., 2004; Heidinger-Pauli et al., 2008). However, whether Rec8 loaded after DNA replication can support SC formation is not known.
To generate a REC8 allele that can be induced at will, we placed REC8 under the control of the GAL1 promoter and introduced this allele into strains that carried a fusion between the Gal4 protein and the estrogen receptor (Gal4-ER; Benjamin et al., 2003). Addition of β-estradiol (βE) to cells of this background causes transport of the Gal4-ER fusion into the nucleus, where it is able to bind to the GAL1 promoter and to induce transcription of REC8. To ensure that cells did not progress past a meiotic stage when SCs can form, the GAL1-REC8 strain also carried a deletion of NDT80. In the absence of this transcription factor, cells arrest in pachytene with fully assembled Zip1 (Xu et al., 1995).
ndt80Δ spo11Δ GAL4-ER cells carrying the pGAL1-REC8 fusion as the sole source of Rec8 were induced to enter meiosis in the absence of βE and incubated for 4.5 h. This allowed cells to enter the meiotic program and progress through meiotic DNA replication in the absence of Rec8-mediated cohesion (Figure 4, A and B). We then induced REC8 expression, and 90 min thereafter initiated DSBs with 20 Krad γ-irradiation (γIR). Zip1 assembly was assayed 3 h thereafter (Figure 4A). Immunofluorescence of cells that were treated with βE revealed that, as expected, Rec8 was expressed in these cells and associated with chromosomes (Figure 4F). This was not true of cells in which βE was not added (data not shown). The analysis of Rad51 foci indicated that 20 Krad γ-irradiation–induced DSBs in these cells (Figure 4F).
Zip1 associated with chromosomes at a very low level in the absence of Rec8 induction (Figure 4C; −βE +IR). When we induced Rec8 in cells in the absence of DSBs, we were also able to observe only a low level of Zip1 assembly (Figure 4C; +βE −IR). In contrast, when we exposed cells to both βE and γIR, 49% of cells were able to assemble Zip1 (Figure 4C; +βE +IR). Similar results were obtained in NDT80 cells expressing stable Rec8, indicating that the Zip1 assembly we observed was not an artifact of the ndt80Δ-induced arrest. Thirty-five percent of βE-treated, γ-irradiated cells assembled Zip1 onto chromosomes under these conditions, compared with <10% of cells that were only βE-treated or γ-irradiated (data not shown). Furthermore, the SC tracts depended on Rec8, as expression of an unstable version of Rec8 (rec8-degron) led to much lower levels of Zip1 polymerization (Figure 4, D and E). We conclude that in contrast to Rec8's cohesion function, Rec8 can support Zip1 assembly when it is loaded onto chromosomes after DNA replication.
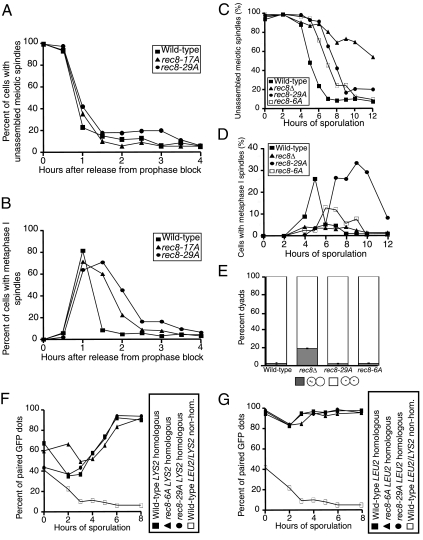
rec8-17A and rec8-29A, But Not rec8-6A Are Defective in Anaphase I Entry
Our studies indicate that Rec8 plays multiple roles during meiosis. The comparison between strains deleted for REC8 and strains lacking sister chromatids but containing functional Rec8, furthermore suggests that Rec8 may mediate its prophase functions through mechanisms distinct from its cohesin function. If this is the case, Rec8 mutants should exist that affect one function of Rec8 but not others.
We previously identified a number of sites in Rec8 that were phosphorylated in vivo (Brar et al., 2006). We mutated various combinations of these sites to the nonphosphorylatable residue, alanine, to examine the importance of these sites to Rec8 cleavage. This analysis revealed that 17 phospho-sites had to be mutated at once to interfere with Rec8 cleavage and anaphase I entry. We also noted that this rec8-17A mutant showed a delay in prophase exit that was dependent on SPO11. A mutant with fewer sites mutated (rec8-6A) also showed a prophase delay, but no metaphase I delay (Brar et al., 2006). A mutant version of REC8 with 29 phosphorylation sites mutated to alanine (rec8-29A) showed a severe prophase delay (Brar et al., 2006).
The effect of mutating 17 phosphorylated amino acids to alanines on anaphase entry was relatively mild (Brar et al., 2006). Alleles with more sites mutated, such as the rec8-29A mutant, could not be examined because of their severe prophase delay. Recently, we developed a protocol to arrest cells in prophase and release them synchronously into the meiotic divisions (Carlile and Amon, 2008). We reasoned that arresting cells in prophase and releasing them may allow us to eliminate the prophase delay of the phospho-mutants and examine the effect on anaphase I entry of these mutants independently of their prophase defect. This was indeed the case. Using this synchronization protocol, we were also able to examine the effects of the rec8-29A mutant on anaphase entry. We detected a substantial metaphase I delay in rec8-17A and rec8-29A cells (1 and 2 h, respectively; Figure 5, A and B). We conclude that rec8-17A and rec8-29A cells are defective in anaphase I entry and that this defect can be separated from the prophase delay that these cells also exhibit.
