Abstract
Hydroxyurea, a well-known DNA replication inhibitor, induces cell cycle arrest and intact checkpoint functions are required to survive DNA replication stress induced by this genotoxic agent. Perturbed DNA synthesis also results in elevated levels of DNA damage. It is unclear how organisms prevent accumulation of this type of DNA damage that coincides with hampered DNA synthesis. Here, we report the identification of stonewall (stwl) as a novel hydroxyurea-hypersensitive mutant. We demonstrate that Stwl is required to prevent accumulation of DNA damage induced by hydroxyurea; yet, Stwl is not involved in S/M checkpoint regulation. We show that Stwl is a heterochromatin-associated protein with transcription-repressing capacities. In stwl mutants, levels of trimethylated H3K27 and H3K9 (two hallmarks of silent chromatin) are decreased. Our data provide evidence for a Stwl-dependent epigenetic mechanism that is involved in the maintenance of the normal balance between euchromatin and heterochromatin and that is required to prevent accumulation of DNA damage in the presence of DNA replication stress.
INTRODUCTION
Cell cycle checkpoint pathways and DNA damage response pathways are essential mechanisms that control the order and timing of all cell cycle transitions and that ensure that critical events such as DNA replication and chromosome segregation are performed with high fidelity (Hartwell and Weinert, 1989;Elledge, 1996;Hurley and Bunz, 2007). In case these mechanisms fail, genetic abnormalities could be passed on to the following generations of cells, and this could lead to genomic instability and diseases such as cancer (Dasika et al., 1999; Houtgraaf et al., 2006). Cell cycle checkpoint and DNA repair genes have initially been identified using forward genetic screens in budding yeast (Saccharomyces cerevisiae) and fission yeast (Schizosaccharomyces pombe) (reviewed in O'Connell et al., 2000; Carr, 2002). Subsequently, forward genetic screens were also performed in Drosophila melanogaster and resulted in the identification of >30 mutagen-hypersensitive (mus) genes (Boyd et al., 1976, 1981; Henderson et al., 1987). Several of these mus genes have been cloned, and their function has been assigned to checkpoint regulation or DNA damage repair (Harris et al., 1996; Oshige et al., 1999; Brodsky et al., 2000; Yamamoto et al., 2000). More recently, using additional screens or by comparing Drosophila homologues with other species (Sibon et al., 1997, 1999; Price et al., 2000; LaRocque et al., 2007; McVey et al., 2007; Wei and Rong, 2007; Klovstad et al., 2008), several other genes were identified that are implicated in proper cell cycle checkpoint function or DNA damage repair. Although discrepancies in survival pathways among different organisms do exist, it can be concluded that many genes involved in responses to DNA damage, and their pathways, are highly conserved (Henderson, 1999; Rhind and Russell, 2000; Sekelsky et al., 2000).
Hydroxyurea (HU) is a compound that inhibits ribonucleoside dephosphate reductase and thereby blocks DNA synthesis (Hendricks and Mathews, 1998). This “classic” feature of HU has been widely used to induce intra-S and S/M checkpoints that delay progression through S phase and prevent mitotic entry due to the presence of incompletely replicated DNA. Using this characteristic effect of HU, various intra-S and S/M checkpoint mutants in yeast were identified based on their hypersensitivity to HU (Al Khodairy and Carr, 1992; Enoch et al., 1992). However, it is often overseen that exposure to HU can also result in modifications of DNA or histones. Prolonged HU incubation results in increased levels of 5-methylcytosine in proliferating tissue culture cells (Nyce et al., 1986; Nyce, 1989). HU also induces phosphorylation of the histone variant H2AX in an ataxia-telangiectasia mutated and Rad3-related (ATR)-dependent way in mammalian cells (Ward and Chen, 2001; Kurose et al., 2006a; Cowell et al., 2007). H2AX is also rapidly phosphorylated in response to exposure of cells to DNA double-strand break (DSB)-inducing agents (Rogakou et al., 1998). These data indicate that HU alters the DNA and inflicts a DNA damage response. This is in accordance with the observation that HU-induced stalled replication forks caused double-strand breaks in specific mutant strains of Escherichia coli (Guarino et al., 2007) and Schizosaccharomyces pombe (Froget et al., 2008). Currently, factors (other than ATR) that influence this specific type of HU-induced DNA damage remain largely unknown.
In a forward genetic Drosophila screen, aimed to identify novel genes involved in survival responses to HU, we identified stonewall (stwl) as an HU-hypersensitive mutant. Stwl was described previously as a female sterile mutant in Drosophila (Clark and McKearin, 1996; Akiyama, 2002; Maines et al., 2007). In contrast to previously identified Drosophila HU-hypersensitive cell cycle checkpoint mutants (grp/Dchk1, Dwee 1, mei-41/Datm; Hari et al., 1995; Sibon et al., 1997, 1999; Price et al., 2000), stwl mutants show intact S/M checkpoint function in response to DNA replication inhibition. We demonstrate that HU induces increased levels of phosphorylated H2Av (Drosophila functional homologue of H2AX; Madigan et al., 2002) in stwl mutants compared with wild type. Light and electron microscopic analysis revealed that Stwl is a nuclear protein associated with heterochromatin and colocalizes with heterochromatin protein 1 (HP1). Stwl displays transcription-repressing activity, and stwl is a dominant suppressor of position-effect variegation. Moreover, in stwl mutants, levels of trimethylated H3K27 and H3K9 (histone modifications associated with heterochromatin) are decreased compared with wild-type controls. Together, these data suggest that Stwl is a protein able to modify chromatin and that is required to maintaining DNA integrity in response to DNA damage induced by perturbed replication.
MATERIALS AND METHODS
Fly Stocks and Genetic Crosses
All fly stocks were maintained at 22°C using standard culture conditions (1.7% agar; Caldic Ingredients, Oudewater, The Netherlands), 3.2% yeast (Desimo, Leeuwarden, The Netherlands), 5.4% sugar (Desimo), and 0.1 mg of methylis parahydroxybenzoas per 100 ml (Spruyt-Hillen, Uitgeest, The Netherlands). The stwl allele 84 was isolated from a collection of P[lacW, ry+] P-element insertion stocks kindly provided by Prof. R. Scott Hawley (Stowers Institute for Medical Research, Kansas City, MO). The stwl null mutant (stwl Δ95, ry/TM3, Sb e ry) and a transgenic stock carrying a stwl minigene (w; P[mini(15a,w+]) were kindly provided by Prof. D. McKearin (Howard Hughes Medical Institute, Washington, DC) and have been described previously (Cooley et al., 1989). The E685 stock balanced over transmembrane ™ 3 was obtained from a female sterile collection generated by Prof. A. Ephrussi (European Molecular Biology Laboratory, Heidelberg, Germany). The Oregon-R wild type was used as a control in all experiments. P-elements were mobilized using a P[Δ2-3; ry+], Sb chromosome. In(1)wm4 flies were kindly provided by Prof. S. Elgin (Washington University, St. Louis, MO).
Screen to Test for Sensitivity to HU and Infrared (IR)
Sensitivity to HU was quantified as follows: five to 10 groups, each containing seven females and three males, were crossed, and the resulting embryos were collected for a period of 48 h. Adult flies were then removed. After 48 h, HU (Sigma-Aldrich, St. Louis, MO) was freshly dissolved in distilled H2O, and 0.5 ml of indicated concentrations of HU was added to the vials containing 7 ml of food. Controls received only 0.5 ml of distilled H2O. When irradiation was applied to rad54 mutants, 20-Gy IR was given to 48- to 96-h-old larvae from a 137Cs source in an IBL 637 irradiator (CIS Biointernational, Saclay, France). Doses were given at a rate of 2.2 Gy/min. After 2 wk, all classes of adult progeny were scored.
Cell Culture
Drosophila Schneider's S2 cells (S2 cells) were cultured in Schneider's Drosophila medium (Invitrogen, Paisley, United Kingdom) supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich), 100 U/ml penicillin, and 100 μg/ml streptomycin in T-25 flasks at 22°C. O23 hamster fibroblasts were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum.
Western Blot Analysis
For SDS-polyacrylamide gel electrophoresis (PAGE), Western blot analysis, the samples of S2 cells and ovaries of female adult flies were prepared as described previously (de Vries et al., 2005). Stwl was detected using the anti-Stonewall rat polyclonal antibody (generously provided by D. McKearin), 1:1000 in 3% bovine serum albumin (BSA)/0.05% Tween/phosphate-buffered saline (PBS). As a loading control, γ-tubulin (T6557; Sigma-Aldrich) or α-tubulin (T5168; Sigma-Aldrich) protein levels were detected. Both antibodies were diluted (1:2000) in phosphate-buffered saline, 0.3% Triton X-100 (PBST). The secondary antibodies anti-rat horseradish peroxidase (HRP) and anti-mouse HRP and the enhanced chemiluminescence (ECL) detection reagents were obtained from GE Healthcare (Little Chalfont, Buckinghamshire, United Kingdom). All standard chemicals were provided by Sigma-Aldrich. Western blot analysis of Cdc25Stg and Cdc2 was performed as described previously (Edgar et al., 1994) by using 12.5% polyacrylamide gels (Protean II; Bio-Rad, Hercules, CA). Cdc25Stg and Cdc2 antibodies were a generous gift from B. A. Edgar (Fred Hutchinson Cancer Research Center, Seattle, WA).
