Protein toxins are among the oldest and most exotic warfare agents.[1] Among these, ribosome-inactivating proteins (RIPs), particularly ricin, represent some of the most poisonous toxins known.[2,3] Ricin is a heterodimeric protein known to be toxic by inhalation and oral and intravenous ingestion. Although it is less lethal than the botulinum toxin, its accessibility and relatively simple isolation from castor beans mean it has potential to become a weapon of choice for bioterrorism.[4] The catastrophic impact of RIPs on eukaryotic cells results from their action on the ribosome, the machinery for protein biosynthesis. Ricin and saporin catalyze the depurination of a specific nucleotide residue on a conserved purine-rich ribosomal RNA sequence known as the α-sarcin/ricin loop (Figure 1).[2,5,6] This N-deglycosylation reaction reduces the affinity of the ribosome for elongation factors that are critical for protein synthesis, thereby resulting in disrupted protein production and ultimately cell death.[7]
Figure 1.
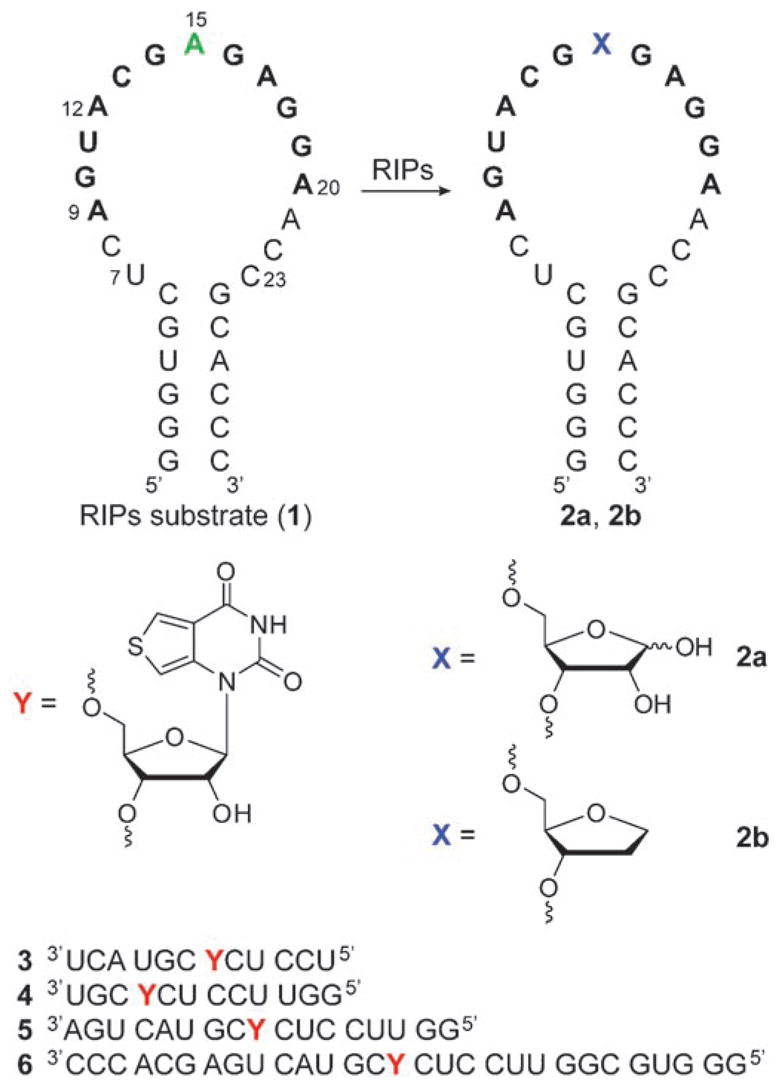
Ribosome inactivating protein mediated depurination of the α-sarcin/ricin hairpin RNA substrate 1 at position A15 (corresponding to A4324 of rat 28S rRNA) yields an RNA product 2 a containing an abasic site. Highlighted residues 9–20 represent the longest conserved RNA sequence in the large ribosomal subunit.[6] RNA 2 b represents a model depurinated RNA product that contains a stable THF residue as an abasic site mimic. Also shown are the chemically synthesized fluorescent probes 3–6, which contain the emissive nucleobase analogue 7, used in this study.
The simple isolation of RIPs from natural sources has alerted the community to the lack of useful field-operable detection tools, as well as the absence of effective antidotes or therapeutics for these proteins. Current approaches to RIP detection rely on polyclonal or monoclonal antibodies that are employed in enzyme-linked immunoadsorbent assays (ELISA).[8,9] These tools are not easily translated to portable devices, and cannot be straightforwardly used for the discovery of inhibitors. We hypothesized that an effective approach, amenable to high-throughput screening and possibly sensor development, could rely on detecting the specific depurinating activity associated with RIPs, which leads to the formation of RNA abasic sites regardless of the identity of the protein.