Figure 5.
rec8-6A and rec8-29A cells form sister chromatid cohesion. (A and B) Wild-type (A21230, ■), rec8-17A (A21234, ▴), and rec8-29A (21232, ·), all carrying GAL-NDT80 and GAL4-ER fusions, were induced to sporulate in the absence of βE. βE was added after 6 h, and samples were taken to determine the percentage of cells with unassembled meiotic spindles (A) and of cells with metaphase I spindles (B). n = 200 cells counted per time point per strain. (C and D) Wild-type (A1972, ■), rec8Δ (A3528, ▴), rec8-29A (A14385, ·), and rec8-6A (A15042, □) cells were induced to sporulate. At the indicated times, samples were taken to determine the percentage of cells with unassembled spindles (A) and of cells with metaphase I spindles (B). n = 200 cells counted per strain per time point. (E) spo11Δ spo13Δ (A21466, A21467, and A21468), spo11Δ spo13Δ rec8Δ (A21472, A21473, and A21474), spo11Δ spo13Δ rec8-29A (A21469, A21470, and A21471), and spo11Δ spo13Δ rec8-6A (A21463, A21464, and A21465) cells carrying a tet repressor-GFP fusion construct and one heterozygous tandem tet operator array inserted at the LEU2 locus were induced to sporulate on plates. After 24 h of sporulation, dyads were scored for sister chromatid segregation by fluorescence microscopy. Cells in which sister chromatids properly segregated apart are represented by the white portion of the bar, whereas cells in which sister chromatids segregate together are represented by the gray portion of the bar. n = 100 cells counted per strain in three independent isogenic diploid strains. Error bars, SD. (F) Wild-type cells with homologous LYS2 dots (A9828, ■), rec8-6A cells with homologous LYS2 dots (A21669, ▴, rec8-29A cells with homologous LYS2 dots (A16412, ·), and wild-type cells with nonhomologous LEU2/LYS2 dots (A11474, □), all deleted for NDT80, assayed for pairing as described in Figure 1A. Note that the same nonhomologous dot control strain is shown in F and G, because the experiments were performed in parallel. (G) Wild-type cells with homologous LEU2 dots (A5111, ■), rec8-6A cells with homologous LEU2 dots (A21670, ▴), rec8-29A cells with homologous LEU2 dots (A21668, ·), and wild-type cells with nonhomologous LEU2/LYS2 dots (A11474, □), all deleted for NDT80, were assayed for pairing as described in Figure 1A.
rec8-6A and rec8-29A Mutants Support Sister Chromatid Cohesion
Next we examined the basis for the prophase defects of the REC8 phospho-mutants. We focused our analyses on the rec8-6A and rec8-29A mutants. Both proteins are produced at wild-type levels and associated with chromosomes (Brar et al., 2006) but cells expressing the rec8-29A mutant as the sole source of REC8 exhibit a 2–4-h prophase delay (the extent of delay varies somewhat with day-to-day variations in meiotic conditions; Brar et al., 2006; Figure 5C). The prophase delay of the rec8-6A mutant is between 1 and 2 h (Brar et al., 2006; Figure 5C). First we determined whether the two mutants were able to support cohesion between sister chromatids. Functional cohesion is necessary for homologous chromosomes, linked by chiasmata, to stably align on the metaphase I spindle. In the absence of sister chromatid cohesion, anaphase I spindle elongation occurs as soon as chromosomes attach on the meiosis I spindle (Klein et al., 1999; Watanabe and Nurse, 1999). The rec8-29A and rec8-6A mutants formed stable metaphase I spindles (Figure 5, B and D), indicating that cohesion is functional in the mutants. In contrast, few cells with metaphase I spindles accumulated in rec8Δ cultures (Figure 5D).
To further examine the cohesive abilities of the various REC8 mutants independently of their ability to support recombination, we investigated whether the rec8-6A and rec8-29A mutants support the equational segregation of spo11Δ spo13Δ mutants. spo11Δ spo13Δ mutants undergo a single meiotic division, during which sister chromatids segregate (Klapholz et al., 1985). This segregation relies on cohesion between sister chromatids. The behavior of chromosomes was tracked in such cells with GFP dots located at the LEU2 locus. When only one of the two copies of chromosome 3 carries these GFP dots, the degree of proper equational segregation in spo11Δ spo13Δ mutants can be assessed. The rec8-6A and rec8-29A mutants supported the full equational segregation of spo11Δ spo13Δ mutants (Figure 5E), indicating that REC8's sister chromatid cohesion function is not affected by the mutations.
It is important to note that sister chromatid cohesion was not abolished in cells lacking REC8. The complete absence of sister chromatid cohesion is expected to result in random segregation, with 50% of sister chromatids segregating to opposite poles and 50% of sister chromatids segregating to the same pole. In rec8Δ spo11Δ spo13Δ cultures, sister chromatids segregated to the same pole in only 20% of cells (Figure 5E). These results indicate that not all meiotic cohesion between sister chromatids depends on REC8.
rec8-6A and rec8-29A Mutants Are Not Defective in Homolog Pairing
Do the rec8-6A and rec8-29A alleles support homolog pairing? To address this question, we examined pairing at LEU2 and LYS2 in rec8-6A and rec8-29A mutant cells. We found that at both loci, replacement of wild-type REC8 with either rec8-6A or rec8-29A had no effect on homolog pairing, with cells capable of pairing efficiently and to wild-type levels (Figure 5, F and G). We conclude that the prophase progression defect seen in rec8-6A and rec8-29A cells is not due to defects in homolog pairing.
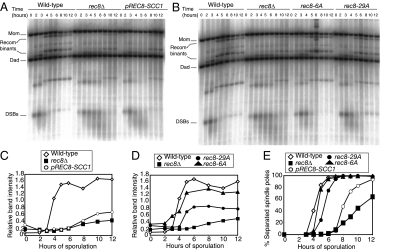
rec8-29A But Not rec8-6A Mutants Are Delayed in Forming Mature Recombinants
Previous studies showed that deletion of REC8 led to a severe recombination defect. rec8Δ cells form DSBs and resect them, but formation of mature recombination products is greatly reduced (Figure 6, A and C; Klein et al., 1999). Using a Southern blot strategy designed to follow recombination status of the artificial HIS4/LEU2 DSB hotspot (Hunter and Kleckner, 2001), we found that cells expressing Scc1/Mcd1 instead of Rec8 during meiosis exhibited a similar recombination defect (Figure 6, A and C), indicating that only the meiotic form of cohesin can support interhomolog recombination. Interestingly, in this experiment, pREC8-SCC1 cells underwent the meiotic divisions with more efficiency than rec8Δ cells (data not shown). The reasons for this are at present unclear.
