Abstract
The Tim9–Tim10 complex plays an essential role in mitochondrial protein import by chaperoning select hydrophobic precursor proteins across the intermembrane space. How the complex interacts with precursors is not clear, although it has been proposed that Tim10 acts in substrate recognition, whereas Tim9 acts in complex stabilization. In this study, we report the structure of the yeast Tim9–Tim10 hexameric assembly determined to 2.5 Å and have performed mutational analysis in yeast to evaluate the specific roles of Tim9 and Tim10. Like the human counterparts, each Tim9 and Tim10 subunit contains a central loop flanked by disulfide bonds that separate two extended N- and C-terminal tentacle-like helices. Buried salt-bridges between highly conserved lysine and glutamate residues connect alternating subunits. Mutation of these residues destabilizes the complex, causes defective import of precursor substrates, and results in yeast growth defects. Truncation analysis revealed that in the absence of the N-terminal region of Tim9, the hexameric complex is no longer able to efficiently trap incoming substrates even though contacts with Tim10 are still made. We conclude that Tim9 plays an important functional role that includes facilitating the initial steps in translocating precursor substrates into the intermembrane space.
INTRODUCTION
Mitochondria harbor 1000–1500 different proteins involved in various cellular processes, including oxidative phosphorylation, apoptosis, Fe-S biogenesis, and ion homeostasis (Ryan and Hoogenraad, 2007; Meisinger et al., 2008; Pagliarini et al., 2008). Most of these proteins are first synthesized as precursors in the cytosol before being imported into mitochondria and sorted to the appropriate subcompartment: outer membrane, intermembrane space (IMS), inner membrane, and matrix. All precursor proteins that cross the outer membrane use the translocase complex of the outer membrane (TOM), before diverging along specific sorting pathways (Baker et al., 2007; Neupert and Herrmann, 2007; Bolender et al., 2008). The inner membrane contains two translocases of the inner membrane (TIM). Precursors with matrix-targeting signals are sorted to the TIM23 complex of the inner membrane. Although most of these precursors translocate into the matrix, a subset contain hydrophobic stop-sorting signals and are laterally released from the TIM23 complex into the inner membrane (Baker et al., 2007; Neupert and Herrmann, 2007; Bolender et al., 2008). Direct contacts between components of the TOM and TIM23 complexes may prevent these hydrophobic precursors from accessing the aqueous IMS, thereby preventing their aggregation (Chacinska et al., 2005; Mokranjac et al., 2005). However, many membrane protein precursors that contain internal targeting signals transit the IMS by using distinct mechanisms. For example, precursors of the metabolite carrier family, such as the ADP/ATP carrier (AAC), translocate the outer membrane TOM complex and are then directed to the TIM22 complex for their integration into the inner membrane (Rehling et al., 2004). Transit of these precursors across the IMS and to the TIM22 complex is accomplished through the chaperone-like action of the small TIM family members. Members of the small TIM family also assist in the transit of β-barrel precursor proteins from the TOM complex to the IMS side of the outer membrane sorting and assembly machinery (SAM) (Bolender et al., 2008; Hoppins and Nargang, 2004; Wiedemann et al., 2004).
All five members of the small TIM family of IMS space proteins (Tim8, Tim9, Tim10, Tim12, and Tim13) contain a “twin CX3C” motif with four conserved cysteines separated by 11–17 residues (Koehler, 2004). Tim9, Tim10, and Tim12 are all essential for yeast cell viability. The majority of Tim9 and Tim10 are found together in a soluble hexameric complex, with the remainder associating with Tim12 at the TIM22 complex (Koehler, 2004). The 3.5-Å structure of the human Tim9–Tim10 hexameric complex revealed that it forms an α-propeller–like structure with alternating subunits (Webb et al., 2006). Both Tim9 and Tim10 adopt an α-hairpin conformation stabilized by two intramolecular disulfide bonds formed between the conserved cysteines of the twin CX3C motif.
Although the report by Webb et al. (2006) provided important structural insight into the overall assembly, it is still not clear how the Tim9–Tim10 complex functions at the molecular level. Studies of the yeast small TIM proteins, mainly using in vitro approaches, have led to the proposal that the N-terminal region of Tim10 is involved in substrate binding, whereas Tim9 principally acts to stabilize the Tim9–Tim10 complex (Gentle et al., 2007; Vergnolle et al., 2005, 2007). However, analysis of the human Tim9–Tim10 crystal structure reveals the importance of both subunits in stabilizing the complex via intermolecular contacts, whereas cross-linking analysis has found that Tim9 also makes close contacts with precursor substrates (Davis et al., 2000). To address this in more detail, we solved the crystal structure of the yeast Tim9–Tim10 complex and used the structural information to address function in vivo. Our results confirm the importance of the Tim9–Tim10 complex in protein import and also uncover a crucial role for the N-terminal tentacle region of Tim9 in facilitating the translocation of precursor substrates into the IMS.
MATERIALS AND METHODS
Crystal Structure Determination of Recombinant the Yeast Tim9–Tim10 Complex
The yeast Tim9 and Tim10 open reading frames were cloned into pGEX-4T2 (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) to produce glutathione transferase (GST)-fusion constructs. Proteins were expressed in Escherichia coli strain Origami DE3 pLysS (Novagen, Madison, WI) and purified (Webb et al., 2006). The Tim9–Tim10 complex in 20 mM Tris-HCl, pH 8.0, 150 mM NaCl was crystallized using 24-well hanging-drop trays, and those used for data collection grew to ∼100 × 50 × 40 μm within 1 wk in the precipitant 0.1 M 2-(N-morpholino)ethanesulfonic acid, pH 6.5, and 3 M sodium formate. Data were collected from a single crystal belonging to the trigonal space group R32 (hexagonal setting) (cell dimensions a = 58.2, c = 243.7 Å) on Beamline ID-23 (European Synchrotron Radiation Facility [ESRF], Grenoble, France) (Table 1). The asymmetric unit contains one subunit each of Tim9 and Tim10, and the biological hexamer was generated by the crystallographic threefold operator. Intensity data were processed and scaled using HKL2000 (Otwinowski and Minor, 1997).
Table 1.
Data collection statistics for the yeast Tim9-Tim10 complex
| Statistic | Value |
|---|---|
| Space group | R32 |
| Cell dimensions (Å) | a = 58.2, c = 243.7 Å |
| Wavelength (Å) | 0.9762 (peak) |
| Resolution range (Å) | 50.0–2.5 |
| Total no. of reflections | 187152 |
| No. of unique reflections | 5723 |
| Rmerge (overall/outer shell) | 0.087/0.4 |
| I/σI (overall/outer shell) | 18.9/5.5 |
| Completeness (%) (overall/outer shell) | 97.8/99.7 |
| Redundancy (overall/outer shell) | 7.0/7.1 |
The electron density was phased by molecular replacement, using PHASER as implemented in the CCP4i suite (McCoy et al., 2007). The search model, derived from the human Tim9-Tim10 structure (2BSK), included residues 13–70 of hTim9 and 14–71 of hTim10 (Webb et al., 2006). A single solution (Z-scores are 6.1 [rotation] and 8.7 [translation]) was used to phase initial electron density maps. Solvent flattening and histogram matching protocols implemented in DM (Cowtan and Zhang, 1999) were used to improve the electron density maps. Model building was carried out using COOT (Emsley and Cowtan, 2004) and O (Jones et al., 1991), and refinements carried out using CNS (Brunger et al., 1998). Atomic coordinates were refined by maximum likelihood and simulated annealing procedures, alternating with cycles of individual B-factor refinement. Side chains of the following residues were truncated at Cβ due to positional disorder: Tim9 Phe12, Gln13, Gln18, Lys19, Gln20, Met21, Lys22, Asp23, Arg26, Lys65, Gln77 and Tim10 Ser15, Gln16, Gln17, Lys18, Glu23, Glu25, Asp27, Lys56, Gln81, Met83. Refinement of 1029 nonhydrogen atoms and 43 water molecules against 5427 data (an additional 293 terms were excluded from refinement and retained as a reference set) in the resolution range 50.0–2.5 Å converged at an Rf of 0.275, Rw 0.245. There are no outliers in the Ramachandran plot (96.2% of residues in the most favored regions), and root mean square deviations (r.m.s.d.) of bond lengths and angles are 0.006 Å and 1.11°, respectively. The average B-factor is 78 Å2. The structure has been deposited in the Protein Structure Database (PDB; accession 3DXR).
