Abstract
ER quality control (ERQC) prevents the exit of misfolded secretory and membrane proteins from the ER. A critical aspect of ERQC is a transcriptional response called the unfolded protein response (UPR), which up-regulates genes that enable cells to cope with misfolded, ER-retained proteins. In this study, we compare the transcriptional responses in yeast resulting from the acute expression of misfolded proteins residing in three different cellular compartments (the ER lumen, membrane, and cytosol), and find that each elicits a distinct transcriptional response. The classical UPR response, here-designated UPR-L, is induced by the ER lumenal misfolded protein, CPY*. The UPR-Cyto response is induced by the cytosolic protein, VHL-L158P, and is characterized by a rapid, transient induction of cytosolic chaperones similar to the heat-shock response. In contrast, the misfolded membrane protein with a cystolic lesion, Ste6p*, elicits a unique response designated UPR-M/C, characterized by the modest induction of >20 genes regulated by Rpn4p, an activator of proteasomal genes. Independently, we identified several genes required for yeast viability during UPR-M/C stress, but not UPR-L or UPR-Cyto stress. Among these is RPN4, highlighting the importance of the Rpn4p-dependent response in tolerating UPR-M/C stress. Further analysis suggests the requirement for Rpn4p reflects severe impairment of the proteasome by UPR-M/C stress.
INTRODUCTION
Protein misfolding plays a critical role in numerous human diseases (i.e., cystic fibrosis, Parkinson's disease, hereditary emphysema, Alzheimer's disease; Otsu and Sitia, 2007; Lin et al., 2008) and is monitored by a variety of cellular “quality control” systems. One such system, endoplasmic reticulum (ER) quality control (ERQC), prevents the exit of misfolded secretory and membrane proteins from the ER. ERQC can be divided into two separate processes: 1) the unfolded protein response (UPR), which refers to the transcriptional up-regulation of genes that are thought to enable the cell to cope with and fold misfolded proteins, and 2) ER-associated degradation (ERAD), whereby misfolded, ER-retained proteins are degraded via the ubiquitin–proteasome system.
The UPR transcriptional pathway activated in response to misfolded ER lumenal proteins has been well characterized in Saccharomyces cerevisiae. The presence of misfolded proteins in the ER results in activation of an ER transmembrane kinase/endoribonuclease, Ire1p, which splices the pre-mRNA of HAC1 (Cox et al., 1993; Cox and Walter, 1996; Mori et al., 1996). Splicing ultimately allows translation of HAC1 into a potent transcriptional activator that binds to the promoters of genes containing UPR elements (UPRE-1, -2, and -3 and others, still not well defined) and activates their transcription (Mori et al., 1992; Kohno et al., 1993; Patil et al., 2004). The full scope of the UPR response was characterized in an elegant study using microarray analysis to identify genes induced in an IRE1- and HAC1-dependent manner by the drugs dithiothreitol (DTT) and tunicamycin (Tm; Travers et al., 2000). DTT and Tm cause widespread protein misfolding in the ER, due to their inhibition of disulfide bond formation and N-linked glycosylation, respectively, and strongly induce the UPR response. More than 381 ORFs were identified as UPR target genes and fell into diverse categories of function, such as protein translocation, folding, glycosylation, vesicle trafficking, and ERAD. Autophagy genes were also recently shown to be induced by ER stress, but in an IRE1- and HAC1-independent manner (Bernales et al., 2006; Yorimitsu et al., 2006). Analysis of the UPR response has revealed considerable insight into how cells cope with the presence of misfolded proteins in the ER lumen without compromising viability.
In addition to the transcriptional targets induced by misfolded ER luminal proteins, much is also known about the ERAD pathway for this type of misfolded protein (Romisch, 2005; Sayeed and Ng, 2005). Recently, the ERAD pathways for misfolded membrane proteins have also been examined, leading to the designation of three classes of ERAD substrates based on the topological location of their misfolded lesion: either lumenal (ERAD-L), cytosolic (ERAD-C), or in a membrane span (ERAD-M; Taxis et al., 2003; Huyer et al., 2004b; Vashist and Ng, 2004; Carvalho et al., 2006). Model substrates for ERAD-L include soluble proteins in the ER lumen (CPY*, KHN) and membrane proteins with misfolded luminal domains (KWW, CT*, and CTG*). ERAD-C and ERAD-M substrates are membrane proteins with misfolded cytosolic (Ste6p*, KSS, and KWS) or membrane domains (Hmg2p, Pdr5p*, and Sec61-2), respectively (Hampton et al., 1996; Bordallo et al., 1998; Taxis et al., 2003; Huyer et al., 2004b; Vashist and Ng, 2004; Carvalho et al., 2006). Although all three ERAD pathways converge post-ubiquitination and at the proteasome, the chaperone and ubiquitination requirements for these three pathways appear to be largely distinct (Nishikawa et al., 2005; Brodsky, 2007).
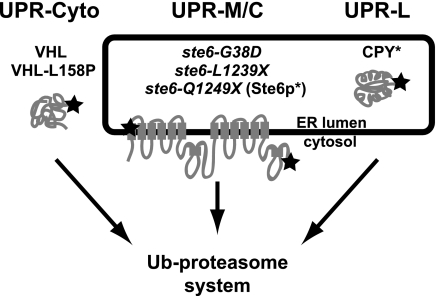
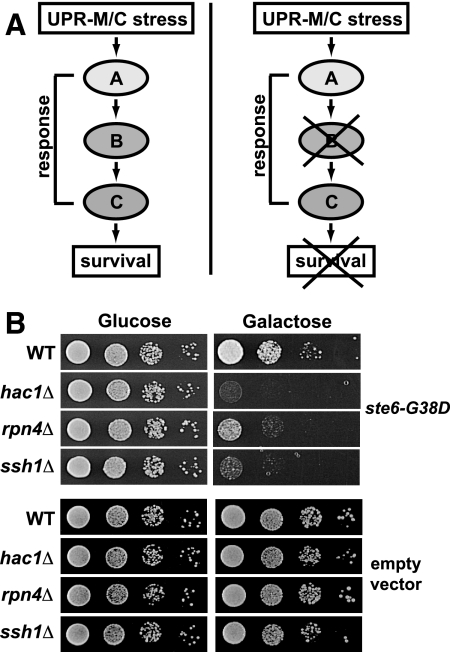
An extension of the delineation of discrete ERAD pathways is that misfolded proteins in different cellular compartments or with distinct topological sites of mutation may also elicit mechanistically unique UPR transcriptional responses. A working model depicting this hypothesis is shown in Figure 1. The “classical” UPR, here called UPR-L (right), is induced by substrates whose misfolded domains are in the lumen of the ER. UPR-M/C (middle) is posited to be induced by misfolded membrane proteins with lesions in a membrane span or a cytosolic domain, and UPR-Cyto (left), by misfolded cytosolic proteins that do not enter the secretory pathway at all.
Figure 1.
Working model for three compartmentally distinct branches of the UPR. In this study, we ask if three classes of misfolded proteins with different cellular locations (ER lumen, membrane, and cytosol) induce distinct patterns of transcriptional responses. Misfolded proteins in the ER lumen, such as CPY*, induce the well-documented UPR-L transcriptional response (generally referred to simply as UPR); misfolded ER membrane proteins with membrane or cytosolic lesions, such as ste6-Q1249X (Ste6p*) and other Ste6p alleles (ste6-L1239X and ste6-G38D), could induce a UPR-M/C response; and misfolded proteins in the cytosol, such as VHL and VHL-L158P, could induce a UPR-Cyto response. All three classes of misfolded proteins are known to be degraded via the ubiquitin–proteasome system (Hiller et al., 1996; Loayza et al., 1998; McClellan et al., 2005). The star in the first transmembrane domain of Ste6p represents the site of the G38D mutation and the star in the C-terminal cytosolic domain represents the sites of the L1239X and Q1249X mutations.
In contrast to the well-defined UPR-L pathway (see Figure 1, right), surprisingly little is known about transcriptional responses induced by misfolded proteins in the ER membrane (the proposed UPR-M/C response; see Figure 1, middle) or cytosol (the proposed UPR-Cyto response; see Figure 1, left). It is currently even unclear whether misfolded proteins in these cellular compartments induce global transcriptional responses. Additionally, despite residing in different cellular compartments (ER membrane and cytosol), both of these classes of misfolded proteins can contain cytosolic misfolded domains, and it is unknown whether they would up-regulate similar or disparate gene targets. The heat-shock response is thought to be a cytosolic response to the thermal misfolding of proteins and their subsequent aggregation (Pinto et al., 1991; Parsell and Lindquist, 1993). The proposed UPR-M/C and UPR-Cyto responses have not been examined to determine what similarities there may be to the heat-shock response or to the well-defined UPR-L response.
To assess whether the unfolded protein response differs in separate cellular compartments, we compared the transcriptional responses resulting from the acute, galactose-induced expression of single misfolded proteins with distinct cellular localizations (ER lumen, membrane, or cytosol, respectively). We find that misfolded proteins residing in these three cellular compartments elicit distinct patterns of gene induction. Using five defined transcriptional markers, we find that the ER lumenal misfolded protein, CPY*, induces the well-characterized UPR-L response, a hallmark of which is the induction of YFR026C (here designated as ULI1), whereas the cytosolic misfolded VHL alleles induce the UPR-Cyto response, which resembles a pattern of gene induction characteristic of heat shock. The UPR-M/C stressors (misfolded Ste6p alleles) do not induce these five transcriptional markers, and we carried out microarray analysis to define the UPR-M/C response profile of gene induction. Among 67 UPR-M/C stress-induced genes are >20 genes known to controlled by Rpn4p, a transcriptional activator of proteasomal genes, suggesting that Rpn4p-mediated transcription is a key aspect of the UPR-M/C response. In agreement with these results, we also identified RPN4 in a synthetic lethality screen designed to identify genes required for viability during UPR-M/C stress, but not UPR-L or UPR-Cyto stresses. Further, UPR-M/C stress was found to severely impair the proteasome. Thus, Rpn4p may be essential for cellular survival in the presence of misfolded membrane proteins due to the critical need for additional proteasomes upon UPR-M/C stress.