Figure 6.
rec8-29A mutant cells exhibit defects in homologous recombination. (A) Wild-type (A1556), rec8Δ (A18933), and pREC8-SCC1 rec8Δ (A16132) were induced to sporulate. At the indicated times, cells were harvested and assayed by Southern blot for DSBs and recombination products at HIS4/LEU2. (B) Wild-type (A1556), rec8Δ (A18933), rec8-29A (A21618), and rec8-6A (A18936) cells were induced to sporulate. At the indicated times, cells were harvested and assayed by Southern blot for DSBs and recombination products at HIS4/LEU2. (C and D) Blots from A and B were subjected to densitometric analysis to quantitate the intensity of the bands representing the upper recombinant band. Values were normalized to the “Mom” parental band for each strain and each time point. (E) Meiotic progression of the strains assayed for recombination shown in A and B. Wild-type (A1556, ◇), rec8Δ (A18933, ■), pREC8-SCC1 rec8Δ (A16132, ○), rec8-29A (A21618, ·), and rec8-6A (A18936, ▴) cells were induced to sporulate. At the indicated times, samples were taken and subjected to α-tubulin immunofluorescence to determine the percentage of cells with unassembled spindles. n = 200 cells counted per strain per time point.
Next we examined whether the rec8-6A and rec8-29A alleles were able to support homologous recombination. Although rec8-6A cells exhibited no detectable defect in DSB formation or formation of mature recombination products, rec8-29A cells made DSBs at normal levels and with normal timing, but exhibited a significant delay in the formation of recombinants and only formed half the number of mature cross-over recombination products observed in wild-type cells (Figure 6, B, D, and E). Consistent with homologous recombination and sister chromatid cohesion occurring normally in rec8-6A mutants is the observation that spore viability is high in this mutant (84%; Brar et al., 2006). Even rec8-29A mutants exhibit a relatively high spore viability (68%; Brar et al., 2006), which is consistent with the observation that cross-overs do form, albeit at a reduced level and with a delay. We cannot determine based on these data whether the defect in mature cross-over formation in rec8-29A cells is due to a requirement for wild-type Rec8 in the completion of cross-over recombination or the resolution of joint molecules. Nonetheless, we conclude that although the rec8-29A mutant supports sister chromatid cohesion, it is defective in efficient DSB repair through cross-over formation.
rec8-6A and rec8-29A Mutants Do Not Affect Chromosome Axis Formation But Disrupt Homolog Synapsis
REC8 is not only essential for homologous recombination but also meiotic chromosome axis morphogenesis and synapsis (Figure 3B; Klein et al., 1999). One of the components of meiotic chromosomes axes is Hop1 (Hollingsworth et al., 1990). Unlike in rec8Δ cells, its association with chromosomes was not altered in rec8-6A or rec8-29A mutants as judged by the ability to assemble partial and full Hop1 ribbons (Figure 7, A–C; Supplemental Figure S7). In contrast, Zip1 assembly was significantly impaired in rec8-6A and rec8-29A cells as judged by the ability of cells to form partial or full Zip1 ribbons (Figure 7D; Supplemental Figure S8). This defect was even present when rec8-6A and rec8-29A cells were given additional time to assemble Zip1 through an ndt80Δ-induced prophase arrest (data not shown). Full synapsis, that is Zip1 assembled along the entire length of all chromosomes, did not occur at all (Figure 7E; Supplemental Figure S8). As is true of analysis of meiotic progression, we observe some variability in the severity of the defect in Zip1 assembly in these mutants, although a consistent defect is observed (Supplemental Figure S9). None of the REC8 alleles with fewer than six mutated phosphorylation sites exhibited a consistent prophase defect (data not shown). We conclude that rec8-6A and rec8-29A mutants are partially defective in Zip1 assembly onto chromosomes. This is consistent with work in maize that has identified alleles of the REC8 homolog AFD1, which show specific defects in Zip1 assembly, but not axis formation (Golubovskaya et al., 2006).
Figure 7.
Axis assembly occurs in rec8-6A and rec8-29A cells, but Zip1 assembly does not. (A) Examples of meiotic cells with various degrees of chromosome axis assembly as judged by Hop1 staining on chromosome spreads. Cells carry a Rec8-3HA construct. α-Hop1 is shown in green, α-HA in red, and DNA in blue. (B–E) Wild-type (A20066, ■), rec8Δ (A3528, ▴), rec8-29A (A14385, ·), and rec8-6A (A15042, □) cells were induced to sporulate. At the indicated times the percentage of cells with unassembled spindles (B) and chromosome spreads were assayed for Hop1 staining (C), Zip1 staining (D), and full Zip1 staining (E). The percentage of cells in the individual Zip1 categories is shown in Supplemental Figure S8. Note that cells were scored as having Hop1 assembled when they showed partial or full Hop1 staining according to the categories shown (A). n = 200 cells counted per strain per time point. Also note that in E the lines for rec8Δ, rec8-29A, and rec8-6A cells all overlap with each other and the x-axis.
