Abstract
Human Bre1, an E3 ligase for H2B monoubiquitination, binds p53 and enhances activator-dependent transcription. Ebp1, an ErbB3 receptor-binding protein, inhibits cell proliferation and acts as a tumor suppressor. Here, we show that hBre1 acts as an E3 ubiquitin ligase for Ebp1 tumor suppressor and promotes its polyubiquitination and degradation. Ebp1 is polyubiquitinated in cancer cells, which is regulated by its phosphorylation. We identified hBre1 acting as an E3 ligase for Ebp1 and increasing its polyubiquitination. Depletion of hBre1 blocks Ebp1's polyubiquitination and elevates its protein level, preventing cancer proliferation. hBre1 binds Ebp1 and suppresses its repressive effect on E2F-1. Moreover, Ebp1 protein level is substantially diminished in human cancers. It is robustly phosphorylated and localized in the nucleus of primary gliomas, correlating with hBre1 subcellular residency. Thus, hBre1 inhibits Ebp1's tumor suppressive activity through mediating its polyubiquitination and degradation.
INTRODUCTION
In yeast, histone H2B K123 monoubiquitination is regulated by the E3 ligase Bre1 and E2-conjugating enzyme RAD6 (Robzyk et al., 2000; Hwang et al., 2003; Wood et al., 2003a). This process is a prerequisite for the subsequent histone H3-K4 and K79 methylation (Sun and Allis, 2002; Wood et al., 2003b), which marks actively transcribed genes (Santos-Rosa et al., 2002). Additionally, PAF complex is also required for H2B ubiquitination (Ng et al., 2003; Wood et al., 2003b). In humans, the homologue of yeast Bre1 (RNF20), designated as hBre1, acts as an E3 ligase for H2B and promotes its ubiquitination at K120, the equivalent of yeast H2B K123 (Kim et al., 2005; Zhu et al., 2005). Moreover, it has a coactivator function in activator-dependent transcription of several genes. Interestingly, hBre1 directly binds p53 and is recruited to the MDM2 promoter, regulating p53-responsive gene transcription in a p53-dependent manner. Overexpression and depletion of hBre1 increases and decreases global H2B ubiquitination, respectively. By contrast, ectopic hRAD6A and hRAD6B, human homologues of yRAD6, do not affect H2B ubiquitination or H3 methylation, suggesting that hRAD6 proteins are not cognate E2 enzymes for hBre1 (Kim et al., 2005). In addition to transcriptional coactivation of specific genes (Osley, 2004), Bre1 in Drosophila is required for Notch signaling and histone modification (Bray et al., 2005).
Ebp1 was originally identified as an ErbB3 receptor-binding protein (Yoo et al., 2000), and it is the human homologue of a previously identified cell cycle-regulated mouse protein p38-2G4 (Radomski and Jost, 1995). PA2G4 gene encodes two Ebp1 isoforms, p48 and p42, which differentially regulate PC12 cell survival and differentiation (Ahn et al., 2006; Liu et al., 2006). P48 is 54 amino acids longer than p42 at its N terminus. The longer form p48 localizes in both the cytoplasm and the nucleolus and suppresses apoptosis, whereas the shorter form p42 predominantly resides in the cytoplasm and promotes cell differentiation (Liu et al., 2006). Ebp1 binds the ribosome and double-stranded RNA, and it is also a component of cytoplasmic bcl-2 messenger ribonucleoprotein (Squatrito et al., 2004; Bose et al., 2006). Recently, we showed that Ebp1 p48 associates with nuclear Akt and prevents apoptosis, which is mediated by protein kinase C (PKC)-δ-triggered phosphorylation on S360 (Ahn et al., 2006). Ebp1's growth regulatory activity has also recently been demonstrated in plants. Elevating or decreasing stEBP1 (homologue to mammalian p48) levels in transgenic plants results in a dose-dependent increase or reduction in organ growth, respectively. During early stages of organ development, stEBP1 promotes cell proliferation, influences cell size threshold for division, and shortens the period of meristematic activity. In postmitotic cells, it enhances cell expansion (Horvath et al., 2006). In contrast, tamoxifen treatment of MCF-7 human breast cancer cells substantially decreases both Ebp1 p48 transcription and protein levels. Patients with breast cancer expressing high levels of PA2G4 have poor clinical outcomes, suggesting it may promote aggressive behavior (Ou et al., 2006). Treatment of serum-starved human breast cancer cells with the ErbB3/4 ligand heregulin (HRG) induced translocation of Ebp1 p42 from the cytoplasm to the nucleus (Lessor et al., 2000). The regulated nuclear accumulation of Ebp1 suggested that Ebp1 might act as a transcription factor or transcriptional coregulator. Indeed, Ebp1 represses transcription of both E2F1 (Xia et al., 2001) and androgen receptor-regulated genes (Zhang et al., 2005a; Zhang and Hamburger, 2005). Ebp1 binds to the E2F-1 promoter in a complex with the E2F-1 transcription factor and retinoblastoma (Rb) (Zhang et al., 2002). Moreover, the activity of both exogenous and endogenous androgen receptor-regulated promoters was inhibited by ectopic expression of Ebp1 independent of prostate cell type (Zhang et al., 2005a). Ebp1 contains an autonomous C-terminal transcriptional repression domain that binds histone deacetylase 2 (Zhang et al., 2003). It strongly suppresses both androgen receptor-mediated gene transcription and tumorigenesis of prostate cancer cells and salivary adenoid carcinoma cell metastasis in mice (Zhang et al., 2005b; Akinmade et al., 2007; Yu et al., 2007). Collectively, these observations suggest that Ebp1 p42 acts as a potent tumor suppressor in various human cancers, whereas p48 might function as an oncogene, promoting cell survival and proliferation. In this report, we show that Ebp1 p42 but not p48 isoform is polyubiquitinated that is regulated by S360 phosphorylation. Moreover, hBre1 acts as an E3 ligase for Ebp1 p42 and accelerates its polyubiquitination, resulting in its degradation in human cancers.
MATERIALS AND METHODS
Cells and Reagents
Anti-FLAG monoclonal antibody (mAb) (M2), anti-hemagglutinin (HA)-horseradish peroxidase (HRP), anti-Myc, and anti-glutathione transferase (GST)-HRP monoclonal antibodies were from Sigma-Aldrich (St. Louis, MO); anti-green fluorescent protein (GFP) mAb was from Roche Diagnostics (Indianapolis, IN). Anti-Ebp1 N was raised against the N-terminal 23 residues of Ebp1 p48 and purified via affinity column. Anti-Ebp1 C antibody was from Millipore (Billerica, MA). Anti-phospho-S360 has been described previously (Ahn et al., 2006). Primer sequences for reverse transcription-polymerase chain reaction were as follows: p42, 5′ primer, GTGACCAAGTATAAGATG and 3′ primer, CTTCAAAGGGGAGAAGTG; and p48, 5′ primer, GTCGTGACCAAGGGTACTTC, and the 3′ primer is the same as p42. Human primary gliomas and normal brain samples were kindly supplied by Dr. Charlie Hao (Emory University). All chemicals were from Sigma-Aldrich.