Cloning and Construction of Yeast Strains
Yeast TIM9 and TIM10 genes were cloned into the single-copy, centromeric pRS415(LEU2) and pRS413(HIS3) yeast shuttle-vectors, respectively (New England Biolabs, Ipswich, MA). The expression of all proteins was under the control of the endogenous TIM9 or TIM10 promoters. Point mutations were incorporated using overlap polymerase chain reaction (PCR), whereas inverse PCR was used for generating truncation mutants. The Saccharomyces cerevisiae strains constructed and used in this study are detailed in Table 2. The pRS415(LEU2) plasmid, encoding wild-type or mutant Tim9, was transformed into GB090 cells. The same procedure was used to generate Tim10 mutant yeast strains except that the pRS413(HIS3) plasmid encoding wild-type or mutant Tim10 was transformed into GB100 cells. Transformants were passaged onto minimal glucose media supplemented with 650 mg/l 5-fluoroorotic acid (5-FOA) to select for loss of the plasmid containing the URA3 gene and wild-type TIM9 or TIM10. Viable strains were passaged to 5-FOA plates and then to glycerol-containing agar plates.
Table 2.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| YPH499 | MATaade2-101 his3-D200 leu2-D1 ura3-52 trp1-D63 lys2-801 | Sikorski and Heiter (1989) |
| GB090 | MATaade2-101 his3-D200 leu2-D1 ura3-52 trp1-D63 lys2-801 tim9::ADE2 pGB5174A3)-TIM9 | This study |
| GB100 | MATaade2-101 his3-D200 leu2-D1 ura3-52 trp1-D63 lys2-801 tim10::ADE2 pGB5184(URA3)-TIM10 | Truscott et al. (2002) |
| Tim9WT | MATaade2-101 his3-D200 leu2-D1 ura3-52 trp1-D63 lys2-801 tim9::ADE2 pRS415(LEU2)-Tim9WT | This study |
| tim9ΔC10 | MATaade2-101 his3-D200 leu2-D1 ura3-52 trp1-D63 lys2-801 tim9::ADE2 pRS415(LEU2)-tim9ΔC10 | This study |
| tim9ΔC13 | MATaade2-101 his3-D200 leu2-D1 ura3-52 trp1-D63 lys2-801 tim9::ADE2 pRS415(LEU2)-tim9ΔC13 | This study |
| tim9ΔC14 | MATaade2-101 his3-D200 leu2-D1 ura3-52 trp1-D63 lys2-801 tim9::ADE2 pRS415(LEU2)-tim9ΔC14 | This study |
| tim9ΔN10 | MATaade2-101 his3-D200 leu2-D1 ura3-52 trp1-D63 lys2-801 tim9::ADE2 pRS415(LEU2)-tim9ΔN10 | This study |
| tim9ΔN15 | MATaade2-101 his3-D200 leu2-D1 ura3-52 trp1-D63 lys2-801 tim9::ADE2 pRS415(LEU2)-tim9ΔN15 | This study |
| tim9E52K | MATaade2-101 his3-D200 leu2-D1 ura3-52 trp1-D63 lys2-801 tim9::ADE2 pRS415(LEU2)-tim9E52K | This study |
| Tim10WT | MATaade2-101 his3-D200 leu2-D1 ura3-52 trp1-D63 lys2-801 tim10::ADE2 pRS413(HIS3)-Tim10WT | This study |
| tim10ΔC10 | MATaade2-101 his3-D200 leu2-D1 ura3-52 trp1-D63 lys2-801 tim10::ADE2 pRS413(HIS3)-tim10ΔC10 | This study |
| tim10ΔN12 | MATaade2-101 his3-D200 leu2-D1 ura3-52 trp1-D63 lys2-801 tim10::ADE2 pRS413(HIS3)-tim10ΔN12 | This study |
| tim10ΔN21 | MATaade2-101 his3-D200 leu2-D1 ura3-52 trp1-D63 lys2-801 tim10::ADE2 pRS413(HIS3)-tim10ΔN21 | This study |
| tim10K68E | MATaade2-101 his3-D200 leu2-D1 ura3-52 trp1-D63 lys2-801 tim10::ADE2 pRS413(HIS3)-tim10K68E | This study |
Mitochondrial In Vitro Import Assays
Yeast mitochondria were isolated according to Daum et al. (1982). In vitro import assays were performed at 24°C according to Stojanovski et al. (2007). Heat-shocking of mitochondria before import was performed as described previously (Kang et al., 1990). Arrest of 35S-AAC-dihydrofolate reductase (DHFR) at the TOM complex, cross-linking, and immunoprecipitation analysis was performed according to Ryan et al. (1999). Cross-linking was performed using 0.8 mM m-maleimidobenzoyl-N-hydroxy succinimide ester (Pierce Chemical, Rockford, IL). After import, mitochondrial proteins were solubilized in 1% (wt/vol) digitonin (Calbiochem, San Diego, CA), 50 mM NaCl, 10% (vol/vol) glycerol, and 20 mM Bis-Tris, pH 7.0, for 15 min on ice. Samples were subjected to blue native-polyacrylamide gel electrophoresis (BN-PAGE) and second-dimensional PAGE according to Lazarou et al. (2007). Fractions for SDS-PAGE analysis were first precipitated with trichloroacetic acid (TCA). Radiolabeled proteins were detected using phosphorimaging. For Western blot analysis, gels were transferred to membranes and immunodetection was achieved using ECL reagents (GE Healthcare) and a gel documentation system (Syngene, Frederick, MD).
RESULTS
Molecular Architecture of the Yeast Tim9–Tim10 Complex
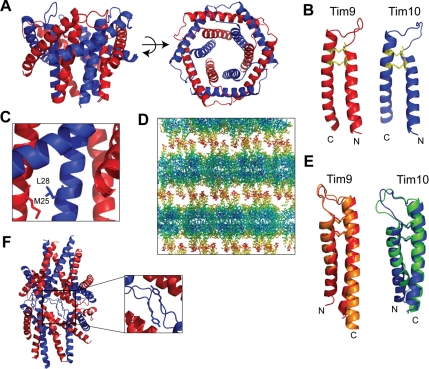
The yeast Tim9–Tim10 complex is composed of alternating subunits arranged into a six-bladed α-propeller (Figure 1A). Residues 12–80 of Tim9 and 15–83 of Tim10 were used to refine the structure to a considerably higher resolution (2.5 Å) than the human assembly (3.5 Å), thus providing a more detailed picture of the inter- and intramolecular contacts. The topology of the yeast Tim9–Tim10 complex is similar to that of its human counterpart, and the peptide backbone superimposes closely in the well-ordered core region of the structure (r.m.s.d. of 37 Cα atoms is 0.87 Å for Tim9 and 0.77 Å for Tim10). The molecular curvature at each subunit interface in the hexamer creates a central channel 10 Å wide at the narrowest point and 14 Å at the widest, and lined predominantly with hydrophilic residues. Six central cysteine-loops form a relatively flat face normal to the molecular axis, whereas 12 tentacle-like projections emanate from the opposite face. Because the N- and C-terminal extremities of Tim9 and Tim10 are highly mobile, 10–15 residues at each terminus could not be accurately modeled into the electron density. Individual subunits have a helix-turn-helix α-hairpin -fold supported by intramolecular disulfide bonds arising from cysteines of the twin CX3C motif (Figure 1B). Disulfide bond formation in the IMS is due to a novel disulfide trapping and release relay (Chacinska et al., 2004; Naoe et al., 2004; Allen et al., 2005; Mesecke et al., 2005; Koehler et al., 2006). The steric bulk of side chains adjacent to the signature cysteine residues causes the flanking helices to splay apart in a bent hairpin topology. Two rings of helices, inner and outer, comprise the basic modular scaffold of the assembly. The inner N-terminal helices make few intermolecular contacts within the ordered core region, but hydrophobic side chains closer to the N-terminal regions cluster adjacent subunits. In one cluster, Val15 and Val16 of Tim9 interact with Leu26 of Tim10, and in the other cluster, Met21 and Met25 of Tim9 connect with Ala24 and Leu28 of Tim10 (Figure 1C). In contrast, the outer C-terminal helices are aligned ∼60° to the molecular axis, creating the blades of the propeller, and they make substantial contacts with adjacent subunits only in the core region.