While the generation of abasic sites in DNA is well documented,[10,11] little is known about their formation, significance, and repair in RNA.[12] Their occurrence appears rather uncommon in cellular RNAs and is almost exclusively associated with the action of RIPs. To advance an effective method to detect RNA depurination by these toxic proteins, we rely on: a) short oligonucleotides comprising the α-sarcin/ricin loop that provide authentic substrates for in vitro applications (Figure 1),[13] and b) a new isomorphic responsive fluorescente nucleoside analogue that signals changes in its microenvironment, particularly the presence of abasic sites (Figure 1). Here we report the use of synthetically modified emissive RNA constructs, complementary to the α-sarcin/ricin loop, that signal the presence of abasic RNA sites by enhanced emission intensity upon hybridization to the depurinated RNA product. We demonstrate the utility of such oligonucleotides to effectively follow the enzymatic activity of RIPs through the use of common fluorescence spectrometric techniques.
As part of our research into the development of fluorescent nucleosides that signal the presence of nucleic acid lesions,[11j] we found that 7 (Scheme 1), a highly emissive analogue based on a thieno[3,4-d]pyrimidine core (φF = 0.48,[14] see Figure S1 and Table S1 in the Supporting Information), signals the presence of abasic RNA sites with significantly enhanced emission. Model studies showed that the short emissive oligonucleotide 11 displayed significant emission quenching upon hybridization to perfect complements, as well as substantial fluorescence enhancement upon hybridization to complementary RNA and DNA oligonucleotides that contain an abasic site opposite the reporter nucleobase (Figure 2). Notably, the probe reports the presence of an abasic site in an RNA–RNA duplex with higher signal enhancement than the corresponding RNA–DNA duplex (Figure 2).[15] This key observation inspired the development of the approach reported here for monitoring the depurination activity of RIPs using fluorescent RNA probes.
Scheme 1.
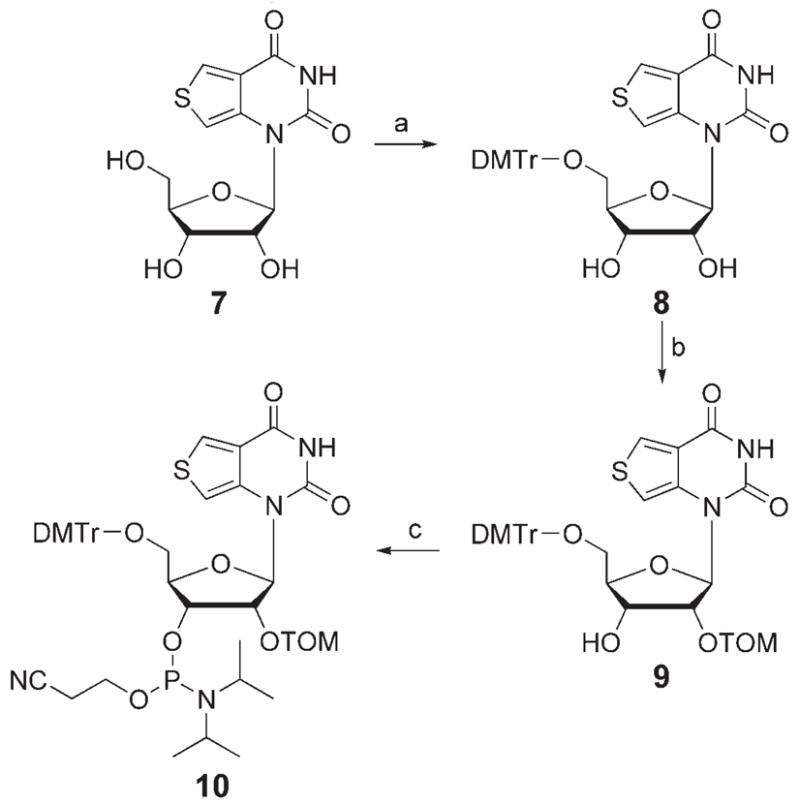
Synthesis of phosphoramidite 10 for solid-phase RNA synthesis. Reagents and conditions: a) DMTrCl, pyridine, RT, 62%; b) 1. nBu2SnCl2, iPr2NEt, DCE, RT; 2. TOMCl, 80 °C; 3. NaHCO3, 39%; c) iPr2NEt, CH2Cl2, iPr2NP(Cl)OEtCN, RT, 67%. TOM = CH2OSi(iPr)3; DMTr =4,4′-dimethoxytrityl; DCE=1,2-dichloroethane.[18] See Ref. [14] for the synthesis of 7.
Figure 2.
