Abstract
The Saccharomyces cerevisiae chromosomal passenger proteins Ipl1 (Aurora B) and Sli15 (INCENP) are required for the tension checkpoint, but the role of the third passenger, Bir1, is controversial. We have isolated a temperature-sensitive mutant (bir1-107) in the essential C-terminal region of Bir1 known to be required for binding to Sli15. This allele reveals a checkpoint function for Bir1. The mutant displays a biorientation defect, a defective checkpoint response to lack of tension, and an inability to detach mutant kinetochores. Ipl1 localizes to aberrant foci when Bir1 localization is disrupted in the bir1-107 mutant. Thus, one checkpoint role of Bir1 is to properly localize Ipl1 and allow detachment of kinetochores. Quantitative analysis indicates that the chromosomal passengers colocalize with kinetochores in G1 but localize between kinetochores that are under tension. Bir1 localization to kinetochores is maintained in an mcd1-1 mutant in the absence of tension. Our results suggest that the establishment of tension removes Ipl1, Bir1, and Sli15, and their kinetochore detachment activity, from the vicinity of kinetochores and allows cells to proceed through the tension checkpoint.
INTRODUCTION
Chromosomal passenger proteins coordinate cell cycle events that are required for proper chromosome segregation (Ruchaud et al., 2007). In metazoans, four proteins form the core chromosomal passenger complex: Aurora B, INCENP, survivin, and borealin. One helix from each of the latter three passengers twines together to form a three-helix bundle, which then recruits Aurora B, explaining why the localization of each passenger depends on all the others (Jeyaprakash et al., 2007). The passengers have a dynamic localization throughout the cell cycle as they move from centromeres to the spindle at anaphase and concentrate at the spindle midzone before cytokinesis. The cell cycle-dependent changes in chromosomal passenger localization reflect their functions at the different stages of the cell cycle. These functions include activation of the spindle checkpoint in response to loss of tension and cytokinesis (Ruchaud et al., 2007).
Budding yeast have three chromosomal passengers, Ipl1 (Aurora B), Sli15 (INCENP), and Bir1 (survivin). The roles of Ipl1 and Sli15 have been well described. Early in mitosis, Ipl1 localizes to kinetochores and is required for the tension checkpoint (Biggins and Murray, 2001). Ipl1 phosphorylates targets at the kinetochore and is required to detach mono-oriented or improperly attached chromosomes that do not generate tension between sister chromatids (Biggins and Murray, 2001; Tanaka et al., 2002; Pinsky et al., 2003, 2006). At telophase, Ipl1 localizes to the spindle midzone and is necessary for timely spindle breakdown (Buvelot et al., 2003). Sli15 seems to be a partner of Ipl1 at these stages and has been shown to stimulate its kinase activity (Kim et al., 1999; Kang et al., 2001; Tanaka et al., 2002). Accordingly, Sli15 mutants have phenotypes that are very similar to the phenotypes of Ipl1 mutants (Kim et al., 1999).
Bir1 also plays a role in chromosome segregation (Li et al., 1998; Uren et al., 1999; Yoon and Carbon, 1999). Bir1 contains two baculovirus inhibitor of apoptosis (IAP) repeat (BIR) domains similar to the BIR domain found in survivin, and an additional 700 amino acids. It copurifies with Sli15 (Kim et al., 1999; Kang et al., 2001; Cheeseman et al., 2002), and all three chromosomal passengers colocalize throughout the cell cycle (Kang et al., 2001; Widlund et al., 2006).
Recently, Sandall and coworkers proposed that a complex of Bir1 and Sli15 senses tension and regulates Ipl1 detachment of improper kinetochore attachments (Sandall et al., 2006). Their evidence was multifold. First, in an in vitro assay, a complex of Bir1 and Sli15 mediates the attachment of centromeres to microtubules. The binding depends on Bir1, the microtubule-binding domain of Sli15, and the CBF3 complex bound to the centromere. Phosphorylation by Ipl1 inhibits binding. In vivo, they showed that the microtubule-binding domain of Sli15 is required for the tension checkpoint, but they did not test the role of Bir1 in the tension checkpoint. In a more recent study, Thomas and Kaplan (2007) constructed mutants of Bir1 that did not bind Sli15 or Ipl1 as tested by immunoprecipitation. Their mutants did not alter anaphase localization of Sli15 or Ipl1 in yeast cells and did not affect the tension checkpoint. Thus, the checkpoint role of Bir1 has not been defined in vivo.
Here, we characterize a temperature-sensitive mutant of Bir1 that reveals its checkpoint function and shows that Bir1 is required for proper localization of Ipl1. The three chromosomal passengers localize to kinetochores in G1 and to the spindle in anaphase as shown previously. Although previous reports disagree about the metaphase localization of Ipl1, we find all three chromosomal passengers exhibit metaphase localizations that are distinct from kinetochores and instead localize between kinetochores that are under tension. Kinetochore localization of Bir1 can be restored by the absence of tension in a cohesin mutant. Our results suggest a model in which tension displaces the chromosomal passengers from kinetochores, thus limiting the ability of Ipl1 to phosphorylate and detach kinetochores that are under tension.
MATERIALS AND METHODS
Media
YPD medium and SD medium were made as described previously (Sherman et al., 1986). SD complete is SD medium supplemented with 50 μg/ml adenine, 25 μg/ml uracil, 100 μg/ml tryptophan, and 0.1% casamino acids. SD-uracil low adenine is SD complete lacking uracil and containing 5 μg/ml adenine. NP-40 buffer was described previously (Hazbun et al., 2003).
Plasmids and Strains
The plasmids used in this study are listed in Table 1. The BIR1 open reading frame (ORF), including 560 base pairs 5′ and 548 base pairs 3′ of the ORF, was cloned into pRS316 to create plasmid pPW02. The plasmid used for the plasmid shuffle is plasmid pPW06, which contains ADE3 (from pLI831) and BIR1 in plasmid pTD29 (Geiser et al., 1993; Muller, 1996). The BIR1 plasmid shuffle strain PWY16-4D was made as described previously (Widlund et al., 2006).
Table 1.
