Abstract
Fission yeast expresses two kinesin-8s, previously identified and characterized as products of the klp5+ and klp6+ genes. These polypeptides colocalize throughout the vegetative cell cycle as they bind cytoplasmic microtubules during interphase, spindle microtubules, and/or kinetochores during early mitosis, and the interpolar spindle as it elongates in anaphase B. Here, we describe in vitro properties of these motor proteins and some truncated versions expressed in either bacteria or Sf9 cells. The motor-plus-neck domain of Klp6p formed soluble dimers that cross-linked microtubules and showed both microtubule-activated ATPase and plus-end–directed motor activities. Full-length Klp5p and Klp6p, coexpressed in Sf9 cells, formed soluble heterodimers with the same activities. The latter recombinant protein could also couple microbeads to the ends of shortening microtubules and use energy from tubulin depolymerization to pull a load in the minus end direction. These results, together with the spindle localizations of these proteins in vivo and their requirement for cell viability in the absence of the Dam1/DASH kinetochore complex, support the hypothesis that fission yeast kinesin-8 contributes both to chromosome congression to the metaphase plate and to the coupling of spindle microtubules to kinetochores during anaphase A.
INTRODUCTION
Kinesin-8 was first discovered in budding yeast as the product of a microtubule-interacting gene, KIP3 (Cottingham and Hoyt, 1997; DeZwaan et al., 1997). Initial work suggested that this motor destabilized microtubules (MTs) in vivo and helped to define nuclear position, especially in the absence of dynein. Subsequent studies showed that yeast Kip3p worked inside the nucleus as well as in the cytoplasm, cooperating with Kar3p to promote the depolymerization of spindle MTs (Cottingham et al., 1999). A similar kinesin was then found in fission yeast (West et al., 2001; Garcia et al., 2002a,b; West et al., 2002), where two genes, klp5+ and klp6+, showed significant sequence similarity to the motor domain of KIP3. The products of these genes had identical and complex patterns of localization: they associated with cytoplasmic MTs during interphase, kinetochores, and/or kinetochore MTs during early mitosis, the midregion of the interpolar spindle during anaphase B, and then they returned to the cytoplasm as the cell reentered interphase. The phenotypes of deletion alleles of klp5+ and klp6+ are very similar during vegetative growth, showing elongated cytoplasmic MTs and spindles, a delay in anaphase onset, and an increase in the frequency of chromosome loss. Subsequent work has suggested that the products of these genes might form heterodimers (Garcia et al., 2002b; Li and Chang, 2003) and have interdependent functions in the regulation of MT dynamics (Unsworth et al., 2008). Interestingly, the meiotic phenotypes of deletions of these motors are quite different (our unpublished data), but this observation has not yet been explored.
Kinesin-8s have now been identified in a wide range of organisms (Lawrence et al., 2004; Wickstead and Gull, 2006), including Drosophila (Pereira et al., 1997), Aspergillus (Rischitor et al., 2004), and humans (Zhu et al., 2005; Mayr et al., 2007). Phenotypic analyses of mutants in metazoans have confirmed a role for this motor family in organized chromosome motion (Goshima and Vale, 2003; Gandhi et al., 2004; Savoian et al., 2004; Gatt et al., 2005), but exactly what these enzymes do to help achieve accurate chromosome motion is still a matter of debate. Studies on budding yeast kinesin-8 expressed in Sf9 cells have confirmed that it is a plus-end–directed motor and abets MT depolymerization with the intriguing properties that it favors the MT plus end (Gupta et al., 2006) and works faster on long MTs than short MTs (Varga et al., 2006). It has been suggested that these properties result from the motor's self-transport to the MT plus end, where it promotes tubulin dissociation. A study of kinesin-8 mutants in fission yeast has recently suggested, however, that this enzyme alters the frequencies of transitions between MT growth and shrinkage, not the rate at which MTs change length (Unsworth et al., 2008). One of the kinesin-8s from human cells (KIF18A) shows a mitotic distribution that is consistent with the in vitro findings, and a reduction in its amount affects both chromosome congression to the spindle equator in prometaphase and the rate of anaphase chromosome movement (Stumpff et al., 2008).
The current article describes in vitro properties of kinesin-8s expressed from the genes of Schizosaccharomyces pombe. Our biochemical results are consistent with much of the work on Kip3p from budding yeast, but they differ in several important ways, drawing attention to the fact that unlike budding yeasts, the genomes of most organisms encode two or more kinesin-8 genes. In these instances, different kinesin-8s may interact in ways that are not anticipated by the work on homodimeric Kip3. We show that although the fission yeast kinesin-8 lacks detectable depolymerization activity for guanylyl (α,β)-methylene diphosphonate (GMPCPP) or Taxol-stabilized MTs in vitro, it can support processive, minus-end–directed motion of microbeads with the shortening ends of dynamic MTs. Thus, kinesin-8s in different cells may achieve their mitotic roles by distinct means.
MATERIALS AND METHODS
Plasmid Construction
Plasmids for protein expression were constructed by standard methods (Sambrook and Russell, 2001), by using the polymerase chain reaction (PCR) and the primers described in Methods in Supplemental Materials. Klp5 and Klp6 full-length (FL) proteins were expressed in insect cells by subcloning their cDNAs from the corresponding bacterial vectors (Klp5 or 6 TopoBlunt II) into the baculovirus expression vectors: pVL1393-6xHis for Klp5 and pFastBac Dual for Klp6 (both from Invitrogen Carlsbad, CA) by using standard methods, as described in Supplemental Materials.
Protein Expression and Purification
For the bacterial expression and purification of both full-length and various truncated forms of these kinesins, we transformed the Escherichia coli strain Rosetta (DE3)pLysS (Novagen, EMD Biosciences, San Diego, CA) with appropriate plasmids, and bacteria were grown by standard procedures (Sambrook and Russell, 2001). Our best success in protein expression and purification from bacteria was with Klp6 motor domain-plus-neck (MDN): amino acids (aa 1-440), tagged at the C terminus with six histidines (His; see Figure 1A). Cultures (2 liters) were induced with isopropyl-β-d-1-thiogalactopyranoside for 4 h at 18°C, and then cells were collected, lysed, and processed as described in Supplemental Materials. Constructs encoding the full-length Klp5p and Klp6p did not produce soluble protein in any of several different bacterial expression systems. In Sf9 cells, however, Klp5FL and Klp6FL were expressed as soluble proteins, so long as they were coexpressed at approximately the same levels.
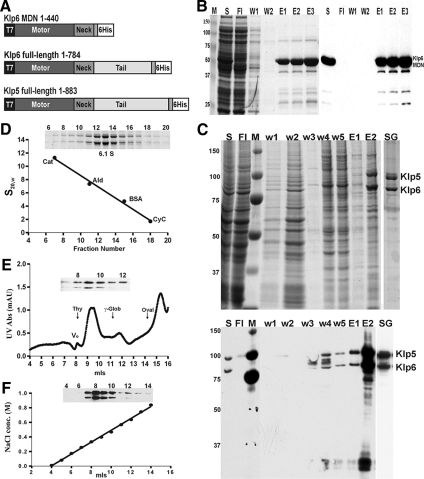
Figure 1.