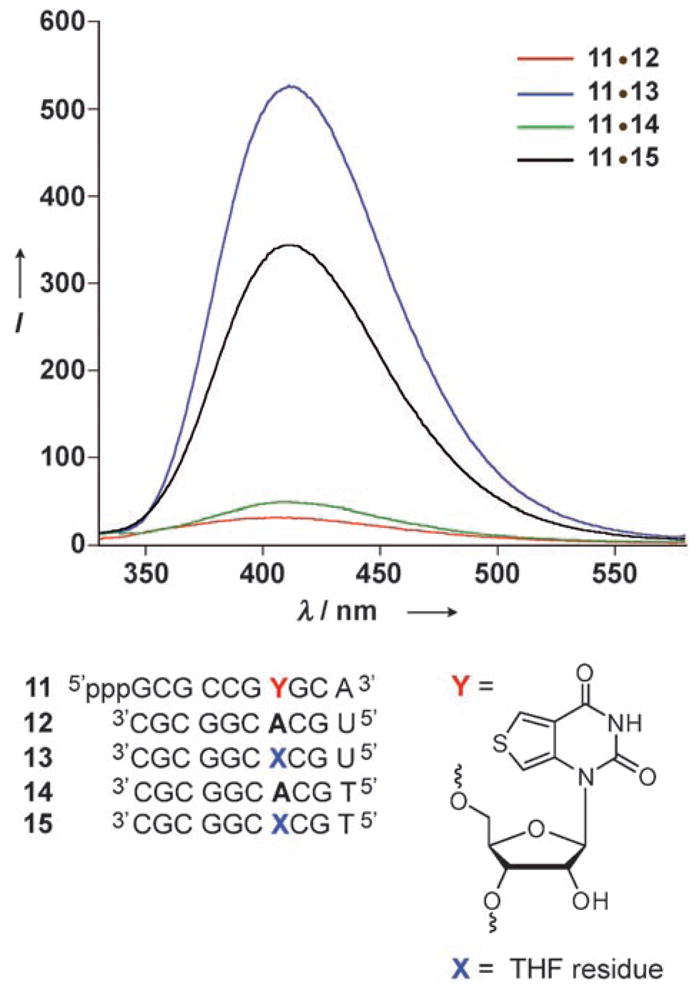
Emission spectra of duplexes obtained upon hybridization of RNA 11[14] to its perfect RNA and DNA complements 12 and 14, respectively, as well as to the corresponding constructs 13 and 15 containing abasic sites. While both the perfect homo- and hetero-duplexes (11·12 and 11·14, respectively) are highly quenched, the duplexes containing a THF residue (a stable abasic-site analogue) are highly emissive, with the RNA·RNA duplex 11·13 showing the highest intensity. Conditions: duplex (1 μM) in cacodylate buffer (20 mM, pH 7.0), NaCl (500 mM), EDTA (0.5 mM), 25 °C, λex = 304 nm. EDTA = ethylenediaminetetraacetic acid.[18]
Four complementary oligoribonucleotides (3–6) that target the conserved α-sarcin/ricin stem–loop domain of ribosomal RNA (rRNA) and place the fluorescent ribonucleoside (7) opposite the common depurination site at A15 were initially designed (Figure 1).[16] Oligonucleotide 3 complements the longest universally conserved sequence (nucleotides 9–20),[16] 4 targets the 3′ end of the loop (12–23), 5 targets the loop region (7–23), and 6 matches the entire stem–loop region (Figure 1). To facilitate the site-specific incorporation of the emissive nucleoside into these singly labeled oligonucleotides 2′-O-TOM-protected phosphoramidite 10 was synthesized and employed in standard solid-phase-synthesis protocols (Scheme 1).[17,18] The integrity of the full-length RNAs and the presence of the fluorescent nucleoside were confirmed by MALDI mass spectrometry (see Figures S2–S5 in the Supporting Information).[18]
To evaluate the effectiveness of the oligonucleotides 3–6 in detecting the presence of abasic sites they were first hybridized to the hairpin RNAs 1 and 2b. RNA 2b serves as a stable model of the actual depurination reaction product 2a, containing a tetrahydrofuran spacer as a surrogate for a D-ribose residue that would be generated upon N-deglycosylation by RIP toxins (Figure 1). When excited at 304 nm, a strong emission at 408 nm is observed for all single-stranded probes 3–6 (Figure 3).[18] Significant quenching is observed upon hybridization to the RIPs substrate 1, with the greatest quenching seen for 1·6.[19] Rewardingly, when probes 3–6 are hybridized to 2b, considerable emission enhancements of 3.5, 1.3, 4.7, and 6.5 times respectively are observed compared to the corresponding perfect duplexes (Figure 3). Thorough biophysical analyses, including thermal denaturation and gel shift experiments,[18] have indicated that oligonucleotide 6, which encompasses the entire stem–loop structure, displayed complete hybridization to both the RIP substrate 1 and its depurinated analogue 2b (see Table S2 and Figure S6 in the Supporting Information). This is in contrast to oligonucleotides 3, 4, and 5, which showed complex hybridization events and multiple product formation.[20] Taken together, these results clearly indicate that the fluorescence enhancement observed for the depurinated duplex 2b·6 over the perfect duplex 1·6 arises from differences in the microenvironment of the fluorescent nucleosides. The emissive oligonucleotide 6 was then selected to probe the enzymatic activity of RIPs.