Plasmids used in this study
| Plasmid | Relevant markers | Source or reference |
|---|---|---|
| pBS4 | CFP::hphMX3 | Yeast Resource Center |
| pBS7 | Venus::kanMX6 | Yeast Resource Center |
| pBS35 | mCherry::hphMX3 | Yeast Resource Center |
| pDH3 | CFP::kanMX6 | Yeast Resource Center |
| pFA6-GFP(S65T)-HIS3MX6 | GFP(S65T)::HIS3MX6 | Wach et al. (1997) |
| pPW02 | BIR1 in pRS316 | Widlund et al. (2006) |
| pPW06 | BIR1 ADE3 in 2 μm vector | Widlund et al. (2006) |
| pPW100 | bir1-107 in pRS306 | This study |
| pPW106 | bir1-110 in pRS306 | This study |
| pPW108 | bir1-111 in pRS306 | This study |
| pRS306 | URA3, f1 origin | Sikorski and Hieter (1989) |
| pRS316 | CEN6, ARSH4, URA3, f1 origin | Sikorski and Hieter (1989) |
The yeast strains used in this study are listed in Table 2. All strains were derived from W303. C-terminal cyan fluorescent protein (CFP), Venus, mCherry, and green fluorescent protein (GFP) fusions were created by amplifying the cyan fluorescent protein (CFP)-hphMX3, CFP-kanMX6, Venus-kanMX6, mCherry-hphMX3, or GFP-HIS3MX6 cassettes from plasmids pBS4, pDH3, pBS7, pBS35, or pFA6-GFP(S65T)::HIS3MX6, respectively (all gifts from Yeast Resource Center, University of Washington, Seattle, WA). Cassettes were integrated in frame at the 3′ end of the target ORF.
Table 2.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| BGY13-6D | MATa, ade3Δ NUF2-Venus::kanMX6 SPC97-CFP::hphMX3 DAM1-myc::kanMX6 cyh2r lys2Δ::HIS3 | Shimogawa et al. (2006) |
| CRY1 | MATa | Geiser et al. (1993) |
| CRY2 | MATα | Geiser et al. (1993) |
| PWY4-2B | MATa, ade3Δ NDC10-CFP::hphMX3 SLI15-Venus::kanMX6 | This study |
| PWY5-5A | MATa, ade3Δ BIR1-Venus::kanMX6 NDC10-CFP::kanMX6 | This study |
| PWY16-4D | MATa, ade3Δ bir1Δ::hphMX3 lys2Δ::HIS3 (with autonomous plasmid pPW06) | Widlund et al. (2006) |
| PWY167-13D | MATa, bir1-107 | This study |
| PWY170-11C | MATa, bir1-110 | This study |
| PWY171-13B | MATa, bir1-111 | This study |
| PWY177-15A | MATa, ade3Δ bir1-107 NUF2-Venus::kanMX6 SPC97-CFP::hphMX3 | This study |
| PWY200-17A | MATa, mcd1-1 PDS1-3xHA::URA3 his3-11::pCUP1-GFP12-lacI12::HIS3 trp1-1::URA3::lacO::TRP1 | Widlund et al. (2006) |
| PWY247-30C | MATa, his3-11::pCUP1-GFP12-lacI12::HIS3 SPC110-mCherry::hphMX3 Chr8::CEN-lacO::TRP1 | This study |
| PWY262-40D | MATa, bir1-107 his3-11::pCUP1-GFP12-lacI12::HIS3 SPC110-mCherry::hphMX3 Chr8::CEN-lacO::TRP1 | This study |
| PWY286-23A | MATa, his3-11::pCUP1-GFP12-lacI12::HIS3 ipl1-321 SPC110-mCherry::hphMX3 Chr8::CEN-lacO::TRP1 | This study |
| PWY303-10D | MATa, ade3Δ bir1-107 IPL1-Venus::kanMX6 NDC10-CFP::hphMX3 | This study |
| PWY303-11A | MATa, ade3Δ IPL1-Venus::kanMX6 NDC10-CFP::hphMX3 | This study |
| PWY304-5A | MATa, ade3Δ BIR1-Venus::kanMX6 ipl1-321 NDC10-CFP::hphMX3 | This study |
| PWY314-7C | MATa, bir1-107 mcd1-1 PDS1-3xHA::URA3 trp1-1::URA3::lacO::TRP1 | This study |
| PWY322-3A | MATa, ade3Δ bir1-107 NDC10-CFP::hphMX3 SLI15-Venus::kanMX6 | This study |
| PWY329-16D | MATa, ade3Δ bir1-107 MTW1-Venus::kanMX6 ndc80-1 SPC97-CFP::hphMX3 | This study |
| PWY329-19C | MATα, ade3Δ MTW1-Venus::kanMX6 ndc80-1 SPC97-CFP::hphMX3 | This study |
| PWY334-1B | MATα, IPL1-3xGFP::HIS3 SPC110-mCherry::hphMX3 | This study |
| PWY335-1B | MATα, NUF2-3xGFP::HIS3 SPC110-mCherry::hphMX3 | This study |
| PWY339-18A | MATα, bir1-107-Venus::kanMX6 lys2Δ::HIS3 | This study |
| PWY341-12D | MATa, BIR1-3xGFP::HIS3 SPC110-mCherry::hphMX3 | This study |
| PWY345 | MATa, ipl1-321 mcd1-1 PDS1-3xHA::URA3 | This study |
| PWY346-1A | MATa, ade3Δ NDC10-CFP::hphMX3 NUF2-Venus::kanMX6 | This study |
| PWY347-1A | MATa, ade3Δ BIR1-Venus::kanMX6 NDC10-CFP::kanMX6 mcd1-1 | This study |
| SFY233-2D | MATa, ipl1-321 | This study |
| TDY188-2A | MATa, ade3Δ BIR1-Venus::kanMX6 NDC10-CFP::kanMX6 pGAL1-HA3-SLI15::kanMX6 | This study |
| TDY189-12A | MATa, ade3Δ IPL1-Venus::kanMX6 NDC10-CFP::hphMX3 pGAL1-HA3-SLI15::kanMX6 | This study |
| W303 | ade2-1oc can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 |
All strains have the same markers as W303 except as shown.
Temperature-sensitive Mutants
Temperature-sensitive mutants of BIR1 were generated by using a plasmid shuffle strategy as described previously (Sundberg et al., 1996). Briefly, plasmid pPW02, containing full-length BIR1 was digested with SphI and SnaBI, which creates a gap in the 3′ end of the BIR1 ORF. The gapped plasmid and a polymerase chain reaction (PCR) mutagenized BIR1 3′ fragment were cotransformed into the plasmid shuffle strain for BIR1, PWY16-4D. Transformants were selected on SD-uracil low adenine plates for 4 d at 37°C. Solid red colonies were restreaked in duplicate on SD-uracil low adenine plates and incubated at 25 and 37°C. Plasmids were rescued from those isolates that sectored white at 25°C and remained red at 37°C. The isolated alleles were sequenced and subcloned into integrating vectors. Temperature-sensitive alleles were introduced as the sole copy at the BIR1 locus by gene replacement using plasmids pPW101, pPW106, and pPW108 as described previously (Widlund and Davis, 2005).