MATERIALS AND METHODS
Yeast Strains, Media, and Growth Conditions
The S. cerevisiae strains and plasmids used in this study are listed in Tables 1 and 2, respectively. Strain and plasmid constructions can be found in the Supplemental Materials and Methods. Solid and liquid drop-out or complete media with glucose as a carbon source were prepared as described previously (Michaelis and Herskowitz, 1988). Media containing galactose as a carbon source lacked glucose and instead contained 4% raffinose and 4% galactose (Sigma-Aldrich, St. Louis, MO; <0.01% glucose; Mumberg et al., 1994). Yeast strains and cultures were grown at 30°C, except for temperature-sensitive strains, which were grown at room temperature (24°C). Yeast transformations were performed by the lithium acetate method (Ito et al., 1983).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| WCG4a | MATaura3-52 leu2-3,112 his3-11,15 PRE1 PRE2 | Heinemeyer et al. (1991) |
| WCG4a-11/21a | MATaura3-52 leu2-3,112 his3-11,15 pre1-1 pre2-1 | Heinemeyer et al. (1991) |
| SM4460a | MATahis3 leu2 met15 ura3 | Open Biosystems |
| SM4464 | MATahis3 leu2 met15 ura3 erg6::kanMX | Open Biosystems |
| SM4817 | MATahis3 leu2 met15 ura3 pep4::kanMX | Open Biosystems |
| SM5295 | MATaleu2-98 ade2-101 ura3-52 lys2-801 YFR026c::HA | Open Biosystems |
| SM5360 | MATahis3 leu2 met15 ura3 doa10::natMX hrd1::kanMX | This study |
| SM5362 | MATahis3 leu2 met15 ura3 ubc6::HIS3 ubc7::natMX | Metzger et al. (2008) |
| SM5382 | MATahis3 leu2 met15 ura3 hac1::kanMX | Open Biosystems |
| SM5383 | MATahis3 leu2 met15 ura3 rpn4::kanMX | Open Biosystems |
| SM5384 | MATahis3 leu2 met15 ura3 ssh1::kanMX | Open Biosystems |
| SM5476 | MATahis3 leu2 met15 ura3 yfr026c::kanMX | Open Biosystems |
a SM4460 is our laboratory's name for BY4741.
Table 2.
Plasmids used in this study
| Plasmid | Genotype | Reference or source |
|---|---|---|
| pESC-VHL | [2μ URA3 PGAL1 VHL] | McClellan et al. (2005) |
| pESC-L158P | [2μ URA3 PGAL1 VHL-L158P] | McClellan et al. (2005) |
| pPW344 | [2μ URA3 4 × UPRE-LacZ] | Patil et al. (2004) |
| pSM922 | [2μ URA3 PGAL1] | Mumberg et al. (1994) |
| pSM1897 | [2μ URA3 PGAL1 STE6::GFPc] | This study |
| pSM1898 | [2μ URA3 PGAL1ste6-G38D::GFPc] | This study |
| PSM2212 | [2μ URA3 PGAL1ste6-L1239X] | This study |
| pSM2213 | [2μ URA3 PGAL1ste6-Q1249X] | This study |
| pSM2215 | [CEN URA3 PGAL1 CPY*] | This study |
| pSM2216 | [CEN LEU2 PPGK Ub-Pro-LacZ] | This study |
| pSM2217 | [2μ URA3 PGAL1pma1-D378S] | This study |
Galactose Induction
Cells containing galactose-inducible plasmids were grown at 30°C in synthetic media containing the appropriate amino acids and 4% raffinose for 2 d to saturation. Cells were diluted back in fresh media containing 4% raffinose and grown to early log phase (OD600 = 0.2) before induction by the addition of 4% galactose to liquid media for indicated periods of time or plating to solid media containing 4% galactose.
Yeast Total RNA Preparation and Northern Blot Analysis
For each time point after galactose induction, 10 OD600 units of cells were harvested. Cells were washed in 1 ml of diethyl pyrocarbonate (DEPC)-treated dH2O, and cell pellets were frozen in liquid nitrogen and stored at −80°C until RNA preparation. For harvesting of yeast total RNA, cells were lysed by vortexing with acid-washed glass beads in the presence of the RNA isolation reagent RNA STAT-60 (Tel-Test, Friendswood, TX), and RNA was extracted from the homogenate by chloroform extraction and centrifugation. The aqueous layer containing RNA was precipitated with isopropanol, pelleted, and then washed using 75% ethanol. RNA pellets were air-dried before resuspending in DEPC dH2O and storage at −20°C. RNA concentration was determined from OD260.
For Northern blots, 5 μg of RNA was mixed with 2 volumes of RNA sample loading buffer (Sigma-Aldrich) and run on a 1% agarose gel containing 1× MOPS (Quality Biological, Gaithersburg, MD), ethidium bromide, and 5% formaldehyde (37% w/w) at 70 V for 3 h in 1× MOPS running buffer. The gel was washed twice for 15 min in 2× SSC (Roche Applied Science, Indianapolis, IN), and RNA was transferred to Nytran SuPerCharge nylon membrane (Whatman Schleicher & Schuell, Keene, NH) by capillary transfer in 10× SSC for 18 h. RNA was cross-linked to the membrane using a UV Stratalinker at 1200 μJ ×100. Membranes were prehybridized in 10 ml of rapid hybridization buffer (Amersham Biosciences, Piscataway, NJ) for 1 h at 60°C. DNA Northern blot probes labeled with 32P were created by first PCR amplifying an ∼500-base pair PCR product from the coding region of the gene of interest and purifying the PCR product using a PCR Purification kit (Qiagen, Chatsworth, CA). The probe was radiolabeled using the Megaprime DNA labeling kit (Amersham) and 30 μCi of [α-32P]dCTP (GE Healthcare, Waukesha, WI). Hot probe was counted using a scintillation counter. Probes were boiled at 100°C for 10 min to denature, added at a concentration of 1 × 106 cpm/ml of hybridization buffer, and incubated with the prehybridized membrane for 2 h at 60°C. After hybridization, the membrane was washed for 5 min in 2× SSC, 10 min in 0.5× SSC/0.1% SDS, and 10 min in 0.1× SSC/0.1% SDS. Blots were visualized and quantitated by Phosphor-Imager and Quantity One software (Bio-Rad, Hercules, CA). For reprobing with a different probe, blots were stripped by incubating twice with boiling 0.1% SDS until cooled and then prehybridized and processed as above. HAC1 splicing was graphed as percentage of HAC1 spliced and was calculated as [lower band]/[lower band + upper band]. Quantitations of blots were normalized to the levels of ACT1 transcript. For galactose-inducible constructs, samples were prepared at various time points after galactose addition. Induction of expression was calculated based on the RNA levels at “zero” time points (just before galactose addition), and each experiment was performed multiple times with different misfolded proteins in each class: UPR-L (CPY*), UPR-M/C (ste6-G38D, -L1239X, or -Q1249X), or UPR-Cyto (VHL or VHL-L158P). All experiments were repeated at least twice with each mutant protein with similar results; a representative experiment is shown in each case.
Microarray Hybridization and Analysis
Three biological replicates of wild-type (SM4460) cells expressing either empty vector (pSM922), ste6-Q1249X (pSM2213), STE6 (pSM1897), or VHL-L158P (pESC-L158P) were harvested after 1 h of galactose induction for RNA preparation (as described above). Processing of the RNA, hybridization to Affymetrix GeneChip yeast Genome 2.0 arrays (Santa Clara, CA), and data analysis are described in the Supplemental Materials and Methods.
Diploid-based Synthetic Lethality Analysis on Microarrays
Diploid-based synthetic lethality analysis on microarrays (dSLAM) was performed essentially as previously described (Pan et al., 2004; Warren et al., 2004) except that pSM1898 (2μ URA3 PGAL1 ste6-G38D::GFP; experimental pool) or pSM922 (2μ URA3 PGAL1; control pool) was transformed into the heterozygous diploid deletion collection pool (a generous gift of Jef Boeke, Johns Hopkins University). After sporulation, MATa haploid yeast cells harboring pSM1898 or pSM922 were selected on haploid selection media containing galactose to induce expression of ste6-G38D::GFPc. Colonies were pooled and yeast genomic DNA was prepared from each pool. To determine mutants underrepresented upon overexpression of ste6-G38D, the barcode tags from the control and experimental pools were PCR-amplified using biotinylated primers and hybridized to Tag3 barcode microarrays, and the microarray data were analyzed as previously described (Lee and Spencer, 2004). Synthetic lethal phenotypes were confirmed individually using the MATa haploid deletion collection (Open Biosystems, Huntsville, AL) by retransforming pSM922 and pSM1898 and assaying growth on media containing galactose. Deletions of the following genes were found to have a synthetic phenotype with ste6-G38D::GFPc: RPN4, SSH1, HAC1, ALF1, STB2, YKL077W, VPS66, YBR095C, ITR1, HSD1, ARR4, CIK1, PHO84, and IDH1. Deletions of RPN4, SSH1, and HAC1 had the strongest synthetic phenotype and are discussed in detail in Results.
Spot Growth Assay
Log phase cultures were diluted to an OD600 of 0.1 and four serial 10-fold dilutions were made in 96-well plates to yield a dilution series in adjacent wells. Ten microliters of each dilution was spotted onto the appropriate media using a multichannel pipetman. Plates containing glucose as a carbon source were incubated for 2 d at 30°C, and plates containing galactose were incubated for 3 d at 30°C.
Preparation of Cell Extracts and Immunoblotting
Cell extracts and immunoblotting were prepared essentially as described previously (Fujimura-Kamada et al., 1997). Briefly, 2.5 OD600 units of cells were grown logarithmically in synthetic dropout media and lysed by the addition of β-mercaptoethanol/NaOH. Proteins were precipitated using 5% trichloroacetic acid, and protein pellets were resuspended in sample buffer (3.5% SDS, 0.5 M DTT, 80 mM Tris, 8 mM EDTA, 15% glycerol, and 4 mg bromophenol blue). YFR026Cp::HA was detected using the 12CA5 mouse anti-hemagglutinin (HA) mAb (Roche Applied Science) diluted 1:10,000 in TBST containing 5% blocking reagent (Roche Applied Science) for 1 h at room temperature. A loading control was done using the anti-Hexokinase Ab (a generous gift of Rob Jensen, Johns Hopkins University) diluted 1:200,000 in TBST containing 5% blocking reagent for 1 h at room temperature.
β-Galactosidase Assays
Cultures were assayed for β-galactosidase activity as previously described (Guarente, 1983). β-Galactosidase activity is expressed in Miller units as 1000 × (A420)/[(tmin)(Vml) × (A600)].