Preparation of Klp5/6 and its active fragment. (A) Constructs from klp5+ and klp6+ that were used to express the proteins studied here. Klp5pFL and Klp6pFL included the entire open reading frames of these genes (West et al., 2001), plus sequences to encode a 6-His C-terminal extension. Klp6pMDN included the motor domain (amino acids 1-403) plus the 36 amino acid putative α-helical segment. (B) Purification of Klp6MDN. Coomassie-stained SDS gel (left) and Western blot with penta-His antibodies (right) of samples collected during various stages of Klp6MDN purification on a Ni-NTA column. M, molecular weight standards; S, whole protein extract; Fl, flow-through; W1, W2, wash fractions (20 and 40 mM imidazole); and E1–E3, Klp6MDN elution fractions. (C) Purification of Klp5FL/Klp6FL. Coomassie-stained SDS gel (top) and Western blot with penta-His antibodies (bottom) of the purification of Klp5FL and Klp6FL after their coexpression from baculovirus in Sf9 cells. S, clarified supernatant from whole cell lysate; Fl, flow-through; M, molecular weight standards; w1–w5, wash-fractions; E1 and E2, Ni-NTA elution fractions; and SG, sucrose gradient-purified Klp5FL and Klp6FL. (D–F) Hydrodynamic characterization of Klp5FL/Klp6FL. (D) Sucrose gradient fractionation of protein standards and Klp5FL/Klp6FL Ni-NTA elution fraction E2 (as shown in C); blot of gradient fractions 6–20 (inset). (E) Gel filtration chromatography of Klp5FL/Klp6FL on a Superdex 200 column. Blot of fractions 7–12 (inset) showing the coelution of Klp5FL and Klp6FL. The elution positions of protein calibration standards are marked by arrows. (F) Ion exchange chromatography of Klp5FL/Klp6FL on a SP Sepharose XL cation exchange column with a blot of fractions 4–14 (inset). Klp5FL and Klp6FL coeluted during a 0–1 M NaCl gradient.
Bacterial protein extracts were allowed to adsorb to nickel-nitrilotriacetic acid (Ni-NTA)-agarose (QIAGEN, Valencia, CA) and then rinsed and eluted with imidazole, following a modification of the manufacturer's instructions (see Supplemental Materials). Klp5FL and Klp6FL were purified from 500-ml cultures of Sf9 insect cells that had been grown and infected with engineered baculovirus by the Tissue Culture Core Facility, University of Colorado Cancer Center (Fitzsimons, Aurora, CO) for 48 h, using a multiplicity of infection of 2 for the virus encoding Klp5 and 1 for the virus with Klp6. Cells were harvested, washed once with phosphate-buffered saline (PBS), and the pellets were frozen in liquid nitrogen. Cells were thawed and lysed, and then protein was isolated as described in Methods of Supplemental Materials.
Biochemical Methods
Hydrodynamic Characterization.
Sucrose gradient assays of sedimentation velocity and gel filtration measures of hydrodynamic size were carried out by standard methods, as described in Supplemental Materials.
Steady-State Enzyme Assays.
The ATPase activities of both full-length Klp5/6 from insect cells and Klp6MDN from bacteria were characterized with the coupled enzyme ATPase assay described on the kinesin home page http://www.proweb.org/kinesin/Methods/ATPase_assay.html, as modified from Huang and Hackney (1994).
Tubulin Isolation and MT Preparation.
Tubulin was isolated from bovine brains by two cycles of polymerization-depolymerization, followed by phosphocellulose chromatography and two additional cycles of polymerization/depolymerization (Williams and Detrich, 1979). Fluorescein (Fl) and rhodamine (Rh) tubulin were prepared from phosphocellulose-purified tubulin as described at http://mitchison.med.harvard.edu/protocols.html by using the fluorophore derivatives 5- (and -6) carboxyfluorescein succinimidyl ester (Fl) or 5- (and -6) carboxy-tetramethylrhodamine succinimidyl ester (Rh) (Invitrogen, Carlsbad, CA). For pelleting assays, Taxol-stabilized MTs were made by polymerizing unlabeled tubulin in BRB80 buffer [80 mM piperazine-N,N'-bis(2-ethanesulfonic acid), 1 mM MgCl2, and 1 mM EGTA] with 1 mM dithiothreitol (DTT) and 1 mM guanosine triphosphate (GTP) at 37°C, and then adding Taxol at 10–15 μM for 5 min before harvesting the polymers by centrifugation through a cushion of 50% glycerol in BRB80 plus 10 μM Taxol, using an Airfuge (Beckman Coulter, Fullerton, CA) at 18 psi for 5 min. For bundling assays, rhodamine-labeled, Taxol-stabilized MTs were prepared from a mixture of labeled and unlabeled tubulin at a ratio of 1:7 and prepared as described above. Motility and depolymerization assays were performed using polarity-marked, GMPCPP MTs that were prepared using highly fluorescent seeds, made using a ratio of Fl-labeled to nonlabeled tubulin of 2:1, polymerized in BRB80 with 1 mM GMPCPP at 37°C. These polymers were collected by centrifugation, as noted above, and extended with a 4 μM mixture of Rh- and unlabeled tubulin (1:8) that had been treated with N-ethylmaleimide (NEM) in BRB80 buffer plus 0.5 mM GMPCPP, as described on http://mitchison.med.harvard.edu/protocols.html. NEM was used to enhance plus end elongation from the seed and inhibit minus end growth. Polarity-marked Taxol-stabilized MTs (Fl-seed and Rh-ends) were also prepared as described above, except that the fluorescent GMPCPP seeds were allowed to elongate in the NEM-Rh-tubulin mixture plus 1 mM MgGTP followed by stabilization with 15 μM Taxol.
MT-dependent Motility
MT gliding assays were carried out in flow chambers built from a glass slide, two stripes of double-sided sticky tape, and an 18- by 18-mm coverslip, sealed with 1:1:1 Vaseline, lanolin, and paraffin. The resulting chamber had an internal volume of ∼20 μl. For motility assays with Klp6MDN, an anti-His antibody (Penta His-Ab; QIAGEN) was flowed into the chamber and incubated for 10 min. The chamber was rinsed with BRB80 plus 0.2–0.5 mg/ml casein. Motor proteins (∼100 μg/ml) were then flowed into the chamber and incubated for 10 min. Motility assays with Klp5FL and Klp6FL were performed as described above or in the absence of antibody but following three applications of motor protein (∼100 μg/ml) to increase the number of bound enzymes. To remove unattached motors, the chamber was washed twice with BRB80 buffer. GMPCPP or Taxol-stabilized MTs were then allowed to bind for 10 min. The chamber was washed twice with BRB80 buffer to remove unbound MTs, followed by a wash with motility buffer (BRB80 buffer supplemented with 0.2 mg/ml casein, 20 mM d-glucose, 0.02 mg/ml catalase, 0.1 mg/ml glucose oxidase, 1% β-mercaptoethanol, and 2 mM MgATP) to induce motor activity. Samples were viewed at room temperature with an Axiophot II microscope (Carl Zeiss, Jena, Germany), using a 100× apochromatic lens (1.4 numerical aperture [n.a.]) or a Nikon Eclipse 80i scope with a 100× Plan Fluor (1.3 n.a.) objective. At the start of each motility experiment, a digital image was taken with both fluorescein and Texas Red fillers to verify MT polarity, and then all subsequent images were taken with a Texas Red filter every 30–60 s for 15–30 min, using a Photometrics Cascade 650 change-coupled device (CCD) camera (Roper Scientific, Ternton, NJ) or a CoolSNAP HQ2 CCD camera (Photometrics, Tucson, AZ). MT movements and length changes were analyzed with MetaMorph imaging software (Molecular Devices, Sunnyvale, CA) by using only MTs that fulfilled the following conditions: continuous movement in one direction for >5 min; no contact with other MTs; and persistence in the field of view for the entire experiment.