Western Blot Analysis
Protein samples from time-course experiments were prepared by trichloroacetic acid precipitation (Wright et al., 1989). Samples were run on 10% SDS polyacrylamide gels and transferred to polyvinylidene difluoride membranes. The anti-hemagglutinin (HA) antibody was mouse monoclonal 12CA5 (Roche Diagnostics, Indianapolis, IN). The rat monoclonal anti-tubulin antibody YOL1/34 (Abcam, Cambridge, MA) was used as a loading control. Antibodies were used at a 1:1000 dilution. The secondary antibodies were goat anti-mouse Alexa-fluor 680 (Invitrogen, Carlsbad, CA) and goat anti-rat IRDye 800 (Rockland Immunochemicals, Gilbertsville, PA), both used at a dilution of 1:8000. Membranes were scanned and quantified using the Odyssey system (Li-Cor Biosciences, Lincoln, NE).
Flow Cytometry
Cells were fixed with 2 volumes of 100% ethanol at 22°C overnight and stained with propidium iodide. Fluorescence was analyzed using an FACScan flow cytometer (BD Biosciences, San Jose, CA), and data were collected with CellQuest software (BD Biosciences). The fraction of cells with 2N DNA content was determined using FlowJo software (BD Biosciences). Briefly, the total number of cells is equal to the number of cells from the left edge of the 1N DNA peak to the right edge of the 2N DNA peak. The fluorescence value midway between the centers of the 1N and 2N peaks was found, and the fraction of total cells with DNA contents greater than this value were considered to have a 2N content of DNA.
Fluorescence Microscopy
Live cell imaging was performed with a DeltaVision microscopy system from Applied Precision (Issaquah, WA). The system incorporates an Olympus IL-70 microscope, a u-plan-apo 100× oil objective (1.35 numerical aperture), a CoolSNAP HQ digital camera from Roper Scientific (Trenton, NJ), and optical filter sets from Omega Optical (Battleboro, VT), Semrock (Rochester, NY), and Chroma Technology (Rockingham, VT). Cells were mounted on a 1% agarose pad containing SD-complete medium (Muller et al., 2005). Single sections were taken with 2 × 2 binning unless otherwise noted. Temperature-sensitive strains were incubated at the nonpermissive temperature for 30 min before imaging, unless otherwise noted.
Conversion of SoftWoRx Image Data
A software tool (R3DConverter) was developed using the Java programming language to convert 12-bit grayscale SoftWoRx data to either 8- or 16-bit TIFF images, for use in our suite of image analysis and quantification tools. R3DConverter allows for the conversion of large numbers of files at a time, while applying universal scaling parameters to the entire set of images. To facilitate visual screening, the original 12-bit image data were converted to an 8-bit TIFF. For quantification, the original 12-bit image data were converted to a 16-bit grayscale TIFF, and the original 12-bit pixel values were retained.
Semiautomated Image Analysis and Quantification of Fluorescence
Metaphase Distribution of Chromosomal Passengers.
A MatLab program, cutup.m, was developed to select cells with mitotic spindles from images containing cells at many different stages of the cell cycle. Cutup identifies pairs of peaks likely to be mitotic spindles in images of cells labeled with the kinetochore marker Ndc10-CFP or the spindle pole body marker Spc110-mCherry. Cutup then “cuts up” the starting 8-bit and 16-bit TIFF images (512 × 512) and produces smaller TIFF images (∼50 × 50), each containing one pair of peaks. The smaller 8-bit files were visually screened, yielding a collection of small image files in TIFF format, each containing an image of a yeast cell with a spindle in the mother cell.
The fluorescence distributions in the corresponding small 16-bit TIFF images were analyzed with another MatLab program, calc.m, based on the method used by Sprague et al. (2003). Briefly, the two brightest 3 × 3 peaks of CFP or mCherry fluorescence were automatically located to define the spindle axis, and the fluorescence at each pixel along the spindle axis was measured. Because the spindle is in a random orientation in each image, the MatLab function “improfile” was used to interpolate the values along the spindle axis by using bilinear interpolation. For the results presented here, the calc routine was adapted to the analysis of 2 × 2 binned images, with fluorescence signal measured five pixels above and below the spindle axis for each spindle position. Background was calculated at each spindle position by averaging values 1–2 pixels above and below the region of signal about the spindle axis. The pushout factor multiplied by the peak separation determined how far outside the peaks data were collected. For example, for two peaks separated by 6 pixels, a pushout factor of 0.5 would result in the measurement of fluorescence for 12 pixels along the spindle axis, starting 3 pixels outside of one peak and extending 3 pixels beyond the other peak. Peak separations of 6–9 pixels (0.76–1.15 μm) between Ndc10 peaks and 11–12 pixels (1.40–1.53 μm) between Spc110 peaks were considered metaphase. A pushout factor of 0.75 was used for Ndc10-labeled spindles, and a pushout factor of 0.3 was used for Spc110-labeled spindles. To compare spindles of variable lengths, data were normalized to a spindle length of 24 pixels. Raw data were normalized by photosensor values to correct for differences in the intensity of the mercury lamp on different days.
Quantification of Total Fluorescence across Metaphase Spindles.
The raw fluorescence data were normalized by photosensor values as described above but not normalized to a constant length. The total fluorescence across each spindle was calculated using a perl script that summed the area under each fluorescence distribution and above a baseline drawn between the first and last data points.
G1 Quantification of Chromosomal Passengers.
The cutup routine was modified to cut up large tiffs into small tiffs each containing a single CFP peak, and unbudded cells were selected. A MatLab program, intens.m, was developed to measure the intensity of each peak of fluorescence in a TIFF image (whether or not it is part of a spindle). Then, 3 × 3 peaks of fluorescence were located in the CFP channel, and fluorescence intensity was measured in a 7 × 7 box about the center of the peak. The fluorescence intensity in the Venus channel was measured in a 7 × 7 box around the brightest 3 × 3 peak within 3 pixels of the center of each CFP peak. Background was calculated from a concentric ring six to seven pixels outside of each 7 × 7 signal box and subtracted. The data were normalized by photosensor values as described above.