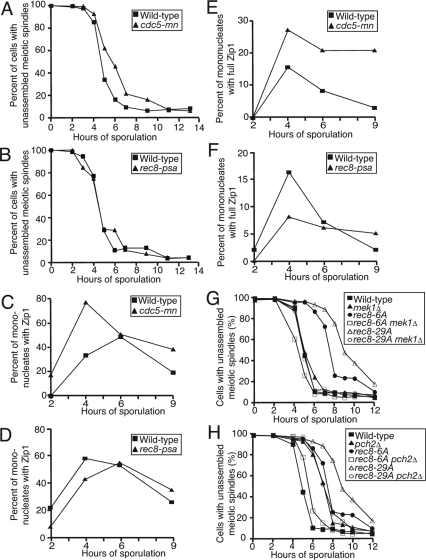
Which protein kinase, if any, is required for Rec8 to promote Zip1 assembly? Rec8 is phosphorylated by the Polo kinase Cdc5, as well as other unidentified kinases (Clyne et al., 2003; Lee and Amon, 2003; Brar et al., 2006). Cdc5-depleted cells have been shown to exhibit a delay in exit from prophase (Clyne et al., 2003), so we wanted to determine whether Cdc5 phosphorylation contributed to the prophase defect seen in Rec8 phospho-mutants. This was not the case. Cells depleted for Cdc5 (cdc5-mn; Lee and Amon, 2003) exhibited a delay in exit from prophase, but Zip1 assembly was largely unaffected (Figure 8, A, C, and E; Supplemental Figure S10, A and B). To the contrary, these cells display a higher level of Zip1 staining than wild-type cells, consistent with a role for Cdc5 in SC disassembly (Sourirajan and Lichten, 2008). Furthermore, a REC8 mutant in which all 11 identified Cdc5-dependent sites on Rec8 were mutated to alanine (rec8-psa; Brar et al., 2006), exhibited only a very mild Zip1 assembly defect and did not display a delay in prophase exit (Figure 8, B, D, and F; Supplemental Figure S10, C and D). Taken together, the analysis of cdc5-mn and rec8-psa cells indicates that Cdc5 phosphorylation cannot account for the significant defect in Zip1 assembly seen in rec8-6A and rec8-29A cells. This is consistent with findings that Cdc5 is induced late in prophase, after SC assembly (Clyne et al., 2003). We conclude that although several Rec8 residues that are phosphorylated play a role in assembly of the SC, Cdc5-dependent phosphorylation does not. We have further excluded the protein kinases Cdc28, Ime2, Ipl1, Mek1, Cdc15, Mec1, and Rad53 through either meiotic depletion or treatment of cells with specific kinase inhibitors, as playing a role in Zip1 assembly (data not shown).
Figure 8.
Effects of Cdc5 phosphorylation on Zip1 assembly and response of rec8-6A and rec8-29A cells to recombination checkpoint defects. (A, C, and E) Wild-type (A20066, ■) and cdc5-mn (A5844, ▴) cells were induced to sporulate. At the indicated times the percentage of cells with unassembled spindles (A) and chromosome spreads were assayed for Zip1 staining (C) and full Zip1 staining (E). The percentage of cells in the individual Zip1 categories is shown in Supplemental Figure S10, A and B. Note that these data are from the same experiment as is presented in Figure 7, so the wild-type control is identical in both figures. (B, D, and F) Wild-type (A14655, ■) and rec8-psa (A15364, ▴) were induced to sporulate. At the indicated times the percentage of cells with unassembled spindles (B) and chromosome spreads were assayed for Zip1 staining (D) and full Zip1 staining (F). The percentage of cells in the individual Zip1 categories is shown in Supplemental Figure S10, C and D. (G) Wild-type (A1972, ■), mek1Δ (A20156, ▴), rec8-6A (A15042, ·), rec8-6A mek1Δ (A20154, □), rec8-29A (A14385, ▵), and rec8-29A mek1Δ (A20157, ○) cells were induced to sporulate. At the indicated times the percentage of cells with unassembled spindles. Note that this experiment was performed concomitantly with that shown in H. Hence the controls are identical. (H) Wild-type (A1972, ■), pch2Δ (A21053, ▴), rec8-6A (A15042, ·), rec8-6A pch2Δ (A20151, □), rec8-29A (A14385, ▵), and rec8-29A pch2Δ (A20164, ○) cells were induced to sporulate. At the indicated times the percentage of cells with unassembled spindles.
Multiple kinases could act in concert to promote Zip1 assembly and recombination. It is also possible, however, that the prophase defects that we observe in specific Rec8 phospho-mutants are due to structural changes in the protein and not actual phosphorylation events on the residues that we mutated. To attempt to address this issue, we generated phospho-mimetic mutants. When the residues mutated to alanine in the rec8-6A mutant were changed to glutamates or aspartates (rec8-6E, rec8-6D), Rec8 was no longer detectable by Western blot analysis (Supplemental Figure S11), indicating that these mutations result in an unstable protein. It is therefore not clear whether phosphorylation or a structural role of these residues is required for Rec8's function in SC formation. It is however certain that the rec8-6A mutant is partially defective in Zip1 assembly but not recombination nor meiotic chromosome axis formation. We conclude that Rec8's functions during prophase can be genetically separated.
The Different Roles of REC8 in Recombination and Synapsis Are Revealed by a Differential Response to Recombination Checkpoint Inactivation
Neither the rec8-6A nor rec8-29A mutant protein supports SC formation. In contrast, cells expressing rec8-6A appear to undergo recombination with wild-type kinetics, whereas cells expressing rec8-29A do not. To probe this difference further, we examined the consequences of inactivating different branches of the recombination checkpoint in the two mutants. The recombination checkpoint is thought to sense the presence of incomplete recombination products or improper SC formation and to delay entry into the meiotic divisions until the defect is repaired (reviewed in Hochwagen and Amon, 2006). The protein kinase Mek1 is a key component of this checkpoint, as well as a component of the meiotic machinery that drives repair from the homolog rather than the sister during recombination (Xu et al., 1997; Wan et al., 2004; Niu et al., 2005). Deletion of MEK1 in rec8-6A and rec8-29A cells suppressed the prophase delay in both mutants (Figure 8G), indicating that the recombination and/or synapsis defects in the two mutants are responsible for the prophase I delay.
Pch2 is a nucleolar protein thought to be primarily responsible for delaying meiosis I entry in response to SC defects, as its deletion is able to suppress the prophase I delay of mutants defective in synapsis but unable to rescue delays caused by recombination defects (San-Segundo and Roeder, 1999; Hochwagen and Amon, 2006; Mitra and Roeder, 2007). Deletion of PCH2 suppressed the prophase delay of rec8-6A mutants but not that of rec8-29A cells. The absolute amount of suppression was the same in both mutants (2 h), but the prophase delay of rec8-29A mutants is significantly greater than that of rec8-6A mutants (Figure 8H). This result is consistent with our observation that rec8-29A mutants show recombination defects in addition to SC formation defects, whereas rec8-6A cells primarily exhibit Zip1 assembly defects. Cells deleted for PCH2 themselves show a mild prophase delay that is likely due to a role for Pch2 in chromosome morphogenesis (Figure 8H; Borner et al., 2008). It is interesting to note that replacement of REC8 by rec8-6A in cells deleted for PCH2 also suppresses this mild prophase delay for reasons that we cannot explain at this time (Figure 8H).