In Vitro Polyubiquitination of p42 Ebp1
The reaction was carried out at 30°C for 1 h in a 15-μl reaction buffer (40 mM Tris-HCl, pH 7.5, 2 mM dithiothreitol, and 5 mM MgCl2) containing the following components: 10 μg of ubiquitin (U-100; Boston Biochem, Cambridge, MA), 20 nM human E1 (E302; Boston Biochem), 500 nM recombinant UbcH7, 2 μM ubiquitin aldehyde (U-201; Boston Biochem), 5 mM ATP, and 150 ng of GST-p42. HeLa S-100 extracts were added as E3 source as indicated in individual experiments. The reaction was terminated by adding 0.5 ml of pull-down buffer (20 mM Tris-HCl, pH 7.5, 500 mM NaCl, 1% Triton X-100, 0.02% bovine serum albumin, and 5 mM β-mercaptoethanol). After addition of 20 μl of glutathione-Sepharose, the samples were rotated at room temperature for 60 min. After washing, the proteins bound to beads were released by boiling in 30 μl of 2× SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer for 5 min. The samples were then resolved by 10% SDS-PAGE followed by immunoblotting with anti-ubiquitin antibody.
Coimmunoprecipitation and In Vitro Binding Assay
Ten-centimeter dishes of human embryonic kidney (HEK) 293 cells were cotransfected with 5 μg of GST-p42 and p48 constructs and 5 μg of HA-hBre1 (Nova Factor, Memphis, TN). In 24 h, the transfected cells were washed once in phosphate-buffered saline (PBS), lysed in 1 ml of lysis buffer (50 mM Tris, pH 7.4, 40 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 1.5 mM Na3VO4, 50 mM NaF, 10 mM sodium pyrophosphate, 10 mM sodium β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, 5 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin A), and centrifuged for 10 min at 14,000 × g at 4°C. After normalizing the protein concentration, 40 μl of 50% slurry glutathione beads were added to the supernatant and incubated with rotation at 4°C for 2 h. The pellet was washed three times with 1000 μl of lysis buffer each time and then resuspended in 30 μl of sample buffer. The coprecipitated proteins were separated by SDS-PAGE, followed by immunoblotting using anti-HA antibody. For in vitro binding, purified GST-recombinant proteins were incubated the lysates from HEK293 cells, transfected with indicated constructs. The following steps are exactly as described previously (Ye et al., 2000).
Immunocytochemistry
Cells were fixed with either methanol at −20°C for 20 min or 10% Formalin at room temperature for 20 min. The fixed cells were blocked by 10% normal goat serum in PBS for 1 h and incubated with primary antibodies for 1 h. Cells were then incubated with secondary antibodies (Alexa Fluor 594-tagged goat anti-mouse immunoglobulin G [IgG], Alexa Fluor 488-tagged goat anti-rabbit IgG, or Alexa Fluor 594-tagged goat anti-mouse IgG antibodies; Invitrogen, Carlsbad, CA) for 1 h, and counterstained for DNA with 4′, 6-diamidino-2-phenylindole. After incubation with antibodies, cells were washed extensively in PBS. Cells were examined under a fluorescence microscope.
5-Bromo-2′-Deoxyuridine (BrdU) Incorporation Assay
Cells were seeded into six-well plates at 1 × 105 cells/well, cultured overnight, and transfected with control or hBre1 small interfering RNA (siRNA). The transfected cells were pulse labeled with 10 μM BrdU (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). The incorporation was continued for 10 min. After washing with cold PBS, the cells were fixed with 4% paraformaldehyde for 15 min and treated with 2 M HCl at 37°C for 30 min. The coverslips were blocked with 2% fetal bovine serum/PBS, and 0.4% Triton X-100 at room temperature for 10 min. For immunostaining, mouse mAb against BrdU (1:200) and anti-GST-fluorescein isothiocyanate-conjugated antibody were used.
Immunohistochemistry Analysis of Human Primary Gliomas
Immunohistochemistry was performed on Formalin-fixed, paraffin-embedded human glioblastoma specimens (n = 9; thickness, 10 μm) for anti-Ebp1-N (1:100 dilution), anti-Ebp1-C (1:100 dilution), anti-phospho-S360 (1:100 dilution), and anti-hBre1 (1:200 dilution). Glioblastoma specimens contained infiltrative neoplastic cells of variable density as well as adjacent nonneoplastic white matter and cortex. The staining procedures were performed using the protocols recommended by the manufacturers.
RESULTS
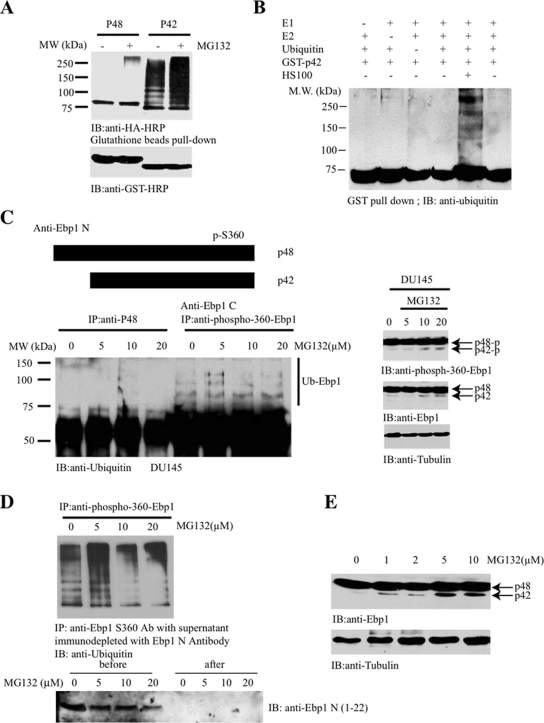
Ebp1 p42 but Not p48 Isoform Is Ubiquitinated
Tumor suppressors are frequently degraded in cancer cells through polyubiquitination-mediated mechanism, including p53 (Maki et al., 1996), PTEN (Trotman et al., 2007; Wang et al., 2007), and merlin (Tang et al., 2007). To investigate whether Ebp1 can also be modified by polyubiquitination, we cotransfected HA-ubiquitin into HEK293T cells with GST-tagged p42 and p48, followed by a proteasome inhibitor MG132 treatment for 4 h. GST-pull-down assay revealed that p42 but not p48 was potently polyubiquitinated, which was enhanced by MG132 treatment (Figure 1A). To investigate whether p42 can be polyubiquitinated in vitro, we developed a biochemical assay. We used recombinant GST-p42 as a substrate. After incubating with recombinant ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme UbcH7 (E2), ubiquitin, ATP, and HeLa cellular extract (S-100), we found that p42 was polyubiquitinated in the presence of S-100 but not control buffer (Figure 1B), indicating that S-100 contains an E3 ligase activity for p42. To explore whether this modification occurs to endogenous proteins, we treated the prostate cancer cell line DU145 with various concentrations of MG132. The endogenous Ebp1 was then immunoprecipitated with either a p48-specific antibody or phospho-S360–specific antibody, which recognizes both of the phosphorylated isoforms. The coprecipitated proteins were analyzed with anti-ubiquitin antibody. p48 was barely ubiquitinated regardless of MG132 treatment; in contrast, the polyubiquitination of phosphorylated Ebp1 was increased after MG132 treatment (Figure 1C, left), suggesting that phosphorylated p42 is selectively polyubiquitinated. Ebp1 p48 phosphorylation was not increased after MG132 treatment, whereas p42 phosphorylation was increased, although p42 total expression was faintly detectable in the cancer cells (Figure 1C, right). Immunoprecipitation with anti-Ebp1-C antibody, which recognizes both p42 and p48 proteins, also displayed evident polyubiquitination upon MG132 treatment (data not shown). To ascertain that p48 was not implicated in anti-phospho-S360 immunocomplex, we immunodepleted p48 from cellular extract with anti-Ebp1 N antibody, which selectively recognizes the N terminus of p48. The resulting supernatant was immunoprecipitated with phospho-S360 antibody, and immunoblotting revealed an MG132-dependent polyubiquitination (Figure 1D), underscoring that p42 but not p48 was polyubiquitinated in cancer cells. Similar results were obtained from experiments on other human cancer cells (data not shown). MG132 titration revealed that p42 protein levels increased in a dose-dependent manner (Figure 1E). Thus, these data support that Ebp1 p42 but not p48 can be selectively polyubiquitinated in human cancer cells.