Quantification of Bir1 Fluorescence in mcd1-1 Cells.
Asynchronous cultures of PWY5–5A (MCD1) and PWY347-1A (mcd1-1) were grown to early log phase in YPD and shifted to the nonpermissive temperature of 37°C. Live cells were mounted on slides and imaged at 0 (22°C), and 60–90 min after the shift. The images were taken with 1 s (Venus) and 0.4 s (CFP) exposures for the samples at 22°C and 0.4 s (Venus and CFP) exposures for the samples at 37°C. All images taken at 22°C and images of the wild-type strain (MCD1) at 37°C were processed with a pushout factor of 0.75 and Ndc10 peak separations of six to nine pixels (0.76–1.15 μm). mcd1-1 spindles at 37°C with longer Ndc10 peak separations of 10–15 pixels (1.28–1.91 μm) were considered to have inactivated cohesin and were processed with a pushout factor of 0.25. These pushout factors were chosen so that an equal range of distances (16–23 pixels) was measured along the spindle axis for both strains and temperatures. For the calculation of total Bir1 fluorescence, the pushout factor was increased to 0.5 for mcd1-1 spindles at 37°C to ensure inclusion of all the fluorescence. Total fluorescence was calculated as described above for the quantification of metaphase spindle fluorescence.
RESULTS
The C Terminus of Bir1 Is Required for Proper Localization
Temperature-sensitive mutants of BIR1 were generated to characterize its essential function and for comparison with the phenotypes described for ipl1 and sli15 mutants. Mutagenesis was targeted to the C-terminal region shown previously to be necessary and sufficient for cell survival, proper localization, and interaction with Sli15 (Widlund et al., 2006). Three mutants that could function as a single copy at the permissive temperature were isolated, bir1-107, bir1-110, and bir1-111 (Figure 1). In several different tests, the bir1-107 allele conferred the most severe growth defect at the restrictive temperature and was therefore chosen for further characterization.
Figure 1.
Characterization of a temperature-sensitive allele of BIR1. (A) Growth phenotype of three bir1 mutants at increasing temperature (PWY167-13D, PWY170-11C, PWY171-13B) compared with a BIR1 strain (CRY1). Cultures were grown in YPD at 22°C and 10-fold serial dilutions were plated on YPD and incubated at the indicated temperatures. (B) The bir1 temperature-sensitive mutants all contain point mutations in the C-terminal Sli15 binding region. The mutations in bir1-107 are marked. The point mutations are as follows: bir1-107: M740K, F760S, A823T, K910E, E936D, M951K; bir1-110: S908P L924S; bir1-111: N876I, E892D, L924R, E939D, M951L. (C) Localization of Bir1-107. Bir1-107-Venus (PWY339-18A) localizes normally (kinetochore and the anaphase spindle) at the permissive temperature of 22°C and mislocalizes throughout the nucleus after 30 min at the restrictive temperature of 37°C. Intensities of the images are scaled equally. Bar, 1 μm.
Bir1-107 was tagged with the yellow fluorescent protein (YFP) variant Venus to determine whether its loss of function was due to a change in localization. At the permissive temperature, Bir1-107-Venus localized like wild-type Bir1-Venus to kinetochores and the anaphase spindle. After shifting to the restrictive temperature for 30 min, Bir1-107-Venus localized diffusely in the nucleus (Figure 1C), similarly to an inactive C-terminal truncation, Bir1-876stop (Widlund et al., 2006).
bir1-107 Cells Display a Biorientation Defect
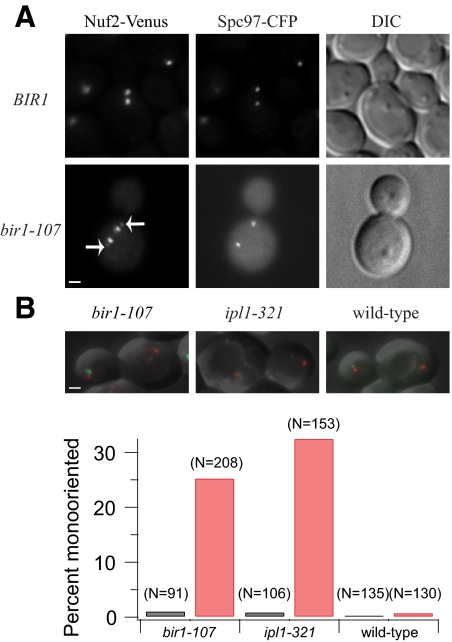
We examined the consequence of inactivating the C-terminus of Bir1 at metaphase. Wild-type kinetochores form bipolar attachments at metaphase, which can be visualized by tagging a kinetochore component, such as Nuf2, with a fluorescent marker. Wild-type cells expressing Nuf2-Venus show two strong peaks at the quarter-spindle, with each peak representing 16 kinetochores under tension (Figure 2A). However, in a bir1-107 mutant at the nonpermissive temperature, a fraction of the Nuf2-Venus signal was seen near or at the spindle pole body in >50% of cells (Figure 2A). The pole localization of these kinetochores is suggestive of monopolar attachments, which are not under tension. Flow cytometry of bir1-107 mutant cells revealed a defect in chromosome segregation, similar to but not as severe as that conferred by an ipl1-321 mutant (Supplemental Figure S1).
Figure 2.
bir1-107 displays a biorientation defect at the restrictive temperature. (A) Wild-type kinetochores display a bilobed distribution of kinetochore fluorescence (Nuf2-Venus) between spindle pole bodies (Spc97-CFP). Asynchronous wild-type cells (BGY13-6D) were imaged live with 1 × 1 binning. PWY177-15A carrying bir1-107, Nuf2-Venus and Spc97-CFP was synchronized with α-factor and released at the restrictive temperature of 37°C. Cells were fixed with 3.7% formaldehyde and imaged with 1 × 1 binning. Arrows mark the Nuf2-Venus signal at the spindle poles corresponding to mono-oriented kinetochores. Bar, 1 μm. (B) bir1-107 (PWY262-40D), ipl1-321 (PWY286-23A) and wild-type (PWY247-30C) cells carrying a LacO array integrated on chromosome 8 and LacI-GFP were shifted to 37°C for 60 min and imaged in 10 Z-sections, spaced 0.4 μm apart, with 1 × 1 binning. Anaphase cells with one spindle pole body (marked with Spc110-mCherry) in the mother and one in the daughter were scored as bioriented if a GFP dot was visible in both mother and daughter and mono-oriented if a GFP dot was visible in only mother or daughter. Black, 22°C; Red, 37°C.