Figure 3.
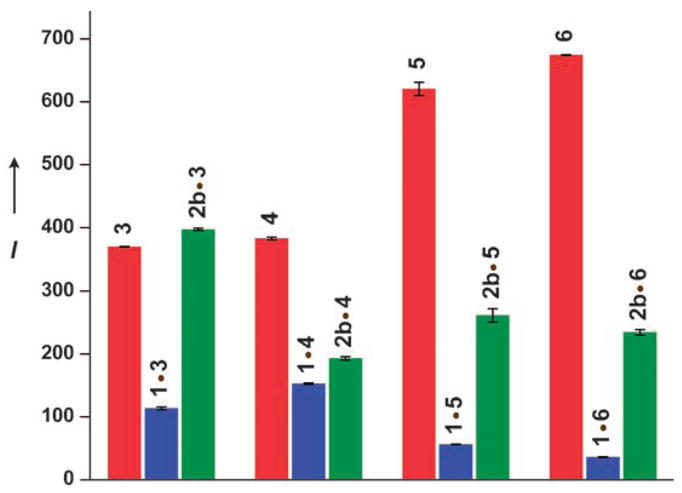
Relative emission intensities of RNA probes 3–6 (red), and when hybridized to RNA 1 (blue) and RNA 2 b (green) in cacodylate buffer (20 mM, pH 7.0), NaCl (100 mM), EDTA (0.5 mM), 25 °C, λex = 304 nm, λem = 408 nm. [18]
We selected saporin as a representative toxin to investigate the N-glycosidase activity of the RIPs, since it exhibits potent depurination activity on synthetic RNAs that contain the α-sarcin/ricin rRNA domain.[21] To identify experimental conditions for fluorescence-based monitoring of toxin-generated RNA abasic sites, saporin-catalyzed depurination reactions were first analyzed by polyacrylamide gel electrophoresis (PAGE). A solution of RIP substrate 1, spiked with 5′-end-radiolabeled 1, was thermally refolded and incubated with saporin in tris(hydroxymethyl)aminomethane (Tris) buffer (30 mM, pH 6.0) at 37°C.[18] Small aliquots of the reaction mixture were taken at short time intervals, quenched, treated with aniline acetate at pH 4.5 (to induce strand cleavage at abasic sites) and resolved by PAGE (Figure 4). The toxin-mediated reaction followed time-dependent saturation kinetics, with the consumption of the substrate RNA 1 and concomitant formation of spliced products (Figure 4, lanes 3–9). Control experiments (lanes 2 and 10) indicate that the enzymatically generated cleaved products are exclusively associated with the formation of abasic sites, which are known to be susceptible to strand scission in the presence of aniline acetate.[22] Sequencing illustrates that saporin activity largely results in the depurination of A15 to predominantly give RNA 2a, thus suggesting that probe 6 would correctly place the reporter nucleoside opposite the main enzymatically generated deglycosylated position (Figure 4).[23]
Figure 4.
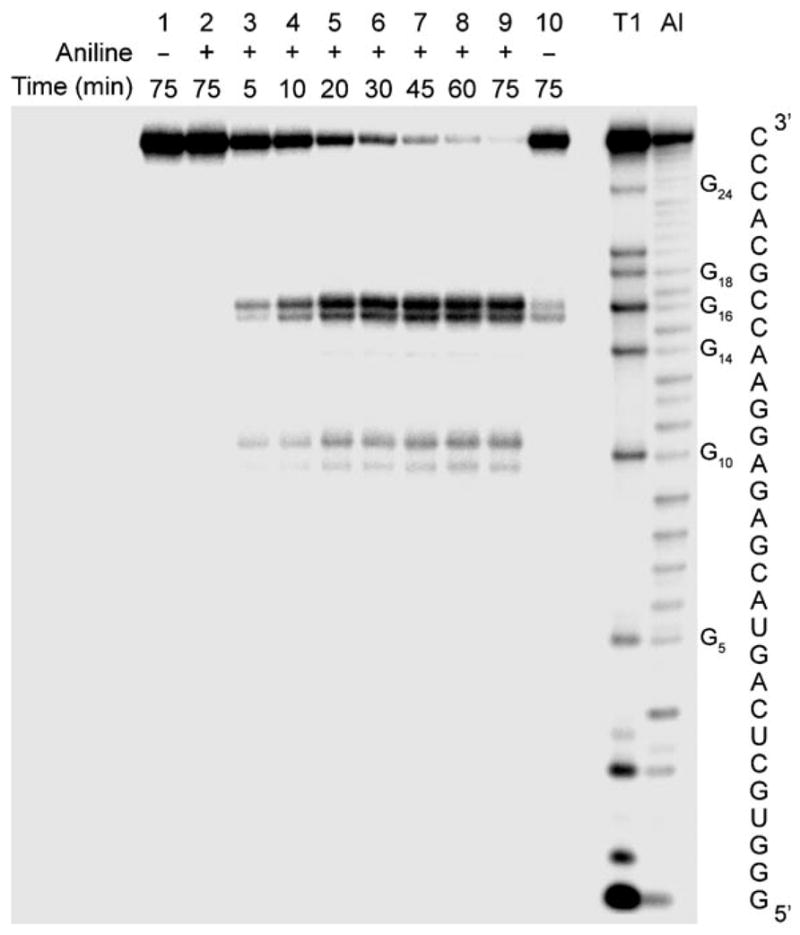
Saporin-catalyzed depurination of RNA 1 (containing trace amounts of 5′-32P-radiolabeled 1) in Tris buffer (30 mM, pH 6.0), NaCl (25 mM), MgCl2 (2 mM), 37 °C. Lanes 1 and 2: RNA substrate 1 with and without aniline acetate treatment, respectively; lanes 3–9: reaction mixtures sampled between 5 and 75 min and treated with aniline acetate; lane 10: reaction duration 75 min without aniline treatment; lanes T1 and Al, RNase T1 and alkaline hydrolysis ladder, respectively. Strand scission with aniline gives rise to two closely migrating products for each of the generated abasic sites, most likely from the generation of the β-eliminated aldehyde and its aniline adduct.[12f, 18]