Bead Movement Induced by MT Depolymerization
Streptavidin-coated polystyrene beads (0.5 μm; Bangs Laboratories, Fishers, IN) were incubated with biotin-anti penta-His antibody (QIAGEN), washed, and then mixed with His-tagged Klp5FL/Klp6FL as described previously (Grishchuk et al., 2008b). Chlamydomonas axonemes (a gift from Dr. M. E. Porter, University of Minnesota) were elongated at 32°C in a chamber with GTP-tubulin, followed by Rh-tubulin in the presence of GMPCPP to create stable, photosensitive caps. After washing out soluble tubulins and all nucleotides, Klp5/6-coated beads were flowed into the chamber in motility buffer (BRB80 with 4 mM MgCl2, 2 mM DTT, 0.5 mg/ml casein, and 5 mg/ml bovine serum albumin) and allowed to bind these segmented MTs. The caps of individual MTs were then removed by illumination with green light, which induced the break-up of the Rh-MT segment, allowing the subsequent depolymerization of the underlying guanosine diphosphate (GDP)-MTs from their plus ends. Images were taken every second, by using low light differential interference contrast (DIC) on an Axiophot II microscope (Carl Zeiss) as described previously (Grishchuk et al., 2008b).
Electron Microscopy
Taxol-stabilized MTs (2.5 μM total tubulin) were mixed with Klp6 MDN (7.5 μM) in BRB80 buffer plus 2 mM AMP-PNP, 10 mM ADP, 1 mM GTP, and 10 μM Taxol and then incubated for 10 min at 37°C and used for negative staining or cryo-electron microscopy (EM). Klp5FL/Klp6FL (0.9 μM) was treated as above but using 0.2 μM Taxol-stabilized MTs.
Negative Staining.
Five microliters of motor-decorated MTs were applied to glow-discharged, carbon-coated grids. After adsorption for ∼1 min the solution was blotted with filter paper from one side of the grid then rinsed with ∼10 μl of 2% uranyl acetate. After ∼30 s, the stain was absorbed with filter paper, and the grid was allowed to dry before examination in a Tecnai F20 transmission electron microscope (FEI, Hillsboro, OR).
Cryo-Electron Microscopy.
A 3- to 4-μl sample of MTs or motor-decorated MTs was applied to glow-discharged grids coated with carbon support films that contained many, regularly spaced 2-μm holes (CU 100/400 mesh R2/2; Quantifoil MicroTools, Jena, Germany). After ∼20 s, excess fluid was blotted with filter paper, and the grid was rapidly plunged into liquid ethane, by using an automatic, home-made guillotine device (Templeton et al., 1997). Frozen grids were transferred under liquid nitrogen to a cryo-transfer holder (Gatan, Pleasanton, CA) and imaged under low-dose conditions at an operating voltage of 300 kV, by using a Tecnai F30 transmission electron microscope (FEI) equipped with a field-emission gun, a high-tilt stage, a sensitive CCD camera (Gatan), and a postcolumn energy filter (Gatan). The grids were scanned with the Navigator module of the microscope control program Serial EM (Mastronarde, 2005), and appropriate regions were imaged as single views.
Cryo-Electron Tomography.
Plunge-freezing was performed as described above except that in addition to the 3-μl sample, 1 μl of 10-nm colloidal gold (Sigma-Aldrich, St. Louis, MO) was applied to the grids before blotting for later use as fiducial markers in the tilt series alignment process. Several single-axis tilt series were recorded digitally on the Tecnai F30 microscope under low-dose conditions, by using the Serial EM software over an angular range of about ±60° with an increment of 2–4° and the energy filter in the zero-loss mode (slit width 20 eV). The total electron dose was ∼80 e−/A2, and the defocus ranged from −4 μm to −8 μm. The tilt series images were aligned, built into tomograms, and analyzed using the IMOD software package (Kremer et al., 1996).
RESULTS
Both a Truncated Allele of Klp6 and Full-Length Klps 5 and 6 Form Dimers In Vitro
To examine in vitro properties of the fission yeast kinesin-8s and their truncated versions, we prepared several DNA constructs and expressed them in bacteria, but only one of these constructs yielded sufficient amounts of soluble protein for further study: the Klp6 motor domain plus the “neck” domain, a putative α-helical coiled-coil of 36 amino acids (Klp6MDN, Figure 1, A and B). The full-length allele of Klp6+ expressed at levels that were too low to be useful, and plasmids containing full length klp5+ or any part thereof were highly unstable in bacteria; they rearranged frequently, and no useful amounts of expressed protein could be detected. We therefore turned to different expression systems. Previous work had shown that overexpression of Klp5p or Klp6p in fission yeast was lethal (West et al., 2001), so we used baculovirus vectors to transfect the relevant DNA into Sf9 cells. Expression of Klp6FL alone resulted in low amounts of soluble protein, but its purification was possible (Supplemental Figure S1). Klp5FL alone did not produce detectable amounts of soluble protein, but a coinfection of insect cells by the two recombinant viruses, each containing the complete cDNA for one motor, gave useful results (Figure 1C).
When His-tagged alleles of Klp5FL and Klp6FL were coexpressed and purified on Ni-NTA-agarose, the eluted polypeptides cosedimented on sucrose gradients with S20,w = 6.1 (Figure 1D), suggesting that they were forming heterodimers. This inference was supported by their coelution after both gel filtration (Figure 1E) and ion-exchange chromatography (Figure 1F). The diffusion coefficient and sedimentation velocity for this complex allowed an estimate of its solution molecular mass as 184 kDa, in good agreement with the value calculated from the predicted amino acid sequences of the two polypeptides (187 kDa). This dimer will hereafter be called Klp5/6FL. A similar characterization of Klp6MDN (Supplemental Figure S2) indicated a molecular mass of 118 kDa. Because this construct has a predicted molecular mass of 54 kDa, Klp6MDN, too, seems to form dimers in vitro. Perhaps the strong, dissimilar charges of the tails on these kinesins (theoretical isoelectric points for Klp5p and Klp6p tails = 5.6 and 10.6, respectively (West et al., 2001) explain both their preference for heterodimer formation and the efficient homodimerization by Klp6MDN, from which the tail has been removed.