To look more carefully at biorientation in bir1-107, individual chromosomes were labeled using a LacO array in cells expressing LacI-GFP. The phenotype of bir1-107 was compared with an ipl1-321 mutant, which is defective in biorientation, after 60 min at the restrictive temperature of 37°C. Anaphase cells were scored for proper segregation of the two sister GFP-labeled chromosomes to opposite cells. In both mutants at the permissive temperature, and in wild-type cells, <1% of cells exhibited a biorientation defect. In contrast, 25% of bir1-107 cells and 32% of ipl1-321 cells missegregated both sisters to the same cell at the restrictive temperature (Figure 2B). Like ipl1-321, bir1-107 confers defects in biorientation.
Bir1 Is Required for the Tension Checkpoint
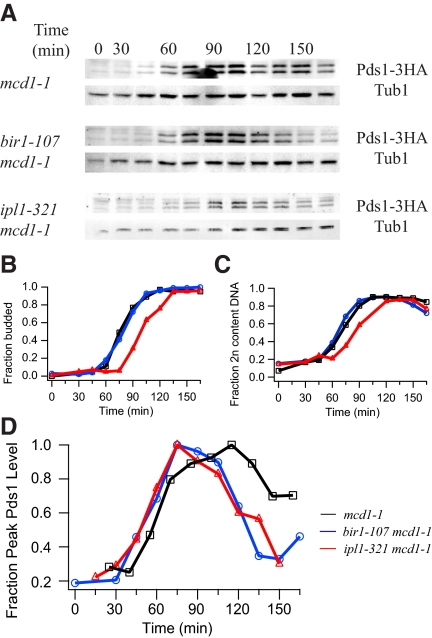
The presence of a biorientation defect in the bir1-107 mutant prompted us to test whether Bir1 is required for the tension checkpoint. The cohesin mutant mcd1-1 lacks sister chromatid cohesion at the restrictive temperature and activates the tension checkpoint. Cells delay at metaphase with high securin (Pds1) levels (Figure 3, A and D). This checkpoint response depends on Ipl1, as an mcd1-1 ipl1-321 double mutant degrades Pds1 despite the lack of tension (Figure 3, A and D; Biggins and Murray, 2001; Stern and Murray, 2001; Pinsky et al., 2003). A bir1-107 mcd1-1 double mutant also degraded Pds1 despite loss of cohesion at the restrictive temperature (Figure 3, A and D). We therefore conclude Bir1 is required for the tension checkpoint.
Figure 3.
Bir1 is required for the tension checkpoint. (A) Cells carrying the temperature-sensitive cohesin allele mcd1-1 were arrested with α-factor and released to the restrictive temperature of 37°C. HA-tagged securin (Pds1) levels were monitored by immunoblot along with tubulin loading controls at each time point. (B) Budding index indicating the fraction of budded cells in mcd1-1 (black squares; PWY200-17A), bir1-107 (blue circles; PWY314-7C) and ipl1-321 (red triangles; PWY345). (C) Fraction of cells with 2N DNA content determined by flow cytometry. (D) For comparison between Western blots, Pds1 levels were normalized to tubulin at each time point and the average Pds1 profiles are plotted as a fraction of peak Pds1 for each strain. For this graph, time courses were aligned based on the flow cytometry profiles. In an mcd1-1 control strain, Pds1 levels remain high (black squares). In contrast, an mcd1-1 strain carrying either the bir1-107 (blue circles) or ipl1-321 allele (red triangles) degrades Pds1.
Bir1 Is Required to Detach Kinetochores
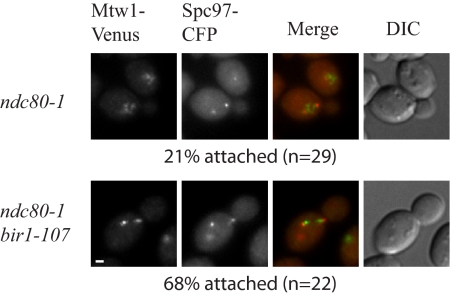
Our results indicate that Bir1, like Ipl1, is required for the tension checkpoint. Because the role of Ipl1 is to detach improperly attached kinetochores, we examined whether Bir1 is required for kinetochore detachment in a mutant with a defective kinetochore. In an ndc80-1 mutant at the restrictive temperature, weak kinetochore microtubule attachments are present unless Ipl1 is active to detach them (Pinsky et al., 2006). Similarly, in the presence of the ndc80-1 mutation, kinetochores were attached to the spindle when Bir1 was inactivated and detached from the spindle when Bir1 was active, suggesting Bir1 plays a similar role to Ipl1 in detaching improper attachments (Figure 4).
Figure 4.
Inactivation of Bir1 stabilizes kinetochore attachments. Kinetochores (Mtw1-Venus, green) are detached from the spindle axis (spindle pole bodies marked by Spc97-CFP, red) in the ndc80-1 mutant at the nonpermissive temperature (PWY329-19C). In a bir1-107, ndc80-1 double mutant, bipolar attachment is restored (PWY329-16D). Cells were shifted asynchronously to 37°C for 60 min. Localization of kinetochores was quantified when both spindle pole bodies were in focus and both spindle pole bodies were still in the mother cell. Spindles were scored as attached if no Mtw1-Venus signal was seen outside of the spindle axis. Images are scaled equally. Bar, 1 μm.
A Checkpoint Role of Bir1 Is to Localize Ipl1
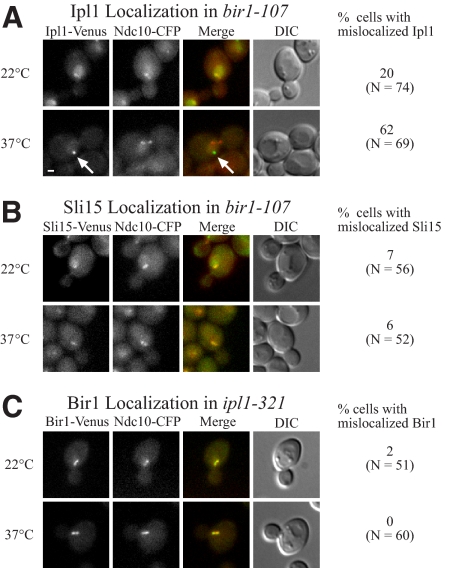
Because Bir1 and Ipl1 are both required for the detachment of kinetochores, we tested whether Ipl1 localization is dependent on functional Bir1. In a bir1-107 mutant shifted to the nonpermissive temperature for 30 min, Ipl1-Venus mislocalized to distinct foci near the edge of the nucleus, away from the spindle and kinetochores (Figure 5A). These Ipl1 foci occurred in 62% of preanaphase cells at the restrictive temperature compared with 20% of cells at the permissive temperature. Under the same conditions, Sli15 did not form aberrant foci in preanaphase cells (Figure 5B). Therefore, Bir1 is likely required for the tension checkpoint because it is required to properly localize Ipl1.