To test the ability of RNA 6 to monitor RIP-mediated depurination, reactions were set up with unlabeled RNA substrate 1 and saporin as described above. At given time intervals, aliquots were removed and immediately quenched by adding complementary RNA 6 followed by rapid thermal denaturation (90°C) and flash-cooling renaturation (0°C). The emission spectrum of each sample was then recorded (Figure 5).[18] A time-dependent fluorescence increase is clearly observed, which reaches a plateau after 30 minutes. The emissive ribonucleoside 7, when placed opposite a developing abasic site, signals the depurinating activity of saporin with an approximately sixfold overall fluorescence enhancement.[24]
Figure 5.
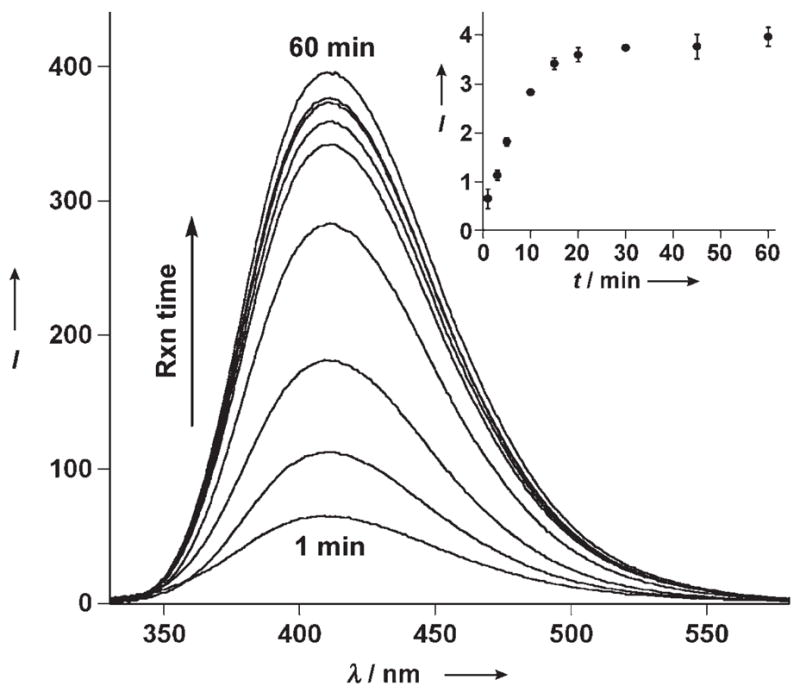
Saporin-catalyzed depurination reaction monitored by fluorescence as a function of time. At regular time intervals, aliquots of the toxin-mediated depurination reaction of RNA 1 were hybridized to RNA 6 (1:0.9 ratio) in Tris buffer (30 mM, pH 6.0), NaCl (100 mM), MgCl2 (2 mM), and emission spectra were recorded (λex = 304 nm). Inset: A plot of intensity at 410 nm versus time depicting saturation kinetics from the formation of the highly emissive 2 a·6.[18]
To confirm that the fluorescence-monitored data represents the same enzymatic transformation directly detected using the radiolabeled RNA substrate, the two data sets were normalized and compared to one another (Figure 6). Apparent rate constants (kap) were determined by fitting the two curves to an exponential rate equation, which gave (2.1 ± 0.3 × 10−3) s−1 and (1.7 ± 0.2 × 10−3) s−1 for the fluorescence-and PAGE-determined depurination reactions, respectively.[25] Despite the distinctly different nature of the two techniques, excellent agreement between the two kinetic profiles and parameters is observed. This correlation clearly indicates that the indirect fluorescence-based sensing of saporin activity detects the same molecular event of RNA depurination as PAGE. Importantly, the fluorescence hybridization method is precise, safer, and far less time consuming than the use of radiolabeled substrates, and approaches a real-time detection of depurination activity.