Figure 1.
Crystal structure of the yeast Tim9–Tim10 hexameric complex. (A) Ribbon diagram of the Tim9–Tim10 complex with a side view (left) and an aerial view (right). Tim9 is in red and Tim10 is in blue. (B) Ribbon diagrams depicting the individual Tim9 and Tim10 subunits forming hairpin-like structures braced by two intramolecular disulfide bonds (yellow). (C) Ribbon diagram depicting the interaction between Met25 of Tim9 and Leu28 of Tim10. (D) Crystal lattice packing of the Tim9–Tim10 complex. Residues are colored according to their individual B-factor. Blue corresponds to residues with low thermal mobility and red corresponds to residues with high thermal mobility. The B-factors ranges from 35 to 153 Å2. (E) -Fold comparison of the human and yeast subunits within the Tim9–Tim10 complex. hTim9 (orange) is superimposed on yTim9 (red), and hTim10 (green) has been superimposed on yTim10 (blue). (F) Ribbon diagram depicting Tyr49 of Tim10 packing into the core of an adjacent Tim9–Tim10 complex.
In the trigonal crystal lattice, layers of hexamers alternate such that the flat faces are closely packed head-to-head, whereas the tentacles loosely intertwine with others in adjacent layers (Figure 1D). The relatively high mobility of the unresolved tentacle regions is indicated by significantly increased thermal parameters of the extremities. The Cα backbone over the central cysteine-loop superimposes closely in the human and yeast structures (Figure 1E), and the angle between inner and outer helices of individual Tim9 or Tim10 subunits is conserved. In the yeast structure, the side chain of Tyr49 in Tim10 flips out of the hexamer core, swapping with a symmetry-related residue from a neighboring assembly (Figure 1F). Critical molecular contacts influencing the conformation of the central cysteine-loops, preserved between the human and yeast structures, include two buried ion-pairs (Tim9K62-Tim10E58 and Tim10K68-Tim9E52) that connect the six subunits into a ring. These side chains of these residues also make main chain contacts, placing an array of molecular constraints on the conformation of the central loop.
Disruption of a Conserved Intermolecular Salt-Bridge Impairs Tim9–Tim10 Complex Assembly and Is Deleterious to Yeast
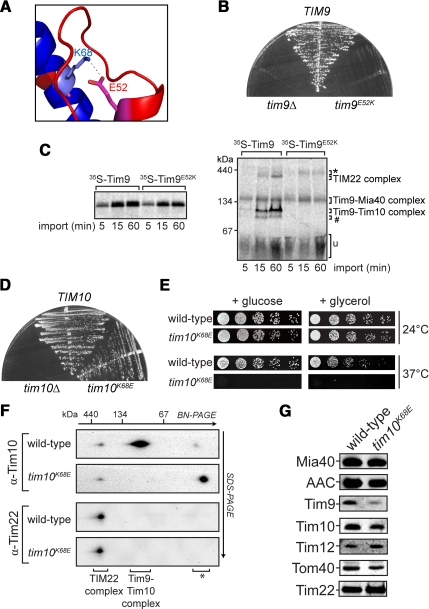
It has been debated previously as to whether the essential function of Tim9 and Tim10 is dependent on the presence of the soluble hexameric complex (Murphy et al., 2001; Truscott et al., 2002). Both studies used temperature-sensitive yeast strains harboring a mutant Tim10 that may contain several point mutations that can impair Tim10 function in multiple ways (e.g., complex assembly and substrate binding), hence complicating analysis. We therefore sought to disrupt the Tim9–Tim10 complex through a single mutation. Glu52 in yeast Tim9 was substituted with Lys (Tim9E52K), to break the ion-pair formed with Lys68 in Tim10 (Figure 2A). However, complementation analysis revealed that transformation of tim9Δ yeast cells with a plasmid expressing Tim9E52K did not support growth (Figure 2B). As expected, a plasmid harboring wild-type Tim9 restored cell growth, whereas tim9Δ cells were not viable. This is the first report where a single mutation in a small TIM protein causes lethality. Tim9E52K was still able to be imported into mitochondria with similar kinetics to the wild-type Tim9 precursor (Figure 2C, left); however, BN-PAGE analysis revealed that it was unable to assemble into either the hexameric or the TIM22 complex (Figure 2C, right). Instead Tim9E52K was arrested at previously defined intermediate complexes that contain Mia40 (Chacinska et al., 2004; Stojanovski et al., 2008). Thus, the assembly defect observed for Tim9E52K may account for the lethality observed.
Figure 2.
Disruption of a conserved salt-bridge between Tim9 and Tim10. (A) Ribbon diagram showing the formation of the Tim9E52–Tim10K68 salt bridge. (B) Growth of yeast strains expressing wild-type Tim9, Tim9E52K, or lacking Tim9 plated on minimal glucose media supplemented with 5-FOA at 24°C. (C) 35S-Tim9 and 35S-Tim9E52K were imported into yeast mitochondria at 24°C for the times indicated. Mitochondria were reisolated and subjected to SDS-PAGE (left) or BN-PAGE (right) followed by phosphorimaging (*, large Tim9–Mia40 complex; #, Tim9–Tim12 intermediate complex; u, unassembled protein). (D) Growth of yeast strains expressing wild-type Tim10, Tim10K68E, or lacking Tim10 plated on minimal glucose media supplemented with 5-FOA at 24°C. (E) Serial dilutions of yeast strains expressing wild-type Tim10 or Tim10K68E were generated and spotted onto nonfermentable (+ glycerol) or fermentable (+ glucose) media and incubated at 24°C or 37°C as indicated. (F) Mitochondria from yeast strains expressing wild-type Tim10 or Tim10K68E were solubilized and run on BN-PAGE in the first dimension and SDS-PAGE in the second dimension before Western blot analysis (*, nonassembled protein). (G) Equal amounts of wild-type or tim10K68E mitochondria were subjected to SDS-PAGE and subsequent Western blot analysis using marker antibodies, as indicated.
The complementary charge reversal mutation in Tim10 was also generated, mutating Lys68 to Glu. In this case, however, cells expressing Tim10K68E were viable (Figure 2D). Plating serial dilutions of this strain showed that tim10K68E cells were temperature sensitive for growth on both fermentable and nonfermentable media (Figure 2E). However, when incubated at lower temperatures (24°C), tim10K68E cells grew like the corresponding wild-type strain. Mitochondria were isolated from cells grown at 24°C and the presence of the Tim9–Tim10 complex was assessed using BN-PAGE and SDS-PAGE in the second dimension (2D BN-PAGE). Tim10K68E was not found in the hexameric assembly but was still present in the TIM22 complex (Figure 2F, top). Western analysis confirmed that the TIM22 complex was intact in tim10K68E mitochondria (Figure 2F, bottom), and the steady-state levels of marker proteins tested in tim10K68E mitochondria seemed normal, although Tim9 levels were reduced (Figure 2G). A possibility is that impaired assembly of the soluble Tim9–Tim10 complex leads to degradation of nonassembled subunits. We conclude that Lys68 of Tim10 is directly involved in stabilizing the soluble Tim9–Tim10 complex but is not required for assembly of Tim10 into the TIM22 complex.