Figure 5.
A bir1-107 mutant mislocalizes Ipl1 but not Sli15, whereas Bir1 localizes properly in ipl1-321. (A) Ipl1-Venus localization in a bir1-107 mutant (PWY303-10D) at the permissive temperature (22°C) and after 30 min at the restrictive temperature (37°C). Ndc10-CFP serves as a marker for kinetochores. The merged images show Ipl1 in green and Ndc10 in red. Note the aberrant localization of Ipl1 indicated by the white arrow at the restrictive temperature. The table on the right shows quantification of Ipl1 localization in preanaphase cells. Bar, 1 μm. (B) Sli15-Venus localizes properly in bir1-107 at both the permissive (22°C) and restrictive (37°C) temperatures (PWY322-3A). The merged images show Sli15 in green and Ndc10 in red. The table on the right shows quantification of Sli15 localization in preanaphase cells. Bar, 1 μm. (C) Bir1-Venus localization in an ipl1-321 mutant (PWY304-5A) at the permissive temperature (22°C) and restrictive temperature (37°C). The merged image shows Bir1 in green and Ndc10 in red. Bir1 shows normal localization at both temperatures. The table on the right shows quantification of Bir1 localization in preanaphase cells. Bar, 1 μm.
Ipl1 Is Not Required for Localization of Bir1
Although Bir1 is required for proper Ipl1 localization, Ipl1 is not required to maintain localization of Bir1. In the presence of the ipl1-321 allele, Bir1 localized similarly at the permissive and restrictive temperatures and was never seen to form aberrant foci at the restrictive temperature (Figure 5C). Localization of Sli15 was likewise unaffected by the ipl1-321 allele (unpublished data) in agreement with previous results with ipl1-2 (Kang et al., 2001). In contrast, pGAL-SLI15 cells grown on glucose to shut off Sli15 expression showed very little Bir1 or Ipl1 localization (Supplemental Figure S2). Our results suggest a model in which Sli15 is required to localize both Ipl1 and Bir1, Bir1 is required to localize Ipl1, and Ipl1 is not required to localize either Bir1 or Sli15.
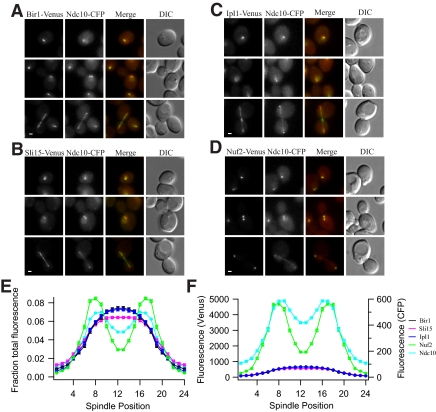
The Chromosomal Passengers Do Not Colocalize with Kinetochores at Metaphase
The localization of Ipl1 in metaphase yeast cells is controversial. It has been reported to localize punctately to kinetochores, which form a bilobed distribution in metaphase, or described as diffuse and distinct from kinetochores (Kim et al., 1999; Kang et al., 2001; Tanaka et al., 2002; Buvelot et al., 2003). We performed the first quantitative analysis of chromosomal passenger localization at different stages of the cell cycle by comparing the localization of the passengers and a known outer kinetochore protein (Nuf2) to the localization of the inner kinetochore protein Ndc10. In unbudded cells, which are in G1, the chromosomal passengers and Nuf2 colocalized with Ndc10 (Figure 6, A–D). However, the total fluorescence of each chromosomal passenger was almost fourfold less than Nuf2 (Table 3). At metaphase, there are eight Nuf2 molecules per kinetochore (Joglekar et al., 2006). From this number and our measurements of Nuf2 fluorescence at metaphase, we calculate that only one to two molecules of each chromosomal passenger are present at each kinetochore before mitosis.
Figure 6.
Chromosomal passengers are not localized to kinetochores under tension. (A) Bir1-Venus (PWY5-5A), (B) Sli15-Venus (PWY4-2B), (C) Ipl1-Venus (PWY303-11A), and (D) Nuf2-Venus (PWY346-1A) localization at metaphase compared with Ndc10-CFP (see Supplemental Figure S4). Cells were grown at 30°C. In the merged images, Venus is shown in green and CFP is shown in red. Images of the three chromosomal passengers are scaled equally, but they are scaled differently than images of Nuf2, which are much brighter. Bar, 1 μm. (E and F) Quantification of the distribution of chromosomal passengers relative to Ndc10 and Nuf2 kinetochores. Fluorescence intensity was measured across ≥70 half-spindles for each strain. Background was subtracted, and the average half-spindle distributions were reflected about the spindle midpoint to produce the graphs. Fluorescence distribution across the spindle is shown as fraction of total fluorescence (E) and as raw fluorescence intensity across the spindle (F). The SE of the mean is approximately the same size as the symbols.
Table 3.
Quantification of chromosomal passengers
| Protein | G1 |
Metaphase |
||
|---|---|---|---|---|
| Total fluorescence (a.u.) | Molecules/kinetochorea | Total fluorescence (a.u.) | Molecules/kinetochoreb | |
| Nuf2 | 21,100 ± 400 | 5 | 34,200 ± 900 | 8c |
| Bir1 | 5600 ± 100 | 1-2 | 4800 ± 300 | 1 |
| Ipl1 | 5200 ± 200 | 1-2 | 4800 ± 300 | 1 |
| Sli15 | 5700 ± 200 | 1-2 | 4200 ± 300 | 1 |
a Calculated relative to metaphase Nuf2 fluorescence.
b Calculated relative to metaphase Nuf2 fluorescence.
c Joglekar, A. P., Bouck, D. C., Molk, J. N., Bloom, K. S., and Salmon, E. D. (2006). Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 8, 581–585.