Colony Viability Assay
Cultures were grown to log phase at 30°C in dropout media containing 4% raffinose. Galactose (4%) was added to induce expression of misfolded proteins, and cultures were split and treated with either 50 μM MG132 (EMD Chemicals, Gibbstown, NJ) in DMSO or an equal volume of DMSO alone, and incubated for 8 h at 30°C. Both cultures were then diluted 1:1500 in dH2O, and 200 μl was plated on dropout media containing galactose and incubated for 3 d at 30°C. Colony forming units (CFUs) were counted for DMSO- and MG132-treated cells and calculated as CFUs of MG132-treated cells as a percentage of CFUs of DMSO-treated cells.
RESULTS
Examining Cellular Transcriptional Responses Using Northern Blot Analysis
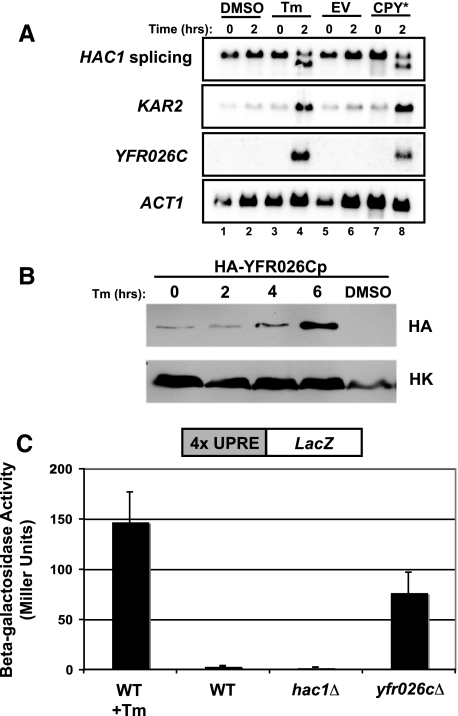
To begin to compare the UPR-L and proposed UPR-M/C and UPR-Cyto transcriptional responses (Figure 1), we examined the induction of five genes, falling into two classes (cytosolic chaperones and well-characterized markers for the UPR-L), by Northern blot analysis. The cytosolic chaperone genes are SSA4, which encodes the main stress-inducible Hsp70 chaperone in the cytosol (Boorstein and Craig, 1990), and STI1, which encodes a cytosolic cochaperone that facilitates interactions between Hsp70 and Hsp90 (Wegele et al., 2003). To assay the UPR-L, we examined HAC1 pre-mRNA splicing, an early hallmark of UPR-L induction, and up-regulation of KAR2, a well-characterized UPR-L target gene (Mori et al., 1992; Kohno et al., 1993). Additionally, we followed a novel marker for UPR-L, YFR026C, which we identified by microarray analysis as the gene most highly induced by the expression of CPY* (our unpublished data). We found that the YFR026C transcript is strongly induced by tunicamycin (Tm) treatment (in agreement with Travers et al., 2000) or expression of CPY*, representing induction of greater than 25-fold (Figure 2A, lanes 4 and 8). The levels of HA-tagged YFR026C protein are also increased in response to Tm treatment (Figure 2B), and deletion of YFR026C induces the UPR-L, as measured by a UPRE-LacZ reporter (Figure 2C), suggesting a role for YFR026C in alleviating or preventing ER stress. The dramatic increase in the levels of YFR026C transcript serves as a robust reporter of the UPR-L response and presumably reflects the UPRE-2 and -3 elements (Patil et al., 2004) within its upstream region.
Figure 2.
YFR026C is a novel gene (designated ULI1, see discussion), highly induced by UPR-L stress. (A) Northern blot to examine UPR-L gene induction by the ER stressors, tunicamycin (Tm; lanes 1–4) and CPY* (lanes 5–8). In lanes 1–4, wild-type cells (SM4460) were treated with either DMSO or 10 μg/ml Tm. Samples were harvested before (0 h) and after (2 h) treatment. In lanes 5–8, wild-type cells (SM4460) expressing empty vector (EV; pSM922) or CPY* (pSM2215) were harvested before (0 h) and after (2 h) galactose induction. Blots were probed to examine HAC1 splicing, KAR2 induction, and YFR026C induction. The ACT1 levels served as a loading control. (B) A Western blot for YFR026Cp protein levels was performed using extracts prepared from cells expressing genomic HA-tagged YFR026Cp (SM5295) under control of its endogenous promoter. Cultures were either untreated (0 h) or treated with 10 μg/ml Tm for 2, 4, and 6 h or DMSO for 6 h and harvested for protein at each time point. Blots were probed with anti-HA and anti-Hexokinase (HK) antibodies. (C) To examine whether lack of YFR026C causes induction of the UPR-L, yfr026cΔ (SM5476) cells expressing a UPRE-LacZ reporter (pPW344) were assayed for β-galactosidase activity. For comparison, the activity of wild-type (SM4460) cells alone (WT) or treated with 10 μg/ml Tm for 2 h before processing (WT + Tm), and hac1Δ cells (SM5382), all expressing a UPRE-LacZ reporter (pPW344), are included as controls. The data reflect an average of three independent experiments; error bars, 1 SD.
To confirm that the five genes we have chosen as transcriptional markers respond characteristically to stress, we assessed their induction over a 5-h time course in response to the well-established global stresses, l-azetidine-2-carboxylic acid (AZC), heat shock, and Tm. As expected, the toxic proline amino acid analogue, AZC, and heat shock, both of which have been used previously to assess cytosolic misfolding (Trotter et al., 2002), quickly and robustly induce the cytosolic chaperones SSA4 and STI1 (Supplemental Figure S1B; column 2, AZC, and column 3, heat shock), though the induction by heat shock is transient in nature, as has been reported previously (Gasch et al., 2000). Also as expected, Tm robustly activates the UPR-L response (Supplemental Figure S1A, column 5, Tm). Interestingly, we observed a mild induction of UPR-L genes by AZC and of the cytosolic chaperone SSA4 by Tm (Supplemental Figure S1A, column 2, AZC, and Supplemental Figure S1B, column 5, Tm). Taken together, we can conclude that the two classes of Northern blot probes act as expected in response to global stresses, but these stresses can also ultimately lead to stress in compartments other than the one that they are targeting.
Analyzing the Cellular Responses to Three Classes of Single Misfolded Protein Stressors
Global stressors, such as AZC or Tm, affect numerous proteins within one or several cellular compartments and may also have off-target effects. The expression of a single misfolded protein can be expected to elicit a more specific and acute cellular response. We have used single misfolded protein stressors that reside in discrete subcellular locations (membrane, lumen, and cytosol) to examine the existence of unique transcriptional responses in these compartments.
To provide a UPR-L stress, we used CPY*, a misfolded allele of the soluble protein carboxypeptidase Y that is retained in the ER lumen and has been used as a model substrate to define the ERAD-L pathway (Finger et al., 1993; Hiller et al., 1996; Taxis et al., 2003; Huyer et al., 2004b). CPY* induction of a UPRE-LacZ reporter has been demonstrated (Spear and Ng, 2003, 2005), but the temporal transcriptional response to CPY* has not been examined in detail.
To assess the UPR-M/C response, several misfolded alleles of the ABC transporter Ste6p were examined. These mutant proteins are retained in the ER and have lesions in either the membrane or cytosolic domains of Ste6p: ste6-G38D has a mutation in the first transmembrane span of Ste6p, and ste6-L1239X and ste6-Q1249X (also known as Ste6p*) are prematurely truncated within a cytosolic nucleotide-binding domain (Loayza et al., 1998; Huyer et al., 2004a; Figure 1 indicates the sites of these mutations with a star). The rates of turnover of these proteins vary, with ste6-G38D and ste6-L1239X having half-lives of ∼45 min and ste6-Q1249X (Ste6p*) having a half-life of <10 min (Nijbroek, 1998; Huyer et al., 2004b).
Cytosolic quality control substrates have been only minimally characterized. Here, we used wild-type and mutant forms of the von Hippel Lindau tumor-suppressor proteins (VHL and VHL-L158P) to evaluate UPR-Cyto stress. VHL is a mammalian, cytosolic protein normally found complexed with the elongins B and C (Feldman et al., 1999). When expressed in yeast, both VHL and VHL-L158P are degraded by the ubiquitin–proteasome pathway, with half-lives of ∼1 h and 15 min, respectively (McClellan et al., 2005).
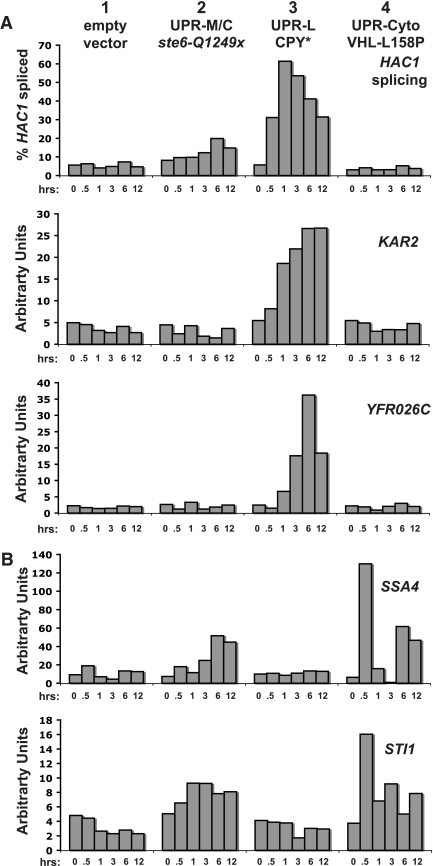
We expressed each of the proteins discussed above from the galactose-inducible GAL1 promoter, and RNA samples were harvested at multiple time points after induction and analyzed by Northern blot analysis for HAC1 splicing or transcriptional induction of KAR2 and YFR026C (Figure 3A) or induction of SSA4 and STI1 (Figure 3B). Figure 3 shows a representative, normalized quantitation of Northern blots with one example from each class of stress. We observed similar patterns of induction for all of the misfolded protein alleles in a particular class of stress (our unpublished data). Below, we first discuss the induction of the UPR-L genes and then the induction of the cytosolic chaperone genes.
Figure 3.
Three distinct classes of misfolded proteins induce unique patterns of transcription. Cells expressing galactose-inducible constructs were harvesting for RNA processing and Northern blotting at 0, 0.5, 1, 3, 6, and 12 h after galactose induction. Shown is a representative experiment using the wild-type strain (SM4460) expressing empty vector (pSM922), ste6-Q1249X (pSM2213), CPY* (pSM2215), and VHL-L158P (pESC-L158P). (A) The indicated probes detect the UPR-L genes, KAR2, YFR026C, and HAC1, splicing. (B) Probes detect the cytosolic chaperones, SSA4 and STI1. Northern blots were performed in duplicate and shown is a representative experiment in which quantitated, normalized values, determined as described in Materials and Methods, are graphed as percent of HAC1 spliced or arbitrary units.