Carrier and β-Barrel Import Pathways Are Defective in Mitochondria with Impaired Soluble Tim9–Tim10 Complex
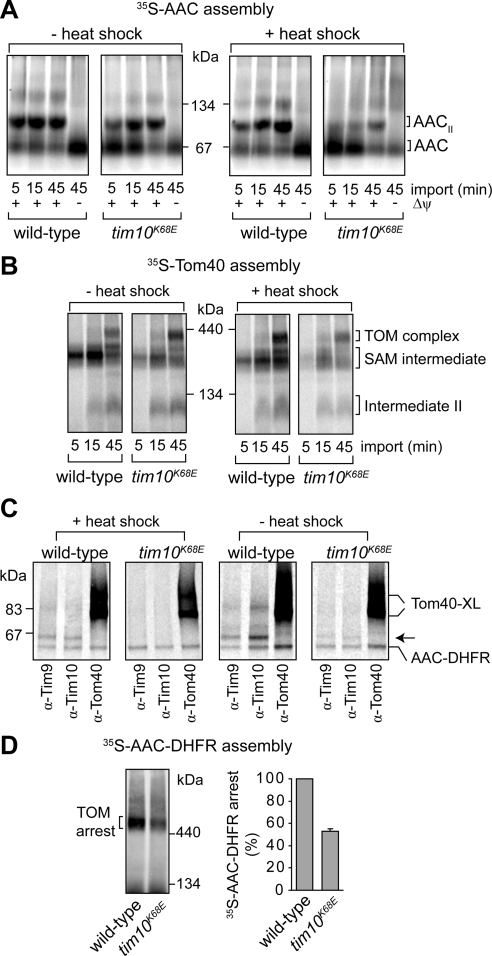
The import and assembly of the model small TIM substrate, AAC, was analyzed to test the effect of the salt-bridge mutation in Tim10. 35S-AAC import and assembly can be monitored by BN-PAGE, because it forms a dimeric assembly in the inner membrane (AACII; Figure 3A). Assembly of 35S-AAC at 24°C was mildly impaired in tim10K68E mitochondria in comparison with the corresponding wild-type mitochondria (Figure 3A, left). Because tim10K68E cells are temperature sensitive for growth, mitochondria were subjected to a 37°C heat-shock pretreatment before the import assay at 24°C. Such a treatment did not affect the stability of the TIM22 complex (data not shown). However, in this case, the import and assembly of 35S-AAC into tim10K68E mitochondria was strongly reduced (Figure 3A, right). Import of matrix-targeted Su9-DHFR, which is not mediated by members of the small TIM family, was not impaired relative to wild-type mitochondria (Supplemental Figure S1A and S1B), indicating that neither matrix import nor the membrane potential (Δψ) are defective.
Figure 3.
Analysis of protein import in tim10K68E mitochondria. (A) 35S-AAC was imported into wild-type or tim10K68E mitochondria preincubated at 24°C (− heat shock) or 37°C (+ heat-shock) before BN-PAGE analysis. Dissipation of the Δψ blocks AAC insertion into the inner membrane (AACII, AAC dimer). (B) 35S-Tom40 was imported into wild-type and tim10K68E mitochondria preincubated at 24°C (− heat shock) or 37°C (+ heat shock). Tom40 assembly intermediates were analyzed by BN-PAGE and phosphorimaging (SAM intermediate, Tom40 precursor at the SAM complex; intermediate II, Tom40 intermediate integrated into the outer membrane). (C) Wild-type or tim10K68E mitochondria were either pretreated with a heat shock or not subjected to a heat shock before incubation with 35S-AAC-DHFR in the presence of methotrexate. After cross-linking, mitochondria were lysed and subjected to immunoprecipitation with antibodies against Tim9, Tim10, and Tom40 before SDS-PAGE and phosphorimaging. Some AAC-DHFR is nonspecifically pulled down (Tom40-XL, cross-links formed between Tom40 and AAC-DHFR; arrow, Tim9 or Tim10 cross-links with AAC-DHFR). (D) 35S-AAC-DHFR arrested at the TOM complex in wild-type and tim10K68E mitochondria was analyzed using BN-PAGE and phosphorimaging (left). Level of arrested AAC-DHFR was quantified (right; error bars represent SD; n = 3).
Any mutation in Tim10 has the potential to affect both the function of the Tim9–Tim10 complex and the TIM22 complex, and it is not possible to discriminate where the AAC import defect resides. We therefore sought to determine the effect of mutations on the specific function of the soluble Tim9–Tim10 complex by using two additional approaches. We first tested the assembly of the precursor Tom40, which follows the outer-membrane β-barrel pathway. It has been established that β-barrel precursors translocate the TOM complex and are then inserted into the outer membrane from the IMS side via SAM (Model et al., 2001; Paschen et al., 2003; Wiedemann et al., 2003). This sorting pathway uses the action of members of the small TIM family and is exclusive of the TIM22 complex (Wiedemann et al., 2004). The import pathway of 35S-Tom40 can be resolved using BN-PAGE into consecutive assembly intermediates. This includes its association with the SAM complex (SAM intermediate), outer membrane integration (intermediate II), and its final assembly into the TOM complex (Model et al., 2001; Wiedemann et al., 2004). The accumulation of 35S-Tom40 at the SAM complex was reduced in tim10K68E mitochondria (with and without heat-shock treatments), compared with wild type (Figure 3B). These results suggest that the impaired passage of Tom40 precursors through the IMS is due to defects in the soluble Tim9–Tim10 complex. In the second approach, we tested whether the chimeric precursor substrate AAC-DHFR can contact Tim9 and/or Tim10K68E in tim10K68E mitochondria using chemical cross-linking. In the presence of methotrexate, 35S-AAC-DHFR arrests at the TOM complex but can still contact the Tim9-Tim10 complex (Ryan et al., 1999; van Wilpe et al., 1999; Wiedemann et al., 2001). Isolated mitochondria were subjected to a heat-shock pretreatment before accumulation of 35S-AAC-DHFR at the TOM complex (at 24°C). In wild-type mitochondria, AAC-DHFR was cross-linked to Tim9, Tim10, and Tom40 as expected (Figure 3C). However, in tim10K68E mitochondria only cross-links to Tom40 were seen, consistent with the absence of the soluble Tim9–Tim10 complex. Because the import of AAC, and to a lesser extent Tom40, was more defective in tim10K68E mitochondria that were subjected to a heat-shock, it is possible that some soluble Tim9–Tim10K68E complex may in fact be present in nonheat-shocked mitochondria. Indeed, cross-linking analysis using mitochondria not treated to a heat-shock revealed that the arrested AAC-DHFR precursor can contact both Tim9 and Tim10K68E (Figure 3C). This suggests that some Tim9–Tim10K68E complex may form; yet, the assembly is labile as evidenced by heat treatment, separation by BN-PAGE, and reduced steady-state protein levels.
The level of AAC-DHFR accumulation at the TOM complex was reduced to 50% in tim10K68E mitochondria compared with wild type (Figure 3D). This reduction was also observed when mitochondria were not subjected to a heat-shock treatment (data not shown), consistent with the overall reduction in the level of Tim9 and Tim10K68E. These results therefore support a role for the soluble Tim9–Tim10 complex in aiding precursor translocation into the IMS and demonstrate its functional importance in both β-barrel and carrier protein import pathways.