In metaphase, when the Ndc10 distribution was bilobed, Nuf2 fluorescence was also bilobed (Figure 6; Supplemental Figures S3 and S4). The bilobed appearance of Ndc10 and Nuf2 indicates that the kinetochores are bioriented and under tension. However, Bir1, Sli15, and Ipl1 did not show bilobed distributions at this stage of the cell cycle, and they were instead localized between kinetochores, often similar to the clouds of Ip1l observed by Tanaka et al. (2002) (Figure 6 and Supplemental Figure S4), with only slight concentration on the mitotic spindle. Thus, the localizations of the chromosomal passengers are distinct from kinetochores at metaphase (Figure 6E). The total fluorescence across metaphase spindles (most of which is not associated with kinetochores) was similar for Bir1, Ipl1, and Sli15 and was only 15% of the amount of Nuf2 (Figure 6F and Table 3). This suggests that the passengers are displaced from kinetochores under tension, as proposed by Tanaka et al. (2002). Because Ipl1 and Bir1 are required for kinetochore detachment, this displacement may be a mechanism by which bioriented kinetochores are stabilized.
Bir1 Colocalizes with Kinetochores That Are Not under Tension
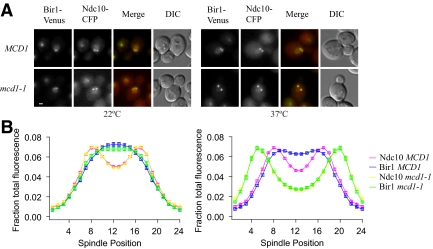
Our metaphase analyses suggested that the chromosomal passengers are physically displaced from kinetochores that are under tension. We tested whether the absence of tension would restore localization to kinetochores. mcd1-1 cells were shifted asynchronously to the restrictive temperature for 60–90 min to inactivate cohesin. Because cohesion is necessary to oppose the force of interpolar microtubules pulling outward, long spindles were considered to contain kinetochores that were not under tension. In the absence of tension, Bir1 was found to colocalize with the kinetochore marker Ndc10 (Figure 7, A and B, and Table 4), suggesting that tension between kinetochores regulates the localization of Bir1 to kinetochores.
Figure 7.
Bir1 localizes to kinetochores that are not under tension. A temperature-sensitive cohesin mutant (mcd1-1) carrying Bir1-Venus and Ndc10-CFP (PWY347-1A) was shifted to the restrictive temperature of 37°C as described in Materials and Methods, along with a wild-type control (MCD1; PWY5-5A). (A) At the permissive temperature (22°C), Bir1 localizes between Ndc10 kinetochores in both MCD1 (Bir1, blue and Ndc10, pink) and mcd1-1 (Bir1, green and Ndc10, yellow). Tension between sister kinetochores is lost in mcd1-1 cells at the restrictive temperature (37°C), as indicated by the increase in spindle length. Average Ndc10 separations were as follows (mean ± SD): MCD1, 22°C (0.89 μm ± 0.13 μm); mcd1-1, 22°C (0.91 ± 0.14 μm); MCD1, 37°C (0.94 ± 0.13 μm); and mcd1-1, 37°C (1.56 ± 0.19 μm). Bir1 localizes between Ndc10 kinetochores in MCD1 (Bir1, blue and Ndc10, pink), but it colocalizes with kinetochores that are not under tension in mcd1-1 (Bir1, green and Ndc10, yellow). Images are scaled equally at each temperature. Bar, 1 μm. (B) Quantification of the distribution of Bir1 and Ndc10. Fluorescence was measured across ≥70 half-spindles for each strain shown. Background was subtracted, and the average half-spindle distributions were reflected about the spindle midpoint. The SE of the mean is approximately the same size as the symbols.
Table 4.
Quantification of Bir1 in the absence of tension
| Strain | Total Bir1 fluorescence (a.u.)a at 22°C | Total Bir1 fluorescence (a.u.)b at 37°C |
|---|---|---|
| MCD1 | 7000 ± 400 | 4000 ± 100 |
| mcd1-1 | 6600 ± 200 | 6900 ± 300 |
a Images taken with 1-s exposures.
b Images taken with 0.4-s exposures.
DISCUSSION
The Ipl1/Aurora B kinase is required for the tension checkpoint in yeast and other eukaryotic cells. Yeast Sli15 and its vertebrate homologue INCENP seem to be required for targeting and activating the kinase (Kim et al., 1999; Kang et al., 2001). Survivin is a third chromosomal passenger that possesses both antiapoptotic and mitotic functions, which include roles in the spindle checkpoint and cytokinesis (Altieri, 2006; Lens et al., 2006). The yeast homologue Bir1 shares the IAP repeats, but it also contains a long C-terminal region not found in survivin. This region has been proposed to be the yeast counterpart to the fourth metazoan chromosomal passenger borealin (Jeyaprakash et al., 2007).
Recent work using a combination of temperature-sensitive alleles and C-terminal point mutants of Bir1 suggests Bir1 is not required for checkpoint function, but it is instead required for proper septin ring formation and cytokinesis (Thomas and Kaplan, 2007). Here, we isolated a temperature-sensitive allele of BIR1, which reveals the checkpoint function of Bir1. Our results demonstrate that alleles with different phenotypes can be isolated, highlighting the multiple functions for Bir1. Using our allele, we show that Bir1 is required for the tension checkpoint and for the proper localization of Ipl1. In contrast, we show that inactivation of Ipl1 has no effect on the localization of Bir1.
How is Ipl1 detachment activity regulated by tension? A Bir1-Sli15 kinetochore complex has been proposed to undergo either tension-dependent conformational changes or detachment, allowing it to regulate Ipl1 kinase activity in response to tension (Sandall et al., 2006). However, previous studies present conflicting data regarding the localization of Ipl1 at metaphase kinetochores (Tanaka et al., 2002; Buvelot et al., 2003). Consistent with the model proposed by Sandall et al. (2006), we do see a tension checkpoint deficiency in our temperature-sensitive mutant of Bir1. However, their model stopped short of determining whether tension regulates the activity or localization of the Bir1-Sli15 complex. Although we see the chromosomal passengers on kinetochores during G1, we find the chromosomal passengers are not at kinetochores once tension is established. Instead, the chromosomal passengers localize between the kinetochores. Although we cannot exclude the possibility of tension-dependent changes in the activity of the complex, our results favor a model in which tension regulates Ipl1 activity by affecting the proximity of the passenger complex to targets at the kinetochore. It is unclear whether the observed metaphase localization reflects binding of the chromosomal passengers to interpolar microtubules or to the extended centromeres formed by intramolecular looping of pericentromeric DNA (Yeh et al., 2008). The latter is appealing because it suggests displacement of Ipl1 from kinetochores is directly coupled to chromatin stretching, and detachment of individual kinetochores is regulated by tension in the adjacent chromatin.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sue Biggins for helpful advice and strains. This work was funded by National Institutes of Health grant R01 GM-40506 and P41RR011823. P.O.W. and M.M.S. were supported by National Institute of General Medical Sciences grant T32GM07270.