Figure 1.
Ebp1 p42 but not p48 isoform can be polyubiquitinated. (A) Ebp1 p42 but not p48 isoform can be ubiquitinated. GST-p42 and p48 were transfected into HEK293 cells with HA-ubiquitin, followed by treatment with 10 μM MG132 for 4 h. Ebp1 was pulled down with glutathione beads and analyzed with anti-HA antibody (top). Equal level of p42 and p48 was expressed in transfected cells (bottom). (B) EBP1 p42 can be ubiquitinated in vitro. The in vitro assay was performed as described in Materials and Methods with individual components added as indicated. GST-p42 was the substrate and HeLa cells S-100 cellular extract 10 μg was used for the E3 ubiquitin ligases. (C) Endogenous p42 can be ubiquitinated in cancer cells. DU145 prostate cancer cells were treated with different doses of MG132, and Ebp1 was immunoprecipitated by anti-p48 or anti-phospho S360 antibody and analyzed with anti-ubiquitin antibody. Phospho-S360 antibody but not p48-specific antibody pulled down polyubiquitinated Ebp1 p42 (left). MG132 slightly enhanced p42 phosphorylation (right). (D) Immunodepletion of p48 does not affect phospho-S360 immunocomplex ubiquitination. Cell lysates from DU145 cells, treated with MG132, were immunodepleted with anti-p48 antibody. The resulting supernatant was further immunoprecipitated with anti-phospho S360. The immunocomplex was analyzed by immunoblotting with Ubiquitin antibody (top). P48 was selectively depleted from the cellular extracts (bottom). (E) MG132 treatment enhances p42 protein levels in DU145 cells in a dose-dependent manner.
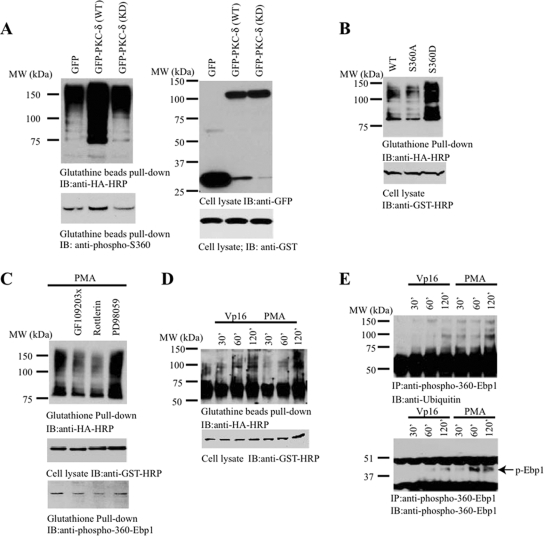
Ebp1 S360 Phosphorylation Enhances Its Polyubiquitination
Ubiquitination is often regulated by substrate protein phosphorylation (Gao and Karin, 2005). We have previously shown that PKC-δ phosphorylates Ebp1 S360 and regulates its nucleolar localization (Liu et al., 2006). To determine whether Ebp1 S360 phosphorylation plays any role in mediating its ubiquitination, we cotransfected GST-p42 and HA-ubiquitin into HEK293 cells with wild-type or kinase-dead PKC-δ. Compared with control and kinase-dead PKC-δ–transfected cells, GFP-PKC-δ substantially enhanced p42 polyubiquitination (Figure 2A, top). P42 S360 phosphorylation activity correlated with its polyubiquitination effect (Figure 2A, bottom). To further explore whether p42 phosphorylation plays any role in its ubiquitination, we used a phosphorylation mimetic mutant S360D and a nonphosphorylate mutant S360A. Compared with wild-type p42 and S360A, p42 S360D was strongly polyubiquitinated (Figure 2B). Using PKC activator phorbol 12-myristate 13-acetate (PMA) and various pharmacologic inhibitory agents, we found that the PKC inhibitor GF109203X and rotterlin but not mitogen-activated protein kinase kinase (MEK)1 inhibitor PD98059 significantly reduced PMA-triggered p42 polyubiquitination (Figure 2C, top). p42 S360 phosphorylation correlated with its polyubiquitination levels (Figure 2C, bottom); supporting the conclusion that p42 phosphorylation by PKC mediates its polyubiquitination. The genotoxic agent VP16 activates PKC-δ and triggers its nuclear translocation before caspase-3 cleavage (Blass et al., 2002). We found that VP16 and PMA treatment also increased p42 polyubiquitination in transfected HEK293 cells in a time-dependent manner (Figure 2D). We made similar observations with endogenous p42 Ebp1 in human cancer cells. p42 ubiquitination correlated with its S360 phosphorylation status (Figure 2E). Hence, Ebp1 S360 phosphorylation regulates p42 polyubiquitination.
Figure 2.
Ebp1 phosphorylation mediates its ubiquitination. (A) PKC-δ enhances p42 ubiquitination. Wild-type or kinase-dead GFP-PKC-δ was cotransfected with GST-p42 and HA-ubiquitin. Transfected p42 was pulled down with glutathione beads and analyzed by immunoblotting with ubiquitin antibody. PKC-δ wild type (wt) enhanced p42 ubiquitination (top left). S360 phosphorylation of GST-p42 in transfected cells was monitored by immunoblotting (bottom left). Similar expression level of transfected construct was confirmed (right). (B) Ebp1 phosphorylation mimetic mutant is potently ubiquitinated. HA-ubiquitin was cotransfected with various GST-p42 constructs. Ebp1 was pulled down with glutathione beads and analyzed by HA antibody. Compared with wild-type p42, S360D, a phosphorylation mimetic mutant, was strongly ubiquitinated. Unphosphorylate mutant S360A displayed a reduced activity (top). Equal level of Ebp1 proteins was expressed (bottom). (C) PKC inhibitors blocked PMA-triggered ubiquitination. GST-p42 and HA-ubiquitin cotransfected cells were pretreated with PKC inhibitor 10 μM GF109203X or 60 nM rotterlin or 10 μM MEK1 inhibitor PD98059, followed by 10 nM PMA for 30 min. PKC inhibitors decreased Ebp1 ubiquitination (top). Verification of transfected constructs and Ebp1 phosphorylation status (middle and bottom). (D and E) VP16 and PMA promote Ebp1 ubiquitination in HEK293 cells and cancer cells. GST-p42 was cotransfected with HA-ubiquitin into HEK293 cells, followed by VP16 or PMA treatment for various times. Ebp1 was pulled down with glutathione beads and analyzed with anti-HA-HRP. Both VP16 and PMA stimulated p42 ubiquitination (D, top). Verification of transfected GST-p42 (D, bottom). DU145 cells were treated with VP16 or PMA and endogenous p42 was immunoprecipitated by anti-phospho-S360 antibody and analyzed by anti-ubiquitin (E, top). Verification of immunoprecipitated Ebp1 phosphorylation (E, bottom).