Figure 6.
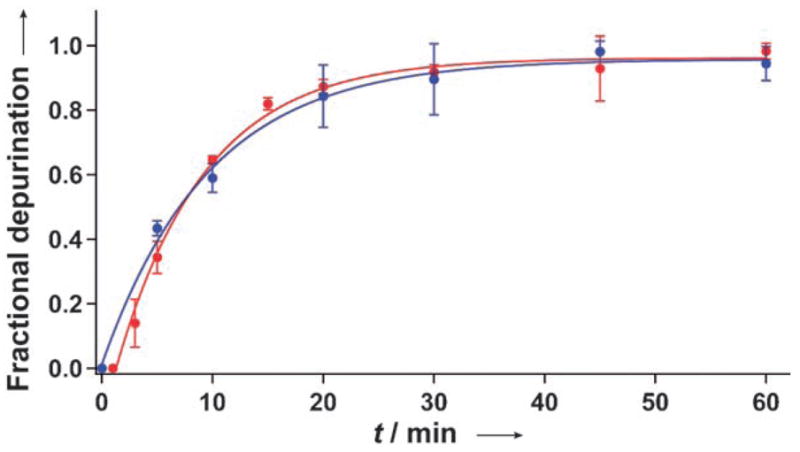
Kinetic profiles of the saporin-mediated depurination reaction of RNA 1 monitored by fluorescence spectroscopy (red) and gel electrophoresis analysis (blue).[18]
In summary, we have developed a simple hybridization assay that can detect the enzymatic activity of toxic ribosome-inactivating proteins. It utilizes an emissive RNA oligonucleotide, complementary to the highly conserved α-sarcin/ricin loop in ribosomal RNA, that contains a new fluorescent nucleobase analogue capable of signaling the presence of abasic RNA sites with considerably enhanced emission. Toxin-mediated RNA depurination is, therefore, directly translated into an increased emission signal in the visible range (410 nm). These results demonstrate the utility of new emissive nucleobase analogues that sense specific nucleic acid lesions. The approach reported here could be extended to the detection of other ribosome-inactivating proteins that act on specific RNA substrates, and could potentially be used for the discovery of RIP inhibitors and antidotes.
Supplementary Material
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.200802199.
Footnotes
We are grateful to the National Institutes of Health (grant number GM069773) for support.
In memory of Tuvia Bercovici
References
- 1.Christopher GW, Cieslak TJ, Pavlin JA, Eitzen EM., Jr J Am Med Assoc. 1997;278:412–417. [PubMed] [Google Scholar]
- 2.a) Nielsen K, Boston RS. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:785–816. doi: 10.1146/annurev.arplant.52.1.785. [DOI] [PubMed] [Google Scholar]; b) Peumans WJ, Hao Q, Van Damme EJM. FASEB J. 2001;15:1493–1506. doi: 10.1096/fj.00-0751rev. [DOI] [PubMed] [Google Scholar]
- 3.No antidotes, inhibitors, therapeutics, or vaccines are currently available to combat ricin poisoning; see: Doan LG. J Toxicol Clin Toxicol. 2004;42:201–208. doi: 10.1081/clt-120030945.Carra JH, Wannemacher RW, Tammariello RF, Lindsey CY, Dinterman RE, Schokman RD, Smith LA. Vaccine. 2007;25:4149–4158. doi: 10.1016/j.vaccine.2007.03.011.Sturm MB, Roday S, Schramm VL. J Am Chem Soc. 2007;129:5544–5550. doi: 10.1021/ja068054h.
- 4.A summary of the NIAID Ricin Expert Panel Workshop can be found at: www3.niaid.nih.gov/Biodefense/ricin_meeting.pdf, April 1 – 2, 2004 Bethesda, MD.
- 5.α-Sarcin inhibits ribosome activity by selectively catalyzing the hydrolysis of a specific phosphodiester linkage on the same conserved sequence.