N- and C-Terminal Tentacle Regions of Tim9 Are Required for Yeast Cell Viability
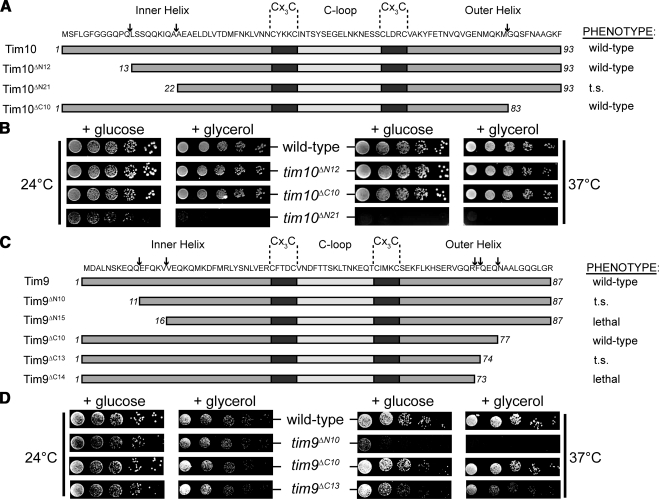
Thermal parameters indicate that all 12 helices projecting from the ventral face of Tim9-Tim10 increase in mobility with distance from the core of the assembly. The extensive plasticity of these tentacles may be a mechanism for interaction with a variety of substrates. It has been proposed that the N-terminal tentacle of Tim10 acts as a substrate-sensing domain, whereas Tim9 is not required for substrate binding but stabilizes the hexameric complex (Gentle et al., 2007; Vergnolle et al., 2005, 2007). However, previous studies used very large N- and C-terminal truncations of both Tim9 and Tim10 that, based on structural considerations, are likely to prevent assembly of the Tim9–Tim10 complex. Here, the importance of the N- and C-terminal regions of Tim9 and Tim10 was analyzed by truncating regions predicted not to disrupt any stabilizing intermolecular contacts. Initially, 10 to 12 residues were removed from each end of Tim9 and Tim10 (termed tim9ΔN10, tim9ΔC10, tim10ΔN12, and tim10ΔC10) (Figure 4, A and C). In all cases, these mutant constructs were able to complement growth of cells lacking the wild-type small TIM, indicating that the termini are not essential for yeast cell viability. Removal of 10–12 residues from the N or C terminus of Tim10 had no effect on yeast growth under the conditions tested (Figure 4, A and B). This was surprising given the previously ascribed role of the N-terminal domain of Tim10 as a substrate-sensing region for carrier precursors (Vergnolle et al., 2005, 2007; Gentle et al., 2007). Because Tim10 or its mutant variants are under the control of the wild-type TIM10 promoter in the plasmid, it is unlikely that there is overexpression to mask any growth phenotype. In addition, Western analysis indicated that the levels of plasmid-borne Tim10 are similar to that found in a yeast strain containing a genomic copy of TIM10 (Supplemental Figure S2A). Tim10ΔN12 levels were somewhat reduced even though a growth phenotype was not observed (Supplemental Figure S2A and S3A). Expression of Tim10 lacking 21 N-terminal residues (tim10ΔN21) was still able to complement growth of yeast cells, although severe growth phenotypes were observed (Figure 4, A and B). In vitro import of Tim10ΔN21 did not seem to be defective (Supplemental Figure S2D). When analyzing the Tim9 truncation mutants, it was found that Tim9 lacking only 10 N-terminal residues displayed a strong growth defect (Figure 4, C and D). Furthermore, expression of Tim9ΔN15 was lethal in yeast cells (Figure 4C and Supplemental S2B). Because expression of the N-terminal truncation mutants in Tim9 resulted in unexpectedly strong growth defects in comparison with Tim10 truncation mutants, we also addressed the requirements for the C-terminal tentacle of Tim9. Yeast cells expressing Tim9ΔC10 were able to grow on fermentable and nonfermentable media like the corresponding wild-type yeast cells that harbor a plasmid with a normal copy of TIM9. However, deletion of three additional C-terminal residues of Tim9 (tim9ΔC13) caused a moderate temperature-sensitive phenotype (Figure 4, C and D), and truncation of an additional residue (tim9ΔC14) was lethal (Figure 4C and Supplemental S2C). The lethality observed for both tim9ΔN15 and tim9ΔC14 does not seem to be due to a defect in the import of these mutant proteins (Supplemental Figure S2D). Thus, we conclude that both the N- and C-terminal tentacle regions of Tim9 have essential functions in yeast.
Figure 4.
Generation of Tim9 and Tim10 truncation mutants. (A) Schematic representation of Tim10 truncation mutants. Arrows point to the most N- or C-terminal residue remaining in the truncation mutants. Growth phenotypes are noted. (B) The growth of yeast strains expressing Tim10 and truncation mutants was analyzed at 24 or 37°C by serially diluting and spotting yeast onto glucose or glycerol containing media. (C) Schematic representation of Tim9 truncation mutants. Arrows point to the most N- or C-terminal residue remaining in the truncation mutants. Growth phenotypes are noted. (D) The growth of yeast strains expressing Tim9 and truncation mutants was analyzed at 24 or 37°C by serially diluting and spotting yeast onto glucose- or glycerol-containing media.
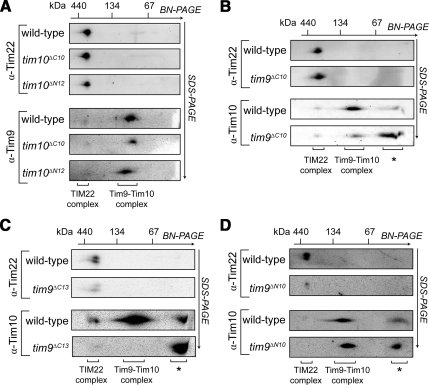
The presence and stability of complexes containing Tim9 and Tim10 truncation mutants was next assessed. In all cases, mitochondria were isolated from strains grown at 24°C. For both N- and C-terminal truncation mutants of Tim10, the levels and the stability of the soluble hexamer and the TIM22 complex were unaffected (Figure 5A) (hexamer levels in Tim10 truncation mutants were analyzed by assessing Tim9, because the Tim10 antibody did not recognize the C-terminally truncated forms of Tim10; Supplemental Figure S3A). The steady-state levels of marker proteins tested in the Tim10 truncation mutants were largely unaffected (Supplemental Figure S3A). Although the level and stability of the TIM22 complex in mitochondria containing Tim9ΔC10 seemed normal (Figure 5B, top), the Tim9ΔC10–Tim10 complex displayed some structural instability and its decreased levels were concomitant with an increase in free Tim10 (Figure 5B, bottom) (in this case the hexameric complex was analyzed with Tim10 antibodies because the Tim9 antibody failed to recognize the C-terminally truncated Tim9). Removal of three additional C-terminal residues in Tim9 caused a decrease in the levels of the TIM22 complex (Figure 5C, top). Furthermore, yeast cells expressing Tim9ΔC13 showed no detectable Tim9ΔC13–Tim10 hexamer, although a large amount of nonassembled Tim10 was present, perhaps suggesting that the complex was further destabilized under the conditions of BN-PAGE (Figure 5C, bottom). Cross-linking analysis further supported the presence of a Tim9ΔC13–Tim10 complex within mitochondria (data not shown). In tim9ΔN10 mitochondria, levels of the TIM22 complex were strongly reduced, yet the levels of the soluble Tim9ΔN10–Tim10 complex seemed normal (Figure 5D). The steady-state levels of most marker proteins tested in the Tim9 truncation mutants, including Tim22, were also similar to wild-type mitochondria (the exception being decreased levels of Tim12 in tim9ΔC13 mitochondria; Supplemental Figure S3B and S3C). Unassembled Tim22 was not observed on the 2D BN-PAGE, presumably due to aggregating and not entering the native gel or by broadly smearing along the gel strip preventing antibody detection. Our results implicate both the extreme N- and C-terminal regions of Tim9 in the assembly and/or stability of the TIM22 complex.
Figure 5.
Analysis of TIM22 and Tim9–Tim10 complexes in truncation mutants. (A–D) Mitochondria isolated from the yeast strains indicated were subjected to BN-PAGE before SDS-PAGE in the second dimension followed by Western blot analysis. The positions of the TIM22 and Tim9–Tim10 complexes are indicated. (*, nonassembled forms of the proteins).