We conclude that rec8-6A mutants primarily exhibit a Zip1 assembly defect. rec8-29A mutants show a Zip1 assembly and recombination defect, and rec8Δ cells display a sister chromatid cohesion, pairing, recombination, axis morphogenesis, and Zip1 assembly defect. The finding that different rec8 mutants affect the various functions of Rec8 to distinct degrees, argues that cohesion, pairing, Zip1 assembly and recombination are mediated by Rec8 through at least partially genetically separable mechanisms.
DISCUSSION
The Multiple Roles of REC8 in Meiotic Chromosome Morphogenesis
The analysis of REC8 deletions and two REC8 alleles revealed that the protein is essential for several key aspects of meiotic chromosome structure and function. The fact that some alleles exhibit only a subset of the phenotypes observed in rec8Δ cells, furthermore indicates that these functions are genetically separable and thus require different functions of REC8 and/or different quantities of functional cohesin.
REC8 and Sister Chromatid Cohesion
Rec8 as part of the cohesin complex is essential to hold sister chromatids together on the metaphase I and metaphase II spindles. This function was revealed by the mis-segregation of sister chromatids in the few rec8Δ cells that enter the meiotic divisions (Klein et al., 1999) and the 20% nondisjunction of sister chromatids in spo11Δ spo13Δ rec8Δ mutants. Although it is clear that sister chromatid cohesion is impaired in the absence of REC8, it was not eliminated. Rec8-containing cohesin complexes are therefore probably not the only factors holding sister chromatids together during meiosis. Low levels of Scc1/Mcd1 present during meiosis could support sister chromatid cohesion (Kateneva et al., 2005). Catenation (Aguilar et al., 2005) and coorientation factors (Monje-Casas et al., 2007) are also capable of holding sister chromatids together and could contribute to their association in the absence of Rec8-containing cohesins.
Rec8 Phosphorylation and Cohesin Removal
Previous studies suggested that phosphorylation of Rec8 by Cdc5 was important for its cleavage at the metaphase-to-anaphase transition. Mutation of many Rec8 phosphorylation sites, however, revealed only a modest delay in cohesin cleavage and anaphase I entry (Brar et al., 2006). All REC8 mutants with 20 or more phosphorylation sites mutated to alanine exhibited a severe prophase delay precluding us from analyzing their effects on anaphase I entry. The development of a method that synchronizes meiotic cells by arresting them in prophase allowed us to assess the anaphase I entry defect of these phospho-mutants because it eliminated the metaphase I entry delay of the mutants, presumably because cells had time to complete most steps of recombination while arrested in the prophase block. In the synchronized cultures, the rec8-17A and rec8-29A cells exhibited a significant anaphase I entry delay, confirming our previous conclusion that Rec8 phosphorylation is important for the timely onset of anaphase I. This role is likely mediated at the level of cohesin removal and not due to an indirect effect of activating the DNA damage and/or recombination checkpoint as rec8–17A and rec8–29A cells release from the prophase block without delay (Figure 5A).
REC8 and Pairing
We developed an assay that allowed us to follow pairing in live cells. It utilizes an array of tet-operators that can be integrated at various loci in the genome and that are visualized using a tetR-GFP fusion (Straight et al., 1996; Michaelis et al., 1997). Pairing of the GFP dots depends on the same events as pairing examined with more traditional assays such as FISH. Cells lacking DSBs were essentially unable to pair. Using this assay, we could show that cells without REC8 showed a partial pairing defect. Furthermore, the requirement for REC8 in sister chromatid cohesion does not contribute to this pairing role, as cells without sister chromatids (Cdc6-depleted cells) pair normally. This finding additionally indicates that the presence of a sister chromatid is not important for homolog recognition.
REC8 and Recombination
The fact that cells lacking REC8 are defective in sister chromatid cohesion and recombination led to the simple hypothesis that linked sister chromatids are a prerequisite for chromosome axis formation and hence the formation of mature recombinants. Our data indicate that this is not the case. Instead it appears that Rec8 promotes recombination independently of its sister chromatid cohesion function. Elimination of a sister chromatid does not dramatically interfere with recombination but deletion of REC8 does (Klein et al., 1999; Hochwagen et al., 2005). Furthermore, the mitotic form of Rec8, Scc1/Mcd1, can support sister chromatid cohesion during meiosis but not recombination. Finally, we isolated a mutant in REC8 (rec8-29A) that supports sister chromatid cohesion but in which recombination is impaired.
As judged by Southern blot analysis of the HIS4/LEU2 hotspot, rec8-29A mutants are delayed in cross-over formation and produce fewer recombinants compared with wild-type cells. Spore viability is high in the rec8-29A mutant, which is consistent with the idea that recombination occurs faithfully but at a reduced efficiency and with a delay. Which aspect of recombination requires cohesin? rec8-29A and rec8Δ mutants form DSBs with wild-type efficiency, but formation of mature recombinants is affected (Klein et al., 1999). Strand invasion or later aspects of double Holliday junction formation could require cohesin function, possibly as a result of Rec8's role in chromosome axis formation.