- 6.a) Endo Y, Wool IG. J Biol Chem. 1982;257:9054–9560. [PubMed] [Google Scholar]; b) Gutell RR, Gray MW, Schnare MN. Nucleic Acids Res. 1993;21:3055–3074. doi: 10.1093/nar/21.13.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Moazed D, Robertson JM, Noller HF. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]; b) Endo Y, Gluck A, Chan YL, Tsurugi K, Wool IG. J Biol Chem. 1990;265:2216–2222. [PubMed] [Google Scholar]; c) Barbieri L, Battelli MG, Stirpe F. Biochim Biophys Acta Rev Biomembr. 1993;1154:237–282. doi: 10.1016/0304-4157(93)90002-6. [DOI] [PubMed] [Google Scholar]
- 8.a) Poli MA, Rivera VR, Hewetson JF, Merrill GA. Toxicon. 1994;32:1371–1377. doi: 10.1016/0041-0101(94)90409-x. [DOI] [PubMed] [Google Scholar]; b) Shyu HF, Chiao DJ, Liu HW, Tang SS. Hybridoma Hybridomics. 2002;21:69–73. doi: 10.1089/15368590252917665. [DOI] [PubMed] [Google Scholar]; c) Dertzbaugh MT, Rossi CA, Paddle BM, Hale M, Poretski M, Alderton MR. Hybridoma. 2005;24:236–243. doi: 10.1089/hyb.2005.24.236. [DOI] [PubMed] [Google Scholar]; d) Guo J, Shen B, Sun Y, Yu M, Hu M. Hybridoma. 2006;25:225–229. doi: 10.1089/hyb.2006.25.225. [DOI] [PubMed] [Google Scholar]; e) Mei Q, Fredrickson CK, Lian W, Jin S, Fan ZH. Anal Chem. 2006;78:7659–7664. doi: 10.1021/ac0610006. [DOI] [PubMed] [Google Scholar]; f) Ler SG, Lee FK, Gopalakrishnakone P. J Chromatogr A. 2006;1133:1–12. doi: 10.1016/j.chroma.2006.08.078. [DOI] [PubMed] [Google Scholar]; g) Lubelli C, Chatgilialoglu A, Bolognesi A, Strocchi P, Colombatti M, Stirpe F. Anal Biochem. 2006;355:102–109. doi: 10.1016/j.ab.2006.05.003. [DOI] [PubMed] [Google Scholar]; h) Fulton RE, Thompson HG. J Immunoassay Immunochem. 2007;28:227–241. doi: 10.1080/15321810701454730. [DOI] [PubMed] [Google Scholar]; i) Keener WK, Rivera WR, Cho CY, Hale ML, Garber EAE, Poli MA. Anal Biochem. 2008;378:87–89. doi: 10.1016/j.ab.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 9.A fluorescence-based activity assay for the formation of abasic site by ricin was recently demonstrated using a short RNA substrate and DNA beacons: Roday S, Sturm MB, Blakaj D, Schramm VL. J Biochem Biophys Methods. 2008;70:945–953. doi: 10.1016/j.jprot.2007.12.010.
- 10.For reviews see: Lindahl T. Nature. 1993;362:709–715. doi: 10.1038/362709a0.Lhomme J, Constant JF, Demeunynck M. Biopolymers. 1999;52:65–83. doi: 10.1002/1097-0282(1999)52:2<65::AID-BIP1>3.0.CO;2-U.Gates KS, Nooner T, Dutta S. Chem Res Toxicol. 2004;17:839–856. doi: 10.1021/tx049965c.Huffman JL, Sundheim O, Tainer JA. Mutant Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012.
- 11.Abasic site detection in DNA: Matray JT, Kool ET. Nature. 1999;399:704–708. doi: 10.1038/21453.Brotschi C, Häberli A, Leumann CJ. Angew Chem. 2001;113:3101–3103. doi: 10.1002/1521-3773(20010817)40:16<3012::AID-ANIE3012>3.0.CO;2-Y.Angew Chem Int Ed. 2001;40:3012–3014. doi: 10.1002/1521-3773(20010817)40:16<3012::AID-ANIE3012>3.0.CO;2-Y.Rachofsky EL, Seibert E, Stivers JT, Osman R, Alexander Ross JB. Biochemistry. 2001;40:957–967. doi: 10.1021/bi001665g.Georgakilas AG, Bennett PV, Sutherland BM. Nucleic Acids Res. 2002;30:2800–2808. doi: 10.1093/nar/gkf393.Hirose T, Ohtani T, Muramatsu H, Tanaka A. Photochem Photobiol. 2002;76:123–126. doi: 10.1562/0031-8655(2002)076<0123:dvoaso>2.0.co;2.JongMin K, Hiroshi M, HeaYeon L, Tomoji K.FEBS Lett 2003555611–615.14675783Yoshimoto K, Nishizawa S, Minagawa M, Teramae N. J Am Chem Soc. 2003;125:8982–8983. doi: 10.1021/ja029786m.Sato K, Greenberg MM. J Am Chem Soc. 2005;127:2806–2807. doi: 10.1021/ja0426185.Valis L, Amann N, Wagenknecht HA. Org Biomol Chem. 2005;3:36–38. doi: 10.1039/b414672g.Greco NJ, Tor Y. J Am Chem Soc. 2005;127:10784–10785. doi: 10.1021/ja052000a.Morita K, Sato Y, Seino T, Nishizawa S, Teramae N. Org Biomol Chem. 2008;6:266–268. doi: 10.1039/b716682f.