Down-Regulation of the Stwl Protein in S2 Cells by RNA Interference (RNAi)
RNA interference was performed as described previously (Clemens et al., 2000). A stwl DNA fragment of 776 base pairs containing a coding sequence of the stwl cDNA (GenBank accession no. U41367, 1464-2240 base pairs) was amplified using polymerase chain reaction (PCR). Each primer used in the PCR contained a 5′-T7 RNA polymerase binding site (GAATTAATACGACTCACTATAGGGAGA) followed by a sequence specific for stwl. The PCR products were purified using the High Pure PCR purification kit (Roche Molecular Biochemicals, Mannheim, Germany). The purified PCR products were used as templates by using a MEGASCRIPT T7 transcription kit (Ambion, Austin, TX) to produce double-stranded (ds)RNA. The dsRNA product was ethanol precipitated and resuspended in distilled H2O. dsRNA was annealed by incubation at 65°C for 30 min followed by slow cooling to room temperature (RT), and then they were analyzed by agarose gel electrophoresis and stored at −20°C. Primer sequences used to generate stwl dsRNA were obtained as follows: stwl, GenBank accession no. U41367, sense primer 1467–1480 and antisense primer 2223-2240. S2 cells were diluted to a final concentration of 1 × 106 cells/ml in Drosophila SFM (Invitrogen). One milliliter of cells was plated in 35-mm dishes, and 10 μg of dsRNA was added to each dish. The cells were incubated for 60 min at 22°C followed by addition of 2.0 ml of complete Schneider's medium. Cells were incubated for 96 h to down-regulate stwl protein to below detection level. Down-regulation of Stwl protein by RNAi was always controlled by Western blot analysis during the course of every experiment.
Immunofluorescence Analysis
Cells were plated on coverslips in six-well plates or in 35-mm dishes. Cells were washed twice with PBS, fixed for 15 min with 3.7% (vol/vol) formaldehyde/PBS, and then washed three times for 5 min each with PBS and incubated for 15 min with 0.2% (vol/vol) Triton X-100 in PBS, followed by 10-min incubation in 100 mM glycine in PBS. Coverslips were blocked for 30 min with 3% (wt/vol) BSA in PBS, and cells were incubated overnight at 4°C with a 1:100 dilution of anti-stwl antibody or with a 1:5000 dilution of Phospho-Histone-H-3 (PH3) (Cell Signaling Technology, Beverly, MA) or with a 1:200 dilution of monoclonal anti-HP1 antibody (monoclonal antibody C1A9 developed by L. L. Wallrath was obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, The University of Iowa, Iowa City, IA) in 5% BSA/0.1% Tween 20/PBS. Coverslips were washed 3 × 10 min in 0.1% Tween 20/PBS and incubated for 1 h at room temperature with secondary antibodies. To visualize stwl, 1:1000 dilution of a Cy3-conjugated anti-rat antibody (Jackson Immunoresearch Laboratories, West Grove. PA) was used. To visualize PH3, a Cy3-conjugated anti-mouse antibody, diluted 1:500 (GE Healthcare) was used. To visualize HP1, Goat-anti-Mouse-FITC (Jackson Immunoresearch Laboratories) diluted 1:200 was used. After three washes in 0.1% Tween 20/PBS, the DNA was stained for 10 min with 0.2 μg/ml 4,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich). Coverslips were washed three times with PBS and mounted with Citifluor AF1 (Agar Scientific, Essex, United Kingdom). The fluorescence was detected with a confocal laser scanning microscope.
Electron Microscopic Analysis
S2 cells were harvested in medium, pelleted, and washed in PBS. Cells were fixed for 15 min in 3.7% (vol/vol) formaldehyde/0.1 M phosphate buffer, pH 7.4. Fixed cells were washed twice in 6.8% sucrose in 0.1 M phosphate buffer, pH 7.4. Cells were embedded by centrifugation in 5% gelatin in 0.05 M phosphate buffer, pH 7.4. Immunolabeling was performed on 0.2-mm-thick gelatin sections by using anti-Stwl antibody incubated overnight (1:40 diluted), followed by biotinylated Rab-anti Rat antibodies (Vector Laboratories, Burlingame, CA) (1:200 diluted) for 2 h. Immunoreactivity was visualized with the ATP-binding cassette method by using a Vectastain kit (Vector Laboratories). The standard gold-substituted silver-intensified peroxidase (GSSP) technique was used in which the diaminobenzidine reaction product is first intensified with silver and then substituted for gold as described previously (van den Pol and Gorcs, 1986; Morara et al., 2001). The GSSP reaction product, at sites where Stwl antibody is localized, is visible as dense black precipitates with sharply defined contours. Immunolabeled gelatin sections were osmicated in 0.5% OsO4, supplemented with 1.5% potassium hexacyanoferrate in cacodylate buffer (0.1 M), pH 7.4, for 15 min, dehydrated in a graded series of ethanol, and embedded in Epon. Semithin sections (90 nm) were cut on an LKB ultratome, and sections were counterstained with uranyl acetate and lead citrate, and finally examined in a Philips CM 100 transmission electron microscope.
Developmental Analysis
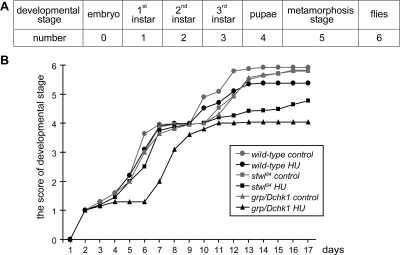
Adult flies were kept on standard apple juice plates, and larvae or embryos of specific age were collected. For the developmental analysis presented in Figure 3, 0- to 4-h-old embryos were harvested. After 16 h, embryos were dechorionated by 50% bleach solution, and after 2 h of recovery, embryos of different genotypes were sorted using a COPAS Select automated sorter according to previously published protocols (Furlong et al., 2001), based on the presence or absence of green fluorescent protein (GFP) bearing balancers. Embryos were raised on control food or food supplemented with 50 mM HU. The developmental stage of the animals and the amount of animals for that specific stage were examined every 24 h. A score was given every 24 h for various genetic backgrounds under control conditions and after HU treatment. Every developmental stage was defined as a number, and the score of every stage was calculated and projected on the y-axis; for a specific time after egg deposition, it is possible that various developmental stages were present in the vials. The score was calculated as follows: 0, embryonic stage; 1, first instar stage; 2, second instar stage; 3, third instar stage, 4, early pupal stage, 5, late pupal stage (wings and eyes are visible within the pupa case); and 6, adult stage. The score of a specific day is the sum of the amount of animals multiplied by the number given to that developmental stage. For example, at day 7, if 36.6% of animals are third instars (3) and 63.4% of animals are early pupae (4), the score is (36.6 × 3) + (63.4 × 4) = 3.63. This score is given in Figure 3B on the y-axis and on the x-axis; time is given in days.
Figure 3.
HU affects development of grp/Dchk1 and stwl84 mutants. Wild type, stwl84, and grp/Dchk1 heterozygous mutant flies were put on standard apple juice plates, and embryos between 0 and 4 h were collected. After 48 h, wild type, stwl84, and grp/Dchk1 homozygous first instar larvae were sorted and raised on control food or food supplemented with 50 mM HU. The effect of HU on development was scored over time using the following approach (also see Materials and Methods). (A) Every developmental stage was defined as a number. (B) The score of every stage was calculated and projected on the y-axis. For every genotype and condition >300 animals were followed over time. Development of HU treated stwl84 and grp/Dchk1 homozygous mutants is affected and delayed compared with wild type.
Checkpoint Analysis
HU (50 mg/ml) was added to the food of third instar larvae for 5 h. Larval brains were extirpated in PBS, fixed for 40 min in PBST (phosphate buffered saline, 0.3% Triton X-100) containing 5% formaldehyde, and washed three times with PBST. After blocking with 5% BSA in PBST for 30 min, samples were incubated overnight with monoclonal anti-phospho-Histone H3 antibody (pH3) (Cell Signaling, Technology, Beverly, MA, USA) diluted 1:200 in 5% BSA in PBST, washed three times with PBST, and incubated 2 h at RT with goat anti-mouse secondary antibody conjugated with Cy3 (GE Healthcare) diluted 1:200 in 5% BSA in PBST. Samples were washed three times with PBST, stained with 10 μg/ml DAPI in PBST for 15 min, and washed three times with PBST before mounting onto slides with Citifluor AF1 (Agar Scientific). Images were acquired using a confocal laser microscope.
Histone Isolation and Western Blot Analysis
HU (50 mg/ml) was added to the food of late second instar larvae for 24 h. Thirty wild-type or stwl homozygous larvae and 60 grp/Dchk1 homozygous larvae were selected and used to extract histones by using an acid extraction protocol (Gorski et al., 2004). To detect γ-H2Av, affinity-purified rabbit polyclonal anti-histone H2AvD Ps 137 (Rockland Immunochemicals, Gilbertsville, PA) was used (diluted 1:2000) in 1% BSA. To detect H3K27Me3 a polyclonal antibody (1:1000) (17-622; Millipore, Billerica, MA) was used, to detect H3K9M3 a rabbit polyclonal antibody (1:1000) (ab8898; Abcam, Cambridge, United Kingdom) was used. As a loading control, a rabbit polyclonal antibody against H2A (Abcam) (diluted 1:2000) in 5% BSA/PBST was used. Secondary anti-rabbit HRP and ECL detection reagents were obtained from GE Healthcare).
5-Bromo-2′-deoxyuridine (BrdU) Incorporation
Salivary glands were extirpated in Schneider's insect culture medium and incubated for 1 h in 0.8 ml of the same medium containing 0.5 mg/ml BrdU in the absence or presence of 20 mM HU on a nutator. Salivary glands were fixed for 40 min in PBST containing 5% formaldehyde, washed three times with PBST, denatured with 2 N HCl for 2 h, and neutralized with three washes of PBST and blocked in 5% BSA before antibody staining. Samples were incubated on the nutator overnight with the monoclonal anti-BrdU antibody (1:200 in blocking solution; BD Biosciences, San Jose, CA) at 4°C, washed three times for 10 min each with PBST, and incubated for 2 h at RT with goat anti-mouse secondary antibody conjugated with Cy3 (GE Healthcare) diluted 1:200 in blocking solution. Samples were washed three times with PBST, stained with 10 μg/ml DAPI in PBST for 15 min, and washed three times with PBST before mounting onto slides with Citifluor AF1 (Agar Scientific).