REC8 and Zip1 Assembly
SC formation depends on recombination in budding yeast (Giroux et al., 1989; reviewed in Zickler and Kleckner, 1998). The inability of rec8-29A and rec8Δ mutants to assemble Zip1 onto chromosomes could therefore be an indirect consequence thereof. However this cannot be true for rec8-6A mutants. rec8-6A cells form sister chromatid cohesion. They also appear to be proficient in recombination, although it is possible that subtle defects exist that are below the threshold of detection of our assay. In contrast, the Zip1 assembly defect of rec8-6A mutants is substantial. Thus, it appears that Rec8's role in SC formation is genetically separable from its other functions. Although the rec8-6A mutant is defective in Zip1 assembly, it is not a phenocopy of a ZIP1 deletion. Cell lacking ZIP1 exhibit low spore viability (50%; Xu et al., 1995). In contrast rec8-6A mutants produce viable spores at wild-type level (Brar et al., 2006). Whether this difference is due to the rec8-6A mutant exhibiting a less severe synapsis defect or due to a role of ZIP1 in recombination that is independent of its role in SC assembly is at present unclear. rec8-6A mutants exhibit a 1–2 h delay in metaphase I spindle formation (Brar et al., 2006). The finding that rec8-6A mutants do not exhibit a recombination defect but a severe Zip1 assembly defect raises the possibility that the delay observed in rec8-6A mutants is due to activation of surveillance mechanisms that halts meiotic progression in response to synapsis defects. Consistent with this idea is the observation that the meiotic delay of rec8-6A mutants depends on the recombination checkpoint components PCH2, a gene previously implicated in regulating the response to synapsis defects (Wu and Burgess, 2006).
A key question that arises from the characterization of the various Rec8 functions during prophase is why meiotic cells would utilize one protein for several disparate functions. We propose that using the same protein for sister chromatid cohesion and interactions between homologous chromosomes is an efficient way for cells to ensure that the sister chromatid cohesion machinery, which is also essential for mediating homolog connections, and is put in place before or concomitantly with the onset of homolog interactions.
How Does REC8 Facilitate Sister Chromatid Cohesion, Pairing, Recombination, and SC Formation?
Our studies indicate that the meiotic cohesin complex is needed for sister chromatid cohesion, pairing, recombination, and SC formation and that these functions are genetically separable. This raises the question of whether Rec8 functions in these aspects of meiotic chromosome morphogenesis through different mechanisms. For example, it is possible that different domains of Rec8 or different phosphorylation events on the protein facilitate Rec8's various roles during meiotic prophase. Testing this idea would require the identification of the protein kinases responsible for phosphorylating the different sites and determining the consequences of inactivating their function. This is likely not a simple task. Rec8 is phosphorylated on as many as 29 sites, and many site mutants exhibit a prophase progression defect (data not shown; Brar et al., 2006). It is thus likely that multiple kinases are involved in activating Rec8 for its role in prophase progression.
The observation that phospho-mimetic mutation of the phosphorylated residues mutated to alanines in the rec8-6A mutant also leads to destabilization of the protein raises the possibility that the REC8 mutants examined here represent an allelic series of loss-of-function alleles rather than separation of function alleles. If the REC8 mutants studied here indeed represent quantitative rather than qualitative differences in cohesin function, it follows that sister chromatid cohesion requires less cohesin than pairing, which requires less cohesin than recombination, which in turn requires less cohesion than SC formation. The observation that cells require greater Rec8 “function” in prophase than for sister chromatid cohesion is consistent with the observation that greater Rec8 levels are present on chromosomes in mammalian prophase I than in the subsequent meiotic divisions. We speculate that the large number of DSBs initiated in prophase require large amounts of Rec8 to stabilize nearby DNA structures. SC formation may need even higher levels of cohesin function. Cohesin complexes, axial element components, and Zip1 may be needed in stoichiometric amounts for SC formation.
Is the SC Dispensable for Recombination?
This long-standing question is revisited by the characterization of the rec8-6A mutant. Zip1 assembly is severely impaired in the mutant, yet the mutant does not exhibit significant recombination defects in the methods used here and produces viable spores with a similar efficiency as wild-type cells. These findings support the idea that, although there is significant interplay between the processes of recombination and SC assembly, SC assembly is not essential for recombination. This theory is based on diverse data including the fact that cells deleted for ZIP1 show surprisingly high spore viability of near 50% despite the inability to form any complete SCs (Xu et al., 1995). Furthermore, cells deleted for RED1 fail to form SCs but make cross-overs (Rockmill and Roeder, 1990). Finally, several organisms such as S. pombe and the silkworm Bombyx mori have been identified that lack SC structures altogether (reviewed in Zickler and Kleckner, 1998). In most meiotic organisms, however, SCs shows striking structural conservation, indicating an important, though mysterious role for this structure during meiosis.
Are Rec8's Roles in Meiotic Prophase Conserved?
REC8 counterparts exist in most meiotic eukaryotes and several lines of evidence suggest that, in other organisms too, the roles of cohesins in mediating the multiple prophase functions of meiotic cohesin are conserved and also distinct from each other. In Coprinus cinereus, mutating the sister chromatid cohesion establishment factor RAD9 (SCC2 in budding yeast) leads to cohesion defects and homolog synapsis defects. When RAD9's role in sister chromatid cohesion is eliminated (by preventing DNA replication) pairing defects persist (Cummings et al., 2002), indicating that in this organism, too, it is not merely the sister chromatid cohesion function of cohesin that is required for homolog pairing and synapsis. In mammalian cells, the vast majority of Rec8 cohesin complexes are removed from chromosomes in a cleavage-independent manner before the first meiotic division (Sumara et al., 2002). It is thought that this eases the burden on Separase, such that meiotic divisions can occur relatively rapidly once initiated. It is unclear, however, why cells incorporate extra Rec8 onto chromosome just to remove it shortly afterward. As in yeast, high levels of cohesins may be necessary for pairing, chromosome axis formation, recombination and SC formation. Consistent with this idea is the observation that mutations in mammalian REC8 also result in prophase defects (Xu et al., 2005). Thus Rec8-containing cohesins may be key determinants of meiotic chromosome morphogenesis in most eukaryotes that use meiosis to form gametes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Franz Klein and Shirleen Roeder for reagents. We also thank members of the Amon lab for critical reading of and comments on the manuscript. This work was supported by the National Institutes of Health Grant GM62207 to A.A. and a National Science Foundation predoctoral fellowship to G.A.B. A.A is also an investigator of the Howard Hughes Medical Institute.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-06-0637) on December 10, 2008.