- 12.a) Schramm VL. Curr Opin Chem Biol. 1997;1:323–331. doi: 10.1016/s1367-5931(97)80069-5. [DOI] [PubMed] [Google Scholar]; b) Aas PA, Otterlei M, Falnes PO, Vågbø CB, Skorpen F, Akbari M, Sundheim O, Bjørås M, Slupphaug G, Seeberg E, Krokan HE. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]; c) Begley TJ, Samson LD. Nature. 2003;421:795–796. doi: 10.1038/421795a. [DOI] [PubMed] [Google Scholar]; d) Trzupek JD, Sheppard TL. Org Lett. 2005;7:1493–1496. doi: 10.1021/ol050120h. [DOI] [PubMed] [Google Scholar]; e) Küpfer PA, Leumann CJ. ChemBioChem. 2005;6:1970–1973. doi: 10.1002/cbic.200500204. [DOI] [PubMed] [Google Scholar]; f) Küpfer PA, Leumann CJ. Nucleic Acids Res. 2007;35:58–68. doi: 10.1093/nar/gkl948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Glück A, Endo Y, Wool IG. J Mol Biol. 1992;226:411–424. doi: 10.1016/0022-2836(92)90956-k. [DOI] [PubMed] [Google Scholar]; b) Szewczak AA, Moore PB. J Mol Biol. 1995;247:81–98. doi: 10.1006/jmbi.1994.0124. [DOI] [PubMed] [Google Scholar]
- 14.Srivatsan SG, Tor Y. Org Biomol Chem. 2008;6:1334–1338. doi: 10.1039/b801054d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.This observation can potentially be ascribed to the conformation differences between the A form of the RNA–RNA duplex and quasi A form of the RNA–DNA duplex, as the RNA–RNA and RNA–DNA duplexes showed identical stability (Tm = 51.8 and 51.9°C, respectively, in 20 mM cacodylate buffer, 500 mM NaCl, 0.5 mM EDTA, pH 7.0).
- 16.The α-sarcin loop in rRNA is conserved across species from human to bacteria; see: Mehta AD, Boston RS. In: A Look Beyond Transcription: Mechanisms Determining mRNA Stability and Translation in Plants. Bailey-Serres J, Gallie DR, editors. American Society for Plant Physiology; Rockville: 1998. pp. 145–152.
- 17.a) Wu X, Pitsch S. Nucleic Acids Res. 1998;26:4315–4323. doi: 10.1093/nar/26.19.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Pitsch S, Weiss PA. Curr Prot Nucleic Acid Chem. 2001:2.9.1–2.9.14. doi: 10.1002/0471142700.nc0209s07. [DOI] [PubMed] [Google Scholar]; c) Pitsch S, Weiss PA. Curr Prot Nucleic Acid Chem. 2001:3.8.1–3.8.15. doi: 10.1002/0471142700.nc0308s07. [DOI] [PubMed] [Google Scholar]; d) Pitsch S, Weiss PA, Jenny L, Stutz A, Wu X. Helv Chim Acta. 2001;84:3773–3795. [Google Scholar]; e) Micura R. Angew Chem. 2002;114:2369–2373. [Google Scholar]; Angew Chem Int Ed. 2002;41:2265–2269. doi: 10.1002/1521-3773(20020703)41:13<2265::AID-ANIE2265>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 18.See the Supporting Information for experimental details.
- 19.Similar quenching effects in perfect duplexes have been previously reported for various fluorescent nucleobases and can be ascribed to partial stacking of the chromophore with other bases, quenching of fluorescence by neighboring bases in the excited state, or both; see: Jean JM, Hall KB. Proc Natl Acad Sci USA. 2001;98:37–41. doi: 10.1073/pnas.011442198.Liu C, Martin CT. J Mol Biol. 2001;308:465–475. doi: 10.1006/jmbi.2001.4601.
- 20.Hybridization of short oligonucleotides to hairpin structures is complex, and has been investigated; see, for example: Lima WF, Monia BP, Ecker DJ, Freier SM. Biochemistry. 1992;31:12055–12061. doi: 10.1021/bi00163a013.Malygin A, Karpova G, Westermann P. FEBS Lett. 1996;392:114–116. doi: 10.1016/0014-5793(96)00798-3.Petyuk VA, Serikov RN, Vlassov VV, Zenkova MA. Nucleosides Nucleotides Nucleic Acids. 2004;23:895–906. doi: 10.1081/NCN-200026038.
- 21.Tang S, Hu RG, Liu WY, Ruan KC. Biol Chem. 2000;381:769–772. doi: 10.1515/BC.2000.098. [DOI] [PubMed] [Google Scholar]
- 22.Lindahl T, Andersson A. Biochemistry. 1972;11:3618–3623. doi: 10.1021/bi00769a019. [DOI] [PubMed] [Google Scholar]
- 23.A small amount of cleavage is also seen at A9 from depurination at this site. It should not interfere with the fluorescence-monitored hybridization experiments.
- 24.Importantly, saporin did not have any apparent effect on the fluorescence of RNA 6 under the above conditions (see Figure S8 in the Supporting Information).
- 25.Although the reaction was not performed under strictly pseudo first-order conditions, a molar ratio of 2:1 (saporin/RNA substrate) yielded kinetic profiles that were analyzed using an exponential correlation with Igor Pro 6.0 software.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.200802199.