Plasmids, Constructs, Transient Transfections
The plasmids used in this study include pSG424 (Tapia-Ramirez et al., 1997), which contains the GAL4 DNA binding domain driven by the SV40 promoter and enhancer. Full-length stwl cDNA (Clark and McKearin, 1996) was cloned in frame in pSG424. The plasmid E1B-TATA-luc contains five copies of the upstream-activating sequence (UAS), a minimal promoter, and the luciferase gene. The typeII-luc plasmid contains a constitutive active type II sodium channel promoter, preceded by five UAS copies, and it drives a luciferase gene. The plasmid pSG424-VP16 contains the VP16 transactivation domain fused to GAL4. The plasmid pSG424-RD contains a carboxy-terminal repressor domain of REST/NRSF fused to GAL4 (Tapia-Ramirez et al., 1997). Transient transfections were performed using Lipofectamine according to the procedure of the manufacturer (Invitrogen) and mixtures of 1 μg of GAL4 plasmids, 1 μg of the UAS-luc plasmids, and 1 μg of pSVLacZ (included as an internal standard) were transfected. All transfections were repeated several times with at least two different preparations of plasmid DNA. Luciferase activity was determined as described previously (Michels et al., 1995) and normalized with respect to β-galactosidase activity also determined as described previously (Simon and Lis, 1987). All expression vectors used were analyzed for fusion-protein expression by loading 25 μg of total cell lysate on a 12.5% polyacrylamide gel as described previously (de Vries et al., 2005). Fusion-protein expression was detected using an anti-GAL4 antibody (Millipore). Trichostatin A (50 and 100 ng/ml; Sigma-Aldrich) was added 24 h before harvesting.
Acquirement of Images
The fluorescence image of cells, brains, salivary glands, and ovaries were detected with a confocal laser scanning microscope (TCS SP2; Leica Microsystems, Heidelberg, Germany). Images were modified with Jasc Paint Shop Pro9 software (Corel, Ottawa, Canada).
RESULTS
84 Mutants Are Sensitive to HU, Female Sterile, and Carry a Mutation in stonewall
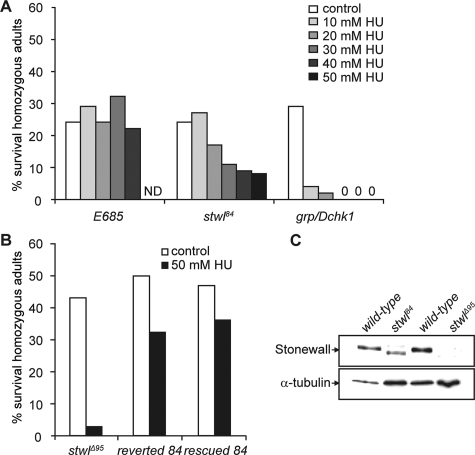
To identify novel genes involved in surviving HU treatment, a collection of Drosophila mutants, generated by P-element insertion (on the third chromosome, balanced over TM3), were screened for enhanced sensitivity to HU. For these studies, heterozygous mutant males were crossed with heterozygous mutant females, and the progeny were raised on food supplemented with various concentrations of HU. Sensitivity is indicated by a preferential loss of homozygous offspring in the presence of HU. We identified a novel HU hypersensitive mutant, initially named “84.” 84 mutants are hypersensitive to HU in a dose-dependent manner (Figure 1A). As a negative control, a random mutation on the third chromosome (E685), balanced over TM3, was used, and this mutant did not show an HU dose-dependent decline in homozygous survivors (Figure 1A). As a positive control the HU-sensitive checkpoint mutant grp/Dchk1 was used (Sibon et al., 1997; de Vries et al., 2005). In contrast to grp/Dchk1 mutants, preferential loss of homozygous 84 offspring was observed after relatively high concentrations of HU (Figure 1A). Another pronounced phenotype of 84 mutants is female sterility marked by a failure to lay eggs. DAPI staining revealed that mutant ovaries are agametic, characterized by a failure of proper germline tissue development and by the absence of nurse cell nuclei (data not shown) (Verheyen and Cooley, 1994; Rodesch et al., 1995).
Figure 1.
Stwl mutants are female sterile and hypersensitive to HU. (A) Zygotic sensitivity of 84 (balanced over TM3) and grp/Dchk1 (balanced over CyO) mutants to different concentrations of HU was assayed. E685 (balanced over TM3) was used as a control. The percentage of homozygous adult survivors is reported and for every condition, >200 surviving (heterozygous and homozygous) flies were counted. The results of a typical experiment are shown. (B) The reverted 84 strain was obtained by excision of the P-element out of the 84 genome. The rescued stock was obtained by crossing a chromosome containing stwl cDNA (stwl-minigene) into the 84 mutant. Pmini/Pmini;84/84 and Pmini/CyO;84/84 flies were counted as homozygous adults, and Pmini/Pmini;84/TM3 and Pmini/CyO;84/TM3 flies were counted as heterozygous adults. Sensitivity of these stocks and sensitivity of stwlΔ95 mutants to HU was measured. (C) Western blot analysis. In wild-type ovaries (lanes 1 and 3), Stwl is detected as a 150-kDa protein. Extracts of ovaries of homozygous 84 females (lane 2) showed a faster migrating band. In ovarian extracts of homozygous stwlΔ95 females (lane 4), no Stwl protein is detected. α-Tubulin was used as a loading control.
To ascertain whether the HU-sensitive and female sterile phenotype of 84 mutants is caused by the P-element insertion, the P-element was removed by reversion analysis and the “reverted” phenotype was investigated. Homozygous reverted females were fertile (data not shown) and homozygous reverted larvae showed significantly reduced hypersensitivity to HU (Figure 1B). Analyzing the insertion site of the P-element revealed that it was integrated in the first intron of the stwl gene, located on the third chromosome, at position 70D7. StwlΔ95 (null) mutants are female sterile and show severe defects in oogenesis (Akiyama, 2002; Clark and McKearin, 1996; Maines et al., 2007). The stwl ovarian defect is characterized by an absence of germline stem cells, and egg chamber numbers decline as females age (Clark and McKearin, 1996; Akiyama, 2002; Maines et al., 2007). The phenotype of the ovaries of homozygous 84 females is similar to these reported stwlΔ95 ovarian defects and is not addressed here in more detail. We tested whether stwlΔ95 mutant larvae were also sensitive to HU. Indeed, treatment with HU resulted in a preferential loss of homozygous offspring (Figure 1B). To confirm that a mutation in the stwl gene is responsible for the HU-hypersensitive and female sterile phenotypes of 84, a stwl minigene (Clark and McKearin, 1996) containing the stwl cDNA was introduced into the 84 genome. HU survival of these “rescued” larvae (Pmini/Pmini; 84/84) was improved (Figure 1B), and rescued homozygous females were fertile (data not shown).
Western blot analysis using an anti-Stwl antibody (Clark and McKearin, 1996) was performed to determine the expression of Stwl protein in extracts from wild-type and homozygous 84 ovaries. As expected, Stwl in ovarian extracts from wild-type females migrated at 150 kDa (Figure 1C, lanes 1 and 3). A protein, recognized by the Stwl antibody with increased electrophoretic mobility, was observed in ovarian extracts from homozygous 84 females (Figure 1C, lane 2). Ovaries of homozygous stwlΔ95 females did not express Stwl (Figure 1C, lane 4) (Clark and McKearin, 1996). These observations indicate that 84 mutants carry a hypomorph allele of Stwl. The 84 mutant will be further designated as stwl84.
Stwl and Grp/DChk1 Are Required for Normal Larval Development and Survival during Prolonged Treatment with HU
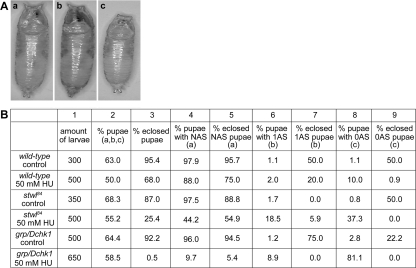
Next, we investigated the consequences of prolonged HU exposure, and the role of Stwl therein, for Drosophila development. First, wild-type, stwl84 and grp/Dchk1 (as a positive HU-hypersensitive control) homozygous larvae were selected using an automated sorter. This device enables the sorting of staged homozygous embryos or larvae, to follow the development of each genotype raised under equal larval-density conditions and to determine the percentage of homozygous survivors after HU treatment. Independent of genotype, addition of HU to the food caused a strong reduction in the number of larvae that were able to reach the pupal stage (Figure 2B, column 2). As expected, a smaller percentage of stwl84 mutant pupae (25.4%) and grp/Dchk1 mutant pupae (0.5%) eclosed after HU treatment compared with wild-type (68.0%) (Figure 2B, column 3). Closer examination of how HU affects development in the various genetic backgrounds revealed that HU induces abnormal pupal anterior spiracles. Pupae were scored for the presence of normal anterior spiracles (NASs) (Figure 2Aa, and B, column 4), the presence of one single spiracle (1AS) (Figure 2Ab, and B, column 6), or the presence of no spiracle (0AS) (Figure 2Ac, and B, column 8). Under control conditions in all genotypes tested, the percentage of the formation of abnormal spiracles (1AS and 0AS) is <3%. In a wild-type background, HU induced abnormal spiracles in only a small percentage of pupae (1AS, 2.0% and 0AS, 10.0%) and 20.0% of 1AS pupae and 0.9% of 0AS pupae developed in adult flies. In stwl84 homozygous mutants, HU caused an increase in the amount of pupae with abnormal anterior spiracles and hardly any of these pupae developed into adult flies compared with wild type (1AS, 18.5% and 0AS, 37.3%, eclosion, respectively, 5.9 and 0.0%). Grp/Dchk1 homozygous mutants show an even more severe phenotype—1AS, 8.9% and 0AS, 81.1%—and none of these abnormal pupae gave rise to viable adult flies. The observed morphological abnormalities are not specific consequences of HU treatment and are not specific characteristics of grp/Dchk1 or stwl84 mutants, because these abnormalities were also observed (although to a lesser extent) in wild types and in repair deficient rad54 mutants after ionizing radiation (data not shown), indicating that these morphological abnormalities can be used as a general marker for the presence of damaged DNA.
Figure 2.