REFERENCES
- Aguilar C., Davidson C., Dix M., Stead K., Zheng K., Hartman T., Guacci V. Topoisomerase II suppresses the temperature sensitivity of Saccharomyces cerevisiae pds5 mutants, but not the defect in sister chromatid cohesion. Cell Cycle. 2005;4:1294–1304. doi: 10.4161/cc.4.9.1997. [DOI] [PubMed] [Google Scholar]
- Alexandru G., Uhlmann F., Mechtler K., Poupart M. A., Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- Benjamin K. R., Zhang C., Shokat K. M., Herskowitz I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 2003;17:1524–1539. doi: 10.1101/gad.1101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. K., Park D., Xu L., Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Blitzblau H. G., Bell G. W., Rodriguez J., Bell S. P., Hochwagen A. Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr. Biol. 2007;17:2003–2012. doi: 10.1016/j.cub.2007.10.066. [DOI] [PubMed] [Google Scholar]
- Borner G. V., Barot A., Kleckner N. Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proc. Natl. Acad. Sci. USA. 2008;105:3327–3332. doi: 10.1073/pnas.0711864105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner G. V., Kleckner N., Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- Brar G. A., Kiburz B. M., Zhang Y., Kim J. E., White F., Amon A. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature. 2006;441:532–536. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- Buonomo S. B., Clyne R. K., Fuchs J., Loidl J., Uhlmann F., Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Cai X., Dong F., Edelmann R. E., Makaroff C. A. The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J. Cell Sci. 2003;116:2999–3007. doi: 10.1242/jcs.00601. [DOI] [PubMed] [Google Scholar]
- Carlile T. M., Amon A. Meiosis I is established through division-specific translational control of a cyclin. Cell. 2008;133:280–291. doi: 10.1016/j.cell.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha R. S., Weiner B. M., Keeney S., Dekker J., Kleckner N. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 2000;14:493–503. [PMC free article] [PubMed] [Google Scholar]
- Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., Brown P. O., Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Clyne R. K., Katis V. L., Jessop L., Benjamin K. R., Herskowitz I., Lichten M., Nasmyth K. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 2003;5:480–485. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O., Peters J. M., Kirschner M. W., Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Cummings W. J., Merino S. T., Young K. G., Li L., Johnson C. W., Sierra E. A., Zolan M. E. The Coprinus cinereus adherin Rad9 functions in Mre11-dependent DNA repair, meiotic sister-chromatid cohesion, and meiotic homolog pairing. Proc. Natl. Acad. Sci. USA. 2002;99:14958–14963. doi: 10.1073/pnas.232316999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick L., Goetsch L., Ammerer G., Byers B. Regulation of meiotic S phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science. 1998;281:1854–1857. doi: 10.1126/science.281.5384.1854. [DOI] [PubMed] [Google Scholar]
- Forsburg S. L. Only connect: linking meiotic DNA replication to chromosome dynamics. Mol. Cell. 2002;9:703–711. doi: 10.1016/s1097-2765(02)00508-7. [DOI] [PubMed] [Google Scholar]
- Gerton J. L., DeRisi J., Shroff R., Lichten M., Brown P. O., Petes T. D. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux C. N., Dresser M. E., Tiano H. F. Genetic control of chromosome synapsis in yeast meiosis. Genome. 1989;31:88–94. doi: 10.1139/g89-017. [DOI] [PubMed] [Google Scholar]
- Golubovskaya I. N., Hamant O., Timofejeva L., Wang C. J., Braun D., Meeley R., Cande W. Z. Alleles of afd1 dissect REC8 functions during meiotic prophase I. J. Cell Sci. 2006;119:3306–3315. doi: 10.1242/jcs.03054. [DOI] [PubMed] [Google Scholar]
- Heidinger-Pauli J. M., Unal E., Guacci V., Koshland D. The kleisin subunit of cohesin dictates damage-induced cohesion. Mol. Cell. 2008;31:47–56. doi: 10.1016/j.molcel.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Henderson K. A., Kee K., Maleki S., Santini P. A., Keeney S. Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell. 2006;125:1321–1332. doi: 10.1016/j.cell.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson K. A., Keeney S. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc. Natl. Acad. Sci. USA. 2004;101:4519–4524. doi: 10.1073/pnas.0400843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochwagen A., Amon A. Checking your breaks: surveillance mechanisms of meiotic recombination. Curr. Biol. 2006;16:R217–R228. doi: 10.1016/j.cub.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Hochwagen A., Tham W. H., Brar G. A., Amon A. The FK506 binding protein Fpr3 counteracts protein phosphatase 1 to maintain meiotic recombination checkpoint activity. Cell. 2005;122:861–873. doi: 10.1016/j.cell.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Hollingsworth N. M., Goetsch L., Byers B. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell. 1990;61:73–84. doi: 10.1016/0092-8674(90)90216-2. [DOI] [PubMed] [Google Scholar]
- Hunter N., Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Kateneva A. V., Konovchenko A. A., Guacci V., Dresser M. E. Recombination protein Tid1p controls resolution of cohesin-dependent linkages in meiosis in Saccharomyces cerevisiae. J. Cell Biol. 2005;171:241–253. doi: 10.1083/jcb.200505020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis V. L., Matos J., Mori S., Shirahige K., Zachariae W., Nasmyth K. Spo13 facilitates monopolin recruitment to kinetochores and regulates maintenance of centromeric cohesion during yeast meiosis. Curr. Biol. 2004;14:2183–2196. doi: 10.1016/j.cub.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Keeney S., Giroux C. N., Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Kitajima T. S., Kawashima S. A., Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Klapholz S., Waddell C. S., Esposito R. E. The role of the SPO11 gene in meiotic recombination in yeast. Genetics. 1985;110:187–216. doi: 10.1093/genetics/110.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F., Mahr P., Galova M., Buonomo S. B., Michaelis C., Nairz K., Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- Lee B., Amon A. Meiosis: how to create a specialized cell cycle. Curr. Opin. Cell Biol. 2001;13:770–777. doi: 10.1016/s0955-0674(00)00282-9. [DOI] [PubMed] [Google Scholar]
- Lee B. H., Amon A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science. 2003;300:482–486. doi: 10.1126/science.1081846. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Marston A. L., Amon A. Meiosis: cell-cycle controls shuffle and deal. Nat. Rev. Mol. Cell Biol. 2004;5:983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- Marston A. L., Lee B. H., Amon A. The Cdc14 phosphatase and the FEAR network control meiotic spindle disassembly and chromosome segregation. Dev. Cell. 2003;4:711–726. doi: 10.1016/s1534-5807(03)00130-8. [DOI] [PubMed] [Google Scholar]