Only the UPR-L Stressor, CPY*, Induces the UPR-L Response
We queried the splicing and induction of the UPR-L genes HAC1, KAR2, and YFR026C (Figure 3A) in response to the three classes of misfolded protein stresses to determine if the proposed UPR-M/C and UPR-Cyto responses are distinct from the UPR-L response. We saw no induction of UPR-L genes in a strain containing empty vector subject to galactose-induction conditions (Figure 3A, column 1, empty vector), confirming that the addition of galactose alone does not induce the UPR-L response. As expected, the UPR-L stress, CPY*, strongly induces HAC1 splicing, followed by transcription of KAR2, and at slightly later time points, a peak of YFR026C transcription (Figure 3A, column 3, CPY*). The levels and timing of this induction are similar to the induction seen with Tm (Supplemental Figure S1A, column 5, Tm). When CPY* is expressed at an even higher level from a high-copy (2μ) plasmid rather than a centromeric plasmid (as shown here), there is also a very similar response with respect to HAC1 splicing and KAR2 and YFR026C induction (our unpublished data).
On the other hand, the UPR-M/C stressor, ste6-Q1249X (Ste6p*), and the UPR-Cyto stressor, VHL-L158P, do not dramatically induce the UPR-L genes (Figure 3A, column 2, ste6-Q1249X, and column 4, VHL-L158P). However, at late time points after the induction of UPR-M/C stressors there is a small amount of HAC1 splicing (Figure 3A, column 2, ste6-Q1249x, top panel, 6 and 12 h), also seen with ste6-G38D and ste6-L1239X (our unpublished data). It is unknown why HAC1 splicing may be induced by UPR-M/C stress, especially because a corresponding induction of KAR2 or YFR026C is absent at these time points. The timing and low level of splicing, however, suggests that this late and modest UPR-L activation is distinct from the acute UPR-L activation induced by CPY* and is not likely to be a primary response to UPR-M/C stress.
Overall, from these results we can conclude that UPR-M/C and UPR-Cyto stresses do not induce an acute UPR-L response comparable to the UPR-L stressor, CPY*. These data support the existence of unique, compartment-specific UPR-M/C and UPR-Cyto transcriptional responses that are different from the UPR-L response.
The Cytosolic Chaperone Genes Are Distinctly Induced by a Misfolded Protein Stressor Residing in the Cytosol
Analysis of the cytosolic chaperone genes, SSA4 and STI1, by Northern blot (Figure 3B) revealed no significant induction when empty vector is expressed (Figure 3B, column 1, empty vector), demonstrating that galactose induction does not affect their transcription. As expected, the UPR-L stressor, CPY*, also does not induce cytosolic chaperone genes (Figure 3B, column 3, CPY*).
Interestingly, the UPR-Cyto stressor, VHL-L158P, induces a unique pattern of induction of the cytosolic chaperone genes: a strong and rapid, but transient, induction, followed by a return of the transcripts to basal levels (Figure 3B, column 4, VHL-L158P, compare 0- and 0.5-h time points). This response is highly reminiscent of the transient induction of SSA4 and STI1 by heat shock (Supplemental Figure S1B, column 3, heat shock), suggesting that the heat-shock response could be a major aspect of the UPR-Cyto response. Interestingly, UPR-Cyto stress induces the cytosolic chaperones again at later time points (Figure 3B, column 4, VHL-L158P, compare 0-h to 6-h and 12-h time points). This secondary induction of SSA4 and STI1 may occur through the same heat-shock-like pathway as the initial induction, or it could result from activation of a secondary pathway controlling these genes.
In response to UPR-M/C stresses, SSA4 is induced, but only modestly and at late time points, and little to no effect on STI1 transcript levels is seen (Figure 3B, column 2, ste6-Q1249X). Because the misfolded domains of the UPR-M/C stressors are in the membrane or cytosol, we might have expected the cytosolic chaperone genes to be rapidly induced as a primary response to these proteins. Instead, only UPR-Cyto stress and heat shock induce these genes, suggesting that the UPR-Cyto and UPR-M/C responses are indeed distinct.
Defining the UPR-M/C Transcriptional Response by Microarray Analysis
Our analysis of the UPR-M/C response by Northern blot did not reveal a rapid induction of either UPR-L or cytosolic chaperone genes, suggesting that a different pattern of gene expression may occur in response to UPR-M/C stress. To examine this further, we performed microarray analysis to determine the scope of genes induced by the UPR-M/C stress, ste6-Q1249X (Ste6p*). Three biological replicates of mRNA were harvested at 1 h after galactose induction from cells expressing ste6-Q1249X, wild-type STE6, or empty vector, and processed for microarray analysis, as described in Materials and Methods and the Supplemental Data (microarray analysis for VHL was also carried out in parallel and is discussed in the next section). After normalization to the empty vector control, the lists of genes induced by the expression of ste6-Q1249X and wild-type STE6 were compared with one another to identify genes uniquely induced by ste6-Q1249X and not simply by overexpression of a membrane protein.
Our analysis revealed 63 genes differentially induced by the UPR-M/C stressor and not by wild-type STE6 (listed in Supplemental Table S1). Using the Gene Ontology (GO) Term Finder Tool in the Saccharomyces Genome Database (SGD, 2008; http://www.yeastgenome.org), we found an enrichment for genes classified by the GO terms “ubiquitin-dependent protein catabolism” (10 genes) and “ubiquitin cycle” (four genes), both related to the ubiquitin–proteasome system. These genes are regulated by the transcription factor, Rpn4p (Mannhaupt et al., 1999; Xie and Varshavsky, 2001; London et al., 2004), and upon further analysis of the full list of 63 UPR-M/C-induced genes we found many additional Rpn4p-target genes (23/63), listed in Table 3. In all, 37% of the genes induced by UPR-M/C stress are known Rpn4p-target genes.
Table 3.
Rpn4p-target genes induced in response to UPR-M/C stress
| ORF | Gene | UPR-M/C ste6-Q1249X (fold induction) | Proteasome inhibitor PS-341 (30 μM) (fold induction)a | Rpn4p target gene |
|---|---|---|---|---|
| YKL195W | MIA40 | 1.99 | 2.50 | PS-341,a chIP-chipb |
| YDR515W | SLF1 | 1.63 | 1.95 | PS-341a |
| YFL044C | OTU1 | 1.43 | 2.11 | PS-341a |
| YOR052C | 1.41 | 1.68 | PS-341,a PACEc | |
| YER142C | MAG1 | 1.39 | 3.49 | PS-341,a predictedb |
| YLL039C | UBI4 | 1.33 | ND | PACE2c |
| YOR259C | RPT4 | 1.32 | 1.52 | PS-341,1a PACEc |
| YER021W | RPN3 | 1.32 | 2.30 | PS-341,1 PACEc |
| YJL036W | SNX4 | 1.30 | 1.51 | PS-3411a |
| YOR007C | SGT2 | 1.29 | 1.73 | PS-341,a chIP-chipb |
| YFR004W | RPN11 | 1.25 | 2.31 | PS-341,a PACEc |
| YOR059C | 1.24 | 1.37 | PS-3411 | |
| YHR027C | RPN1 | 1.23 | 1.75 | PS-341,a PACEc |
| YGR048W | UFD1 | 1.22 | 3.10 | PS-341,a chIP-chipb |
| YML092C | PRE8 | 1.21 | ND | PACE2c |
| YMR314W | PRE5 | 1.21 | 2.06 | PS-341,a chIP-chipb |
| YHL030W | ECM29 | 1.20 | 3.70 | PS-341,a PACEc |
| YJL014W | CCT3 | 1.19 | 1.91 | PS-341a |
| YFR003C | YPI1 | 1.19 | 1.99 | chIP-chipb |
| YIL075C | RPN2 | 1.17 | 1.87 | PS-341,a PACEc |
| YPR103W | PRE2 | 1.17 | 1.79 | PS-341,a chIP-chipb |
| YFR010W | UBP6 | 1.15 | 1.76 | PS-341,a chIP-chipb |
| YEL037C | RAD23 | 1.13 | 2.33 | PS-341,a chIP-chipb |
Rpn4p not only up-regulates 26S proteasome subunits, but also other ubiquitin–proteasome pathway components. Rpn4p target genes have been identified in several ways: through promoter analysis revealing an Rpn4p-binding PACE element, by their Rpn4p-dependent induction in response to the proteasome inhibitor PS-341, or by chromatin immunoprecipitation (ChIP)-CHIP analysis with Rpn4p (Mannhaupt et al., 1999; Fleming et al., 2002; Beyer et al., 2006; Table 3; Rpn4p-target genes). Included in the list of UPR-M/C induced genes are 10 different subunits of the proteasome and several other genes that have known or putative roles in ubiquitin–proteasome-related pathways. Although the induction of the Rpn4p-dependent genes in response to UPR-M/C stress is modest, ranging from 1.13- to 1.99-fold (Table 3), the large number of Rpn4p-target genes suggests that this induction is likely to be significant. Additionally, in previously published studies more than half of these genes were also induced less than twofold by treatment with 30 μM proteasome inhibitor (PS-341) for 1 h, and the highest induction seen under these conditions was 3.7-fold (Fleming et al., 2002; noted here in Table 3).
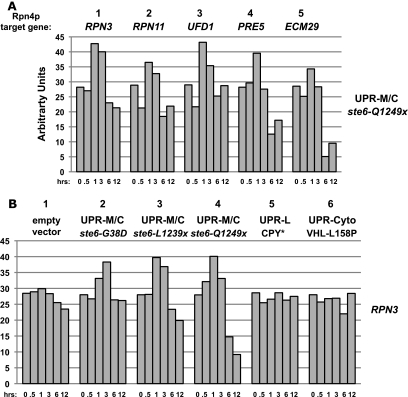
The UPR-M/C-induced transcriptional profile of Rpn4p-regulated genes was confirmed by Northern blot analysis, where, in agreement with our microarray data, the levels of induction for a selection of these genes (RPN3, RPN11, UFD1, PRE5, and ECM29) ranged from 1.2- to 1.5-fold upon expression of ste6-Q1249X (Figure 4A). The pattern of induction elicited by UPR-M/C stress is transient in nature, with the transcript levels falling to or below basal levels by 6 h after expression of UPR-M/C stress. For all of the genes examined, we find that the three UPR-M/C misfolded protein stressors (ste6-G38D, ste6-L1239X, and ste6-Q1249X) show a similar induction profile (Figure 4B with the proteasome subunit, RPN3, and our unpublished data).
