Abstract
Breast carcinogenesis involves genetic and epigenetic alterations that cause aberrant gene function. Recent progress in the knowledge of epigenomics has had a profound impact on the understanding of mechanisms leading to breast cancer, and consequently the development of new strategies for diagnosis and treatment of breast cancer. Epigenetic regulation has been known to involve three mutually interacting events – DNA methylation, histone modifications and nucleosomal remodeling. These processes modulate chromatin structure to form euchromatin or heterochromatin, and in turn activate or silence gene expression. Alteration in expression of key genes through aberrant epigenetic regulation in breast cells can lead to initiation, promotion and maintenance of carcinogenesis, and is even implicated in the generation of drug resistance. We currently review known roles of the epigenetic machinery in the development and recurrence of breast cancer. Furthermore, we highlight the significance of epigenetic alterations as predictive biomarkers and as new targets of anticancer therapy.
Keywords: breast cancer, CpG islands, demethylating agents, DNA methylation, epigenetics, histone deacetylase inhibitors, histone modifications, nucleosomal rem odeling
Breast cancer is among the most frequently diagnosed neoplasias and the second leading cause of cancer death among American women. Generally, cancer has been viewed as a disease that is driven by progressive genetic abnormalities, involving mutations in oncogenes and tumor suppressor genes, and chromosomal abnormalities [1,2]. However, it has been shown that breast cancer, similar to other types of cancer, is also a disease that is driven by epigenetic alterations, which do not affect the primary DNA sequence [3,4]. The outcome of these alterations is aberrant transcriptional regulation that results in a change in expression patterns of genes implicated in cellular proliferation, survival and differentiation [3,5,6]. Epigenetic alterations occur at the chromosomal level in transformed cells. These involve changes in DNA methylation and histone modifications, and altered expression and function of factors implicated in regulating assembly and remodeling of nucleosomes [5-9]. Alterations in DNA methylation include global hypomethyation and focal hypermethylation. Global hypomethylation has been found to increase with age and is linked to genomic instability and activation of oncogene expression [10-12]. By contrast, gene-locus-specific hypermethylation can lead to the transcriptional silencing of tumor suppressor genes [3,5-9]. In addition to DNA methylation, post-translational histone modification is another epigenetically regulated mechanism that can modulate chromatin structure to regulate gene expression [5-8,13]. DNA methylation is often associated with some specific types of histone modifications that can cooperatively affect chromatin structure to silence gene expression [6,8,14,15]. In addition, current work on identifying and studying regulators that control nucleosomal remodeling has deciphered that some of them are also involved in regulation of DNA methylation and histone modifications [5,7-9,13,15]. Therefore, three epigenetic events, DNA methylation, histone modifications and nucleosomal remodeling, mutually interact with each other to regulate gene-expression. The effort to elucidate molecular events in chromatin regulation that initiate and maintain epigenetic gene silencing and oncogene activation in cancer cells, one can envision, is very important for the translation of epigenomics to clinical application. Here, we provide an overview of the current understanding of the contribution of epigenetic alterations to breast tumorigenesis, recent advances in genome-scale technologies aimed at revealing epigenetic alterations in breast cancer, and the current progress in translating this profiling knowledge into diagnosis, prognosis and therapy of breast cancer.
The epigenome in breast cancer
■ DNA methylation
DNA methylation is one of the three known layers of epigenetic control of germline- and tissue-specific gene expression. Hypermethylation plays an integral role in genomic imprinting wherein one of the two parental alleles of a gene is silenced in order to establish monoallelic expression; X-chromosome inactivation in females occurs through a similar imprinting mechanism [16,17]. Stated simply, DNA methylation is a heritable, epigenetic change that alters gene expression, and is confined to the addition of a methyl group to the 5-carbon position of cytosine in a CpG dinucleotide. In the vertebrate genome, CpG dinucleotide sequences have been severely depleted to approximately 20% of the predicted frequency during evolution, and among the remaining CpG dinucleotides, over 70% are methylated [3]. A study of the human genome revealed that the distribution of CpG dinucleotides is not random, and some of them cluster together to form CpG-rich DNA regions called CpG islands. CpG islands are mostly located in the upstream promoter and exon 1 region of over half of human genes [18]. In normal cells, CpG islands in actively expressed genes are unmethylated. However, during neoplastic transformation, DNA methylation in cancer cells exhibits inverse profiles compared with normal cells; focal hypermethylation of CpG islands of 5′-end regions of many genes is observed [5-8]. Thus, a change in DNA methylation profile is a hallmark of almost all human cancers, including breast cancer.