Dimers of These Kinesin-8s Bind and Cross-Link MTs In Vitro
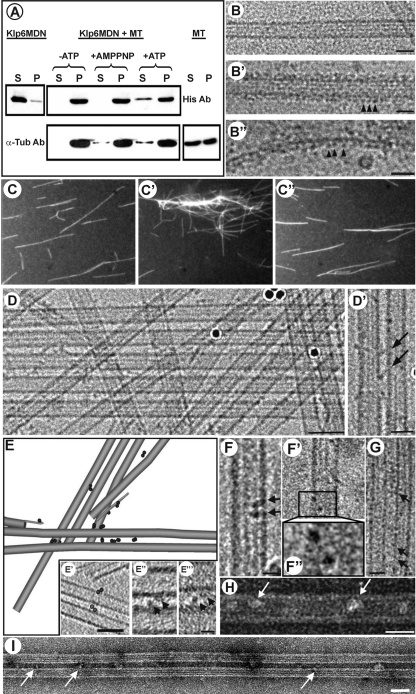
We explored the MT binding of bacterially expressed Klp6MDN in several ways. Assays of protein cosedimentation with MTs in the presence or absence of ATP or its nonhydrolyzable analogue AMPPNP indicated that Klp6MDN bound tubulin polymers in an ATP-sensitive manner (Figure 2A). Cryo-electron microscopy of control MTs showed the expected smooth cylinders, 23–24 nm in diameter, whereas MTs mixed with Klp6MDN were thicker (36–37 nm in diameter) and showed a robust periodicity, even on protofilaments that extended beyond their neighbors at the MT end (Figure 2B′ and 2B″). MTs became fully decorated when the ratio of Klp6MDN dimer to tubulin dimer was ∼1.5:1, suggesting that most of the motors in solution could display active MT binding. Neighboring particle heads were separated along the MT by ∼8 nm, suggesting one motor binding site per tubulin dimer.
Figure 2.
Evidence for Klp6MDN and Klp5/6FL cross-bridging of MTs. (A) Cosedimentation of Klp6MDN with MTs is ATP sensitive. Klp6MDN (2 μM) was incubated with Taxol-stabilized MTs (3 μM total tubulin) for 15 min at room temperature under various conditions: the absence of ATP, in the presence of 2 mM MgAMP-PNP, or in the presence of 2 mM MgATP. Samples were centrifuged for 30 min at 30,000 × g, and the pellets (P) were resuspended in SDS-buffer at a volume equal to the supernatants (S). Equal amounts of pellet and supernatant fractions were loaded on SDS gels and analyzed by Western blots. The top panel was probed with anti-His and the bottom panel with anti α-tubulin antibodies. (B) MT binding of Klp6MDN visualized by cryo-electron microscopy. (B) A rapidly frozen, ice-embedded, Taxol-stabilized MT. (B′) A similar MT after incubation with Klp6MDN for 10 min without ATP; motor-decorations (arrowheads) are evident on MT walls and on single protofilament extensions (B″). Bars, 25 nm. (C) Klp6MDN promotes ATP-sensitive MT bundling. Rhodamine-labeled MTs (C) were incubated with 0.5 μM Klp6MDN in the absence (C′) or presence (C″) of 1 mM MgATP. Samples were taken after 5 min, fixed with 0.2% glutaraldehyde, and then visualized by fluorescence light microscopy. (D) MT bundling in the presence of Klp6MDN visualized by cryo-EM. Without Klp6MDN most MTs were randomly distributed in the ice-layer (data not shown); in the presence of Klp6MDN, many MTs ran parallel to each other, forming bundles of up to ∼10 MTs (D). The distance between neighboring MTs within these bundles ranged from ∼10–13 nm. (D′) In some areas the gap between neighboring MTs was bridged by diagonal linkers (arrows). Dark circles are colloidal gold used for alignment of tilted views. Bars, 50 nm (D); 25 nm (D′). (E) Klp6MDN links between MTs were dimers when seen in 3D reconstructions. A 3D graphical model of a subvolume from a cryo-tomogram of ice-embedded MTs with Klp6MDN. The sample was imaged at 3° intervals over a tilt range of −63° to +72°. MTs and their protofilament extensions were modeled in gray, attached motors are shown as dark 4-nm discs. The majority of Klp6MDN particles occurred in pairs (circled in E′). E′–E‴ show 1-nm-thick tomographic slices of Klp6MDN dimers; E″ and E‴ show two motor heads (arrowheads) that bind adjacent MTs and are connected, most likely by their neck-domain. Bars, 50 nm (E′); 10 nm (E″ and E‴). (F–I) MT binding and bundling by Klp5/6FL visualized by cryo- (F and G) and negative staining (H and I) EM. Taxol-stabilized MTs were incubated with Klp5/6FL at a ratio of 1:2 plus 2 mM AMPPNP for 10 min. Klp5/6FL motors (arrows) bind to MTs (F–F″, H), often as dimers (F–F″). F″ is a 2× magnification of the two motor domains in the boxed area in (F′). Klp5/6FL also promotes MT bundle formation (G and I), whereby neighboring MTs seem to be connected by diagonal linkers (G, arrows). Bars, 50 nm (I); 25 nm (G and H); 12.5 nm (F and F′). Protein is black in B′–B″ and D–G and white in H and I due to different EM techniques used to prepare these images.
MT binding, together with the above-mentioned evidence for dimerization, raised the possibility that Klp6MDN might cross-link MTs in vitro. We prepared Taxol-stabilized, rhodamine-labeled MTs and mixed them with Klp6MDN in the absence of ATP (Figure 2C′). In this mixture, MTs were significantly more bundled than in controls, and the bundling was ATP sensitive (Figure 2, C–C″). Cryo-electron microscopy of these preparations revealed small cross-bridges between adjacent MTs (Figure 2D and 2D′). Closer scrutiny of these cross-bridges showed that they were constructed from two, 4-nm globular domains that associated directly with MT walls plus a slender connection between them (Figure 2, E′–E‴). A three-dimensional (3D) model of a subvolume from this tomogram (Figure 2E, diagram) showed MTs (gray cylinders) and Klp6MDN-dimers (black spheres) positioned so that the majority of Klp6MDN heads occurred in pairs, again indicating that Klp6MDN formed dimers under these conditions.
Examination of Klp5/6FL by analogous methods revealed similar structural patterns. This larger complex again associated with the walls of MTs, as seen by both cryo-electron microscopy (Figure 2, F and G) and negative staining (Figure 2H). In many cases, two roughly spherical structures were closely associated (Figure 2, F–F″); sometimes bridges between neighboring MTs were evident, and bundles formed (Figure 2, G and I). The distance between parallel MTs within these bundles seemed slightly wider (12–16 nm) than in Klp6MDN-linked bundles.