Disruption of stwl and grp/Dchk1 affects metamorphosis in the presence of HU. (A) Wild-types and homozygous stwl84 and grp/Dchk1 mutants were sorted and raised on control food or food supplemented with 50 mM HU. HU induced several morphological abnormalities of the pupae. a, normal pupa. B, pupa with one single anterior spiracle (1AS). c, pupa with no anterior spiracle (0AS). (B) Survival rates and the percentages of abnormal pupae cases of different genotypes in the presence and absence of HU. Amount of larvae = amount of larvae of each genotype selected; % pupae = total % of larvae that reached the pupal stage including; normal morphology (Aa), 1AS morphology (Ab), or with 0AS morphology (Ac). % eclosed pupae = percentage of these pupae that eclosed; % pupae with NAS = percentage of pupae with normal anterior spiracles (Aa); % eclosed NAS pupae = percentage of pupae with an NAS morphology that eclosed; % pupae with 1AS = percentage of pupae with one single anterior spiracle (Ab); % eclosed 1AS pupae = percentage of pupae with a 1AS morphology that eclosed; % pupae with 0AS = percentage of pupae with no single anterior spiracles (Ac); % eclosed NAS pupae = percentage of pupae with an 0AS morphology that eclosed.
HU also induced a developmental delay (Figure 3), which occurred at an earlier time point and resulted in earlier developmental arrest in grp/Dchk1 mutants compared with wild type. Developmental arrest also occurred in stwl84 mutants but at a later stage compared with grp/Dchk1 (Figure 3). Based on these results, we conclude that HU in stwl84 and grp/Dchk1 mutants did not lead to increased larval death compared with wild type, but it led to an increased developmental delay and a higher percentage of morphologically abnormal pupae in which proper metamorphosis did not occur.
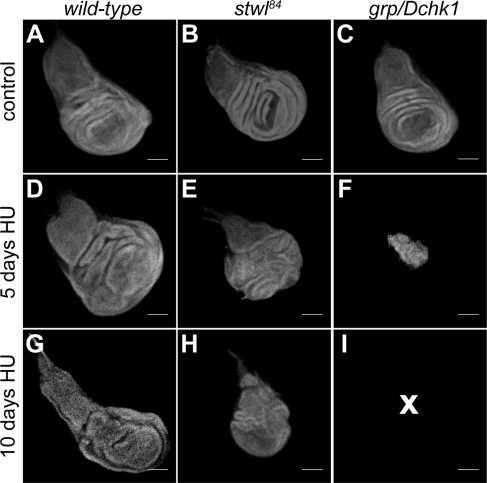
HU Affects Imaginal Disk Morphology in stwl84 and grp/Dchk1 Mutants
Because HU induces a developmental delay and abnormal metamorphosis (Figures 2 and 3), we tested whether differences could be observed in proliferating larval tissues in addition to these gross abnormalities. When 50 mg/ml HU is added to the food of stwl84 late second instar larvae, imaginal discs become progressively disorganized and smaller in size compared with HU-treated wild type (Figure 4). The effect of HU on the imaginal discs of grp/Dchk1 mutants is more dramatic and after 10 d, no imaginal disk structures could be detected in the wandering larvae (Figure 4). These results show that increased developmental delay and pupal abnormalities in HU-sensitive mutants coincide with tissue degeneration in proliferating imaginal discs.
Figure 4.
Integrity of imaginal discs is affected by HU. Imaginal discs were dissected from wild type (A, D, and G), stwl84 (B, E, and H), and grp/Dchk1 (C, F, and I) homozygous mutant larvae that were raised on food supplemented with H2O as a control (A–C) or with 50 mg/ml HU for 5 d (D–F)) or for 10 d (G–I), and tissue morphology was visualized by DAPI staining. Images were made using a confocal scanning microscope. NB, tissue morphology of imaginal discs of grp/Dchk1 mutants (5 and 10 d after HU treatment, presented in F and I) and of stwl84 mutants (10 d after HU treatment, presented in H) is affected in such a way that it is not possible to identify the type of discs. No imaginal disk structures were observed in grp/Dchk1 homozygous larvae after 10-d HU treatment, as indicated by a cross. A, D, and G, wild-type imaginal wing discs; B and E, stwl84 imaginal wing disk; and C, grp/Dchk1 imaginal wing disk. Bar, 100 μm.
Stwl Is Not Required to Prevent Entrance into Mitosis in the Presence of Incompletely Replicated DNA
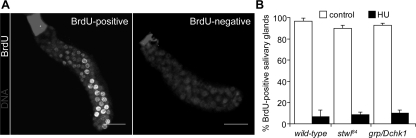
To investigate the cellular mechanism underlying the developmental abnormalities observed in HU treated stwl84 mutants, we first tested whether HU leads to a similar inhibitory effect on DNA synthesis in wild type and mutants. BrdU incorporation in salivary glands was equally reduced in wild type and stwl84 and grp/Dchk1 mutants after HU treatment (Figure 5), implying that differences in HU sensitivity in the various genetic backgrounds are not caused by differences in DNA synthesis inhibition in these backgrounds. Next, the ability to arrest cell cycle progression in response to HU was investigated by visualizing mitotic cells in larval brains in the absence and presence of HU (Figure 6A). Consistent with previous reports (Sibon et al., 1997, 1999, 2000; de Vries et al., 2005), grp/Dchk1 mutant cells enter mitosis in the presence of incompletely replicated DNA (Figure 6A). However, stwl84 mutants showed a normal checkpoint response after HU treatment comparable with wild type (Figure 6A). This was confirmed using Stwl-depleted (by RNAi) cultured Schneider's S2 cells (Figure 6B), in which a strong reduction of mitotic cells was observed in response to HU, similar to control cells (Figure 6C). In line with this, normal posttranslational modifications or changes in expression levels of the cell cycle regulators, Cdc2 and Cdc25Stg (Edgar et al., 1994; de Vries et al., 2005) were observed in Stwl-depleted S2 cells in response to HU (Figure 6D). These data indicated that Stwl is not required to prevent entrance into mitosis in the presence of incompletely replicated DNA.
Figure 5.
DNA synthesis is equally inhibited in salivary glands in wild type, stwl84, and grp/Dchk1 mutants. Larval salivary glands were dissected from wild type, homozygous stwl84, and grp/Dchk1 second instar larvae and were cultured in fresh Schneider's insect culture medium (control) or medium with 20 mM HU for 1 h; subsequently, glands were incubated with 50 mM BrdU. Larval salivary glands were stained with BrdU antibody. The results of a typical experiment are shown. (A) Salivary glands were scored for negative or positive BrdU signal. Bar, 100 μm. (B) Quantification of the percentage of positive and negative salivary glands under control conditions and after HU treatment of wild type, stwl84, and grp/Dchk1 mutants. DNA synthesis is equally inhibited after HU treatment in wild type, stwl84, and grp/Dchk1 mutants.
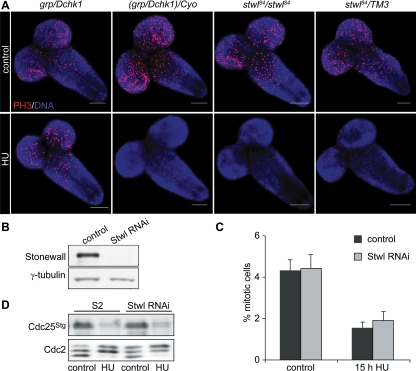
Figure 6.
Grp/Dchk1, but not Stwl, is required to prevent entrance into mitosis in the presence of HU. (A) Larval brains were dissected from homozygous (stwl84) and heterozygous (stwl84/TM3) stwl84 mutants and grp/Dchk1 homozygous (grp/Dchk1) and heterozygous [(grp/Dchk1)/CyO)] third instar mutant larvae after feeding with 50 mg/ml HU for 5 h. Larval brains were stained with an antibody against PH3, used as a marker for mitotic cells. Grp/Dchk1 heterozygous mutants and stwl84 heterozygous and homozygous mutants show intact checkpoint function. Checkpoint function is impaired in grp/Dchk1 homozygous mutants. Bar, 100 μm. (B) Western blot analysis of Stwl in Schneider's S2 cells under control conditions and after RNAi. Lane 1 shows expression of Stwl protein levels in control cells. In cells incubated with dsRNA of stwl (Stwl-depleted cells) for 96 h, Stwl protein was not detected (lane 2). γ-Tubulin was used as a loading control. (C) Control Schneider's S2 cells and Stwl-depleted S2 cells were stained with a PH3 antibody to detect mitotic cells, and percentages were determined using fluorescence-activated cell sorting analysis. Schneider's S2 control cells and Stwl-depleted cells show a comparable percentage of mitotic cells under control conditions. In control cells and in Stwl-depleted cells, a comparable decrease of mitotic cells was observed after incubation with 10 mM HU for 15 h. (D) Schneider's S2 control cells and Stwl-depleted cells were treated with 10 mM HU and protein levels of Cdc25Stg and isoforms of Cdc2 were determined using Western blot analysis. In line with previous results (Sibon et al., 1999; de Vries et al., 2005) in control Schneider's S2 cells, HU treatment induced a decrease in Cdc25Stg and accumulation of inhibitory forms Cdc2. Stwl-depleted cells show comparable Cdc25Stg and Cdc2 modifications in response to HU treatment compared with wild type.
Stwl Is Not Required to Survive DNA Hypermethylation Induced by HU
Previously, it has been demonstrated that prolonged exposure to HU induces hypermethylation in replicating mammalian cells (Nyce et al., 1986; Nyce, 1989). Because stwl84 mutants do not show cell cycle defects, it is possible that stwl84 mutants are hypersensitive to HU because of HU-induced hypermethylation. To test this hypothesis, levels of global methylated DNA were analyzed using chromatography-electrospray ionization-tandem mass spectrometry (Kok et al., 2007) and capillary electrophoresis (Stach et al., 2003). No increased levels of global methylated DNA could be observed using these two methods in untreated stwl84 mutants, in HU-treated wild type or in stwl84 mutants after prolonged HU treatment (data not shown). Based on these experiments, we concluded that the HU hypersensitivity of stwl84 mutants cannot be explained by increased DNA methylation.