Figure 4.
The UPR-M/C response is characterized by the induction of Rpn4p-target genes. (A) Wild-type cells (SM4460) expressing galactose-inducible ste6-Q1249X (pSM2213) were harvested for RNA processing at 0, 0.5, 1, 3, 6, and 12 h after galactose induction, and Northern blotting was performed with probes to the indicated UPR-M/C genes. (B) Northern blot analysis of wild-type cells (SM4460) expressing galactose-inducible empty vector (pSM922), ste6-G38D (pSM1898), ste6-L1239X (pSM2212), ste6-Q1249X (pSM2213), CPY* (pSM2215), or VHL-L158P (pESC-L158P) were harvested for RNA processing and Northern blotting with RPN3 at the indicated times after galactose induction. Northern blots were performed in duplicate and shown is a representative experiment in which quantitated, normalized values, determined as described in Materials and Methods, are graphed as arbitrary units.
The UPR-M/C Response Differs from the UPR-Cyto and UPR-L Responses
To determine whether this transcriptional response is unique to UPR-M/C stress, we analyzed the induction of the Rpn4p-dependent genes discussed above in response to UPR-L and UPR-Cyto stressors. The induction of the UPR-M/C response appears to be largely specific to UPR-M/C stresses (Figure 4B, compare CPY* and VHL-L158P in lanes 5 and 6 with the UPR-M/C stressors in lanes 2–4). To compare the UPR-M/C response to the UPR-Cyto response at a more global level, we performed microarray analysis with VHL-L158P. Three biological replicates of mRNA were harvested at 1 h after galactose induction from cells expressing VHL-L158P, microarray analysis was performed, and the data were normalized to empty vector as described above for ste6-Q1249X. However, unlike the case for ste6-Q1249X, where we could compare to the “isogenic” wild-type STE6, a properly folded cytosolic protein comparable to VHL-L158P is not available. As a result, we are not able to make strong conclusions regarding the precise profile of gene induction the UPR-Cyto response. Additional studies with several other misfolded cytosolic proteins, including misfolded endogenous yeast proteins, will be needed to fully characterize the UPR-Cyto response. Despite these limitations, we are able to make several notable observations from our analysis: first, multiple genes are induced by the expression of all three of the proteins we examined (ste6-Q1249X, STE6, and VHL-L158P), including those involved in generalized metabolic functions, such as PHO84 and ADE17. The induction of these genes may be related to the high level of protein expression from the galactose-inducible promoter. Second, the GO Term Finder Tool did not reveal a significant enrichment of genes with common GO terms in the list of VHL-L158P–induced genes, as was identified in the ste6-Q1249X data set. There also is not a significant induction of heat shock genes at this 1 h time point, which is in agreement with our Northern blot data showing that the cytosolic chaperone genes have already returned to their basal transcript levels by 1 h after induction of VHL-L158P (Figure 3B). Microarray analysis of samples collected at multiple time points after induction of VHL-L158P will be required to fully define the transient UPR-Cyto response characterized thus far by induction of SSA4 and STI1.
Most importantly, from our genomic analysis we can conclude that very few of the Rpn4p-dependent genes induced uniquely by UPR-M/C stress are also induced by VHL-L158P (4/23; our unpublished data). In addition to the UPR-Cyto response, we compared our UPR-M/C microarray data to the previous microarray analysis defining the UPR-L response induced by the drugs DTT and Tm (Travers et al., 2000) to determine any overlap between the UPR-M/C and the UPR-L response. This analysis reveals a mild induction of some of the same genes (11/23) induced by UPR-M/C stress. None of these, however, would be characterized as UPR-L target genes, defined as those dependent on HAC1 and IRE1 for their induction. Many of these overlapping genes, such as UBI4, the gene encoding ubiquitin, or RAD23 and UFD1, both involved in targeting of misfolded proteins from the ER to the proteasome, may play roles in a variety of cellular stress pathways.
The modest induction of a large number of Rpn4p-regulated genes that characterizes the response to UPR-M/C stress appears to be physiologically relevant, because in synthetic lethal analysis (described below) we find RPN4 to be uniquely required for viability in the presence of UPR-M/C stresses, but not UPR-L or UPR-Cyto stresses. Thus, the requirement for Rpn4p may reflect the fact that a functional UPR-M/C transcriptional response is essential for viability in cells experiencing misfolded membrane protein stress.
Synthetic lethality with UPR-M/C stresses reveals unique differences in the cell's ability to cope with misfolded proteins in different cellular compartments
To learn more about the UPR-M/C pathway and identify genes that enable cells to cope with misfolded membrane proteins, we performed a modified synthetic lethality screen. We used dSLAM (Pan et al., 2004; Warren et al., 2004) to identify gene deletions that cannot tolerate UPR-M/C stress. Presumably, deletions of genes required to promote cell survival when cells are challenged by UPR-M/C stress would be inviable upon expression of a UPR-M/C stress (Figure 5A). We performed dSLAM using ste6-G38D expressed from the galactose-inducible promoter and identified 15 genes that show a synthetic lethal or “sick” phenotype with ste6-G38D (see Materials and Methods for the list of these genes).
Figure 5.
Synthetic lethality analysis reveals a requirement for Rpn4p, Hac1p, and Ssh1p for viability during UPR-M/C stress. (A) Model for the use of synthetic lethality to identify genes in a pathway required to cope with a UPR-M/C stress. Hypothetical genes (labeled A–C and here are shown in a single pathway, for simplicity) are hypothesized to be required for survival of a UPR-M/C stress. When any of these genes are deleted, cells are inviable. (B) Serial dilutions of the wild-type (SM4460) and indicated mutant (hac1Δ, rpn4Δ, ssh1Δ; SM5382, SM5383, SM5384, respectively) strains containing the galactose-inducible ste6-G38D plasmid (pSM1898) or empty vector (pSM922) growing on the indicated media.
Among the gene deletions that have the most strongly synthetic phenotypes are rpn4Δ, hac1Δ, and ssh1Δ, all of which show significantly reduced growth when ste6-G38D expression is induced on media containing galactose (Figure 5B, top). Deletions of these genes show no growth defect on media containing galactose when empty vector is expressed, indicating they are not merely inviable on galactose (Figure 5B, bottom). The identification of HAC1 as an essential gene during UPR-M/C stress suggests that the small amount of splicing seen at late time points in response to this class of stress (Figure 3A, column 2, top) is significant for the long-term survival of cells. The UPR-L controls multiple aspects of secretory pathway function, including protein import and trafficking, glycosylation, and membrane production (Travers et al., 2000), and may be required to maintain membrane homeostasis during prolonged UPR-M/C stress (Ron and Hampton, 2004; Federovitch et al., 2005). We do not yet know what specific function the UPR-L provides to cells experiencing chronic UPR-M/C stress.
Another synthetic lethal gene, SSH1, is a homologue of the Sec61p translocon (Finke et al., 1996) and has been suggested to have a role in cotranslational protein import (Finke et al., 1996; Wittke et al., 2002). We previously found that Ssh1p may play a role in the ERAD of ste6-Q1249X (Ste6p*) under certain conditions (Huyer et al., 2004b). Ssh1p is present in a complex containing Sbh2p (Finke et al., 1996), and we also find a mild growth defect in sbh2Δ experiencing a UPR-M/C stress (our unpublished data). Further work will be needed to determine what role Ssh1p is playing in maintaining ER homeostasis during UPR-M/C stress.
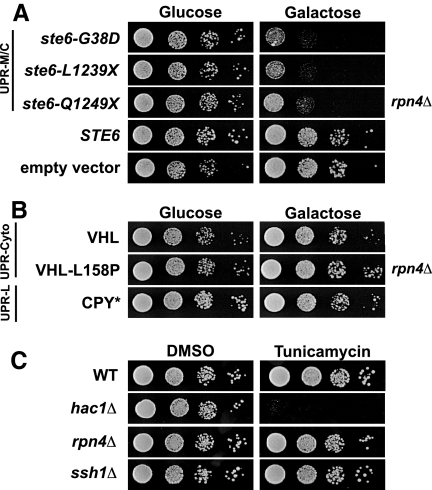
We chose to focus on rpn4Δ because Rpn4p-dependent genes are coordinately induced in response to UPR-M/C stress, as reported above. To determine whether the sensitivity of rpn4Δ to ste6-G38D is allele-specific, we asked whether rpn4Δ was also sensitive to the other UPR-M/C stresses, ste6-L1239X and ste6-Q1249X, or to expression of wild-type STE6. The rpn4Δ strain is sensitive to all of the UPR-M/C stresses we tested, and not to wild-type STE6 or empty vector (Figure 6A, galactose). Thus, Rpn4p is required for the tolerance of UPR-M/C stresses, but not for overexpression of a wild-type secretory protein. The sensitivity of rpn4Δ to UPR-M/C stresses with different rates of turnover and distinct sites of mutations (transmembrane or cytosolic) suggests that the effect is independent of differences in half-life or location of mutation, but correlates with misfolding and ER retention.
Figure 6.
Rpn4p is required to survive UPR-M/C stress, but not UPR-L or UPR-Cyto stress. (A) Serial dilutions of the rpn4Δ mutant (SM5383) containing wild-type STE6 (pSM1897), empty vector (pSM922), or the UPR-M/C stresses ste6-G38D (pSM1898), ste6-L1239X (pSM2212), or ste6-Q1249X (pSM2213) are shown. (B) Serial dilutions of the rpn4Δ mutant (SM5383) containing plasmids that induce the UPR-Cyto stress, VHL (pESC-VHL) or VHL-L158P (pESC-L158P), or the UPR-L stress, CPY* (pSM2215) are shown. (C) Serial dilutions of wild-type (SM4460), rpn4Δ (SM5383), hac1Δ (SM5382), and ssh1Δ (SM5384) cells were grown on media containing 0.1 μg/ml tunicamycin or an equal volume DMSO for 2 d at 30°C.