The ATPase Activity of S. pombe Klp5/6 Is Stimulated by MTs
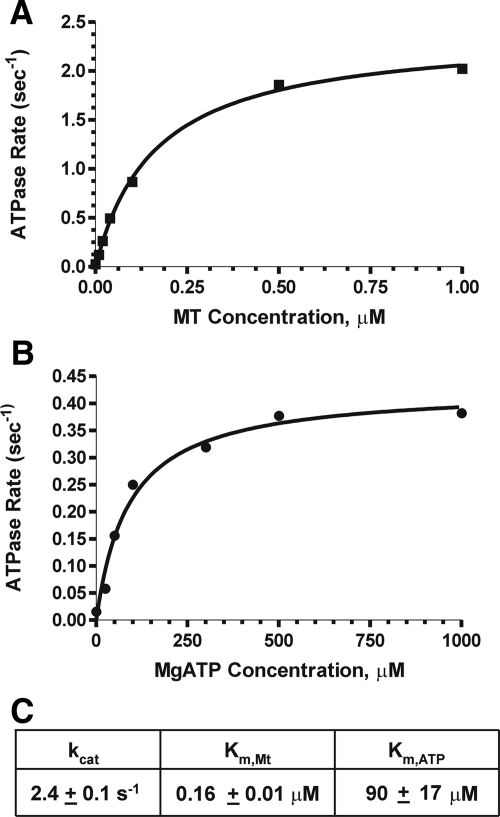
We have examined the enzyme activities of these motor constructs by measuring the activation of their ATPase by Taxol-stabilized MTs or soluble tubulin. Klp5/6FL showed classic Michaelis-Menton kinetics with respect to both MgATP and MTs (Figure 3). The Km, MT value was 0.16 ± 0.01 μM, with a Kcat value of 2.4 ± 0.1 s−1; with saturating MTs, the Km value for ATP was 90 ± 17 μM. These values are comparable though slightly higher than those obtained for another kinesin-8 (Gupta et al., 2006), suggesting that our protein preparations were active, although the exact values for our measured rate constants varied from one preparation to the next. Similar experiments were carried out on Klp6MDN, and again increasing amounts of Taxol-stabilized MTs stimulated a corresponding increase in ATPase activity (Supplemental Figure S3). The Kcat value with saturating concentrations of MTs was 0.93 ± 0.01 s−1, and the Km, MT value was 0.06 ± 0.004 μM.
Figure 3.
ATPase activities of Klp5/6FL. (A) Enzyme activity as a function of MT concentration: 100 nM Klp5/6FL, 1 mM MgATP, and Taxol-stabilized MTs (0–1 μM tubulin dimers). (B) Enzyme activity as a function of MgATP: 100 nM Klp5/6FL, 1 μM tubulin (polymerized into MTs), and 0–1 mM MgATP. The points on graphs A and B are an average of two experiments performed with the same protein preparation. (C) Steady-state ATPase values for Klp5/6FL.
Kinesin-8 from budding yeast is activated by soluble tubulin (Gupta et al., 2006), a property it shares with kinesin-13s (Hunter et al., 2003), so we asked whether Klp5/6FL had this activity. There was no increase in the ATPase activity of Klp5/6FL, even with concentrations of tubulin dimers up to 10 μM (data not shown).
Kinesin-8s Are Plus-End–directed Motors In Vitro
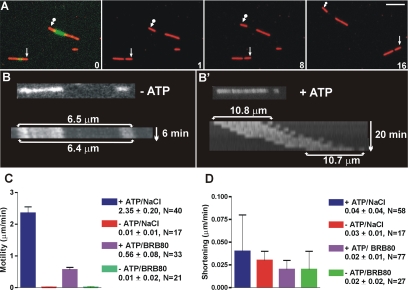
We used fluorescence microscopy to examine the motility of fission yeast kinesin-8s. Segmented MTs that contained a fluorescein-labeled seed and rhodamine-tubulin extensions were prepared in the presence of NEM to minimize minus-end growth, so we could easily determine the direction of enzyme motion (see Materials and Methods). Klp5/6FL was a plus end-directed motor (Figure 4, A and B, and Supplemental Movie S1); its rate of motility was sensitive to the concentration of added salt. At room temperature in BRB80 buffer, MTs moved with an average speed of 0.56 ± 0.08 μm/min (mean ± SD; N = 33), but in BRB80 plus 200 mM NaCl, they moved at 2.35 ± 0.20 μm/min (N = 40) (Figure 4C). Klp6MDN and Klp6FL were also plus-end–directed motors in vitro; they moved MTs at 3.36 ± 0.66 μm/min (N = 23) and 0.12 ± 0.09 μm/min (N = 17), respectively, even when BRB80 buffer was supplemented with 200 mM NaCl.
Figure 4.
Klp5/6FL is a plus end motor in vitro with no MT depolymerization activity. (A) Motility assay showing plus-end–directed motor activity of Klp5/6FL. Images of polarity marked MTs (green, Fl labeled seed with red, Rh extensions). First panel is a superposition of images taken through fluorescein isothiocyanate and Texas Red filter cubes; subsequent images show Texas Red images only with arrows marking the minus end of two MTs. Time between images in minutes. Bar, 5 μm. (B) Kymograph of polarity-marked MTs (Fl seed and Rh ends) visualized in Texas Red channel. MT with Klp5/6FL in the absence of ATP shows no motility or shortening; MT with Klp5/6FL plus 2 mM MgATP shows motility but no depolymerization (B′). (C) Summary bar graph of Klp5/6FL plus-end motility rates. Experiments were performed in the presence of NaCl/BRB80 buffer + ATP (blue bar); in NaCl/BRB80 buffer − ATP (red bar); in BRB80 buffer + ATP (purple bar); in BRB80 with no ATP (green bar). Error bars are standard deviations. (D) Summary bar graph showing the absence of MT shortening by Klp5/6FL. Experiments were performed in NaCl/BRB80 buffer + ATP (blue bar); in NaCl/BRB80 buffer − ATP (red bar); in BRB80 buffer + ATP (purple bar); BRB80 with no ATP (green bar). Error bars are standard deviations. Note difference in graph scales for C and D.
None of these Kinesin-8s Enhanced the Depolymerization of Taxol or GMPCPP Stabilized MTs
While assessing MT motility, we also monitored MT length (Figure 4). In none of our experiments did MT length change significantly, regardless of whether ATP was present or salt was added to enhance motility. This was surprising because kinesin-8 from both budding yeast (Gupta et al., 2006; Varga et al., 2006) and mammalian cells (Mayr et al., 2007) show ATP-dependent MT shortening in vitro. Our fission yeast Klp5/6FL was clearly producing MT movement, so we wondered whether our assay conditions might have inhibited the enzyme's full range of activities. We therefore tried a “tethered MT” assay, similarly to that described previously (Gupta et al., 2006). Biotin antibody was flowed into the chamber, unbound antibody was washed out, and then biotin-labeled MTs were introduced and allowed to attach. Klp5/6 was then flowed into the chamber in the presence of MgATP, and MT length was monitored. MT length did not change detectably, either with full-length heterodimers or with the truncated allele of KLP6 (data not shown).
To examine this issue in a different way, we prepared Taxol-stabilized MTs, added 2 mM MgATP, and mixed them with either Klp5/6FL, a kinesin-13 (MCAK generously given by Claire Walczak, Indiana University, Bloomington, IN), or buffer only. Each preparation was incubated at room temperature for 30 min and then spun in an AirFuge. The amount of tubulin that pelleted remained unchanged in the presence of Klp5/6FL, whereas the kinesin-13, used as a positive control, depolymerized MTs efficiently (Supplemental Figure S4). We concluded that even when fission yeast kinesin-8 motors were interacting with MTs and ATP in solution, they still lacked measurable depolymerization activity for stabilized MTs in vitro, a marked contrast to other members of this family.