Tim9 Truncation Mutants Exhibit Strong Defects in Carrier and β-Barrel Sorting
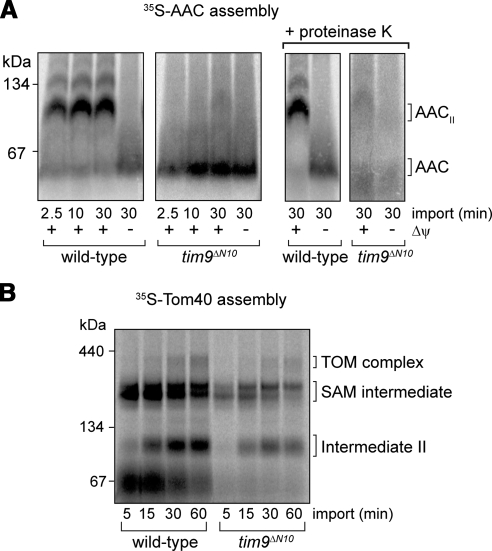
Import and assembly analysis of 35S-AAC and 35S-Tom40 by using mitochondria isolated from tim10ΔN12 or tim10ΔC10 cells revealed no defects (Supplemental Figure S4A and S4B), even after a heat-shock pretreatment (data not shown). Coupled with normal growth phenotypes and the presence of wild-type TIM22 and Tim9–Tim10 complexes, it can be concluded that the N- and C-terminal ends of Tim10 are not critical for substrate protein import under the conditions used. We next tested for import defects in mitochondria expressing Tim9ΔC13 and Tim9ΔN10. Import of the matrix-targeted Su9-DHFR precursor was not impaired in either preparation, even when mitochondria were subjected to a heat-shock pretreatment, indicating that the matrix-sorting pathway and the Δψ were not compromised (Supplemental Figure S4C and S4D). In vitro import and assembly of 35S-AAC in tim9ΔC13 mitochondria was inhibited when mitochondria were subjected to a heat-shock pretreatment (data not shown). However, the TIM22 complex was also found to be destabilized after the heat-shock treatment, making it difficult to elucidate at which stage the import of the AAC precursor was defective. When 35S-AAC was imported into tim9ΔN10 mitochondria, virtually no assembly of AAC was seen, even in the absence of a heat-shock pretreatment (Figure 6A). A large amount of 35S-AAC precursor instead migrated closer to the bottom of the gel. To test whether this represented nonimported AAC or AAC precursor that had translocated the outer membrane but was not inserted into the inner membrane (the stage III intermediate; Ryan et al., 1999), mitochondria were subsequently treated with proteinase K. In contrast to wild-type mitochondria, virtually all 35S-AAC precursor associated with tim9ΔN10 mitochondria was degraded, indicating that the precursor did not translocate across the outer membrane (Figure 6A, right). The assembly of newly imported 35S-Tom40 was also impaired in tim9ΔN10 mitochondria compared with the corresponding wild-type mitochondria (Figure 6B) and its ability to cross the outer membrane was also reduced (data not shown). The level of 35S-Tom40 binding to the SAM complex was decreased in tim9ΔN10 mitochondria, indicating that the soluble Tim9ΔN10–Tim10 complex is defective in substrate translocation and/or binding. The residual import and assembly seen for the 35S-Tom40 precursor is most likely due to the ability of the nonessential Tim8–Tim13 complex to also engage with this substrate (Hoppins and Nargang, 2004). In contrast, the Tim8–13 complex does not interact with the 35S-AAC precursor (Davis et al., 2000; Truscott et al., 2002; Hoppins and Nargang, 2004), and this correlates with the strong import defect observed for AAC.
Figure 6.
Import analysis in tim9ΔN10 mitochondria. (A) 35S-AAC was imported into wild-type and tim9ΔN10 mitochondria (without a heat-shock pretreatment) at 24°C in the presence and absence of Δψ. After treating some samples to proteinase K, mitochondria were isolated, solubilized, and analyzed using BN-PAGE and phosphorimaging. Right, AAC complexes (AAC and AACII) protected from proteinase K treatment of intact mitochondria. (B) 35S-Tom40 was imported into wild-type and tim9ΔN10 mitochondria (without heat shock) at 24°C and analyzed using BN-PAGE and phosphorimaging. (SAM intermediate, Tom40 precursor at the SAM complex; intermediate II, Tom40 intermediate integrated into the outer membrane).
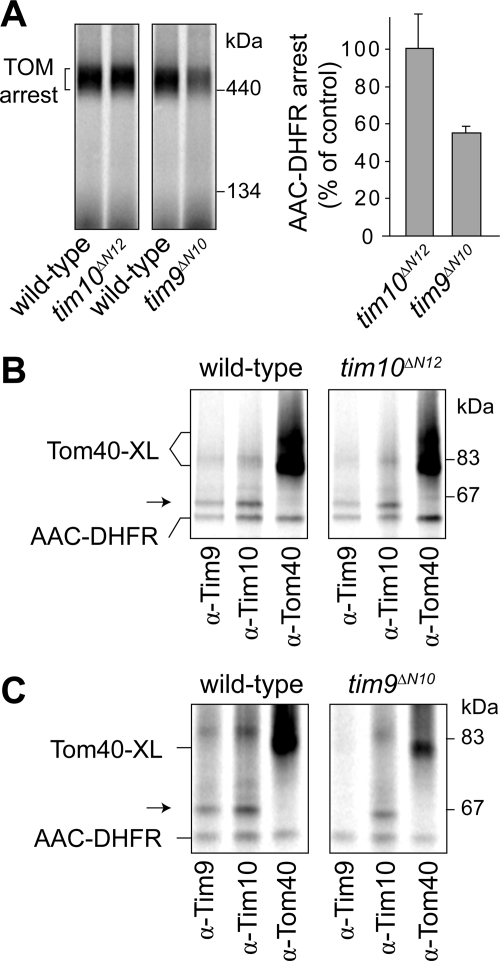
Given the impaired import of 35S-AAC and 35S-Tom40 into tim9ΔN10 mitochondria, we asked whether the Tim9ΔN10–Tim10 complex could still contact the 35S-AAC-DHFR precursor arrested at the TOM complex. Wild-type and tim10ΔN12 mitochondria were used as controls. AAC-DHFR was arrested at the TOM complex in all mitochondrial preparations when analyzed using BN-PAGE (Figure 7A); however, the level of accumulation was strongly reduced in tim9ΔN10 mitochondria. Cross-linking analysis (Figure 7, B and C) showed that in tim10ΔN12 mitochondria, Tim9, Tim10ΔN12, and Tom40 could be cross-linked to 35S-AAC-DHFR (Figure 7B), consistent with the lack of an import defect in this mutant. However, in tim9ΔN10 mitochondria, 35S-AAC-DHFR cross-links were formed between both Tim10 and Tom40 but not with Tim9ΔN10 (Figure 7C). The same results were also observed when mitochondria were not subjected to a heat-shock pretreatment (data not shown). These results, in conjunction with the yeast cell growth analyses and import studies, suggest that although a Tim9ΔN10–Tim10 complex is present and retains the capacity to interact with a substrate at the TOM complex via Tim10, this may not be sufficient for complete translocation. Instead, additional contacts with the N-terminal region of Tim9 seem to be required for driving substrate translocation into the IMS.
Figure 7.
Accumulation and cross-linking analysis of AAC-DHFR at the TOM complex. (A) 35S-AAC-DHFR was incubated with mitochondria isolated from indicated yeast strains at 24°C in the presence of methotrexate and then analyzed using BN-PAGE and phosphorimaging (left). The levels of TOM-arrested AAC-DHFR were quantified (right; error bars represent SD, n = 3). (B) 35S-AAC-DHFR was incubated with wild-type or tim10ΔN12 mitochondria in the presence of methotrexate. Cross-linking to Tim9, Tim10ΔN12, and Tom40 was assessed by immunoprecipitation and SDS-PAGE (Tom40-XL, cross-links formed between Tom40 and AAC-DHFR; arrow, cross-links formed between either Tim9 or Tim10 and AAC-DHFR). Some noncross-linked AAC-DHFR is nonspecifically pulled down. (C) 35S-AAC-DHFR was incubated with tim9ΔN10 mitochondria in the presence of methotrexate and subjected to cross-linking analysis as described in B.