- Marston A. L., Tham W. H., Shah H., Amon A. A genome-wide screen identifies genes required for centromeric cohesion. Science. 2004;303:1367–1370. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- McKee B. D. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim. Biophys. Acta. 2004;1677:165–180. doi: 10.1016/j.bbaexp.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Mitra N., Roeder G. S. A novel nonnull ZIP1 allele triggers meiotic arrest with synapsed chromosomes in Saccharomyces cerevisiae. Genetics. 2007;176:773–787. doi: 10.1534/genetics.107.071100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll T., Tebb G., Surana U., Robitsch H., Nasmyth K. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell. 1991;66:743–758. doi: 10.1016/0092-8674(91)90118-i. [DOI] [PubMed] [Google Scholar]
- Molnar M., Bahler J., Sipiczki M., Kohli J. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics. 1995;141:61–73. doi: 10.1093/genetics/141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje-Casas F., Prabhu V. R., Lee B. H., Boselli M., Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128:477–490. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Niu H., Wan L., Baumgartner B., Schaefer D., Loidl J., Hollingsworth N. M. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell. 2005;16:5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S. L., Hawley R. S. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- Pasierbek P., Jantsch M., Melcher M., Schleiffer A., Schweizer D., Loidl J. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 2001;15:1349–1360. doi: 10.1101/gad.192701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples T. L., Dean E., Gonzalez O., Lambourne L., Burgess S. M. Close, stable homolog juxtaposition during meiosis in budding yeast is dependent on meiotic recombination, occurs independently of synapsis, and is distinct from DSB-independent pairing contacts. Genes Dev. 2002;16:1682–1695. doi: 10.1101/gad.983802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples-Holst T. L., Burgess S. M. Multiple branches of the meiotic recombination pathway contribute independently to homolog pairing and stable juxtaposition during meiosis in budding yeast. Genes Dev. 2005;19:863–874. doi: 10.1101/gad.1293605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila P. J., Skrzynia C. Independent synaptic behavior of sister chromatids in Coprinus cinereus. Can. J. Botany. 1995;73:S215–S220. [Google Scholar]
- Rabitsch K. P., Gregan J., Schleiffer A., Javerzat J. P., Eisenhaber F., Nasmyth K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr. Biol. 2004;14:287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- Rockmill B., Roeder G. S. Meiosis in asynaptic yeast. Genetics. 1990;126:563–574. doi: 10.1093/genetics/126.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Segundo P. A., Roeder G. S. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 1999;97:313–324. doi: 10.1016/s0092-8674(00)80741-2. [DOI] [PubMed] [Google Scholar]
- Shonn M. A., McCarroll R., Murray A. W. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science. 2000;289:300–303. doi: 10.1126/science.289.5477.300. [DOI] [PubMed] [Google Scholar]
- Smith K. N., Penkner A., Ohta K., Klein F., Nicolas A. B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis. Curr. Biol. 2001;11:88–97. doi: 10.1016/s0960-9822(01)00026-4. [DOI] [PubMed] [Google Scholar]
- Sourirajan A., Lichten M. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev. 2008;22:2627–2632. doi: 10.1101/gad.1711408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A., Xu L., Cao L., Kleckner N. Crossover and noncrossover recombination during meiosis: timing and pathway relationships. Proc. Natl. Acad. Sci. USA. 1995;92:8512–8516. doi: 10.1073/pnas.92.18.8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A. F., Belmont A. S., Robinett C. C., Murray A. W. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Strom L., Lindroos H. B., Shirahige K., Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Stuart D., Wittenberg C. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev. 1998;12:2698–2710. doi: 10.1101/gad.12.17.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I., Vorlaufer E., Stukenberg P. T., Kelm O., Redemann N., Nigg E. A., Peters J. M. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol. Cell. 2002;9:515–525. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- Sym M., Roeder G. S. Zip1-induced changes in synaptonemal complex structure and polycomplex assembly. J. Cell Biol. 1995;128:455–466. doi: 10.1083/jcb.128.4.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., Brown A., Oliver S. G., Greenwell P., Petes T. D. Genetic analysis of a meiotic recombination hotspot on chromosome III of Saccharomyces cerevisiae. Genetics. 1991;128:717–727. doi: 10.1093/genetics/128.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A., Rabitsch K. P., Galova M., Schleiffer A., Buonomo S. B., Nasmyth K. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- Uhlmann F. Chromosome cohesion and separation: from men and molecules. Curr. Biol. 2003;13:R104–114. doi: 10.1016/s0960-9822(03)00039-3. [DOI] [PubMed] [Google Scholar]
- Unal E., Arbel-Eden A., Sattler U., Shroff R., Lichten M., Haber J. E., Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Visintin R., Craig K., Hwang E. S., Prinz S., Tyers M., Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Wan L., de los Santos T., Zhang C., Shokat K., Hollingsworth N. M. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic double strand break repair in budding yeast. Mol. Biol. Cell. 2004;15:11–23. doi: 10.1091/mbc.E03-07-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Weiner B. M., Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- Whitby M. C. Making crossovers during meiosis. Biochem. Soc. Trans. 2005;33:1451–1455. doi: 10.1042/BST0331451. [DOI] [PubMed] [Google Scholar]
- Wu H. Y., Burgess S. M. Two distinct surveillance mechanisms monitor meiotic chromosome metabolism in budding yeast. Curr. Biol. 2006;16:2473–2479. doi: 10.1016/j.cub.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Beasley M. D., Warren W. D., van der Horst G. T., McKay M. J. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev. Cell. 2005;8:949–961. doi: 10.1016/j.devcel.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Xu L., Ajimura M., Padmore R., Klein C., Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:6572–6581. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Weiner B. M., Kleckner N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.