KLP5/6FL Can Remain Bound to the Plus End of a Shortening MT and Carry a Load
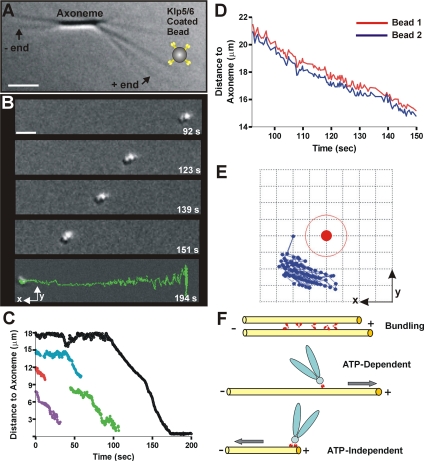
The human kinesin-8, Kif18A, has been proposed to regulate kinetochore MT dynamics and thereby control mitotic chromosome position (Stumpff et al., 2008). Likewise, the S. pombe kinesin-8s are important for efficient prometaphase (West et al., 2002). We therefore asked whether Klp5/6FL might serve as a coupler for harnessing the energy of tubulin depolymerization to do mechanical work by using an assay established previously in our laboratory (Grishchuk et al., 2005). We stably conjugated full-length kinesin-8 heterodimers to polystyrene microbeads and rinsed them free of soluble proteins. These beads were then added to a microchamber with segmented MTs of known polarity (Figure 5A; see Materials and Methods); they bound readily to the walls of the capped MTs, whereas similarly prepared beads that lacked the motor protein failed to make robust attachments. The caps of individual MTs were then removed by illumination with green light, which induces the break-up of Rh-MTs, allowing the subsequent depolymerization of the GDP-MTs from their plus ends. The motor-coated beads followed the depolymerizing plus end (Supplemental Movie S2), just as we have seen previously with some nonmotor proteins, such as Dam1/DASH (Grishchuk et al., 2008b) and NDC80 (McIntosh et al., 2008). In contrast, beads coated with streptavidin or heat-stable MT-associated proteins from brain do not follow shortening MT ends, even though they bind stably to MT polymers grown with biotinylated tubulin (Grishchuk et al., 2005). The processivity of Klp5/6FL was good, because the mean distance for bead travel was 7.2 ± 5.9 μm (Figure 5, B and C). Interestingly, the mean speed of these motions was only 10.3 ± 2.8 μm/min, which is significantly slower than the mean shortening speed of MTs made of pure tubulin under these conditions (22 μm/min). We concluded that this kinetochore-associated motor enzyme is capable of tubulin depolymerization-dependent, ATP-independent motion in the minus direction, an interesting addition to its ATP-dependent motion toward the plus end.
Figure 5.
Klp5/6FL coated beads follow the end of a depolymerizing MT. (A) Experimental setup: Chlamydomonas axonemes (DIC image) have been elongated with tubulin plus GTP to form conventional MTs and then capped with Rh-tubulin in the presence of GMPCPP (arrows point to plus and minus MT ends). Tubulin was then washed out, and Klp5/6FL-coated beads were flowed into the chamber (note that bead is not drawn to scale). A given MT was uncapped by photobleaching its Rh-tubulin cap and images were recorded as the MT depolymerized. Bar, 2 μm. (B) Time-lapse images of a cluster of two or more Klp5/6FL-coated polystyrene beads. The beads move toward the axoneme (MT minus ends) during MT depolymerization (time in seconds). The final image (bottom) displays the trajectory of the entire movement. Bar, 2 μm. (C) Plots of distance versus time for moving Klp5/6FL-coated beads. Sometimes, beads detached from the shortening MTs before reaching the axonemes. Black line corresponds to the most processive example (shown in B). (D) Graph of distance to axoneme for the two attached beads traveling as one cluster. Selected images for this experiment are shown in B and one of these curves is shown in black in C. (E) Graph showing relative positions of the centers of two attached beads that traveled together. The motion of bead 2 (blue symbols) relative to the center of bead 1 (solid red circle) as they followed the shortening MT end. This graph is a view in the image plane with the x- and y-axes oriented as in B; the MT runs along the x-axis. Grid size is 0.2 μm, so the thin red line corresponds to the outer perimeter of bead 1. (F) Model of suggested Klp5/6FL motor activities functions in vivo. The enzyme (small red symbols) can bundle MTs, accomplish ATP-dependent transport of a cargo (drawn here as a chromosome) toward the MT plus end, and ATP-independent transport of a cargo (drawn here as a chromosome) toward the MT minus end as it follows tubulin depolymerization.
KLP5/6FL-coated Beads Follow Shortening MT Ends by a Rotation-free Mechanism
We and others have previously shown that the kinetochore-associated complex, Dam1 (a.k.a. DASH), will conjugate microspheres to MTs and promote their motion with the end of a shortening MT. When rings of Dam1 can form, MT depolymerization is slowed, whereas in the absence of a ring, MTs shorten at their normal speed and the Dam1-coated microspheres roll (Grishchuk et al., 2008a, 2008b). Because rolling is unlikely to be a good model for depolymerization-associated chromosome motion in vivo, we asked whether beads coated with Klp5/6FL were rolling as they followed a shortening MT end. This question was comparatively easy to answer because ∼67% of the beads that moved were in groups of two or more whose rotation could readily be detected by phase microscopy. The bead clusters obviously moved without rotation (Figure 5B), a behavior that is analyzed in Figure 5, D and E. We tracked the trajectories of three sets of two or more beads as they traveled together for ≥60 s over a distance of >6 μm. In every case, the trajectories of the individual beads did not cross but moved in concert, suggesting that the beads did not roll as they moved parallel to the MT axis. This conclusion was verified by plotting the coordinates for the center of one moving bead, using the other as the origin of a coordinate system (Figure 5E). The motions of the first bead (blue symbols on Figure 5E) are confined to one quadrant of the two-dimensional area around the reference bead (red symbol), demonstrating the absence of cluster rotation. KLP5/6FL-coated beads therefore follow shortening MT ends by a rotation-free mechanism. Thus, by both the criterion of rotation and speed of MT shortening, beads coupled to MTs by this kinesin-8 were more similar to those coupled by Dam1 rings than by simple Dam1 binding.
These properties of kinesin-8–coupled beads may be related to the fact that bead clusters were seen to move more frequently than single beads, even though the suspensions of beads usually contained <10% of bead clusters (a result of energetic sonication). Forty percent of MT-associated bead clusters moved with MT shortening, compared with 7% of the single beads; perhaps the attachment of multiple kinesin-8s is required for minus-end–directed movement, and this condition is favored when multiple beads are in close association. Regardless of the mechanism, this is the first motor for which these properties have been described, although antibodies to CENP-E have been shown to inhibit MT depolymerization-driven chromosome motion in vitro (Lombillo et al., 1995a). It remains to be determined whether this motor too can support depolymerization-dependent motion of beads in vitro by a rotation-free mechanism.