DNA methylation is mediated by DNA methyltransferases (DNMTs) that catalyze the transfer of the methyl group from S-adenosyl l-methionine (SAM) to the cytosine in CpG dinucleotide [19]. To date, the known DNMTs are DNMT1, DNMT2, DNMT3a, DNMT3b and DNMT3L. The maintenance of established methylation patterns in hemimethylated genes is mediated by DNMT1 that copies methylation patterns from the parent strand to the daughter [20]. De novo DNA methylation is catalyzed by DNMT3a, DNMT3b and DNMT3L [21,22]. DNMT3L lacks the ability to bind to SAM, and is responsible for increasing the binding of DNMT3a to SAM [22,23]. DNMT2, a small 391-amino-acid protein, is reported to possess weak DNA methyltransferase activity, but its biological function is not yet elucidated [24]. Very recent studies have shown that Dicer-mediated microRNA biogenesis is involved in modulation of DNA methylation by indirectly regulating the expression of DNMT3 genes [25,26]. Dicer belongs to the RNase III family enzymes and is implicated in processing the biosynthesis of small interfering RNAs (siRNAs) and microRNAs (miRNAs) [27]. In Dicer−/− cells, the microRNAs of the miR-290 cluster are depleted and expression levels of their target Rbl2 protein (retinoblastoma-like protein) are increased, leading to downregulation of DNMT3 gene expression through Rbl2-mediated transcriptional repression, and in turn causing the DNA methyl ation defect (global hypomethylation) [25,26]. Regarding the role of DNMTs in breast tumori genesis, it has been reported that DNMT3b mRNA is overexpressed in breast cancer, a finding that correlates well with the hypermethylator phenotype and poor prognosis in breast tumors [28,29].
In normal cells, repetitive genomic sequences (e.g., centromeric satellite α-DNA and juxtacentromeric satellite DNA) are heavily methylated [6,7]. The maintenance of methylation in this repetitive DNA could be important for the protection of chromosomal integrity by preventing chromosomal rearrangements, translocations and gene disruption through the reactivation of transposable elements [7,11,30]. Besides hypermethylation of gene-associated CpG islands, hypomethylation of repetitive genomic DNA has also been identified as a specific feature in human cancers [7,31,32]. Although less well studied than DNA hypermethylation, several lines of investigation indicate that the global DNA hypomethylation identified in cancer cells might contribute to structural changes in chromosomes, loss of imprinting (LOI), micro satellite and chromosome instability through aberrant DNA recombination, aberrant activation of proto-oncogene expression and increased mutagenesis [7,11,33,34]. Global genomic hypomethylation in breast cancer has been known to correlate with some clinical features such as disease stage, tumor size and histological grade [35]. Some proto-oncogenes implicated in proliferation and metastasis (e.g., synuclein γ and urokinase genes) or drug resistance to endocrine therapy (e.g., N-cadherin, ID4, annexin A4, β-catenin and WNT11 genes) have been found to be upregulated in breast cancer through the hypomethylation of their promoters [36-38].
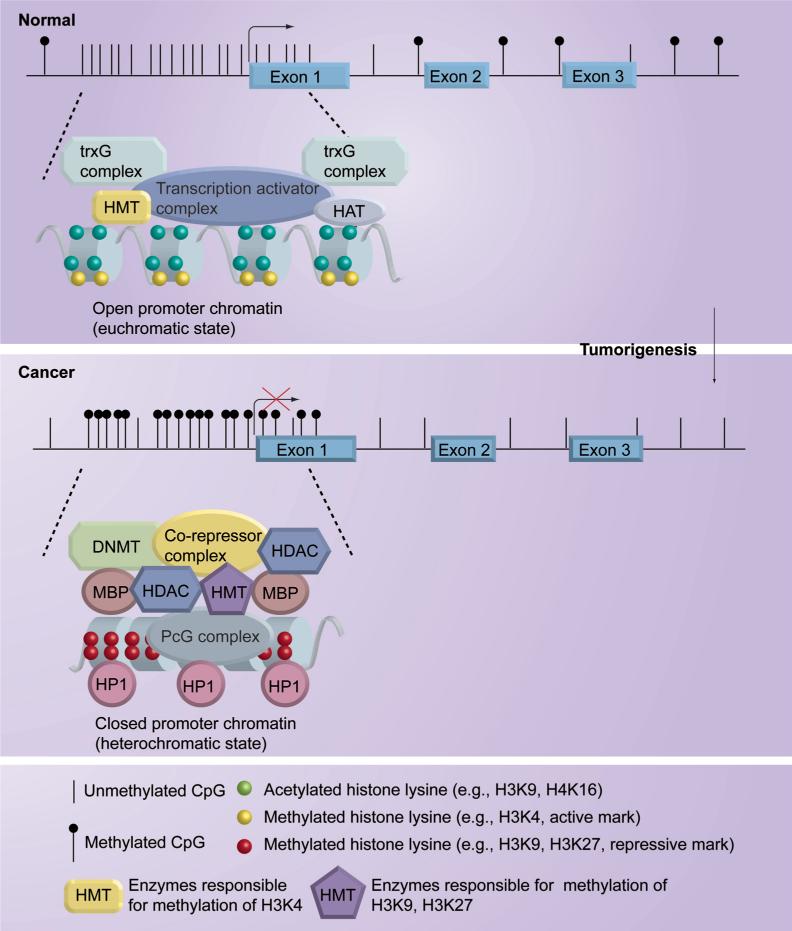
CpG-island-containing gene promoters are usually unmethylated in normal cells to maintain euchromatic structure, which is the transcriptionally active conformation allowing gene expression. However, during cancer development, many of these genes are hypermethylated at their CpG-island-containing promoters to inactivate their expression by changing open euchromatic structure to compact heterochromatic structure (Figure 1) [5-7,9,14,15]. These genes are selectively hypermethylated in tumorigenesis for inactivation owing to their functional involvement in various cellular pathways that prevent cancer formation. Some of the methylated genes identified in human cancers are classic tumor suppressor genes in which one mutationally