DISCUSSION
Targeted Disruption of Structurally Important Residues Blocks Tim9–Tim10 Complex Assembly and Leads to Import Defects
In this study, the structure of the yeast Tim9–Tim10 complex has been determined and used to analyze subunit interactions and to address functional regions of the assembly. The previously reported human Tim9–Tim10 crystal structure revealed several unusual features, which are conserved in the yeast Tim9–Tim10 complex: intramolecular disulfide bridges constricting the neck of fixed-angle helical hairpins, intermolecular salt-bridges buried at subunit interfaces, and thermally labile tentacle-like helices projecting from the ventral face of the assembly. The structure of the yeast Tim9–Tim10 complex reported here has the advantage of substantially higher resolution (at 2.5 Å), with a correspondingly clearer definition of molecular interactions. More importantly, it has direct application to experiments carried out using yeast as a model system.
Intermolecular Salt-Bridge Contacts Stabilize the Tim9–Tim10 Complex
The importance of the hexameric complex was addressed by reversing the polarity of charged residues in a conserved ion-pair (i.e., Tim10K68E and Tim9E52K). The tim9E52K strain proved unconditionally lethal, whereas the tim10K68E strain was temperature sensitive for growth. From import studies and cross-linking analysis, our data suggest that a Tim9–Tim10K68E complex can still assemble and function in the IMS but it is heat-labile, consistent with the temperature-sensitive growth phenotype observed. Our results are in agreement with previous findings that translocation of carrier precursors across the TOM complex requires the presence of the soluble, functional Tim9–Tim10 complex (Truscott et al., 2002; Vasiljev et al., 2004).
Because E52 of Tim9 and K68 of Tim10 form an ion-pair, the question arises as to why tim10K68E cells are viable and tim9E52K cells are not. One explanation is that Tim10K68E still has the capacity to form a hexamer, but the E52K mutation in Tim9 cannot. E52 in Tim9 is not strictly required because an earlier study reported that tim9E52G yeast were viable but displayed a temperature-sensitive phenotype (Leuenberger et al., 2003). These results indicate that the salt-bridge formed by the ion-pair is not essential for the function of the Tim9–Tim10 complex. The E52K mutation in Tim9 is more deleterious, potentially blocking the formation of both the Tim9–Tim10 complex (by disrupting other important core contacts) and the TIM22 complex, adding to the defects in yeast. This is consistent with our in vitro import analysis of Tim9E52K, which accumulated at the Mia40 complex. In contrast, tim10K68E mitochondria contained a fully assembled TIM22 complex that included the mutant form of Tim10. This suggests that the contacts made between subunits within the soluble 70-kDa complex differ from those contacts made within the TIM22 complex. This is supported by the recent findings that Tim9 and Tim10 make distinct interactions with Tim12 and are present in the TIM22 complex in a different stoichiometry than the soluble Tim9–Tim10 complex (Gebert et al., 2008). In addition, we observed defects in the assembly of the TIM22 complex in tim9ΔN10 mitochondria, whereas the Tim9ΔN10–Tim10 complex remained intact.
Tim9 Tentacles Play Important Roles in the Structure and Function of the Tim9–Tim10 Complex
It has been proposed previously that the N- and C-terminal α-helices in the Tim9–Tim10 complex may act to bind to substrates (Webb et al., 2006; Baker et al., 2007). Antiparallel helices in each subunit make contacts with one another in the Tim9–Tim10 core region, whereas the terminal regions veer apart as unrestrained amphiphilic “tentacles.” Deletion of the C-terminal tentacle regions from either yeast Tim9 or Tim10 (Tim9ΔC10 and Tim10ΔC10) did not affect yeast growth or import, indicating that they are dispensable for function. However, deletion of 13 C-terminal residues in Tim9 was deleterious to yeast growth, causing a slight temperature-sensitive phenotype and defects in the stability of both the Tim9–10 complex and the TIM22 complex. Deletion of a further residue in Tim9 (Tim9ΔC14) is lethal and may be explained by the yeast Tim9–Tim10 structure where the 14th residue from the C terminus of Tim9, Phe74, is an essential part of the hydrophobic core of the assembly. It is therefore probable that truncation at Phe74 abolishes hexamer assembly. Deletion of the 13 residues C-terminal to Phe74 may disrupt local helical secondary structure just preceding the truncation, making subunit association less stable and thereby providing an explanation for the temperature sensitivity of the tim9ΔC13 strain. Although our results support previous findings that Tim9 is involved in stabilizing the complex (Vergnolle et al., 2005), it is likely that any sufficiently long truncation in either Tim9 or Tim10 will cause defects in Tim9–Tim10 complex assembly.
Yeast Tim9 and Tim10 contain longer N-terminal helices than their human counterparts (7 additional residues in Tim9 and 12 more in Tim10). In both the human and yeast Tim9–Tim10 complexes, the N-terminal helices reside in the inner layer of the assembly. It has been suggested previously that the N-terminal region of Tim10 is the main subunit involved in binding to precursor substrates such as AAC (Vergnolle et al., 2005; Gentle et al., 2007). However, using our in vivo approaches, it was found that the N-terminal tentacle region of Tim10 is dispensable for substrate recognition. tim10ΔN12 cells grew normally, displayed no protein import defects in vitro, and Tim10ΔN12 could still be cross-linked to a precursor substrate. It remains possible that substrates make contacts with Tim10 further into the N-terminal α-helix. However, we also showed that the region encompassing residues 1-21 are not absolutely essential for yeast cell viability. Instead, we uncovered an important role for the N-terminal helical region of Tim9 in the proper functioning of the Tim9–Tim10 complex in protein import. tim9ΔN10 cells were strongly temperature-sensitive for growth and translocation of the AAC precursor into the IMS was blocked. The levels of AAC-DHFR accumulating at the TOM complex were also reduced and Tim9 contacts were lost. The import of the β-barrel precursor substrate Tom40 was also impaired at the stage of outer membrane translocation and delivery to the SAM complex. Thus, even though the TIM22 complex was also reduced in tim9ΔN10 mitochondria, the defect was observed at an earlier stage in the import pathway.
In conclusion, we have demonstrated that the Tim9–Tim10 complex is required for protein import and sorting. The complex engages unproductively with substrates if the inner tentacles of Tim9 are truncated, indicating that the N-terminal region of Tim9 is crucial for function. Although a substrate can contact Tim10 independently of the action of the Tim9 N-terminal tentacle, substrate-trapping at the TOM complex is reduced and translocation is defective. Our data provides novel information to direct future approaches to address how the N-terminal tentacle of Tim9 directly interacts with a variety of substrate precursors.
Supplementary Material
ACKNOWLEDGMENTS
We thank scientists at Beamline ID-23 ESRF, J. Newman at the Bio21 C3 crystallization facility, M. Kvansakul for assistance in data collection, E. Dodoson for help with crystallographic aspects of the project, and N. Pfanner for reagents and advice. This work was supported by the Association pour la Recherche sur le Cancer (ARC) Center for Coherent X-Ray Science and grants from the ARC and National Health and Medical Research Council (NHMRC). C. W. is supported by an NHMRC Dora Lush postgraduate award, and A. F. is supported by a Human Frontiers Science Program fellowship.
Abbreviations used:
- AAC
ADP/ATP carrier
- DHFR
dihydrofolate reductase
- IMS
intermembrane space
- TIM
translocase of the inner membrane
- TOM
translocase of the outer membrane.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-09-0903) on November 26, 2008.