Stwl Is a Heterochromatin-associated Protein and Colocalizes with HP1
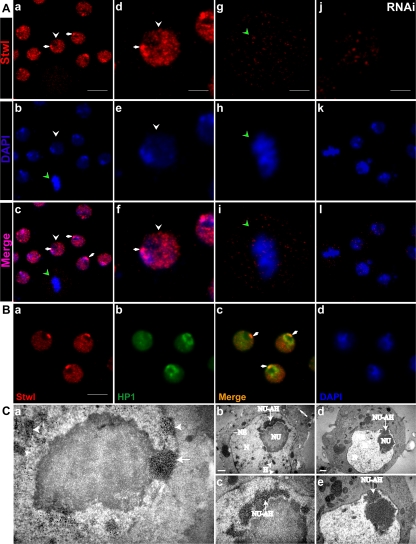
To understand the molecular function of Stwl in more detail, localization studies were performed. Outside ovaries, Stwl in Drosophila somatic cells was hardly detectable when the available anti-Stwl polyclonal antibodies were used for Western blot analysis or immunolocalization studies (Clark and McKearin, 1996) (our data not shown). Although, a specific protein trap line expressing a Stwl-GFP fusion protein (Buszczak et al., 2007) did reveal the presence of low levels of Stwl in interphase nuclei of salivary glands (Supplemental Figure 1), expression levels were too low to investigate specific patterns of Stwl localization at high resolution on polytene chromosome squashes. Fortunately, in S2 cells Stwl is highly expressed (Figure 6B); therefore, S2 cells were used to investigate localization patterns of Stwl in more detail. Consistent with the presence of two nuclear localization signal domains (Clark and McKearin, 1996), Stwl was present in the nucleus of interphase S2 cells (Figure 7). In addition, in most interphase cells, one or more foci with strong Stwl immunoreactivity were observed at the edge of the nucleoli at sites where the nucleolus and the nuclear envelope are in proximity (arrows, Figure 7Ad; also see below and Figure 7Ca). Incubation of S2 cells with dsRNA of Stwl for 96 h reduced Stwl protein levels to below detection by Western blot analysis (Figure 6B, lane 2), and no Stwl signal was observed in Stwl-depleted cells by using immunofluorescence (compare Figure 7, Aa and Aj), demonstrating the specificity of the Stwl antibody. Specificity was further supported by the observation that expression of a myc-stwl fusion protein in S2 cells and the subsequent detection of the fusion construct by using anti-myc antibodies or expressing Stwl-GFP resulted in a comparable pattern in the nucleus (data not shown). During prometaphase, metaphase, anaphase, and telophase, Stwl localization was diffuse and less pronounced. A representative S2 cell in metaphase is shown (green arrowhead, Figure 7, Aa and Ag).
Figure 7.
Stwl is a heterochromatin-associated protein and Stwl colocalizes with HP1. (A) Schneider's S2 cells were grown on coverslips and labeled with the Stwl antibody (a, d, g, and j). DNA was stained using DAPI (b, e, h, and k); overlays of the images are shown in c, f, i, and l. Arrows in Aa point to localization of Stwl protein in close association with the nucleolus. White arrowheads point to an interphase cell enlarged in d, e, and f, and green arrowheads point to a mitotic cell enlarged in g, h, and i. Stwl depletion by RNAi treatment for 96 h strongly reduces the specific Stwl labeling in Schneider's S2 cells (j, k, and l). Bar, 8 μm (a, d, g, and j). (B) Schneider's S2 cells were grown on coverslips and labeled with the Stwl antibody and with an antibody against HP1. Stwl colocalizes with HP1 at sites were the nucleolus (white arrowheads) and the nuclear envelope are in proximity. a, Stwl antibody. b, HP1 labeling. c, merged image of Stwl and HP1 labeling. d, DAPI staining. Bar, 6 μm. (C) Electron microscopic analysis was performed to examine the nuclear morphology of Schneider's S2 cells and to visualize Stwl-associated ultrastructures. a, immunolabeling was performed using Stwl antibodies. Sites where Stwl antibody is localized are visible as dense black precipitates. Stwl colocalizes with a specific structure associated with the nucleolus (arrows); Stwl is also present at heterochromatin-like structures dispersed throughout the nucleus (arrowheads). b–e, Examination of nonlabeled sections revealed that the Stwl-associated structures show a specific electron-dense morphology, previously described as nucleolus-associated heterochromatin. NU, nucleolus; N, nucleus; NU-AH, nucleolus-associated heterochromatin; H, heterochromatin; NE, nuclear envelope. Bars, 500 nm.
To further investigate the association of Stwl with heterochromatin in interphase cells, we tested whether Stwl colocalizes with HP1, a heterochromatin-associated protein (Eissenberg et al., 1990). Our data revealed that Stwl partly colocalizes with HP1 (Figure 7B). Colocalization is in most of the cells observed in the area closely associated with the nucleolus (arrows, Figure 7Ad) that shows strong Stwl immunoreactivity. In 87% of cells in which Stwl is localized in this bright spot, HP1 labeling was also observed in this area (n = 200 cells). However, localization of HP1 and Stwl is not restricted to this area. No obvious colocalization (upon visual inspection) between Stwl and HP1 was observed in areas outside this nucleolus-associated spot. Comparable with the Stwl labeling during prometaphase, metaphase, anaphase and telophase, HP1 localization was diffuse and less pronounced (data not shown). These data demonstrated that heterochromatin-associated proteins such as Stwl and HP1 show a cell cycle-specific distribution and localize to heterochromatin during interphase but not during mitosis. To investigate whether the localization pattern of HP1 depends on the presence of Stwl, HP1 localization was examined in Stwl-depleted (by RNAi) S2 cells and in salivary glands of stwl84 mutants. HP1 localization in Stwl-depleted S2 cells and S2 control cells is comparable (data not shown). In addition, localization of HP1 in nuclei of wild-type salivary glands is comparable with the localization pattern of HP1 in nuclei of salivary glands dissected from stwl84 larvae (Supplemental Figure 2). Together, these data indicate that localization of HP1 does not change in a stwl mutant background or when Stwl protein levels are decreased.
To examine the nuclear localization of Stwl in more detail and to investigate whether Stwl localizes to defined nuclear structures, immunoelectron microscopy was performed (Figure 7C). Ultrastructural analysis of Stwl-labeled sections revealed the presence of Stwl in a structure closely associated with the nucleolus and in heterochromatin-like structures dispersed throughout the nucleus (Figure 7Ca). In nonlabeled sections, these nucleolus-associated structures and heterochromatin structures are visible as distinct electron dense structures (Figure 7C, b–e). An analysis of several sections showed that the size of this Stwl-containing nucleolus-associated structure varied between 250 and 600 nm, most likely depending on the orientation of this structure within the sections. In a previous report using Drosophila KCo cells, this electron-dense structure has been described as “dense fibrillar nucleolus-associated transcriptionally inactive heterochromatin intermingled with fibrillo-granular ramifications extending from the nucleolus toward the nuclear envelope” (Knibiehler et al., 1982).
Next, we investigated whether localization of Stwl alters in response to HU treatment. Various concentrations of HU were tested; however, no change in Stwl localization was observed after HU treatment in S2 cells (data not shown) nor in salivary glands after adding HU to the food of wandering larvae (Supplemental Figure 3). Moreover, no posttranslational modifications of the Stwl protein could be observed after HU treatment examined by Western blot analysis (data not shown). Thus, so far there is no evidence that Stwl itself is a signaling molecule; it rather suggests that Stwl has a structural function.
Stwl Is a Suppressor of Position Effect Variegation and Represses Transcription
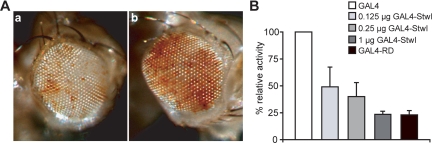
Because Stwl is associated with heterochromatin, we further investigated a possible role of Stwl in heterochromatin organization. Heterochromatin position effect variegation (PEV) is a phenomenon observed when a gene is translocated to a position adjacent to heterochromatin; therefore, this gene experiences inactivation. The inactivation is thought to be a consequence of spreading of a heterochromatin structure into euchromatin (Ebert et al., 2006). Several mutations have been identified that act as suppressors [Su(var)] or enhancers [E(var)] of variegation (Schotta et al., 2003). HP1 is such a heterochromatin-associated protein and reduction of HP1 levels results in a suppression of PEV (Eissenberg et al., 1990). We next investigated whether Stwl, like HP1, acts as Su(var). To address this, the stwlΔ95 allele was crossed into a line in which PEV can be monitored because these flies carry the white-variegating rearrangement In(1)wm4 (Shaffer et al., 2006). ln(1)wm4 flies show a great proportion of nonpigmented eye facets (Figure 8), and the effect of reduced levels of Stwl can be monitored by a change in the proportion of pigmented facets. Indeed the proportion of pigmented facets in ln(1)wm4;stwlΔ95/+ is strongly increased (Figure 8Ab) compared with ln(1)wm4; +/+ flies (Figure 8Aa). These results are in agreement with previous data by using different alleles (Maines et al., 2007) and showed that Stwl act as Su(var) and influences the heterochromatin status.
Figure 8.
Stwl is dominant suppressor of position-effect variegation and Stwl represses transcription. (A) a, eye of an In(1)wm4 fly showing a variegation phenotype. B, Eye of a ln(1)wm4;stwlΔ95/+ fly that shows an increase in the proportion of pigmented facets. (B) Mammalian O23 cells were transiently transfected with the UAS-type II sodium channel luciferase reporter in combination with the indicated expression plasmids: GAL4 (GAL4 DNA binding domain), GAL4-Stwl (GAL4-Stwl fusion construct), and GAL4-RD (carboxy-terminal repressor domain of REST/NRSF fused to GAL4) was used as a positive control. Various concentrations of the GAL4-Stwl fusion construct were used. Stwl suppresses UAS-type II sodium channel luciferase reporter activity in a dose-dependent manner. The normalized luciferase activity is given, error bars indicate SEM of three independent experiments.
To directly investigate whether Stwl is indeed able to repress gene expression, an established GAL4/UAS-luciferase system (Sadowski and Ptashne, 1989) was used in mammalian O23 cells that do not express endogenous Stwl. O23 cells were cotransfected with pSG424-stwl and a constitutively active UAS-type II-luciferase reporter, and the results showed a decrease in reporter activity in a stwl dose-dependent manner (Figure 8B). The above-mentioned studies indicated that, when tethered to the DNA by using a GAL4-UAS system, Stwl is able to repress reporter gene activity.