DISCUSSION
Fission yeast kinesin-8s expressed in insect cells formed a heterodimer that hydrolyzed MgATP in a MT-dependent manner and moved on MTs toward their plus ends, properties it shares with its budding yeast and mammalian homologues. Kinesin-8s from the latter organisms will depolymerize stable MTs in vitro (Gupta et al., 2006; Varga et al., 2006; Mayr et al., 2007), but we were unable to detect ATP-dependent MT shortening with any of the kinesin-8 alleles examined here. This was a surprise because both our group (West et al., 2001; West et al., 2002) and others (Garcia et al., 2002a,b) have shown that deletion of the genes encoding either of these proteins leads to the elongation of cellular MTs. Moreover, in vivo overproduction of either full-length KLP6p or Klp6MDN, the same truncated allele used here, caused cytoplasmic MTs to shorten (West and McIntosh, 2008). However, recent measurements of in vivo MT dynamics in the absence of Klp5/Klp6 have shown that polymer growth and shrinkage rates are not significantly different from those in wild-type cells. Rather catastrophe/rescue frequency and dynamicity are suppressed (Unsworth et al., 2008). Similarly, the deletion of Kip3 reduced the frequencies of both catastrophes and pauses in living budding yeasts (Gupta et al., 2006). Perhaps all kinesin-8s modulate MT dynamicity but only a subset of these enzymes can actually change the rates of MT depolymerization. The effects of Klp5/6 on MT catastrophe and rescue frequencies in vitro will be an interesting question for future studies.
Another difference between our enzyme and the kinesin-8s described previously was its lack of tubulin-activated ATPase activity, a property that other kinesin-8s share with kinesin-13 (Gupta et al., 2006). These differences may be a result of our having lost an activity during enzyme isolation, but the robust ATPase and motility activities argue against this interpretation. Our data are consistent with two hypotheses: 1) kinesin-8 from fission yeast differs from homologous enzymes in other systems; or 2) our preparations lack an essential activity, either because our recombinant proteins lack necessary post-translational modifications or because the native fission yeast enzyme includes an additional component that we have not yet identified.
Several aspects of our biochemical results supported the idea that Klp5FL and Klp6FL prefer to form heterodimers: 1) the heterocomplex expressed much better and was more amenable to purification than either of its components expressed alone; 2) these distinct polypeptides cosedimented and coeluted from both gel filtration and ion exchange columns; and 3) the measured solution molecular mass was a good fit to the mass predicted from the two amino acid sequences. The heterodimeric behavior of fission yeast kinesin-8 in vitro is consistent with evidence from vegetative cells that these proteins heterodimerize in vivo (Garcia et al., 2002b; Li and Chang, 2003). It remains to be seen whether homodimers of Klp5 and Klp6 are formed in vivo and what physiological role they play, if any. The interdependence of these two proteins for vegetative cell growth (Unsworth et al., 2008) suggests that their principal modes of action are cooperative, but the distinct phenotypes of the two motor gene deletions during meiosis remain to be explained. These results emphasize the caution that should be exercised when interpreting work on a single kinesin-8 polypeptide from an organism that possesses two or more genes for this enzyme.
Our results show that fission yeast kinesin-8s can couple a cargo to the end of a shortening MT. This property has not yet been investigated for other kinesin-8s, but it was found for mammalian kinesin-1 (Lombillo et al., 1995b), and it may be a common feature of this enzyme superfamily. Our in vitro observation that a Klp5/6 coated bead can follow a depolymerizing MT in the absence of ATP is different from what has been observed in vivo for Kip3p. The budding yeast kinesin-8 accumulates at the plus ends of growing but not shortening MTs (Gupta et al., 2006; Varga et al., 2006). Consistent with this observation, an increase in the concentration of the human kinesin-8, Kif18A, was observed at one sister kinetochore relative to the other as Kif18A accumulates at MT plus ends (Stumpff et al., 2008). Note that it seems unlikely that a kinesin-8 could enhance tubulin depolymerization without actually being at the shortening MT end, so the previously published observations may be quantitatively correct but not qualitatively accurate. The apparent differences in kinesin-8 motor behaviors may thus be due to assay conditions or subtle variations in enzymatic properties.
A plus-end–directed motor that can follow a shortening MT in the minus end direction may constitute an important cellular mechanism for ensuring the productive attachment of cargo, be it a chromosome or a vesicle, to the end of a depolymerizing MT. Fission yeast kinesin-8 resembled the Dam1 ring in providing rotation-free motion in association with a slowed MT shortening. Our in vitro observations may, therefore, be directly relevant to this motor's function in chromosome motion. Indeed, these properties of Klp5/6 may be at the heart of the interactions between the genes for these motors and for subunits of the Dam1 complex in fission yeast (Sanchez-Perez et al., 2005); although neither of these kinetochore components is essential for vegetative growth, their loss-of-function alleles show synthetic lethality. Perhaps these very different proteins represent parallel but distinct components of the system that ensure attachment between kinetochores and shortening spindle MTs in vivo.
The ability of Klp5/6FL both to move in the plus direction through ATP hydrolysis and to pull a cargo in the minus end direction as tubulin depolymerizes in the absence of ATP is particularly intriguing in the light of this motor's apparent roles in both prometaphase congression to the spindle midplane and successful chromosome attachment to the spindle. If properly tethered to a kinetochore, this motor and appropriate cofactors or posttranslational modifications could help with both plus-end–directed motions toward the spindle equator and minus-end–directed motions toward the spindle poles. The regulation of such an enzyme is obviously an interesting and important subject for future work, and we hope that our systems for measuring both motility and force generation (Grishchuk et al., 2008a) will allow us to unravel the intricacies of this kind of biologically significant regulation.
Fission yeast lacks a kinesin-7, i.e., an orthologue of CENP-E, which is a plus-end–directed, kinetochore-associated kinesin (Kim et al., 2008) that contributes to the ability of isolated mammalian chromosomes to follow the ends of depolymerizing MTs in vitro (Lombillo et al., 1995a). We propose that kinesin-8 in fission yeast may share some properties and functions with kinesin-7s and be an example of the principle that across a broad range of phylogeny, kinetochores preserve functions but not necessarily protein identities (McIntosh et al., 2002). Klps5/6 may be particularly interesting for mitosis, however, because the in vivo activities of this motor-pair include an enhancement of MT depolymerization, analogous to the roles of kinesin-13s. Fission yeast, like budding yeast, lack a gene for kinesin-13, so kinesin-8s may be doing at least double, and perhaps triple, duty in the spindles of these organisms. It is no wonder that their deletion increases the frequency of chromosome loss by 100-fold (Garcia et al., 2002b).
From a mechanistic point of view, these multiple functions, which are suggestive of different members of the kinesin superfamily, may reflect a complexity of protein structure that is not evident from simple amino acid sequence. When the atomic structures of more kinesins are known, it may become possible to associate 3D structural motifs with particular cellular functions and understand how some cells do without an enzyme that can readily be recognized as either a kinesin-7 or -13.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mary Porter and Claire Walczak for the generous gift of reagents and members of the McIntosh and Ataullakhanov laboratories for technical help and useful discussions. This work was supported by National Institutes of Health grants GM-033787 and RR-000592 (to J.R.M.).
Abbreviations used:
- FL
full length
- GMPCPP
guanylyl (α,β)-methylene diphosphonate
- MDN
motor domain-plus-neck
- MT
microtubule.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-09-0979) on November 26, 2008.