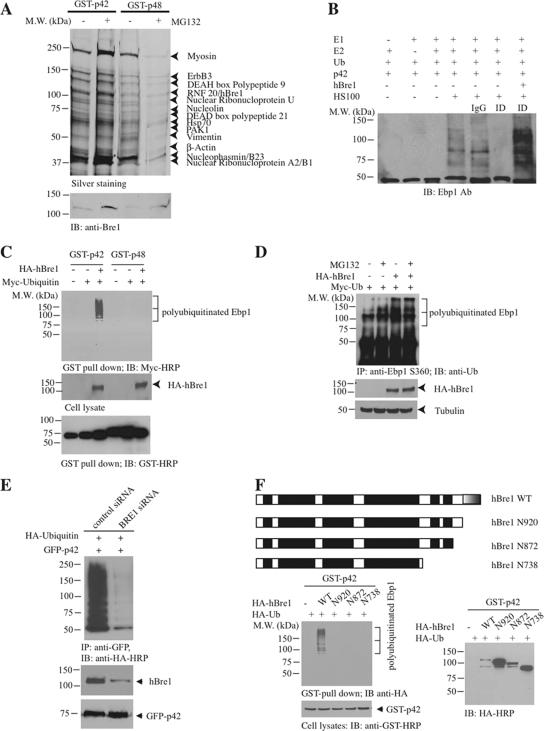
Human Bre1 Is an E3 Ubiquitin Ligase for p42
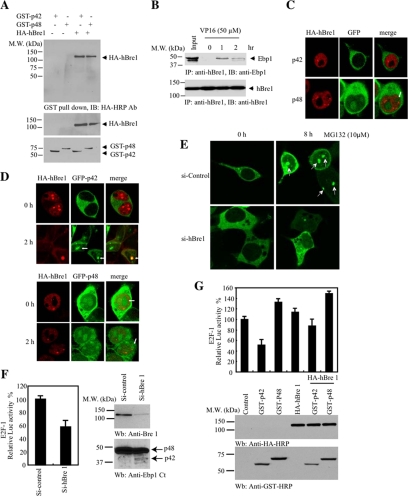
In most eukaryotes, there is only one E1 for ubiquitin, a considerable but limited number of E2s, and a much larger number of E3s (Pickart, 2001; Gao and Karin, 2005). In the polyubiquitination process, the E3s serve the critical role of selecting and targeting specific substrate for ubiquitin conjugation (Hershko and Ciechanover, 1998). To search for p42's E3 ubiquitin ligase, we conducted a GST-pull-down assay with the cellular extract from DU145 cells treated with or without MG132. The bead-bound proteins were eluted and resolved on SDS-PAGE, followed by silver staining. MG132 treatment enhanced more proteins strongly associated with GST-p42 than GST-p48. The prominent protein bands were subjected to proteomic analysis (Figure 3A). As expected, we identified nucleophasmin (NPM)/B23, which confirmed our previous finding that Ebp1 associates with B23 in the nucleolus (Okada et al., 2007). In addition, we identified ribonucleoproteins, verifying previous report (Squatrito et al., 2004). Among these binding partners, we also identified a Ring-finger containing protein hBre1, which serves as an E3 ubiquitin ligase for H2B (Kim et al., 2005). MG132 treatment provoked a prominent binding by RNF20/hBre1 to both p42 and p48 (Figure 3A, bottom). To assess whether hBre1 possesses an E3 ligase activity to p42, we conducted an in vitro ubiquitination assay using HeLa S-100 cellular extract. Compared with control IgG, immunodepletion of hBre1 selectively abolished S-100's E3 ligase activity. Adding back recombinant hBre1 restored p42 polyubiquitination, indicating that hBre1 is required for p42 polyubiquitination in vitro (Figure 3B). To explore whether hBre1 mediates p42 ubiquitination in intact cells, we cotransfected Myc-ubiquitin and HA-hBre1 into HEK293 cells with GST-p42 or GST-p48. GST-pull-down assays revealed that p42 but not p48 was selectively polyubiquitinated (Figure 3C, top), suggesting that hBre1 can facilitate p42 polyubiquitination in intact cells. Endogenous Ebp1 polyubiquitination was evidently increased by hBre1 regardless of MG132 treatment (Figure 3D, lanes 3 and 4). To ascertain that hBre1 is required for p42 polyubiquitination, we depleted endogenous hBre1 with siRNA. Knocking down of hBre1 substantially abolished p42 polyubiquitination (Figure 3E). Truncation assay revealed that the C-terminal Ring-finger domain was indispensable for hBre1's E3 ligase activity (Figure 3F). Together, these data demonstrate that hBre1 acts as an E3 ubiquitin ligase for p42 and stimulates its polyubiquitination.
Figure 3.
hBre1 is an E3 ubiquitin ligase for Ebp1 p42. (A) hBre1 is an Ebp1-binding protein. Recombinant GST-p42 and GST-p48 proteins were incubated with cellular extract from DU145 cells, treated with MG132. After extensive washing, Ebp1-bound proteins were eluted with 1 M NaCl and analyzed by silver staining after SDS-PAGE. The bands selectively bound to Ebp1 were identified by proteomic analysis. The protein identity was labeled on the right side of the SDS-PAGE. RNF20/hBre1 strongly bound to p42 upon MG132 treatment. (B) hBre1 is required for p42 polyubiquitination in vitro. In vitro ubiquitination assay was conducted as described in Materials and Methods. Immunodepletion of hBre1 with its specific antibody but not control IgG abolished S-100's E3 ligase activity. Adding back hBre1 (1 μg) restored the E3 ubiquitin ligase activity. (C) hBre1 enhances transfected p42 polyubiquitination in vivo. GST-p42 and p48 were cotransfected with Myc-ubiquitin into HEK293 cells in the presence or absence of hBre1. Ebp1 was pulled down with glutathione beads and analyzed by immunoblotting with anti-Myc antibody. hBre1 selectively enhanced p42 but not p48 polyubiquitination (top). Verification of transfected proteins (middle and bottom). (D) Human Bre1 promotes endogenous p42 polyubiquitination. HA-ubiquitin and hBre1 were cotransfected into HEK293 cells, treated with 10 μM MG132 for 4 h. Ebp1 was immunoprecipitated with anti-phospho-S360 and analyzed with anti-ubiquitin. MG132 treatment enhanced polyubiquitination, and the maximal effect occurred when both hBre1 and MG132 were used. (E) Knocking down of hBre1 inhibits p42 polyubiquitination. HEK293 cells were transfected with GFP-p42 and Myc-ubiquitin, followed by control siRNA or hBre1 siRNA transfection. Ablation of hBre1 blocked p42 polyubiquitination. (F) hBre1 that contains Ring-finger domain is required for its E3 ligase activity. Schematic diagram of hBre1. Ring finger domain at the C terminus is indicated by half shaded box. The black boxes are the predicted coiled-coil domain. Predicted coiled-coil domains are shown as black boxes (top). Deletion of the Ring-finger domain crippled hBre1's E3 ligase activity (bottom left). Verification of transfected proteins (bottom).