Stwl Is Required for Normal Levels of Trimethylated H3K27 and H3K9
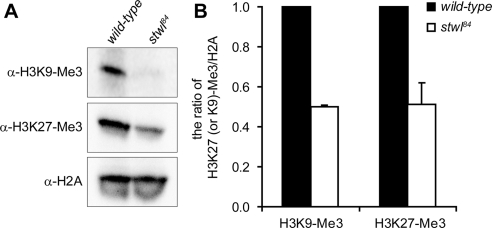
Our results suggested that in the absence of Stwl, chromatin is in a more relaxed state, and to investigate this further we compared levels of histone modifications that are associated with silent heterochromatin (trimethylated H3K27 and H3K9). In line with the observation that Stwl is a heterochromatin-associated protein with repressive capacities and the fact that stwl is a suppressor of PEV, we found that levels of trimethylated H3K27 and H3K9 are decreased in stwl84 mutants (Figure 9). These results are not due to a global decrease of DNA in stwl84 mutants, because no significant differences between mutants and wild-type levels of trimethylated H3K4 (data not shown) and H2A could be observed (Figure 9).
Figure 9.
Levels of trimethylated H3K9 and H3K27 are decreased in stwl84 mutant larvae. (A) Histone fractions were isolated from wild-type and stwl84 third instar larvae and levels of trimethylated H3K9 and H3K27 were analyzed. (B) The average ratios of levels of these specific histone modifications to levels of H2A were quantified using three independent experiments.
Based on our results, we concluded that Stwl is required to maintain chromatin organization, suggesting that this is (in addition to intact cell cycle checkpoint functions) required to survive HU.
Levels of γ-H2Av Are Increased in Stwl84 Mutants after HU Treatment
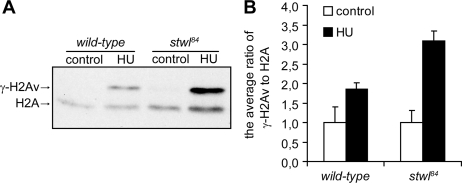
Evidence is accumulating that specific histone modifications are required for proper DNA damage responses (Loizou et al., 2006; Altaf et al., 2007; Escargueil et al., 2008), and thus histone modifications may in part determine an organism's sensitivity to DNA-damaging agents. Because global levels of trimethylated H3K9 and H3K27 were decreased in stwl84 mutants, we tested whether these abnormal levels of specific histone modifications correlate with levels of increased DNA damage after HU treatment. Previously, it was demonstrated in mammalian cells that HU induces elevated levels of γ-H2AX (Kurose et al., 2006b). γ-H2AX is a marker for the presence of damaged DNA (Madigan et al., 2002), and levels of γ-H2AX correlate with the amount of damage present (Rogakou et al., 1998; Madigan et al., 2002), meaning that the more DNA damage, the more γ-H2AX is detected. H2Av is the Drosophila histone variant that is also phosphorylated (γ-H2Av) in response to the induction of DNA damage (Madigan et al., 2002). First, we investigated whether in stwl84 mutants levels of γ-H2Av were spontaneously increased. Western blot analysis showed that in untreated stwl84 mutant larvae, no increase was observed in γ-H2Av, compared with wild type (Figure 10). However, in response to HU, levels of γ-H2Av were increased in wild type and were further increased in stwl84 mutants (Figure 10). These data show that in Drosophila (like in mammalian cells) in response to HU, DNA damage is inflicted. In addition our data demonstrate that in stwl84 mutants abnormal levels of trimethylated H3K9 and H3K27 coincide with increased DNA damage after HU and increased sensitivity to this compound. Together, these data suggest that the abnormal histone modifications in stwl mutants are not required for viability; however, after HU treatment, these abnormalities lead to increased DNA damage, explaining the hypersensitivity of stwl mutants to HU.
Figure 10.
HU induces increased levels of phosphorylation of histone H2Av in stwl84 mutants. (A) An antiserum directed against H2Av phosphorylated at Ser137 (γ-H2Av) on histone isolations was used as a marker for DNA damage induced by HU in wild-type and stwl84 homozygous larvae after 50 mg/ml HU treatment for 24 h. HU induces accumulation of γ-H2Av in wild-types and in stwl84 mutants; however, in stwl84 mutants, a larger increase of γ-H2Av levels was detected compared with wild-types. H2A was used as a loading control. (B) The average ratio of γ-H2Av expression levels to H2A expression levels was quantified using four independent experiments.
DISCUSSION
In this report, we present evidence for a novel function of Stwl, a protein previously described to be involved in oogenesis and stem cell maintenance (Clark and McKearin, 1996; Maines et al., 2007). Our data show that Stwl is required to survive HU-induced replication inhibition. In contrast to other previously identified HU-hypersensitive Drosophila mutants (such as grp/Dchk1, wee1, and mei41), stwl mutants show a normal HU-induced cell cycle delay, demonstrating that HU-induced lethality is not due to aberrant S/M checkpoint function. Stwl localizes to heterochromatin, colocalizes with HP1 and has transcription repressing capacities. In the absence of Stwl, levels of trimethylated H3K9 and H3K27 (marks for silent chromatin) are decreased and stwl is a suppressor of PEV. In addition, in stwl mutants HU causes an increase in accumulation of γ-H2Av. Together these findings show that Stwl plays a role in modifying chromatin to a more heterochromatin-like state, and this configuration is favorable to survive HU-induced DNA damage, linking epigenetic regulation of gene expression to DNA damage responses.
HU was initially used as an anticancer drug, and later it was shown that HU is beneficial for patients with sickle cell disease (Letvin et al., 1984). Despite the widespread clinical use of HU to treat patients, many issues about this chemical compound remain unresolved (Brawley et al., 2008). Among these, the response in patients varies, long-term side-effects are unknown, and mechanisms underlying the effectiveness of HU remain elusive. Thus, the consequences of HU treatment on whole organisms are largely unknown. In contrast, cellular consequences of DNA replication inhibition provoked by HU have been studied in more detail. These studies have been mainly performed in yeast, bacteria, and mammalian tissue culture cells. In the presence of replication stress, cell cycle checkpoints are induced, replication forks stall, and to survive, stalled replication forks need to be stabilized and reactivated (Lambert et al., 2007). Evidence obtained from studies using budding and fission yeast show that cell cycle checkpoint proteins are not only required to halt cell cycle progression but are also necessary to maintain DNA integrity at the stalled replication forks (Meister et al., 2005;Lopes et al., 2001). Because stwl mutants show a normal cell cycle arrest, Stwl may play a role downstream from induction of cell cycle arrest.
In addition to cell cycle arrest, other effects of HU are known, and it has been shown that HU causes an increase in phosphorylation of H2AX, which is indicative for DNA damage (Kurose et al., 2006a;Ward and Chen, 2001). Our data show that in the absence of Stwl and after HU treatment, γ-H2AX accumulates to higher levels compared with wild type. Therefore, it is most likely that HU-induced lethality of stwl mutants is caused by increased levels of DNA damage. Accumulation of DNA damage may be explained by an altered chromatin structure in stwl mutants, leading to more DNA damage compared with wild type in response to the same concentrations of HU. But it may also be possible that Stwl is required to unload γ-H2AX from damaged DNA, and when this unloading is affected in stwl mutants, DNA damage repair is less effective (Chowdhury et al., 2005). The presence of increased levels of DNA damage after HU in stwl mutants is also in line with our findings that in stwl mutants after HU, imaginal discs are more degenerated compared with wild types. Moreover, the morphological abnormalities of stwl pupae after HU treatment, resemble abnormalities observed in irradiated repair-deficient mutants. Together, these results indicate that after treatment with HU, stwl mutants suffer from increased DNA damage compared with wild type.
Stwl mutants are viable but show decreased levels of H3K9 and H3K27, indicating that these altered levels of specific histone modifications are not incompatible with life. However, in the presence of perturbed DNA replication, such as after HU treatment, normal levels of H3K9 and H3K27 may be required to prevent the accumulation of DNA damage. Increased sensitivity to various DNA-damaging agents in association with other histone modifications has been reported in S. cerevisiae and in S. pombe; H3K27 methylation is required for repair of UV-induced DNA damage (Evans et al., 2008), decreased acetylation of H3K56 coincides with increased sensitivity to various DNA synthesis inhibitors (Xhemalce et al., 2007), mutations in specific lysine residues in the histone H3 tail correlate with increased sensitivity to methylmethane sulfonate (Qin and Parthun, 2002), and decreased levels of H3K56 result in decreased survival after HU (Ozdemir et al., 2005). Although the connection between decreased levels of trimethylated H3K9 and H3K27 and HU hypersensitivity of stwl mutants needs to be proven, our data and data published by others strongly suggest that histone modifications are linked to stress responses.
It may be possible that not only alterations in specific histone modifications but also the more euchromatin-like state of chromatin in the stwl mutants induce increased vulnerability to HU. Previous reports describe that DNA damage-induced histone modifications differ between heterochromatin and euchromatin. Cowell et al. (2007) reported that γ-H2AX foci form preferable in euchromatin of non-S-phase mammalian cells after ionizing radiation, and Kim et al. (2007) presented evidence, using yeast and mammalian cells, that in heterochromatin adjacent to a DSB, levels of γ-H2AX are lower compared with levels in euchromatin. These data imply that histone modifications in response to DNA damage differ between heterochromatin and euchromatin and because histone modifications are involved in proper DNA damage responses (Loizou et al., 2006; Altaf et al., 2007; Escargueil et al., 2008), it is likely that heterochromatin and euchromatin are not equally resistant to various DNA-damaging insults, further explaining the HU hypersensitivity of stwl mutants.
Our results suggest that Stwl is a heterochromatin-associated protein able to modify chromatin. These results are in agreement with knowledge obtained from previous studies concerning Stwl and Stwl-like proteins (Clark and McKearin, 1996; Bhaskar and Courey, 2002) as explained below. In addition to two nuclear localization signals, the Stwl protein contains two conserved domains: an N-terminal MADF domain (amino acids [aa] 10-98) and a C-terminal BESS domain (aa 602-641) (Clark and McKearin, 1996). The same architecture (N-terminal MADF domain and C-terminal BESS domain) has been found in 13 other Drosophila proteins (Bhaskar and Courey, 2002). For one of these proteins, Dip3, it has been demonstrated that the MADF domain can directly bind to DNA in a sequence-specific way, whereas the BESS domain is involved in protein–protein interactions. These results, in addition to the results presented in this manuscript strongly suggest that Stwl is a protein able to bind and modify chromatin.