Abbreviations used:
- a.u.
arbitrary units
- BIR
baculovirus inhibitor of apoptosis repeat
- IAP
inhibitor of apoptosis.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-07-0723) on December 3, 2008.
REFERENCES
- Altieri D. C. The case for survivin as a regulator of microtubule dynamics and cell-death decisions. Curr. Opin. Cell Biol. 2006;18:609–615. doi: 10.1016/j.ceb.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Biggins S., Murray A. W. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot S., Tatsutani S. Y., Vermaak D., Biggins S. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 2003;160:329–339. doi: 10.1083/jcb.200209018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Anderson S., Jwa M., Green E. M., Kang J., Yates J. R., 3rd, Chan C. S., Drubin D. G., Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Geiser J. R., Sundberg H. A., Chang B. H., Muller E. G., Davis T. N. The essential mitotic target of calmodulin is the 110-kilodalton component of the spindle pole body in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:7913–7924. doi: 10.1128/mcb.13.12.7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazbun T. R., et al. Assigning function to yeast proteins by integration of technologies. Mol. Cell. 2003;12:1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A. A., Klein U. R., Lindner D., Ebert J., Nigg E. A., Conti E. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell. 2007;131:271–285. doi: 10.1016/j.cell.2007.07.045. [DOI] [PubMed] [Google Scholar]
- Joglekar A. P., Bouck D. C., Molk J. N., Bloom K. S., Salmon E. D. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Cheeseman I. M., Kallstrom G., Velmurugan S., Barnes G., Chan C. S. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 2001;155:763–774. doi: 10.1083/jcb.200105029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Kang J. S., Chan C. S. Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:1381–1394. doi: 10.1083/jcb.145.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lens S. M., Vader G., Medema R. H. The case for Survivin as mitotic regulator. Curr. Opin. Cell Biol. 2006;18:616–622. doi: 10.1016/j.ceb.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Li F., Ambrosini G., Chu E. Y., Plescia J., Tognin S., Marchisio P. C., Altieri D. C. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- Muller E. G. A glutathione reductase mutant of yeast accumulates high levels of oxidized glutathione and requires thioredoxin for growth. Mol. Biol. Cell. 1996;7:1805–1813. doi: 10.1091/mbc.7.11.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller E. G., Snydsman B. E., Novik I., Hailey D. W., Gestaut D. R., Niemann C. A., O'Toole E. T., Giddings T. H., Jr., Sundin B. A., Davis T. N. The organization of the core proteins of the yeast spindle pole body. Mol. Biol. Cell. 2005 doi: 10.1091/mbc.E05-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky B. A., Kung C., Shokat K. M., Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- Pinsky B. A., Tatsutani S. Y., Collins K. A., Biggins S. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev. Cell. 2003;5:735–745. doi: 10.1016/s1534-5807(03)00322-8. [DOI] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W. C. Chromosomal passengers: conducting cell division. Nat. Rev. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Sandall S., Severin F., McLeod I. X., Yates J. R., 3rd, Oegema K., Hyman A., Desai A. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell. 2006;127:1179–1191. doi: 10.1016/j.cell.2006.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Hicks J. B. Cold Spring Harbor Laboratory. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Shimogawa M. M., et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr. Biol. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague B. L., Pearson C. G., Maddox P. S., Bloom K. S., Salmon E. D., Odde D. J. Mechanisms of microtubule-based kinetochore positioning in the yeast metaphase spindle. Biophys. J. 2003;84:3529–3546. doi: 10.1016/S0006-3495(03)75087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern B. M., Murray A. W. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr. Biol. 2001;11:1462–1467. doi: 10.1016/s0960-9822(01)00451-1. [DOI] [PubMed] [Google Scholar]
- Sundberg H. A., Goetsch L., Byers B., Davis T. N. Role of calmodulin and Spc110p interaction in the proper assembly of spindle pole body components. J. Cell Biol. 1996;133:111–124. doi: 10.1083/jcb.133.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M. J., Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Thomas S., Kaplan K. B. A Bir1p Sli15p kinetochore passenger complex regulates septin organization during anaphase. Mol. Biol. Cell. 2007;18:3820–3834. doi: 10.1091/mbc.E07-03-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren A. G., Beilharz T., O'Connell M. J., Bugg S. J., van Driel R., Vaux D. L., Lithgow T. Role for yeast inhibitor of apoptosis (IAP)-like proteins in cell division. Proc Natl. Acad. Sci. USA. 1999;96:10170–10175. doi: 10.1073/pnas.96.18.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat A., Alberti-Segui C., Rebischung C., Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Widlund P. O., Davis T. N. A high-efficiency method to replace essential genes with mutant alleles in yeast. Yeast. 2005;22:769–774. doi: 10.1002/yea.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund P. O., Lyssand J. S., Anderson S., Niessen S., Yates J. R., 3rd, Davis T. N. Phosphorylation of the chromosomal passenger protein Bir1 is required for localization of Ndc10 to the spindle during anaphase and full spindle elongation. Mol. Biol. Cell. 2006;17:1065–1074. doi: 10.1091/mbc.E05-07-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A. P., Bruns M., Hartley B. S. Extraction and rapid inactivation of proteins from Saccharomyces cerevisiae by trichloroacetic acid precipitation. Yeast. 1989;5:51–53. doi: 10.1002/yea.320050107. [DOI] [PubMed] [Google Scholar]
- Yeh E., Haase J., Paliulis L. V., Joglekar A., Bond L., Bouck D., Salmon E. D., Bloom K. S. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H. J., Carbon J. Participation of Bir1p, a member of the inhibitor of apoptosis family, in yeast chromosome segregation events. Proc. Natl. Acad. Sci. USA. 1999;96:13208–13213. doi: 10.1073/pnas.96.23.13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.