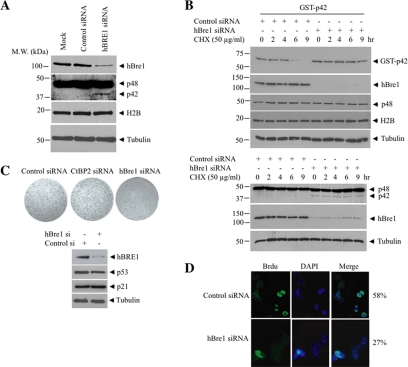
Human Bre1 Regulates p42 Expression and Cancer Cell Growth
To confirm whether hBre1 is a physiological E3 ligase for p42 and negatively regulates its protein levels, we transfected MCF7 cells with its siRNA. Depletion of hBre1 selectively enhanced p42 but not p48 expression. As expected, H2B expression level was not altered, fitting with a previous report that hBre1-promoted monoubiquitination does not elicit H2B degradation (Figure 4A). To explore whether hBre1 regulates p42 steady-state level, we transfected HEK293 cells with control siRNA or hBre1 siRNA. In 16 h, we transfected GST-p42 into these cells, followed by protein translation inhibitor cycloheximide for various times. Immunoblotting analysis revealed that cycloheximide blocked p42 expression in a time-dependent manner. Knocking down of hBre1 prevented p42 degradation, leading to markedly enhanced p42 protein levels (Figure 4B, top, top panel). In contrast, endogenous p48 expression levels were not altered at all. Similar observations were made with the endogenous p42 as well (Figure 4B, bottom). H2B protein levels also remained unchanged (Figure 4B, third and fourth top panels). Thus, hBre1 specifically and negatively regulates p42 expression. Because p42 acts as a tumor suppressor, it might be expected that up-regulation of p42 expression after ablation of hBre1 would inhibit cell proliferation. To explore this hypothesis, we transfected MCF7 cells with control or hBre1 siRNA. Ablation of hBre1 significantly reduced the growth rates of these cells (Figure 4C), suggesting that hBre1 inactivation inhibits cell proliferation. By monitoring BrdU incorporation, we again observed that hBre1 knockdown inhibited cell proliferation in both cell lines (Figure 4D). Thus, hBre1 is a physiological negative regulator of p42 in human cancer cells.
Figure 4.
hBre1 regulates p42 expression and cancer cell growth. (A) Depletion of hBre1 induces p42 expression. MCF7 cells were transfected with control or hBre1 siRNA. In 24 h, cell lysates were analyzed by immunoblotting with anti-hBre1 and anti-Ebp1 antibodies, respectively. Ablation of hBre1 elevated p42 but not H2B protein levels (top, second, and third panels). (B) Knocking down of hBre1 increases p42 steady-state level. HEK293 cells were transfected with control or hBre1 siRNA. In 16 h, the cells were transfected with GST-p42, followed by cycloheximide treatment for various times. Immunoblotting was conducted to monitor GST-p42, hBre1, and Ebp1 expression levels. Depletion of endogenous hBre1 increased p42 steady-state level. (C) Ablation of hBre1 blocks cancer cell proliferation. Endogenous hBre1 was knocked down in MCF7 with control or hBre1 siRNA. The cells were stained with crystal violet 3 d after siRNA treatment. (D) Elimination of hBre1 decreases BrdU incorporation. MCF7 cells were treated with control or hBre1 siRNA. The cells were labeled and stained 1 d after siRNA treatment.
Human Bre1 Interacts with Ebp1 and Inhibits Its Repressive Activity on E2F-1
To explore the interaction between Ebp1 and hBre1, we cotransfected GST-tagged p42 and p48 into HEK293 cells with HA-hBre1. GST-pull-down revealed that both isoforms associated with hBre1 (Figure 5A). To assess whether the interaction occurred between the endogenous proteins, we treated MCF7 cells with DNA damage agent VP16 for different time points, and immunoprecipitated hBre1. Immunoblotting revealed that hBre1 bound to Ebp1 in a time-dependent manner (Figure 5B), indicating that DNA damage regulates the association between Ebp1 and hBre1. To further investigate the interaction between these two proteins, we conducted immunofluorescent staining with HEK293 cells cotransfected with GFP-Ebp1 and HA-hBre1. As expected, GFP-p42 exclusively resided in the cytoplasm, whereas HA-hBre1 located in the nucleus, as indicated in previous reports (Andersen et al., 2005; Hinsby et al., 2006). By contrast, GFP-p48 distributed in both the cytoplasm and the nucleolus (Figure 5C). Surprisingly, VP16 treatment markedly provoked p42 nuclear translocation, colocalizing with hBre1 (Figure 5D, top). P48 remained in both the cytoplasm and the nucleolus irrespective of VP16 treatment (Figure 5D, bottom). Knocking down of hBre1 completely inhibited p42 nucleolar translocation (Figure 5E), demonstrating that endogenous hBre1 is required for this event. Collectively, these data support that Ebp1 colocalizes with hBre1 in the nucleolus, which can be regulated by the proteasome inhibitor MG132.
Figure 5.
hBre1 binds Ebp1 and regulates its repressive effect on E2F-1. (A) Both p42 and p48 bind hBre1. GST-p42 and p48 were transfected into HEK293 cells with or without HA-hBre1. Ebp1 was pulled down with glutathione beads, and coprecipitated proteins were analyzed by immunoblotting with anti-HA antibody. Both p42 and p48 bound to hBre1 (top). Verification of transfected proteins (middle and bottom). (B) VP16 provokes Ebp1 to bind hBre1. MCF7 cells were treated with VP16 for various times. Endogenous hBre1 was immunoprecipitated with agarose-conjugated antibody, and the coprecipitated proteins were analyzed by anti-Ebp1 antibody. VP16 treatment triggered hBre1 to bind Ebp1. (C) GFP-p42 resides in the cytoplasm, whereas GFP-p48 occurs in both the cytoplasm and the nucleolus. (D) VP16 triggers p42 nucleolar translocation. HEK293 cells were cotransfected with GFP-p42 and GFP-p48 and HA-hBre1, followed by treatment with 50 μM VP16 for 2 h. The transfected cells were stained with mouse monoclonal HA antibody. VP16 provoked GFP-p42 nucleolar translocation (white arrowhead) (top), whereas GFP-p48 remained in both the cytoplasm and the nucleolus regardless of VP16 treatment (bottom). (E) hBre1 is required for MG132 to provoke p42 nucleolar translocation. MG132 elicited p42 nucleolar translocation (top). Knocking down of hBre1 inhibited GFP-p42 nucleolar translocation (bottom). (F) Ablation of hBre1 decreases E2F1 promoter activity. MCF7 cells were transfected with E2F1-promoter-luciferase construct, followed by transfection with control or hBre1 siRNA. Elimination of hBre1 reduced E2F-1 promoter activity (left). Depletion of hBre1 enhanced p42 expression (right). (G) hBre1 decreases p42 expression and enhances E2F-1 promoter activity. Overexpression of hBre1 evidently augmented E2F-1 promoter activity in both p42 and p48 transfected cells (top). hBre1 overexpression decreased p42 but not p48 expression levels (compare lanes 2 and 3 with lanes 5 and 6, bottom).
Ebp1 binds to the E2F-1 promoter in a complex with the E2F-1 transcription factor and Rb and represses E2F-1's transcriptional activity (Zhang et al., 2002). To investigate whether hBre1 mediates p42's suppressive effect on E2F-1, we conducted a luciferase assay. Ablation of hBre1 decreased E2F-1 promoter activity, which correlated with the enhanced p42 expression in MCF7 cells (Figure 5F). As predicted, overexpression of p42 alone repressed E2F-1 activity; by contrast, p48 significantly elevated it compared with control. Cotransfection of hBre1 decreased p42 but not p48 expression level, fitting with its E3 ubiquitin ligase activity. In agreement with this observation, E2F-1 activity was markedly up-regulated in hBre1-cotransfected cells (Figure 5G), supporting that hBre1 promotes cell proliferation presumably through suppressing p42 expression.