One possibility to explain the sensitivity of rpn4Δ to UPR-M/C stresses is that all misfolded proteins requiring the proteasome for degradation will cause such inviability. However, this does not appear to be the case, as we find that rpn4Δ is not sensitive to expression of the UPR-Cyto stresses, VHL or VHL-L158P, or to expression of the UPR-L stress, CPY* (Figure 6B, galactose). This indicates, as our transcriptional data also suggests, that Rpn4p is uniquely required for coping with the UPR-M/C class of stress, despite all three classes of misfolded proteins depending on the proteasome for degradation. Previously, Ng et al. (2000) demonstrated that Rpn4p is required for viability in cells lacking UPR-L function (i.e., ire1Δ and hac1Δ). To confirm that cells experiencing UPR-L stress do not require Rpn4p for viability, we asked whether rpn4Δ is sensitive to Tm. The rpn4Δ mutant is viable on media containing Tm, whereas the hac1Δ strain is highly sensitive to this type of stress (Figure 6C, tunicamycin). Like rpn4Δ, the ssh1Δ mutant is not sensitive to Tm. Taken together, it appears that neither a UPR-L–inducing drug nor the UPR-L– or UPR-Cyto–inducing misfolded proteins that we tested require Rpn4p for viability. Instead, only the misfolded membrane proteins require Rpn4p, suggesting this is a unique property of this type of protein. These findings also highlight the importance of the UPR-M/C transcriptional response described above in tolerating misfolded membrane protein stress and are in agreement with the UPR-M/C response being uniquely induced by misfolded membrane proteins.
Functional Proteasomes Are Required for Viability in Cells Experiencing UPR-M/C Stress
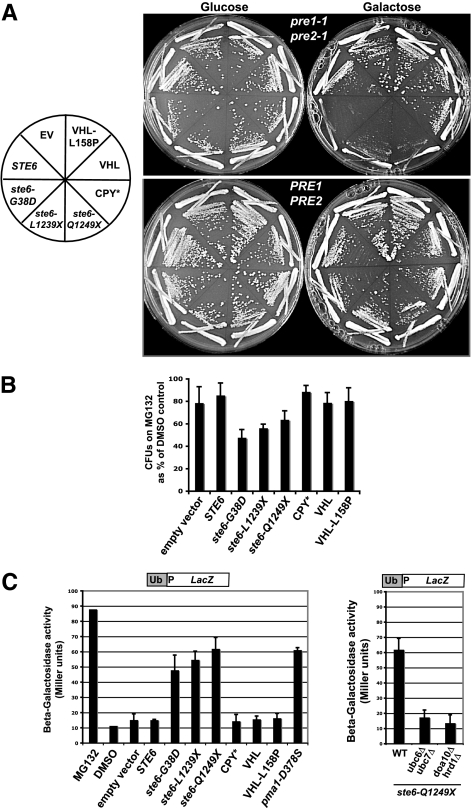
Rpn4p is required by cells to maintain basal levels of proteasome subunits and to induce proteasome subunits under conditions of reduced proteasome function (Xie and Varshavsky, 2001; Ju et al., 2004; London et al., 2004), but it is also responsible for the induction of DNA repair genes and other ubiquitin–proteasome pathway components in response to a variety of stress conditions (Mannhaupt et al., 1999; Jelinsky et al., 2000; Fleming et al., 2002). We asked whether mutations in proteasome subunits showed the same pattern of sensitivity as rpn4Δ, or whether it is the inability to induce other target genes of Rpn4p that causes the sensitivity to a UPR-M/C stress. Because the proteasome is essential, we used the pre1-1 pre2-1 temperature-sensitive mutant, which has conditional defects in the 20S proteasome subunits PRE1 and PRE2 (Richter-Ruoff et al., 1992). The pre1-1 pre2-1 mutant shows strong sensitivity to expression of any of the misfolded ste6 alleles (ste6-G38D, -L1239X, and -Q1249X), with very little growth even at permissive temperature (24°C) when expressing these UPR-M/C stressors (Figure 7A, top panel). In contrast, pre1-1 pre2-1 mutant cells expressing UPR-Cyto stressors (VHL and VHL-L158P) or the UPR-L stressors, CPY*, show no sensitivity. Interestingly, wild-type Ste6p overexpression also has a modest effect on the viability of the pre1-1 pre2-1 mutant, though not nearly as dramatic as a UPR-M/C stressor. This is despite its degradation occurring by vacuolar peptidases and not by the proteasome (Kelm et al., 2004) and may indicate that wild-type Ste6p trafficking may “back up” into the ER when overexpressed for prolonged periods of time and thus require the proteasome for some of it degradation.
Figure 7.
Proteasome function is specifically impaired by the presence of misfolded membrane proteins. (A) Wild-type (PRE1 PRE2; WCG4a) and proteasome mutant strain pre1-1 pre2-1 (WCG4A-11/21A) containing the indicated galactose-inducible misfolded proteins (see Table 2 for plasmids names) were streaked to the indicated media and grown at 24°C for 4 d. (B) A strain sensitized to MG132, erg6::kanMX (SM4464), was transformed with plasmids expressing the indicated galactose-inducible misfolded proteins (see panel A). Transformants were assayed for viability by plating on solid media after an 8 h treatment in liquid with 50 μM MG132, as described in Materials and Methods. Colony-forming units (CFUs) were counted for MG132- and DMSO-treated cells and graphed as CFUs on MG132 as a percentage of CFUs on DMSO. Each experiment was repeated at least three times and graphed as the average; error bars, 1 SD. A paired t test gives p < 0.01 for ste6-G38D and a p < 0.05 for ste6-L1239X and ste6-Q1249X, all compared with empty vector. (C) To examine proteasome function, wild-type cells (SM4460) containing empty vector (pSM922) or the indicated galactose-inducible misfolded proteins and the Ub-Pro-LacZ reporter for proteasome activity (pSM2216) were induced by the addition of 4% galactose for 6 h at 30°C before preparation for β-galactosidase assay. For comparison, cells were treated with 50 μM MG132 or an equal volume of DMSO for 12 h at 30°C. Each experiment was repeated at least three times and graphed as the average; error bars, 1 SD. (D) To examine the affect of ubiquitination mutants on proteasome inhibition by ste6-Q1249X, wild-type (SM4460), ubc6Δ ubc7Δ (SM5362), and doa10Δ hrd1Δ (SM5360) cells expressing galactose-inducible ste6-Q1249X (pSM2213) and Ub-Pro-LacZ (pSM2216) were grown, prepared, and analyzed as in C.
To confirm the above result obtained with the pre1-1 pre2-1 mutant, erg6Δ cells (used to increase permeability to the drug MG132) expressing the three classes of misfolded protein stresses were assayed for viability after 8 h of treatment with the proteasome inhibitor, MG132. Galactose induction of cells containing empty vector or wild-type Ste6p resulted in greater than 80% viability after MG132 treatment, whereas cells expressing the three UPR-M/C stressors were impaired for viability when treated with MG132 (Figure 7B). No effect was seen on cells expressing the UPR-Cyto or UPR-L stressors (VHL, VHL-L158P, or CPY*). The particular sensitivity of proteasome-impaired cells to a UPR-M/C stress mirrors the pattern of sensitivity we see in rpn4Δ cells, supporting the hypothesis that the inviability of rpn4Δ cells experiencing UPR-M/C stress is due to Rpn4p's role in induction of 26S proteasomes. Thus, misfolded membrane proteins appear to provide a particular challenge to cells with reduced levels of the proteasome.
The Proteasome Is Impaired by UPR-M/C Stresses in a Ubiquitin-dependent Manner
Cells expressing UPR-M/C stressors appear to require Rpn4p-dependent gene induction and specifically show sensitivity to impairments of the proteasome by mutation or drug. A possible reason for this could be that misfolded membrane proteins “tie up” or “clog” proteasomes and thus may require the synthesis of additional proteasomes to maintain essential proteasome function in the cell, a situation necessitating upregulation by Rpn4p. To address this possibility, wild-type cells expressing UPR-M/C stressors for 6 h were assayed for proteasome impairment using the ubiquitin fusion degradation substrate, Ub-Pro-LacZ (Bachmair et al., 1986), as a reporter of proteasome function. When the proteasome is functioning normally, Ub-Pro-LacZ is degraded rapidly, resulting in very low β-galactosidase activity. If proteasome function is impaired, however, there is stabilization of Ub-Pro-LacZ, which can be detected as increased β-galactosidase activity (Figure 7C, MG132). A greater than five-fold increase in β-galactosidase activity is seen in cells expressing the UPR-M/C stressors versus cells with vector only, indicating proteasome impairment (Figure 7C). Proteasome impairment occurs with all three misfolded ste6 alleles, as well as with another misfolded membrane protein, pma1-D378S. No proteasome impairment is seen in cells expressing UPR-L or UPR-Cyto stresses. Therefore, the expression of UPR-M/C stresses uniquely impairs proteasome function and may suggest that misfolded membrane proteins “choke” the proteasome. Because UPR-L and UPR-Cyto stresses do not have this effect, the impairment may be directly related to the presence of membrane spans in misfolded Ste6p and Pma1p.
Proteasome impairment by UPR-M/C stresses is dependent on ubiquitination of the misfolded protein. When the ubiquitination of ste6-Q1249X is prevented by deletion of the E2s, Ubc6p and Ubc7p, or the E3s, Doa10p and Hrd1p, required for ste6-Q1249X's degradation, it no longer impairs the proteasome (Figure 7D), indicating that a ubiquitinated form of ste6-Q1249X is responsible for the proteasome impairment.
DISCUSSION
Distinct Transcriptional Responses to Misfolded Proteins in Different Cellular Compartments
A key aspect of protein quality control is the ability to up-regulate genes that help cells to refold, degrade, and otherwise cope with misfolded proteins. Such transcriptional activation is exemplified by the well-studied, classical UPR pathway, induced by ER lumenal stress. Here, we provide evidence that three distinct responses (designated UPR-L, UPR-Cyto, and UPP-M/C in this study) are induced by misfolded proteins residing in different cellular compartments (ER, cytosol, and membrane, respectively). First, by examining a panel of five reporter genes we show that the UPR-L and UPR-Cyto responses induced by single misfolded proteins are different from one another (induction of HAC1 splicing, KAR2, and YFR026C characterizes UPR-L vs. induction of SSA4 and STI1 for UPR-Cyto). A UPR-M/C stressor does not significantly induce either of these responses, but microarray analysis reveals that there is indeed a distinct UPR-M/C response.
The UPR-M/C Response
By microarray, we characterized a UPR-M/C transcriptional response that consists largely (23/63 genes) of Rpn4p-target genes. We also demonstrate that Rpn4p is required for cellular viability in the presence of UPR-M/C stress, in support of this response being functionally significant for coping with misfolded membrane proteins. Rpn4p is crucial for cell survival during proteasome inhibition (Fleming et al., 2002) and accordingly, we find that UPR-M/C stress (but not UPR-L or UPR-Cyto stress) specifically blocks proteasomal degradation. Rpn4p is thus likely required during UPR-M/C stress to generate new proteasomes to meet the essential degradative needs of the cell. Interestingly, the role of Rpn4p in the UPR-M/C response parallels the role of Hac1p in the UPR-L response, where Hac1p is essential to combat the effects of a UPR-L stress by upregulating genes that promote cell survival (Mori et al., 1996; Nikawa et al., 1996).