inactivated allele is inherited. According to Knudson's two-hit model, complete inactivation of a tumor suppressor gene requires loss-of-function of both gene copies [39]. Epigenetic silencing of the remaining wild-type allele of the tumor suppressor gene, thus, can be considered as the second hit in this model. For example, some well-known tumor suppressor genes, such as p16INK4a, APC and BRCA1, that are mutationally inactivated in the germline occasionally lose function of the remaining functional allele in breast epithelial cells through DNA hypermethylation [40-42]. Since the consequence of aberrant DNA methylation is transcriptional silencing, novel tumor suppressor genes can be identified using methylated CpG islands as a marker. Hypermethylated genes identified from breast neoplasms now form a long list (summarized in Table 1). Their biological functions encompass cell cycle regulation (p16INK4a, p14ARF, 14−3−3σ, cyclin D2, p57KIP2), apoptosis (APC, DAPK1, HIC1, HOXA5, TWIST, TMS1), DNA repair (GSTP1, MGMT, BRCA1), hormone regulation (ERα, PR), cell adhesion and invasion (CDH1, APC, TIMP3), angiogenesis (maspin, THBS1), cellular growth-inhibitory signaling (RARβ, RASSF1A, SYK, TGFβRII, HIN1, NES1, SOCS1, SFRP1, WIF1), and so on. In addition to protein-coding genes, recent studies showed that microRNAs with tumor-suppressor function could be silenced in breast cancer cells through DNA methylation [43]. These breast-genome methylation patterns have been developed as biomarkers for early detection and the classification of subtype of breast tumors, as predictors for risk assessment and for monitoring prognosis, and as indicators of susceptibility or response to therapy [3]. These advances in the knowledge of the breast methylome strongly indicate that DNA hypermethylation plays a crucial role in initiation, promotion and maintenance of breast carcinogenesis, which cooperatively and synergistically interact with other genetic alterations to promote the development of breast cancer. For example, human mammary epithelial cells (HMECs) that gained the ability to emerge from the first transient growth plateau lost p16INK4A expression concurrently with hypermethylation of p16INK4A promoter, indicating that loss of tumor-suppressor function of p16INK4A is required for HMECs to gain growth competency by successfully bypassing the stage of cell senescence [3,44]. This finding is consistent with other studies where the life span of stem cells could be extended by germline loss of this gene [45]. Deregulation of cell cycle control by inhibiting the function of the cyclin-dependent kinase inhibitor, p16INK4A, could create a context for facilitating early abnormal clonal expansion of cells at risk for cancer. It is believed that loss of p16INK4A gene is permissive for enabling such expanding cells to develop genomic instability [46] and further epigenetic gene-silencing events [47]. In addition to cell-cycle regulatory genes, DNA methylation-mediated silencing of DNA repair genes, such as BRCA1 and MGMT, could result in further inactivation of tumor suppressor genes or activation of oncogenes, which further drive breast tumorigenesis [48]. More recently, the genes that function as inhibitors of WNT oncogenic pathway, such as SFRP1 and WIF1, have been found to be frequently hypermethylated in primary breast tumors [49,50]. Thus, in addition to the genetic mutation-mediated mechanism, epigenetic gene silencing is another mechanism that fosters malignant transformation of the mammary gland by aberrantly activating oncogenic signaling pathways.
Figure 1. DNA methylation, histone modifications and chromatin remodeling in normal and cancer cells.
DNMT: DNA methyltransferases; HAT: Histone acetyltransferases; HDAC: Histone deacetylases; HMT: histone methyltransferases; HP1: Heterochromatin protein 1; MBP: Methyl-CpG binding proteins; PcG: Polycomb group; trxG: Trithorax group.