Ebp1 p42 Tumor Suppressor Is Inhibited in Cancer Cells
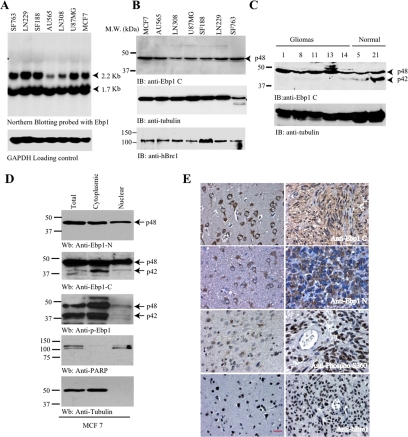
Ebp1 is ubiquitously expressed in all human tissues (Yoo et al., 2000). PA2G4 gene transcribes two mRNAs, which encode p48 and p42 isoforms. The expression levels of 1.7-kb mRNAs were comparable in human cancer cells, although 2.2 kb varied among the samples (Figure 6A). Surprisingly, p42 protein level was almost undetectable in the cancer cells, whereas p48 was strongly expressed. (Figure 6B, top), suggesting that p42 protein is unstable or degraded in human cancer cells. In contrast, hBre1 was demonstrable in all samples with various amounts (Figure 6B, bottom). To explore whether p42 is selectively degraded in malignant human cancers, we analyzed Ebp1 expression in primary gliomas and normal human brain tissues. Immunoblotting revealed that p42 was demonstrable in normal human brain samples, whereas it was barely detectable in malignant gliomas (Figure 6C). We have made the similar observations in normal human breast and lung tissue as well as cancer cells (data not shown). Thus, p42 is selectively degraded in human cancer cells. The malignant cells displayed normal p42 mRNA, but expressed surprisingly low to undetectable level of p42 protein, suggesting that hBre1 might contribute to the low p42 protein levels in these samples. To determine the subcellular distribution of p48 and p42 in malignant cancer cells, we conducted subcellular fractionation assay and used 3 different Ebp1 antibodies: anti-Ebp1-N (specific for p48), anti-Ebp1-C (detects both p48 and p42) and anti-phospho-S360, which recognizes both p48 and p42 phosphorylated forms. P48, which was recognized by Ebp1 N antibody, occurred in both the cytoplasm and the nucleus (Figure 6D, top). Modest p42 was detected in total lysate and distributed in both the cytosolic and nuclear fractions, but it was significantly enriched in the cytosolic fraction (Figure 6D, second panel). Immunoblotting with anti-phospho-S360 demonstrated that both p48 and p42 were strongly phosphorylated and predominantly occurred in the cytosolic fraction with negligible amount in the nuclear fraction (Figure 6D, third panel). We made the similar observation with other human glioblastoma cells (data not shown).
Figure 6.
Ebp1 p42 but not p48 is lost in human cancer cells through ubiquitination. (A) Northern blotting of Ebp1 in human cancer cell lines. Ebp1 encoded two mRNAs in various human cancer cell lines. (B) Ebp1 p42 expression is undetectable in various human cancer cells. Immunoblotting analysis of various human cancer cells with anti-Ebp1 antibody, which recognizes the C termini of both p48 and p42. Only p48 was detected (top). Similar levels of hBre1 and H2B were detected in all cells (third and bottom panels). (C) p42 is demonstrable in normal human brain samples, but it is degraded in human primary gliomas. The number on the top of the gel is the sample ID. (D) Subcellular fractionation of MCF7 cells. Three Ebp1 antibodies were characterized by immunoblotting with subcellular fractionated MCF7 cells (top to third panel). Identities if nuclear and cytosolic fractions were confirmed (fourth and bottom panels). (E) Immunohistochemistry on human primary gliomas. Both anti-Ebp1 N (specific for p48) and anti-Ebp1 C (recognizing both p42 and p48) revealed robust cytoplasmic staining and faint nuclear staining in neurons and astrocytes (left top and second panels). Anti-Ebp1 C staining displayed in both the cytoplasm and the nucleus in gliomas, whereas anti-Ebp1 N exclusively stained the cytoplasm of the neoplastic cells (right top and second panels). Normal neurons and astrocytes showed faint Ebp1 S360 phosphorylation. In contrast, glioblastoma displayed strong nuclear staining by anti-phospho-S360 (third panel). hBre1 specifically distributed in the nucleus of all cells (bottom). Bar, 10 μm.
To explore whether hBre1 is relevant physiologically to human cancers, we conducted immunohistochemistry on a panel of human gliomas with (Figure 6E). Glioblastoma specimens contained infiltrative neoplastic cells of variable density as well as adjacent nonneoplastic white matter and cortex. The normal cerebral cortex showed strong staining for anti-Ebp1-C within the cytoplasm of cortical neurons, with only minimal staining of normal astrocytes, oligodendrocytes and neuropil (Figure 6E, top left). Staining for anti-Ebp1 N showed a similar pattern, but the intensity of immunoreactivity was lower, especially in cortical neurons (Figure 6E, left, second panel). The cellular components of the normal cortex and white matter showed very little nuclear or cytoplasmic staining for phospho-S360. Only minimal cytoplasmic staining was seen in pyramidal neuron and astrocyte cytoplasm. Nuclear staining of phospho-S360 was absent (Figure 6E, left, third panel). Tumor cells showed moderate staining for anti-Ebp1-C within the cytoplasm and weak nuclear staining (Figure 6E, top, right). The neoplastic cells of glioblastoma showed strong immunoreactivity for anti-Ebp1-N within the cytoplasm but did not show any nuclear staining (Figure 6E, right, second panel). Glioblastoma cells showed intense nuclear staining for phospho-S360 in nearly all cells without any cytoplasmic staining. The strong staining of neoplastic nuclei for anti-phospho-S360 was highlighted when contrasted with the negative staining of endothelial cell lining tumoral blood vessels and cortical neurons that were infiltrated by neoplastic cells (Figure 6E, right, third panel). Immunohistochemistry for hBre1 in normal cerebral cortex showed exclusive nuclear staining in neurons, astrocytes, oligodendrocytes, and endothelial cells. In human glioblastoma cells, tumor cell nuclei were uniformly intensely immunoreactive, with little variation based on tumor cell size or tumor microenvironment (Figure 6E, bottom). Thus, hBre1 colocalizes with phospho-Ebp1 in the nucleus of primary human gliomas, fitting with its E3 ligase activity for promoting phosphorylated Ebp1 p42 polyubiquitination in cancers.
DISCUSSION
Previous studies establish that Bre1 is a H2B-specific E3 ligase, which regulates its K120 (K123 in yeast) monoubiquitination, a cotranscriptional event mediating histone H3 methylation at K4 and 79 (Hwang et al., 2003; Wood et al., 2003a). In this report, we identified hBre1 as a physiological E3 ubiquitin ligase for p42 Ebp1, which promoted p42 polyubiquitination and degradation. In contrast, ablation of hBre1 by its siRNA substantially elevated p42 expression level in human cancer cells. In agreement with this observation, depletion of hBre1 blocked MCF7 cell proliferation, supporting the growth suppressive role of p42. Moreover, we showed that polyubiquitination of p42 was up-regulated by its phosphorylation. Interestingly, we found that DNA damage agent VP16 provoked p42 association with hBre1 and nucleolar translocation, for which endogenous hBre1 is required. Collectively, our results demonstrate that hBre1 acts as a negative regulator of p42. Genomic DNA sequence analysis revealed numerous p53 binding motifs in the promoter region of PA2G4 (data not shown), indicating that p53 might regulate p42 transcription. It has been shown before that hBre1 directly interacts with p53 and that it is recruited to the mdm promoter in a p53-dependent manner. Through direct activator interactions, hBre1 functions as a transcriptional coactivator (Kim et al., 2005). Presumably, hBre1 negatively controls p42 expression levels by two means: 1) Interacting with p53 in the nucleoplasm, blocking p42 transcription; and 2) increasing p42 polyubiquitination in the nucleolus and triggering its proteasomal degradation. However, in the nucleoplasm, hBre1 selectively provokes H2B monoubiquitination and binds p53 and mediates gene transcription. At this point, the mechanism dictating hBre1's coactivating or inhibitory effect on p53 has not been defined yet.