The presence of membrane spans, rather than the topological site of the mutation or the half-life of the mutant protein, appears to be the distinguishing characteristic that elicits the UPR-M/C transcriptional profile. Cells expressing any of the three misfolded Ste6p alleles require Rpn4p for viability, and by Northern blot analysis they appear to induce similar Rpn4p-dependent UPR-M/C responses, despite having mutations in different cellular locations. For instance, ste6-Q1249X and -L1239X truncate a cytosolic ATP-binding domain, whereas ste6-G38D lies in a membrane span. Further genome-wide analysis with the ste6-G38D and ste6-L1239X alleles may reveal nuanced differences in the transcriptional profiles for the different alleles.
How Are Rpn4p-dependent Genes Induced by UPR-M/C Stresses?
Although Rpn4p can be a downstream transcriptional target of other transcription factors (Owsianik et al., 2002; Hahn et al., 2006), we see no change in RPN4 transcript levels after UPR-M/C stress (our unpublished data), so this is not likely to be how UPR-M/C stress activates Rpn4p-dependent transcription. In addition to being an activator of the proteasome, Rpn4p is also a rapidly degraded substrate of the proteasome (Xie and Varshavsky, 2001; Ju et al., 2004). Thus, Rpn4p protein levels serve as a sensitive “sensor” of proteasome function, and Rpn4p-target gene induction by UPR-M/C stress may simply reflect the proteasome impairment seen under these conditions (Figure 7). Alternatively, Rpn4p can be modulated post-translationally (Ju and Xie, 2004, 2007), and UPR-M/C stress could be sensed independently of proteasome function and Rpn4p activated post-translationally.
What Role Does the Proteasome Play in Degradation of Misfolded Membrane Proteins?
An important question to answer is why UPR-M/C stress specifically leads to impairment or “choking” of the proteasome, whereas UPR-L or UPR-Cyto stress apparently do not? Although the degradation of UPR-L and UPR-Cyto misfolded proteins is proteasome-dependent (Hiller et al., 1996; McClellan et al., 2005), under “overflow” conditions, CPY* can be degraded in the vacuole (Spear and Ng, 2003). It was also shown that autophagy occurs in response to Tm and DTT (Bernales et al., 2006; Yorimitsu et al., 2006). We find that GFP-Atg8p is cleaved in response to CPY*, although CPY* does not impair growth in autophagy mutants (our unpublished data). Vacuolar routing from the secretory pathway or autophagy followed by Pep4p-dependent degradation may prevent proteasome impairment by CPY*. Interestingly, UPR-M/C stressors do not induce autophagy, nor is their degradation Pep4p-dependent, even after prolonged expression (our unpublished data).
Another possibility to explain why UPR-M/C stressors uniquely cause proteasome impairment could be that multispanning membrane proteins are degraded by the proteasome in a manner unique from soluble proteins. For UPR-M/C substrates, the proteasome may be involved in their membrane extraction. Several studies have concluded that membrane extraction and proteasomal degradation are coupled (Mayer et al., 1998; Xiong et al., 1999; Piwko and Jentsch, 2006; Baker and Tortorella, 2007). There is also precedence for the proteasome directly cleaving and degrading membrane proteins from the ER membrane (Hoppe et al., 2000; Piwko and Jentsch, 2006). The proteasome thus may play a more direct or complicated role in degrading polytopic membrane proteins.
Nakatsukasa et al. (2008) recently demonstrated in vitro that full-length Ste6p* was released into the cytosol post-ubiquitination (Nakatsukasa et al., 2008). If this also occurs in vivo, membrane proteins, perhaps because of strong intramolecular interactions between their spans, could provide a challenge for the proteasome to efficiently degrade. Proteasome impairment is caused by a variety of disease-associated aggregation prone proteins, such as prion protein, Huntingtin, and CFTR-ΔF508 (Bence et al., 2001; Apodaca et al., 2006; Kristiansen et al., 2007). Although some misfolded membrane proteins aggregate, others, such as Ste6p* do not (Huyer et al., 2004a). Future study regarding the mechanisms of proteasomal degradation of misfolded membrane proteins will reveal insight into these interesting possibilities.
The UPR-L Response
The UPR-L transcriptional response induced by the global stressors Tm or DTT has been defined genomically (Travers et al., 2000). In this study we characterized the induction of a novel gene, YFR026C, that is highly induced by CPY*, as assayed by Northern analyses (>15-fold; Figure 2A) and appears among the most highly induced genes by UPR-L genome-wide microarray analysis (Travers et al., 2000). Although YFR026Cp shows no sequence similarity to known proteins, it does have a predicted transmembrane domain and signal sequence, suggesting it could be ER-localized (SGD, 2008). The loss of YFR026C leads to UPR-L induction (Figure 2), indicating it may play a role in preventing or abrogating ER stress. Additionally, yeast two-hybrid studies indicates two interactions for YFR026C: with the E2, Ubc7p, and with the 20S proteasome subunit, Pre10p (S. Fields, The Yeast Resource Center, University of Washington, personal communication). However, YFR026C does not appear to influence the degradation of misfolded ER proteins, because there is no effect on the turnover of CPY* in yfr026cΔ (our unpublished data). Although YFR026C's role in ERQC remains unknown, it is a useful reporter for the UPR-L response because of its very strong induction by UPR-L stress. We propose naming this gene ULI1, for UPR-L–inducible gene.
The UPR-Cyto Response
We found that the UPR-Cyto response, induced by VHL and VHL-L158P, resembles the heat-shock response. It is implicit in quality control literature that the heat-shock response results from misfolded cytosolic proteins, but this has not been analyzed directly. Because we have not compared the genome-wide responses to UPR-Cyto and heat stress in parallel, we cannot definitively conclude that UPR-Cyto stress induces the exact same genes induced by heat stress, but our Northern blot analysis shows that the timing and degree of induction of the cytosolic chaperone genes by the two are highly similar (Figure 3B and Supplemental Figure S1B). At late time points, heat shock and UPR-Cyto stress do differ slightly, where UPR-Cyto stress shows a unique reinduction of the chaperones, suggesting that this stress, unlike heat stress, is persistent and unrepairable. From our analysis we can conclude that the UPR-M/C and UPR-Cyto responses are distinct, and future studies will be required to fully define the UPR-Cyto response and determine the transcription factors mediating this gene induction.
Supplementary Material
ACKNOWLEDGMENTS
We thank Peter Walter (University of California, San Francisco), Davis Ng (National University of Singapore), Amy Chang (University of Michigan), Judith Frydman (Stanford University), Mark Hochstrasser (Yale University), and Rob Jensen for generously supplying plasmids and antibodies. We are grateful to Forrest Spencer, Jef Boeke, Peter Espenshade, Haiping Hao, and Chunfa Jie for technical assistance. We thank the members of the Michaelis lab for helpful discussions and Jemima Barrowman and Jim Mullally for the critical reading of this manuscript. This work was supported by National Institutes of Health Grant GM51508 to S.M.
Abbreviations used:
- AZC
l-azetidine-2-carboxylic acid
- DEPC
diethyl pyrocarbonate
- DTT
dithiothreitol
- ERAD
ER-associated degradation
- ERQC
ER-quality control
- Tm
tunicamycin
- UPR
unfolded protein response
- UPRE
unfolded protein response element
- VHL
von Hippel Lindau protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0140) on December 10, 2008.