Sequence analysis of the genomic region adjacent to the P-element revealed that the P-element is integrated within the first intron (lying in between the first and second exon) of the stwl gene. Because the MADF domain is encoded by the C-terminal part of exon 1 and the N-terminal part of exon 2, most likely the P-element insertion is causing abnormal splicing of the stwl transcript, thereby leading to a disruption of the MADF domain. Loss of Stwl activity in stwl84 mutants therefore may be due to loss of the MADF domain, truncation of the protein, and lower expression levels of the truncated transcript (Figure 1C).
Based on our results, we suggest the following model: Stwl is required for normal compaction of chromatin. The Stwl-dependent configuration of chromatin is not required under normal replicating conditions; however, when replication stress is induced and cell cycle arrest and stalled replication forks arise, Stwl is required to maintain DNA integrity.
In summary, our data indicate that in addition to the genotype, epigenetic modifications strongly influence the potential of an organism to survive externally induced cytotoxic insults. Understanding the mechanisms of survival responses to specific compounds such as HU is not only of fundamental interest but also is beneficial to understanding the clinical consequences of these specific treatments.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lee Fradkin for generously providing plasmids containing Drosophila-specific promotors. We thank Bart Kanon, Jurre Hageman, and Jan Kootstra for technical assistance. Stwl antibodies were a generous gift from Dennis McKearin, and Cdc25Stg and Cdc2 antibodies were a generous gift from Bruce Edgar. This work was supported by a grant from the Dutch Cancer Society KWF (RUG99-1949), by a VIDI grant from the Netherlands Organisation for Scientific Research NWO (917-36-400), and by an NWO middelgroot grant (911-06-001) to O.C.M.S; by a UMCG Bernouilli Bursary grant (809003) to X. Y; by a Ubbo Emmius grant (714010) from the Graduate School GUIDE to K. S.; and by a Topmaster grant from the graduate school GUIDE to A. R.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-06-0639) on December 3, 2008.
REFERENCES
- Akiyama T. Mutations of stonewall disrupt the maintenance of female germline stem cells in Drosophila melanogaster. Dev. Growth Differ. 2002;44:97–102. doi: 10.1046/j.1440-169x.2002.00625.x. [DOI] [PubMed] [Google Scholar]
- Al Khodairy F., Carrm A. M. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altaf M., Saksouk N., Cote J. Histone modifications in response to DNA damage. Mutat. Res. 2007;618:81–90. doi: 10.1016/j.mrfmmm.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Bhaskar V., Courey A. J. The MADF-BESS domain factor Dip3 potentiates synergistic activation by Dorsal and Twist. Gene. 2002;299:173–184. doi: 10.1016/s0378-1119(02)01058-2. [DOI] [PubMed] [Google Scholar]
- Boyd J. B., Golino M. D., Nguyen T. D., Green M. M. Isolation and characterization of X-linked mutants of Drosophila melanogaster which are sensitive to mutagens. Genetics. 1976;84:485–506. doi: 10.1093/genetics/84.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd J. B., Golino M. D., Shaw K. E., Osgood C. J., Green M. M. Third-chromosome mutagen-sensitive mutants of Drosophila melanogaster. Genetics. 1981;97:607–623. doi: 10.1093/genetics/97.3-4.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawley O. W., et al. National Institutes of Health Consensus Development Conference statement: hydroxyurea treatment for sickle cell disease. Ann. Intern. Med. 2008;148:932–938. doi: 10.7326/0003-4819-148-12-200806170-00220. [DOI] [PubMed] [Google Scholar]
- Brodsky M. H., Sekelsky J. J., Tsang G., Hawley R. S., Rubin G. M. mus304 encodes a novel DNA damage checkpoint protein required during Drosophila development. Genes Dev. 2000;14:666–678. [PMC free article] [PubMed] [Google Scholar]
- Buszczak M., et al. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr A. M. DNA structure dependent checkpoints as regulators of DNA repair. DNA Rep. 2002;1:983–994. doi: 10.1016/s1568-7864(02)00165-9. [DOI] [PubMed] [Google Scholar]
- Chowdhury D., Keogh M. C., Ishii H., Peterson C. L., Buratowski S., Lieberman J. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Clark K. A., McKearin D. M. The Drosophila stonewall gene encodes a putative transcription factor essential for germ cell development. Development. 1996;122:937–950. doi: 10.1242/dev.122.3.937. [DOI] [PubMed] [Google Scholar]
- Clemens J. C., Worby C. A., Simonson-Leff N., Muda M., Maehama T., Hemmings B. A., Dixon J. E. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L., Berg C., Kelley R., McKearin D., Spradling A. Identifying and cloning Drosophila genes by single P element insertional mutagenesis. Prog. Nucleic Acid Res. Mol. Biol. 1989;36:99–109. doi: 10.1016/s0079-6603(08)60164-6. [DOI] [PubMed] [Google Scholar]
- Cowell I. G., Sunter N. J., Singh P. B., Austin C. A., Durkacz B. W., Tilby M. J. gammaH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS ONE. 2007;2:e1057. doi: 10.1371/journal.pone.0001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasika G. K., Lin S. C., Zhao S., Sung P., Tomkinson A., Lee E. Y. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene. 1999;18:7883–7899. doi: 10.1038/sj.onc.1203283. [DOI] [PubMed] [Google Scholar]
- de Vries H. I., Uyetake L., Lemstra W., Brunsting J. F., Su T. T., Kampinga H. H., Sibon O. C. Grp/DChk1 is required for G2-M checkpoint activation in Drosophila S2 cells, whereas Dmnk/DChk2 is dispensable. J. Cell Sci. 2005;118:1833–1842. doi: 10.1242/jcs.02309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A., Lein S., Schotta G., Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14:377–392. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- Edgar B. A., Sprenger F., Duronio R. J., Leopold P., O'Farrell P. H. Distinct molecular mechanism regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes Dev. 1994;8:440–452. doi: 10.1101/gad.8.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., James T. C., Foster-Hartnett D. M., Hartnett T., Ngan V., Elgin S. C. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Enoch T., Carr A. M., Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- Escargueil A. E., Soares D. G., Salvador M., Larsen A. K., Henriques J. A. What histone code for DNA repair? Mutat. Res. 2008;658:259–270. doi: 10.1016/j.mrrev.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Evans M. L., Bostelman L. J., Albrecht A. M., Keller A. M., Strande N. T., Thompson J. S. UV sensitive mutations in histone H3 in Saccharomyces cerevisiae that alter specific K79 methylation states genetically act through distinct DNA repair pathways. Curr. Genet. 2008;53:259–274. doi: 10.1007/s00294-008-0182-1. [DOI] [PubMed] [Google Scholar]
- Froget B., Blaisonneau J., Lambert S., Baldacci G. Cleavage of stalled forks by fission yeast mus81/eme1 in absence of DNA replication checkpoint. Mol. Biol. Cell. 2008;19:445–456. doi: 10.1091/mbc.E07-07-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong E. E., Profitt D., Scott M. P. Automated sorting of live transgenic embryos. Nat. Biotechnol. 2001;19:153–156. doi: 10.1038/84422. [DOI] [PubMed] [Google Scholar]
- Gorski M. M., Romeijn R. J., Eeken J. C., de Jong A. W., van Veen B. L., Szuhai K., Mullenders L. H., Ferro W., Pastink A. Disruption of Drosophila Rad50 causes pupal lethality, the accumulation of DNA double-strand breaks and the induction of apoptosis in third instar larvae. DNA Rep. 2004;3:603–615. doi: 10.1016/j.dnarep.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Guarino E., Salguero I., Jimenez-Sanchez A., Guzman E. C. Double-strand break generation under deoxyribonucleotide starvation in Escherichia coli. J. Bacteriol. 2007;189:5782–5786. doi: 10.1128/JB.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari K. L., Santerre A., Sekelsky J. J., McKim K. S., Boyd J. B., Hawley R. S. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell. 1995;82:815–821. doi: 10.1016/0092-8674(95)90478-6. [DOI] [PubMed] [Google Scholar]
- Harris P. V., Mazina O. M., Leonhardt E. A., Case R. B., Boyd J. B., Burtis K. C. Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol. Cell. Biol. 1996;16:5764–5771. doi: 10.1128/mcb.16.10.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Henderson D. S. DNA repair defects and other (mus) takes in Drosophila melanogaster. Methods. 1999;18:377–400. doi: 10.1006/meth.1999.0797. [DOI] [PubMed] [Google Scholar]
- Henderson D. S., Bailey D. A., Sinclair D. A., Grigliatti T. A. Isolation and characterization of second chromosome mutagen-sensitive mutations in Drosophila melanogaster. Mutat. Res. 1987;177:83–93. doi: 10.1016/0027-5107(87)90024-8. [DOI] [PubMed] [Google Scholar]
- Hendricks S. P., Mathews C. K. Differential effects of hydroxyurea upon deoxyribonucleoside triphosphate pools, analyzed with vaccinia virus ribonucleotide reductase. J. Biol. Chem. 1998;273:29519–29523. doi: 10.1074/jbc.273.45.29519. [DOI] [PubMed] [Google Scholar]
- Houtgraaf J. H., Versmissen J., van der Giessen W. J. A concise review of DNA damage checkpoints and repair in mammalian cells. Cardiovasc. Revasc. Med. 2006;7:165–172. doi: 10.1016/j.carrev.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Hurley P. J., Bunz F. ATM and ATR: components of an integrated circuit. Cell Cycle. 2007;6:414–417. doi: 10.4161/cc.6.4.3886. [DOI] [PubMed] [Google Scholar]