Most recently, ARF-BP1, a key factor associated with ARF in vivo, has been identified as an ubiquitin ligase (Chen et al., 2005; Zhong et al., 2005). It acts as a critical mediator of both the p53-independent and p53-depedent tumor suppressor functions of ADP-ribosylation factor (ARF). We found that ARF-BP1 robustly bound p42 and provoked its polyubiquitination (Supplemental Figure 1A); however, depletion of ARF-BP1 failed to trigger p42 expression in human cancer cells (Supplemental Figure 1C), indicating that ARF-BP1 might not act as a physiological E3 ubiquitin ligase for p42. Alternatively, p42 might have numerous E3 ubiquitin ligases, and eliminating a minor E3 ligase, ARF-BP1, is not sufficient to restore p42 expression level.
The retinoblastoma protein (RB) and p53 transcription factor are regulated by two distinct proteins that are encoded by the INK4a/ARF locus. Genes encoding these four tumor suppressors are disrupted, either in whole or in part, in most human cancers (Sherr, 2001). RB restricts cell cycle progression by binding to and restraining E2Fs, which, when untethered from RB, promote S phase entry. INK4 proteins release Cip/Kip proteins from cyclin D–cyclin-dependent kinase (CDK) complexes and result in CDK2 inhibition, leading to RB activation in its hypophosphorylated state. Interestingly, Ebp1 has been shown before to associate with Rb, and up-regulate its transcription suppressive activity on E2F-1. Dephosphorylation of Ebp1 enhances its interaction with Rb (Xia et al., 2001). Conceivably, Ebp1 phosphorylation decreases its association with Rb, leading to E2F-1 activation and cell proliferation. This notion is also consistent with our observation that Ebp1 phosphorylation stimulates its ubiquitination and degradation (Figure 2), resulting in loss of its tumor-suppressive activity. Human Bre1 binds Ebp1 and inhibits its repressive effect on E2F-1 (Figure 5). Consistently, depletion of hBre1 enhances p42 expression and decreases human cancer cell growth (Figure 4).
p53 tumor suppressor plays an essential role in monitoring genomic integrity in response to cellular stress. p53 exerts its antiproliferative functions through regulating a variety of gene transcription, resulting in cell cycle arrest, apoptosis, and cellular senescence. Nucleolar ARF stabilizes nucleoplasmic p53 through binding to MDM2 and sequestering it in the nucleolus (Weber et al., 1999). Nonetheless, nucleoplasmic ARF activates p53 function by directly inhibiting the ubiquitin ligase activity of MDM2 (Midgley et al., 2000; Lin and Lowe, 2001; Llanos et al., 2001). The extent of p53 ubiquitination under different cellular conditions is determined not only by changes in MDM2 and ARF-BP1 activities but also by phosphorylation of the substrate p53 (Gao and Karin, 2005). Phosphorylation of p53 also reduces its affinity to MDM2, thereby inhibiting p53 ubiquitination and degradation (Mayo et al., 1997). Ebp1 can be potently phosphorylated by PKC-δ, which is activated by VP16 or PMA. In human cancer cells, phospho-Ebp1 ubiquitination was increased upon genotoxic VP16 and PMA treatment. In addition, we show that wild-type PKC-δ but not kinase-dead mutant stimulated Ebp1 phosphorylation and ubiquitination (Figure 2). In alignment with this notion, the ubiquitination on unphosphorylate p42 S360A was substantially impaired compared with wild-type or phosphorylation mimetic S360D mutant. These results demonstrate that p42 S360 phosphorylation by PKC-δ is required for its ubiquitination (Figure 2).
The levels of histone H2B-K120 monoubiquitination regulate Hox gene expression, which is required for proper development. Overexpression of HOX genes is associated with acute myeloid leukemia and promotes leukemogenesis (Scholl et al., 2007). Overexpression of hBre1 triggers robust up-regulation of many Hox genes, whereas RNA interference knocking down of hBre1 specifically down-regulates the same set of Hox genes (Zhu et al., 2005). This report suggests that hBre1 possesses proto-oncogenic activity, which fits with our observation that depletion of hBre1 suppresses MCF7 cell proliferation by enhancing p42 tumor suppressor expression, leading to down-regulation of E2F-1 promoter activity (Figure 5). Presumably, in addition to facilitation of p42 expression, repression of HOX gene expression by knocking down of hBre1 might also contribute to cell growth-suppressive effect. Together, these findings disclose a novel molecular mechanism by hBre1 to negatively regulate p42 expression in cancer cells by acting as an E3 ubiquitin ligase.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Robert Roeder at Rockefeller University for Flag-H2B and hBre1 constructs. E1 and E2 constructs were from Dr. Shigetsugu Hatakeyama (Hokkaido University, Japan). This work is supported by National Institute of Health grant R01, NS-045627 (to K.Y.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-09-0983) on November 26, 2008.