REFERENCES
- Cottingham F. R., Gheber L., Miller D. L., Hoyt M. A. Novel roles for saccharomyces cerevisiae mitotic spindle motors. J. Cell Biol. 1999;147:335–350. doi: 10.1083/jcb.147.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham F. R., Hoyt M. A. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan T. M., Ellingson E., Pellman D., Roof D. M. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J. Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R., Bonaccorsi S., Wentworth D., Doxsey S., Gatti M., Pereira A. The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol. Biol. Cell. 2004;15:121–131. doi: 10.1091/mbc.E03-05-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M. A., Koonrugsa N., Toda T. Spindle-kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J. 2002a;21:6015–6024. doi: 10.1093/emboj/cdf611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M. A., Koonrugsa N., Toda T. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr. Biol. 2002b;12:610–621. doi: 10.1016/s0960-9822(02)00761-3. [DOI] [PubMed] [Google Scholar]
- Gatt M. K., Savoian M. S., Riparbelli M. G., Massarelli C., Callaini G., Glover D. M. Klp67A destabilises pre-anaphase microtubules but subsequently is required to stabilise the central spindle. J. Cell Sci. 2005;118:2671–2682. doi: 10.1242/jcs.02410. [DOI] [PubMed] [Google Scholar]
- Goshima G., Vale R. D. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E. L., Efremov A. K., Volkov V. A., Spiridonov I. S., Gudimchuk N., Westermann S., Drubin D., Barnes G., McIntosh J. R., Ataullakhanov F. I. The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proc. Natl. Acad. Sci USA. 2008a;105:15423–15428. doi: 10.1073/pnas.0807859105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E. L., Molodtsov M. I., Ataullakhanov F. I., McIntosh J. R. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- Grishchuk E. L., Spiridonov I. S., Volkov V. A., Efremov A., Westermann S., Drubin D., Barnes G., Ataullakhanov F. I., McIntosh J. R. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc. Natl. Acad. Sci. USA. 2008b;105:6918–6923. doi: 10.1073/pnas.0801811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M. L., Jr, Carvalho P., Roof D. M., Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Huang T. G., Hackney D. D. Drosophila kinesin minimal motor domain expressed in Escherichia coli. Purification and kinetic characterization. J. Biol. Chem. 1994;269:16493–16501. [PubMed] [Google Scholar]
- Hunter A. W., Caplow M., Coy D. L., Hancock W. O., Diez S., Wordeman L., Howard J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Heuser J. E., Waterman C. M., Cleveland D. W. CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J. Cell Biol. 2008;181:411–419. doi: 10.1083/jcb.200802189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J. R., Mastronarde D. N., McIntosh J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lawrence C. J., et al. A standardized kinesin nomenclature. J. Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chang E. C. Schizosaccharomyces pombe Ras1 effector, Scd1, interacts with Klp5 and Klp6 kinesins to mediate cytokinesis. Genetics. 2003;165:477–488. doi: 10.1093/genetics/165.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo V. A., Nislow C., Yen T. J., Gelfand V. I., McIntosh J. R. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J. Cell Biol. 1995a;128:107–115. doi: 10.1083/jcb.128.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo V. A., et al. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature. 1995b;373:161–164. doi: 10.1038/373161a0. [DOI] [PubMed] [Google Scholar]
- Mastronarde D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mayr M. I., Hummer S., Bormann J., Gruner T., Adio S., Woehlke G., Mayer T. U. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr. Biol. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- McIntosh J. R., Grishchuk E. L., West R. R. Chromosome-microtubule interactions during mitosis. Annu. Rev. Cell Dev. Biol. 2002;18:193–219. doi: 10.1146/annurev.cellbio.18.032002.132412. [DOI] [PubMed] [Google Scholar]
- McIntosh J. R., Grishchuk E. L., Morphew M. K., Efremov A. K., Zhudenkov K., Volkov V. A., Cheeseman I. M., Desai A., Mastronarde D. N., Ataullakhanov F. I. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 2008;135:322–333. doi: 10.1016/j.cell.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A. J., Dalby B., Stewart R. J., Doxsey S. J., Goldstein L. S. Mitochondrial association of a plus end-directed microtubule motor expressed during mitosis in Drosophila. J. Cell Biol. 1997;136:1081–1090. doi: 10.1083/jcb.136.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rischitor P. E., Konzack S., Fischer R. The Kip3-like kinesin KipB moves along microtubules and determines spindle position during synchronized mitoses in Aspergillus nidulans hyphae. Eukaryot. Cell. 2004;3:632–645. doi: 10.1128/EC.3.3.632-645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sanchez-Perez I., Renwick S. J., Crawley K., Karig I., Buck V., Meadows J. C., Franco-Sanchez A., Fleig U., Toda T., Millar J. B. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J. 2005;24:2931–2943. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoian M. S., Gatt M. K., Riparbelli M. G., Callaini G., Glover D. M. Drosophila Klp67A is required for proper chromosome congression and segregation during meiosis I. J. Cell Sci. 2004;117:3669–3677. doi: 10.1242/jcs.01213. [DOI] [PubMed] [Google Scholar]
- Stumpff J., von Dassow G., Wagenbach M., Asbury C., Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton N. S., Lasic D. D., Frederik P. M., Strey H. H., Roberts D. D., Pavlakis G. N. Improved DNA: liposome complexes for increased systemic delivery and gene expression. Nat. Biotechnol. 1997;15:647–652. doi: 10.1038/nbt0797-647. [DOI] [PubMed] [Google Scholar]
- Unsworth A., Masuda H., Dhut S., Toda T. Fission yeast kinesin-8 Klp5 and Klp6 are interdependent for mitotic nuclear retention and required for proper microtubule dynamics. Mol. Biol. Cell. 2008;19:5104–5115. doi: 10.1091/mbc.E08-02-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V., Helenius J., Tanaka K., Hyman A. A., Tanaka T. U., Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- West R. R., Malmstrom T., McIntosh J. R. Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J. Cell Sci. 2002;115:931–940. doi: 10.1242/jcs.115.5.931. [DOI] [PubMed] [Google Scholar]
- West R. R., Malmstrom T., Troxell C. L., McIntosh J. R. Two related kinesins, klp5(+) and klp6(+), foster microtubule disassembly and are required for meiosis in fission yeast. Mol. Biol. Cell. 2001;12:3919–3932. doi: 10.1091/mbc.12.12.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R. R., McIntosh J. R. Novel interactions of fission yeast kinesin 8 revealed through in vivo expression of truncation alleles. Cell Motil. Cytoskeleton. 2008;65:626–640. doi: 10.1002/cm.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B., Gull K. A “holistic” kinesin phylogeny reveals new kinesin families and predicts protein functions. Mol. Biol. Cell. 2006;17:1734–1743. doi: 10.1091/mbc.E05-11-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. C., Jr, Detrich H. W., 3rd Separation of tubulin from microtubule-associated proteins on phosphocellulose. Accompanying alterations in concentrations of buffer components. Biochemistry. 1979;18:2499–2503. doi: 10.1021/bi00579a010. [DOI] [PubMed] [Google Scholar]
- Zhu C., Zhao J., Bibikova M., Leverson J. D., Bossy-Wetzel E., Fan J. B., Abraham R. T., Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell. 2005;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.