REFERENCES
- Allen S., Balabanidou V., Sideris D. P., Lisowsky T., Tokatlidis K. Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J. Mol. Biol. 2005;353:937–944. doi: 10.1016/j.jmb.2005.08.049. [DOI] [PubMed] [Google Scholar]
- Baker M. J., Frazier A. E., Gulbis J. M., Ryan M. T. Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol. 2007;17:456–464. doi: 10.1016/j.tcb.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Bolender N., Sickmann A., Wagner R., Meisinger C., Pfanner N. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 2008;9:42–49. doi: 10.1038/sj.embor.7401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A. T., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chacinska A., et al. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 2005;120:817–829. doi: 10.1016/j.cell.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Chacinska A., Pfannschmidt S., Wiedemann N., Kozjak V., Sanjuan Szklarz L. K., Schulze-Specking A., Truscott K. N., Guiard B., Meisinger C., Pfanner N. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23:3735–3746. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. D., Zhang K. Y. Density modification for macromolecular phase improvement. Prog. Biophys. Mol. Biol. 1999;72:245–270. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- Daum G., Bohni P. C., Schatz G. Import of proteins into mitochondria: cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- Davis A. J., Sepuri N. B., Holder J., Johnson A. E., Jensen R. E. Two intermembrane space TIM complexes interact with different domains of Tim23p during its import into mitochondria. J. Cell Biol. 2000;150:1271–1282. doi: 10.1083/jcb.150.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gebert N., Chacinska A., Wagner K., Guiard B., Koehler C. M., Rehling P., Pfanner N., Wiedemann N. Assembly of the three small Tim proteins precedes docking to the mitochondrial carrier translocase. EMBO Rep. 2008;9:548–554. doi: 10.1038/embor.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle I. E., et al. Conserved motifs reveal details of ancestry and structure in the small TIM chaperones of the mitochondrial intermembrane space. Mol. Biol. Evol. 2007;24:1149–1160. doi: 10.1093/molbev/msm031. [DOI] [PubMed] [Google Scholar]
- Hoppins S. C., Nargang F. E. The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J. Biol. Chem. 2004;279:12396–12405. doi: 10.1074/jbc.M313037200. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kang P. J., Ostermann J., Shilling J., Neupert W., Craig E. A., Pfanner N. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990;348:137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- Koehler C. M. The small Tim proteins and the twin Cx3C motif. Trends Biochem. Sci. 2004;29:1–4. doi: 10.1016/j.tibs.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Koehler C. M., Beverly K. N., Leverich E. P. Redox pathways of the mitochondrion. Antioxid. Redox Signal. 2006;8:813–822. doi: 10.1089/ars.2006.8.813. [DOI] [PubMed] [Google Scholar]
- Lazarou M., McKenzie M., Ohtake A., Thorburn D. R., Ryan M. T. Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol. Cell. Biol. 2007;27:4228–4237. doi: 10.1128/MCB.00074-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuenberger D., Curran S. P., Wong D., Koehler C. M. The role of Tim9p in the assembly of the TIM22 import complexes. Traffic. 2003;4:144–152. doi: 10.1034/j.1600-0854.2003.00095.x. [DOI] [PubMed] [Google Scholar]
- McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C., Sickmann A., Pfanner N. The mitochondrial proteome: from inventory to function. Cell. 2008;134:22–24. doi: 10.1016/j.cell.2008.06.043. [DOI] [PubMed] [Google Scholar]
- Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., Herrmann J. M. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Model K., Meisinger C., Prinz T., Wiedemann N., Truscott K. N., Pfanner N., Ryan M. T. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat. Struct. Biol. 2001;8:361–370. doi: 10.1038/86253. [DOI] [PubMed] [Google Scholar]
- Mokranjac D., Popov-Celeketic D., Hell K., Neupert W. Role of Tim21 in mitochondrial translocation contact sites. J. Biol. Chem. 2005;280:23437–23440. doi: 10.1074/jbc.C500135200. [DOI] [PubMed] [Google Scholar]
- Murphy M. P., Leuenberger D., Curran S. P., Oppliger W., Koehler C. M. The essential function of the small Tim proteins in the TIM22 import pathway does not depend on formation of the soluble 70-kilodalton complex. Mol. Cell. Biol. 2001;21:6132–6138. doi: 10.1128/MCB.21.18.6132-6138.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoe M., Ohwa Y., Ishikawa D., Ohshima C., Nishikawa S., Yamamoto H., Endo T. Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J. Biol. Chem. 2004;279:47815–47821. doi: 10.1074/jbc.M410272200. [DOI] [PubMed] [Google Scholar]
- Neupert W., Herrmann J. M. Translocation of Proteins into Mitochondria. Annu. Rev. Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pagliarini D. J., et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen S. A., Waizenegger T., Stan T., Preuss M., Cyrklaff M., Hell K., Rapaport D., Neupert W. Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature. 2003;426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- Rehling P., Brandner K., Pfanner N. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol. 2004;5:519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- Ryan M. T., Hoogenraad N. J. Mitochondrial-nuclear communications. Annu. Rev. Biochem. 2007;76:701–722. doi: 10.1146/annurev.biochem.76.052305.091720. [DOI] [PubMed] [Google Scholar]
- Ryan M. T., Muller H., Pfanner N. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem. 1999;274:20619–20627. doi: 10.1074/jbc.274.29.20619. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Heiter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D., Pfanner N., Wiedemann N. Import of proteins into mitochondria. Methods Cell Biol. 2007;80C:783–806. doi: 10.1016/S0091-679X(06)80036-1. [DOI] [PubMed] [Google Scholar]
- Stojanovski D., Milenkovic D., Müller J. M., Gabriel K., Schulze-Specking A., Baker M. J., Ryan M. T., Guiard B., Pfanner N., Chacinska A. Mitochondrial protein import: precursor oxidation in a ternary complex with disulfide carrier and sulfhydryl oxidase. J. Cell Biol. 2008;183:195–202. doi: 10.1083/jcb.200804095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott K. N., Wiedemann N., Rehling P., Muller H., Meisinger C., Pfanner N., Guiard B. Mitochondrial import of the ADP/ATP carrier: the essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol. Cell. Biol. 2002;22:7780–7789. doi: 10.1128/MCB.22.22.7780-7789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wilpe S., et al. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature. 1999;401:485–489. doi: 10.1038/46802. [DOI] [PubMed] [Google Scholar]
- Vasiljev A., et al. Reconstituted TOM core complex and Tim9/Tim10 complex of mitochondria are sufficient for translocation of the ADP/ATP carrier across membranes. Mol. Biol. Cell. 2004;15:1445–1458. doi: 10.1091/mbc.E03-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnolle M. A., Alcock F. H., Petrakis N., Tokatlidis K. Mutation of conserved charged residues in mitochondrial TIM10 subunits precludes TIM10 complex assembly, but does not abolish growth of yeast cells. J. Mol. Biol. 2007;371:1315–1324. doi: 10.1016/j.jmb.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Vergnolle M. A., Baud C., Golovanov A. P., Alcock F., Luciano P., Lian L. Y., Tokatlidis K. Distinct domains of small Tims involved in subunit interaction and substrate recognition. J. Mol. Biol. 2005;351:839–849. doi: 10.1016/j.jmb.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Webb C. T., Gorman M. A., Lazarou M., Ryan M. T., Gulbis J. M. Crystal structure of the mitochondrial chaperone TIM9.10 reveals a six-bladed alpha-propeller. Mol. Cell. 2006;21:123–133. doi: 10.1016/j.molcel.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Wiedemann N., Kozjak V., Chacinska A., Schonfisch B., Rospert S., Ryan M. T., Pfanner N., Meisinger C. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- Wiedemann N., Pfanner N., Ryan M. T. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 2001;20:951–960. doi: 10.1093/emboj/20.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N., Truscott K. N., Pfannschmidt S., Guiard B., Meisinger C., Pfanner N. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway. J. Biol. Chem. 2004;279:18188–18194. doi: 10.1074/jbc.M400050200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.