REFERENCES
- Ahn J. Y., Liu X., Liu Z., Pereira L., Cheng D., Peng J., Wade P. A., Hamburger A. W., Ye K. Nuclear Akt associates with PKC-phosphorylated Ebp1, preventing DNA fragmentation by inhibition of caspase-activated DNase. EMBO J. 2006;25:2083–2095. doi: 10.1038/sj.emboj.7601111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinmade D., Lee M., Zhang Y., Hamburger A. W. Ebp1-mediated inhibition of cell growth requires serine 363 phosphorylation. Int. J. Oncol. 2007;31:851–858. [PubMed] [Google Scholar]
- Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Blass M., Kronfeld I., Kazimirsky G., Blumberg P. M., Brodie C. Tyrosine phosphorylation of protein kinase Cdelta is essential for its apoptotic effect in response to etoposide. Mol. Cell. Biol. 2002;22:182–195. doi: 10.1128/MCB.22.1.182-195.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S. K., Sengupta T. K., Bandyopadhyay S., Spicer E. K. Identification of Ebp1 as a component of cytoplasmic bcl-2 mRNP complexes. Biochem. J. 2006;396:99–107. doi: 10.1042/BJ20051548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S., Musisi H., Bienz M. Bre1 is required for Notch signaling and histone modification. Dev. Cell. 2005;8:279–286. doi: 10.1016/j.devcel.2004.11.020. [DOI] [PubMed] [Google Scholar]
- Chen D., Kon N., Li M., Zhang W., Qin J., Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Gao M., Karin M. Regulating the regulators: control of protein ubiquitination and ubiquitin-like modifications by extracellular stimuli. Mol. Cell. 2005;19:581–593. doi: 10.1016/j.molcel.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hinsby A. M., Kiemer L., Karlberg E. O., Lage K., Fausboll A., Juncker A. S., Andersen J. S., Mann M., Brunak S. A wiring of the human nucleolus. Mol. Cell. 2006;22:285–295. doi: 10.1016/j.molcel.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Horvath B. M., Magyar Z., Zhang Y., Hamburger A. W., Bako L., Visser R. G., Bachem C. W., Bogre L. EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J. 2006;25:4909–4920. doi: 10.1038/sj.emboj.7601362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. W., Venkatasubrahmanyam S., Ianculescu A. G., Tong A., Boone C., Madhani H. D. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- Kim J., Hake S. B., Roeder R. G. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol. Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Lessor T. J., Yoo J. Y., Xia X., Woodford N., Hamburger A. W. Ectopic expression of the ErbB-3 binding protein ebp1 inhibits growth and induces differentiation of human breast cancer cell lines. J. Cell. Physiol. 2000;183:321–329. doi: 10.1002/(SICI)1097-4652(200006)183:3<321::AID-JCP4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Lin A. W., Lowe S. W. Oncogenic ras activates the ARF-p53 pathway to suppress epithelial cell transformation. Proc. Natl. Acad. Sci. USA. 2001;98:5025–5030. doi: 10.1073/pnas.091100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Ahn J. Y., Liu X., Ye K. Ebp1 isoforms distinctively regulate cell survival and differentiation. Proc. Natl. Acad. Sci. USA. 2006;103:10917–10922. doi: 10.1073/pnas.0602923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llanos S., Clark P. A., Rowe J., Peters G. Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat. Cell Biol. 2001;3:445–452. doi: 10.1038/35074506. [DOI] [PubMed] [Google Scholar]
- Maki C. G., Huibregtse J. M., Howley P. M. In vivo ubiquitination and proteasome-mediated degradation of p53(1) Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- Mayo L. D., Turchi J. J., Berberich S. J. Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Res. 1997;57:5013–5016. [PubMed] [Google Scholar]
- Midgley C. A., Desterro J. M., Saville M. K., Howard S., Sparks A., Hay R. T., Lane D. P. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene. 2000;19:2312–2323. doi: 10.1038/sj.onc.1203593. [DOI] [PubMed] [Google Scholar]
- Ng H. H., Dole S., Struhl K. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 2003;278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- Okada M., Jang S. W., Ye K. Ebp1 association with nucleophosmin/b23 is essential for regulating cell proliferation and suppressing apoptosis. J. Biol. Chem. 2007;282:36744–36754. doi: 10.1074/jbc.M706169200. [DOI] [PubMed] [Google Scholar]
- Osley M. A. H2B ubiquitylation: the end is in sight. Biochim. Biophys. Acta. 2004;1677:74–78. doi: 10.1016/j.bbaexp.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Ou K., et al. Quantitative profiling of drug-associated proteomic alterations by combined 2-nitrobenzenesulfenyl chloride (NBS) isotope labeling and 2DE/MS identification. J. Proteome Res. 2006;5:2194–2206. doi: 10.1021/pr060115n. [DOI] [PubMed] [Google Scholar]
- Pickart C. M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Radomski N., Jost E. Molecular cloning of a murine cDNA encoding a novel protein, p38-2G4, which varies with the cell cycle. Exp. Cell Res. 1995;220:434–445. doi: 10.1006/excr.1995.1335. [DOI] [PubMed] [Google Scholar]
- Robzyk K., Recht J., Osley M. A. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C., Schreiber S. L., Mellor J., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Scholl C., et al. The homeobox gene CDX2 is aberrantly expressed in most cases of acute myeloid leukemia and promotes leukemogenesis. J. Clin. Invest. 2007;117:1037–1048. doi: 10.1172/JCI30182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C. J. The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- Squatrito M., Mancino M., Donzelli M., Areces L. B., Draetta G. F. EBP1 is a nucleolar growth-regulating protein that is part of pre-ribosomal ribonucleoprotein complexes. Oncogene. 2004;23:4454–4465. doi: 10.1038/sj.onc.1207579. [DOI] [PubMed] [Google Scholar]
- Sun Z. W., Allis C. D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Tang X., Jang S. W., Wang X., Liu Z., Bahr S. M., Sun S. Y., Brat D., Gutmann D. H., Ye K. Akt phosphorylation regulates the tumour-suppressor merlin through ubiquitination and degradation. Nat. Cell Biol. 2007;9:1199–1207. doi: 10.1038/ncb1641. [DOI] [PubMed] [Google Scholar]
- Trotman L. C., et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J. D., Taylor L. J., Roussel M. F., Sherr C. J., Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Wood A., et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell. 2003a;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- Wood A., Schneider J., Dover J., Johnston M., Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 2003b;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- Xia X., Cheng A., Lessor T., Zhang Y., Hamburger A. W. Ebp1, an ErbB-3 binding protein, interacts with Rb and affects Rb transcriptional regulation. J. Cell Physiol. 2001;187:209–217. doi: 10.1002/jcp.1075. [DOI] [PubMed] [Google Scholar]
- Ye K., Hurt K. J., Wu F. Y., Fang M., Luo H. R., Hong J. J., Blackshaw S., Ferris C. D., Snyder S. H. Pike. A nuclear GTPase that enhances PI3kinase activity and is regulated by protein 4.1N. Cell. 2000;103:919–930. doi: 10.1016/s0092-8674(00)00195-1. [DOI] [PubMed] [Google Scholar]
- Yoo J. Y., Wang X. W., Rishi A. K., Lessor T., Xia X. M., Gustafson T. A., Hamburger A. W. Interaction of the PA2G4 (EBP1) protein with ErbB-3 and regulation of this binding by heregulin. Br. J. Cancer. 2000;82:683–690. doi: 10.1054/bjoc.1999.0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Chen W., Zhang Y., Hamburger A. W., Pan H., Zhang Z. Suppression of salivary adenoid cystic carcinoma growth and metastasis by ErbB3 binding protein Ebp1 gene transfer. Int. J. Cancer. 2007;120:1909–1913. doi: 10.1002/ijc.22541. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Akinmade D., Hamburger A. W. The ErbB3 binding protein Ebp1 interacts with Sin3A to repress E2F1 and AR-mediated transcription. Nucleic Acids Res. 2005a;33:6024–6033. doi: 10.1093/nar/gki903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Fondell J. D., Wang Q., Xia X., Cheng A., Lu M. L., Hamburger A. W. Repression of androgen receptor mediated transcription by the ErbB-3 binding protein, Ebp1. Oncogene. 2002;21:5609–5618. doi: 10.1038/sj.onc.1205638. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Hamburger A. W. Specificity and heregulin regulation of Ebp1 (ErbB3 binding protein 1) mediated repression of androgen receptor signalling. Br. J. Cancer. 2005;92:140–146. doi: 10.1038/sj.bjc.6602257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang X. W., Jelovac D., Nakanishi T., Yu M. H., Akinmade D., Goloubeva O., Ross D. D., Brodie A., Hamburger A. W. The ErbB3-binding protein Ebp1 suppresses androgen receptor-mediated gene transcription and tumorigenesis of prostate cancer cells. Proc. Natl. Acad. Sci. USA. 2005b;102:9890–9895. doi: 10.1073/pnas.0503829102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Woodford N., Xia X., Hamburger A. W. Repression of E2F1-mediated transcription by the ErbB3 binding protein Ebp1 involves histone deacetylases. Nucleic Acids Res. 2003;31:2168–2177. doi: 10.1093/nar/gkg318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q., Gao W., Du F., Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Zhu B., Zheng Y., Pham A. D., Mandal S. S., Erdjument-Bromage H., Tempst P., Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.