REFERENCES
- Apodaca J., Kim I., Rao H. Cellular tolerance of prion protein PrP in yeast involves proteolysis and the unfolded protein response. Biochem. Biophys. Res. Commun. 2006;347:319–326. doi: 10.1016/j.bbrc.2006.06.078. [DOI] [PubMed] [Google Scholar]
- Bachmair A., Finley D., Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Baker B. M., Tortorella D. Dislocation of an endoplasmic reticulum membrane glycoprotein involves the formation of partially dislocated ubiquitinated polypeptides. J. Biol. Chem. 2007;282:26845–26856. doi: 10.1074/jbc.M704315200. [DOI] [PubMed] [Google Scholar]
- Bence N. F., Sampat R. M., Kopito R. R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Bernales S., McDonald K. L., Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer A., Workman C., Hollunder J., Radke D., Moller U., Wilhelm T., Ideker T. Integrated assessment and prediction of transcription factor binding. PLoS Comput. Biol. 2006;2:e70. doi: 10.1371/journal.pcbi.0020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein W. R., Craig E. A. Structure and regulation of the SSA4 HSP70 gene of Saccharomyces cerevisiae. J. Biol. Chem. 1990;265:18912–18921. [PubMed] [Google Scholar]
- Bordallo J., Plemper R. K., Finger A., Wolf D. H. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol. Biol. Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J. L. The protective and destructive roles played by molecular chaperones during ERAD (endoplasmic-reticulum-associated degradation) Biochem. J. 2007;404:353–363. doi: 10.1042/BJ20061890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Goder V., Rapoport T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Cox J. S., Shamu C. E., Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Cox J. S., Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Federovitch C. M., Ron D., Hampton R. Y. The dynamic ER: experimental approaches and current questions. Curr. Opin. Cell Biol. 2005;17:409–414. doi: 10.1016/j.ceb.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Feldman D. E., Thulasiraman V., Ferreyra R. G., Frydman J. Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol. Cell. 1999;4:1051–1061. doi: 10.1016/s1097-2765(00)80233-6. [DOI] [PubMed] [Google Scholar]
- Finger A., Knop M., Wolf D. H. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur. J. Biochem. 1993;218:565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- Finke K., Plath K., Panzner S., Prehn S., Rapoport T. A., Hartmann E., Sommer T. A second trimeric complex containing homologs of the Sec61p complex functions in protein transport across the ER membrane of S. cerevisiae. EMBO J. 1996;15:1482–1494. [PMC free article] [PubMed] [Google Scholar]
- Fleming J. A., Lightcap E. S., Sadis S., Thoroddsen V., Bulawa C. E., Blackman R. K. Complementary whole-genome technologies reveal the cellular response to proteasome inhibition by PS-341. Proc. Natl. Acad. Sci. USA. 2002;99:1461–1466. doi: 10.1073/pnas.032516399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura-Kamada K., Nouvet F. J., Michaelis S. A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast a-factor precursor. J. Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Hahn J. S., Neef D. W., Thiele D. J. A stress regulatory network for co-ordinated activation of proteasome expression mediated by yeast heat shock transcription factor. Mol. Microbiol. 2006;60:240–251. doi: 10.1111/j.1365-2958.2006.05097.x. [DOI] [PubMed] [Google Scholar]
- Hampton R. Y., Gardner R. G., Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W., Simeon A., Hirsch H. H., Schiffer H. H., Teichert U., Wolf D. H. Lysosomal and non-lysosomal proteolysis in the eukaryotic cell: studies on yeast. Biochem. Soc. Trans. 1991;19:724–725. doi: 10.1042/bst0190724. [DOI] [PubMed] [Google Scholar]
- Hiller M. M., Finger A., Schweiger M., Wolf D. H. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Hoppe T., Matuschewski K., Rape M., Schlenker S., Ulrich H. D., Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- Huyer G., Longsworth G. L., Mason D. L., Mallampalli M. P., McCaffery J. M., Wright R. L., Michaelis S. A striking quality control subcompartment in Saccharomyces cerevisiae: the endoplasmic reticulum-associated compartment. Mol. Biol. Cell. 2004a;15:908–921. doi: 10.1091/mbc.E03-07-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J. Biol. Chem. 2004b;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinsky S. A., Estep P., Church G. M., Samson L. D. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol. 2000;20:8157–8167. doi: 10.1128/mcb.20.21.8157-8167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju D., Wang L., Mao X., Xie Y. Homeostatic regulation of the proteasome via an Rpn4-dependent feedback circuit. Biochem. Biophys. Res. Commun. 2004;321:51–57. doi: 10.1016/j.bbrc.2004.06.105. [DOI] [PubMed] [Google Scholar]
- Ju D., Xie Y. Proteasomal degradation of RPN4 via two distinct mechanisms, ubiquitin-dependent and -independent. J. Biol. Chem. 2004;279:23851–23854. doi: 10.1074/jbc.C400111200. [DOI] [PubMed] [Google Scholar]
- Ju D., Xu H., Wang X., Xie Y. Ubiquitin-mediated degradation of Rpn4 is controlled by a phosphorylation-dependent ubiquitylation signal. Biochim. Biophys. Acta. 2007;1773:1672–1680. doi: 10.1016/j.bbamcr.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Kelm K. B., Huyer G., Huang J. C., Michaelis S. The internalization of yeast Ste6p follows an ordered series of events involving phosphorylation, ubiquitination, recognition and endocytosis. Traffic. 2004;5:165–180. doi: 10.1111/j.1600-0854.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- Kohno K., Normington K., Sambrook J., Gething M. J., Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen M., et al. Disease-associated prion protein oligomers inhibit the 26S proteasome. Mol. Cell. 2007;26:175–188. doi: 10.1016/j.molcel.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Lee M. S., Spencer F. A. Bipolar orientation of chromosomes in Saccharomyces cerevisiae is monitored by Mad1 and Mad2, but not by Mad3. Proc. Natl. Acad. Sci. USA. 2004;101:10655–10660. doi: 10.1073/pnas.0404102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. H., Walter P., Yen T. S. Endoplasmic reticulum stress in disease pathogenesis. Annu. Rev. Pathol. 2008;3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza D., Tam A., Schmidt W. K., Michaelis S. Ste6p mutants defective in exit from the endoplasmic reticulum (ER) reveal aspects of an ER quality control pathway in Saccharomyces cerevisiae. Mol. Biol. Cell. 1998;9:2767–2784. doi: 10.1091/mbc.9.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London M. K., Keck B. I., Ramos P. C., Dohmen R. J. Regulatory mechanisms controlling biogenesis of ubiquitin and the proteasome. FEBS Lett. 2004;567:259–264. doi: 10.1016/j.febslet.2004.04.078. [DOI] [PubMed] [Google Scholar]
- Mannhaupt G., Schnall R., Karpov V., Vetter I., Feldmann H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 1999;450:27–34. doi: 10.1016/s0014-5793(99)00467-6. [DOI] [PubMed] [Google Scholar]
- Mayer T. U., Braun T., Jentsch S. Role of the proteasome in membrane extraction of a short-lived ER-transmembrane protein. EMBO J. 1998;17:3251–3257. doi: 10.1093/emboj/17.12.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan A. J., Scott M. D., Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121:739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- Metzger M. B., Maurer M. J., Dancy B. M., Michaelis S. Degradation of a cytosolic protein requires Endoplasmic Reticulum-associated degradation machinery. J. Biol. Chem. 2008;283:32302–32316. doi: 10.1074/jbc.M806424200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis S., Herskowitz I. The a-factor pheromone of Saccharomyces cerevisiae is essential for mating. Mol. Cell. Biol. 1988;8:1309–1318. doi: 10.1128/mcb.8.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K., Kawahara T., Yoshida H., Yanagi H., Yura T. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- Mori K., Sant A., Kohno K., Normington K., Gething M. J., Sambrook J. F. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K., Huyer G., Michaelis S., Brodsky J. L. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D. T., Spear E. D., Walter P. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijbroek G. Dissertation. Baltimore, MD: Johns Hopkins University; 1998. Distinct Classes of Yeast ste6 Mutants Provide Insights into ER Quality Control. [Google Scholar]
- Nikawa J., Akiyoshi M., Hirata S., Fukuda T. Saccharomyces cerevisiae IRE2/HAC1 is involved in IRE1-mediated KAR2 expression. Nucleic Acids Res. 1996;24:4222–4226. doi: 10.1093/nar/24.21.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S., Brodsky J. L., Nakatsukasa K. Roles of molecular chaperones in endoplasmic reticulum (ER) quality control and ER-associated degradation (ERAD) J. Biochem. 2005;137:551–555. doi: 10.1093/jb/mvi068. [DOI] [PubMed] [Google Scholar]
- Otsu M., Sitia R. Diseases originating from altered protein quality control in the endoplasmic reticulum. Curr. Med. Chem. 2007;14:1639–1652. doi: 10.2174/092986707780830952. [DOI] [PubMed] [Google Scholar]
- Owsianik G., Balzi E., Ghislain M. Control of 26S proteasome expression by transcription factors regulating multidrug resistance in Saccharomyces cerevisiae. Mol. Microbiol. 2002;43:1295–1308. doi: 10.1046/j.1365-2958.2002.02823.x. [DOI] [PubMed] [Google Scholar]
- Pan X., Yuan D. S., Xiang D., Wang X., Sookhai-Mahadeo S., Bader J. S., Hieter P., Spencer F., Boeke J. D. A robust toolkit for functional profiling of the yeast genome. Mol. Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Parsell D. A., Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu. Rev. Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- Patil C. K., Li H., Walter P. Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol. 2004;2:E246. doi: 10.1371/journal.pbio.0020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto M., Morange M., Bensaude O. Denaturation of proteins during heat shock. In vivo recovery of solubility and activity of reporter enzymes. J. Biol. Chem. 1991;266:13941–13946. [PubMed] [Google Scholar]
- Piwko W., Jentsch S. Proteasome-mediated protein processing by bidirectional degradation initiated from an internal site. Nat. Struct. Mol. Biol. 2006;13:691–697. doi: 10.1038/nsmb1122. [DOI] [PubMed] [Google Scholar]
- Richter-Ruoff B., Heinemeyer W., Wolf D. H. The proteasome/multicatalytic-multifunctional proteinase. In vivo function in the ubiquitin-dependent N-end rule pathway of protein degradation in eukaryotes. FEBS Lett. 1992;302:192–196. doi: 10.1016/0014-5793(92)80438-m. [DOI] [PubMed] [Google Scholar]
- Romisch K. Endoplasmic reticulum-associated degradation. Annu. Rev. Cell Dev. Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- Ron D., Hampton R. Y. Membrane biogenesis and the unfolded protein response. J. Cell Biol. 2004;167:23–25. doi: 10.1083/jcb.200408117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayeed A., Ng D. T. Search and destroy: ER quality control and ER-associated protein degradation. Crit. Rev. Biochem. Mol. Biol. 2005;40:75–91. doi: 10.1080/10409230590918685. [DOI] [PubMed] [Google Scholar]
- SGD. Saccharomyces Genome Database. 2008 doi: 10.1093/nar/26.1.73. http://www.yeastgenome.org/ [DOI] [PMC free article] [PubMed]
- Spear E. D., Ng D. T. Stress tolerance of misfolded carboxypeptidase Y requires maintenance of protein trafficking and degradative pathways. Mol. Biol. Cell. 2003;14:2756–2767. doi: 10.1091/mbc.E02-11-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear E. D., Ng D. T. Single, context-specific glycans can target misfolded glycoproteins for ER-associated degradation. J. Cell Biol. 2005;169:73–82. doi: 10.1083/jcb.200411136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C., Hitt R., Park S. H., Deak P. M., Kostova Z., Wolf D. H. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J. Biol. Chem. 2003;278:35903–35913. doi: 10.1074/jbc.M301080200. [DOI] [PubMed] [Google Scholar]
- Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Trotter E. W., Kao C. M., Berenfeld L., Botstein D., Petsko G. A., Gray J. V. Misfolded proteins are competent to mediate a subset of the responses to heat shock in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:44817–44825. doi: 10.1074/jbc.M204686200. [DOI] [PubMed] [Google Scholar]
- Vashist S., Ng D. T. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren C. D., Eckley D. M., Lee M. S., Hanna J. S., Hughes A., Peyser B., Jie C., Irizarry R., Spencer F. A. S-phase checkpoint genes safeguard high-fidelity sister chromatid cohesion. Mol. Biol. Cell. 2004;15:1724–1735. doi: 10.1091/mbc.E03-09-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegele H., Haslbeck M., Reinstein J., Buchner J. Sti1 is a novel activator of the Ssa proteins. J. Biol. Chem. 2003;278:25970–25976. doi: 10.1074/jbc.M301548200. [DOI] [PubMed] [Google Scholar]
- Wittke S., Dunnwald M., Albertsen M., Johnsson N. Recognition of a subset of signal sequences by Ssh1p, a Sec61p-related protein in the membrane of endoplasmic reticulum of yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2223–2232. doi: 10.1091/mbc.01-10-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl. Acad. Sci. USA. 2001;98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X., Chong E., Skach W. R. Evidence that endoplasmic reticulum (ER)-associated degradation of cystic fibrosis transmembrane conductance regulator is linked to retrograde translocation from the ER membrane. J. Biol. Chem. 1999;274:2616–2624. doi: 10.1074/jbc.274.5.2616. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T., Nair U., Yang Z., Klionsky D. J. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.