- Kim J. A., Kruhlak M., Dotiwala F., Nussenzweig A., Haber J. E. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J. Cell Biol. 2007;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klovstad M., Abdu U., Schupbach T. Drosophila brca2 is required for mitotic and meiotic DNA repair and efficient activation of the meiotic recombination checkpoint. PLoS Genet. 2008;4:e31. doi: 10.1371/journal.pgen.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knibiehler B., Mirre C., Rosset R. Nucleolar organizer structure and activity in a nucleolus without fibrillar centres: the nucleolus in an established Drosophila cell line. J. Cell Sci. 1982;57:351–364. doi: 10.1242/jcs.57.1.351. [DOI] [PubMed] [Google Scholar]
- Kok R. M., Smith D. E., Barto R., Spijkerman A. M., Teerlink T., Gellekink H. J., Jakobs C., Smulders Y. M. Global DNA methylation measured by liquid chromatography-tandem mass spectrometry: analytical technique, reference values and determinants in healthy subjects. Clin. Chem. Lab Med. 2007;45:903–911. doi: 10.1515/CCLM.2007.137. [DOI] [PubMed] [Google Scholar]
- Kurose A., Tanaka T., Huang X., Traganos F., Dai W., Darzynkiewicz Z. Effects of hydroxyurea and aphidicolin on phosphorylation of ataxia telangiectasia mutated on Ser 1981 and histone H2AX on Ser 139 in relation to cell cycle phase and induction of apoptosis. Cytometry A. 2006a;69:212–221. doi: 10.1002/cyto.a.20241. [DOI] [PubMed] [Google Scholar]
- Kurose A., Tanaka T., Huang X., Traganos F., Darzynkiewicz Z. Synchronization in the cell cycle by inhibitors of DNA replication induces histone H2AX phosphorylation: an indication of DNA damage. Cell Prolif. 2006b;39:231–240. doi: 10.1111/j.1365-2184.2006.00380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S., Froget B., Carr A. M. Arrested replication fork processing: interplay between checkpoints and recombination. DNA Rep. 2007;6:1042–1061. doi: 10.1016/j.dnarep.2007.02.024. [DOI] [PubMed] [Google Scholar]
- LaRocque J. R., Jaklevic B., Su T. T., Sekelsky J. Drosophila ATR in double-strand break repair. Genetics. 2007;175:1023–1033. doi: 10.1534/genetics.106.067330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letvin N. L., Linch D. C., Beardsley G. P., McIntyre K. W., Nathan D. G. Augmentation of fetal-hemoglobin production in anemic monkeys by hydroxyurea. N. Engl. J. Med. 1984;310:869–873. doi: 10.1056/NEJM198404053101401. [DOI] [PubMed] [Google Scholar]
- Loizou J. I., Murr R., Finkbeiner M. G., Sawan, Wang Z. Q., Herceg Z. Epigenetic information in chromatin: the code of entry for DNA repair. Cell Cycle. 2006;5:696–701. doi: 10.4161/cc.5.7.2616. [DOI] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C. S., Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- Madigan J. P., Chotkowski H. L., Glaser R. L. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 2002;30:3698–3705. doi: 10.1093/nar/gkf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines J. Z., Park J. K., Williams M., McKearin D. M. Stonewalling Drosophila stem cell differentiation by epigenetic controls. Development. 2007;134:1471–1479. doi: 10.1242/dev.02810. [DOI] [PubMed] [Google Scholar]
- McVey M., Andersen S. L., Broze Y., Sekelsky J. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics. 2007;176:1979–1992. doi: 10.1534/genetics.106.070052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P., Taddei A., Vernis L., Poidevin M., Gasser S. M., Baldacci G. Temporal separation of replication and recombination requires the intra-S checkpoint. J. Cell Biol. 2005;168:537–544. doi: 10.1083/jcb.200410006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels A. A., Nguyen V. T., Konings A. W., Kampinga H. H., Bensaude O. Thermostability of a nuclear-targeted luciferase expressed in mammalian cells. Destabilizing influence of the intranuclear microenvironment. Eur. J. Biochem. 1995;234:382–389. doi: 10.1111/j.1432-1033.1995.382_b.x. [DOI] [PubMed] [Google Scholar]
- Morara S., van der Want J. J., de Weerd H., Provini L., Rosina A. Ultrastructural analysis of climbing fiber-Purkinje cell synaptogenesis in the rat cerebellum. Neuroscience. 2001;108:655–671. doi: 10.1016/s0306-4522(01)00433-x. [DOI] [PubMed] [Google Scholar]
- Nyce J. Drug-induced DNA hypermethylation and drug resistance in human tumors. Cancer Res. 1989;49:5829–5836. [PubMed] [Google Scholar]
- Nyce J., Liu L., Jones P. A. Variable effects of DNA-synthesis inhibitors upon DNA methylation in mammalian cells. Nucleic Acids Res. 1986;14:4353–4367. doi: 10.1093/nar/14.10.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell M. J., Walworth N. C., Carr A. M. The G2-phase DNA-damage checkpoint. Trends Cell Biol. 2000;10:296–303. doi: 10.1016/s0962-8924(00)01773-6. [DOI] [PubMed] [Google Scholar]
- Oshige M., Aoyagi N., Harris P. V., Burtis K. C., Sakaguchi K. A new DNA polymerase species from Drosophila melanogaster: a probable mus308 gene product. Mutat. Res. 1999;433:183–192. doi: 10.1016/s0921-8777(99)00005-1. [DOI] [PubMed] [Google Scholar]
- Ozdemir A., Spicuglia S., Lasonder E., Vermeulen M., Campsteijn C., Stunnenberg H. G., Logie C. Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:25949–25952. doi: 10.1074/jbc.C500181200. [DOI] [PubMed] [Google Scholar]
- Price D., Rabinovitch S., O'Farrell P. H., Campbell S. D. Drosophila wee1 has an essential role in the nuclear divisions of early embryogenesis. Genetics. 2000;155:159–166. doi: 10.1093/genetics/155.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S., Parthun M. R. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell. Biol. 2002;22:8353–8365. doi: 10.1128/MCB.22.23.8353-8365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N., Russell P. Chk1 and Cds 1, linchpins of the DNA damage and replication checkpoint pathways. J. Cell Sci. 2000;113:3889–3896. doi: 10.1242/jcs.113.22.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodesch C., Geyer P. K., Patton J. S., Bae E., Nagoshi R. N. Developmental analysis of the ovarian tumor gene during Drosophila oogenesis. Genetics. 1995;141:191–202. doi: 10.1093/genetics/141.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Sadowski I., Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G., Ebert A., Reuter G. SU(VAR)3-9 is a conserved key function in heterochromatic gene silencing. Genetica. 2003;117:149–158. doi: 10.1023/a:1022923508198. [DOI] [PubMed] [Google Scholar]
- Sekelsky J. J., Brodsky M. H., Burtis K. C. DNA repair in Drosophila: insights from the Drosophila genome sequence. J. Cell Biol. 2000;150:F31–F36. doi: 10.1083/jcb.150.2.f31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer C. D., Cenci G., Thompson B., Stephens G. E., Slawson E. E., Adu-Wusu K., Gatti M., Elgin S. C. The large isoform of Drosophila melanogaster heterochromatin protein 2 plays a critical role in gene silencing and chromosome structure. Genetics. 2006;174:1189–1204. doi: 10.1534/genetics.106.057604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibon O. C., Kelkar A., Lemstra W., Theurkauf W. E. DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat. Cell Biol. 2000;2:90–95. doi: 10.1038/35000041. [DOI] [PubMed] [Google Scholar]
- Sibon O. C., Laurencon A., Hawley R., Theurkauf W. E. The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr. Biol. 1999;9:302–312. doi: 10.1016/s0960-9822(99)80138-9. [DOI] [PubMed] [Google Scholar]
- Sibon O. C., Stevenson V. A., Theurkauf W. E. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature. 1997;388:93–97. doi: 10.1038/40439. [DOI] [PubMed] [Google Scholar]
- Simon J. A., Lis J. T. A germline transformation analysis reveals flexibility in the organization of heat shock consensus elements. Nucleic Acids Res. 1987;15:2971–2988. doi: 10.1093/nar/15.7.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stach D., Schmitz O. J., Stilgenbauer S., Benner A., Dohner H., Wiessler M., Lyko F. Capillary electrophoretic analysis of genomic DNA methylation levels. Nucleic Acids Res. 2003;31:E2. doi: 10.1093/nar/gng002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia-Ramirez J., Eggen B. J., Peral-Rubio M. J., Toledo-Aral J. J., Mandel G. A single zinc finger motif in the silencing factor REST represses the neural-specific type II sodium channel promoter. Proc. Natl. Acad. Sci. USA. 1997;94:1177–1182. doi: 10.1073/pnas.94.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Pol A. N., Gorcs T. Synaptic relationships between neurons containing vasopressin, gastrin-releasing peptide, vasoactive intestinal polypeptide, and glutamate decarboxylase immunoreactivity in the suprachiasmatic nucleus: dual ultrastructural immunocytochemistry with gold-substituted silver peroxidase. J. Comp Neurol. 1986;252:507–521. doi: 10.1002/cne.902520407. [DOI] [PubMed] [Google Scholar]
- Verheyen E., Cooley L. Looking at oogenesis. Methods Cell Biol. 1994;44:545–561. [PubMed] [Google Scholar]
- Ward I. M., Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- Wei D. S., Rong Y. S. A genetic screen for DNA double-strand break repair mutations in Drosophila. Genetics. 2007;177:63–77. doi: 10.1534/genetics.107.077693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xhemalce B., Miller K. M., Driscoll R., Masumoto H., Jackson S. P., Kouzarides T., Verreault A., Arcangioli B. Regulation of histone H3 lysine 56 acetylation in Schizosaccharomyces pombe. J. Biol. Chem. 2007;282:15040–15047. doi: 10.1074/jbc.M701197200. [DOI] [PubMed] [Google Scholar]
- Yamamoto R. R., Axton J. M., Yamamoto Y., Saunders R. D., Glover D. M., Henderson D. S. The Drosophila mus101 gene, which links DNA repair, replication and condensation of heterochromatin in mitosis, encodes a protein with seven BRCA1 C-terminus domains. Genetics. 2000;156:711–721. doi: 10.1093/genetics/156.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.