Table 1.
Genes hypermethylated in breast cancer.
| Gene | Description | Function | Ref. |
|---|---|---|---|
|
14−3−3σ |
Stratifin |
Cell cycle regulation |
[195,196] |
|
APC |
Adenomatous polyposis of the colon |
Inhibitor of β-catenin |
[41,197] |
|
B4GALT1 |
β-1,4-galactosyltransferase I |
An enzyme involved in glycoconjugate biosynthesis |
[141] |
|
BRCA1 |
Breast cancer 1 |
DNA repair and recombination, transcriptional regulation |
[42] |
|
CCND2 |
Cyclin D2 |
Cell cycle regulation |
[198] |
|
CDH1 |
E-cadherin |
Cell adhesion |
[199] |
|
CDH13 |
H-cadherin |
Cell adhension |
[200] |
|
CDK10 |
Cyclin-dependent kinase 10 |
Regulates cell cycle progression and signaling transduction |
[122] |
|
DAPK1 |
Death-associated protein kinase 1 |
Involved in induction of apoptosis |
[201] |
|
ER |
Estrogen receptor α and β |
Involved in transduction of estrogen signaling |
[3,119,120] |
|
FHIT |
Fragile histidine triad gene |
Modulates cell proliferation and apoptosis, tumor suppression |
[202] |
|
GPC3 |
Glypican 3 |
Involved in control of cell division and growth regulation |
[203] |
|
GSTP1 |
Glutathione-S-tranferase P1 |
Carcinogen detoxification |
[204] |
|
HIC-1 |
Hypermethylated in cancer 1 |
Transcriptional regulation |
[205] |
|
HIN1 |
Cytokine high in normal-1; secretoglobin |
Suppresses cell growth |
[206] |
|
KIF1A |
Kinesin family member 1A |
An anterograde motor protein that transports membranous organelles along axonal microtubules |
[141] |
|
MGMT |
O-6-methylguanine-DNA methyltransferase |
DNA repair of O6-alkyl-guanine |
[48] |
|
NES1 |
Kallikrein-related peptidase 10 |
Tumor suppression |
[207] |
|
NISCH |
Nischarin; imidazoline receptor candidate |
Candidate for the I1-imidazoline receptor |
[141] |
|
Nm23-H1 |
Metastasis inhibition factor NM23 |
Suppressor of metastasis |
[208] |
|
NOEY2 |
Ras homolog gene family member I |
Suppresses clonogenic growth and regulates cell cycle |
[209] |
|
NORE1 |
Ras association (RalGDS/AF-6) domain family member 5 |
Regulation of lymphocyte adhesion and suppression of cell growth in response to activated Rap1 or Ras |
[210] |
|
OGDHL |
Oxoglutarate dehydrogenase-like |
Conversion of 2-oxoglutarate (α-ketoglutarate) to succinyl-Coenzyme A and CO2 during the Krebs cycle |
[141] |
|
p16INK4a |
Cyclin-dependent kinase inhibitor 2A |
Cell cycle regulation, involved in senescence |
[3] |
|
PAK3 |
p21 protein (Cdc42/Rac)-activated kinase 3 |
Cytoskeleton reorganization and nuclear signaling |
[141] |
|
PR |
Progesterone receptor |
Growth regulation |
[211] |
|
RARβ |
Retinoic acid receptor β |
Regulates apoptosis, proliferation and differentiation |
[157,158] |
|
RASSF1A |
Ras association domain family protein 1 |
Functions as tumor suppressor and inhibits tumor formation |
[212] |
|
RIZ1 |
Retinoblastoma protein-binding zinc finger protein |
Transcriptional regulation |
[213] |
|
RUNX3 |
Runt-related transcription factor 3 |
Transcriptional regulation |
[214] |
|
SERPINB5 |
Serpin peptidase inhibitor; Maspin |
Inhibitor of angiogenesis |
[215] |
|
SFRP1 |
Secreted frizzled-related protein 1 |
Inhibitor of Wnt signaling |
[49] |
|
SOCS1 |
Suppressor of cytokine signaling 1 |
Inhibitor of JAK-STAT signaling pathway |
[3] |
|
SRBC |
Sdr-related gene product that binds to c-kinase |
BRCA1-binding protein |
[216] |
|
SYK |
Spleen tyrosine kinase |
Inhibits tumor growth and metastasis |
[217] |
| TGFβRII |
Transforming growth factor β receptor II |
Cell cycle regulation |
[3] |
|
TIMP3 |
Tissue inhibitor of metalloproteinase 3 |
Suppresses tumor growth, angiogenesis and metastasis |
[218] |
|
TMS1 |
Target of methylation-induced silencing-1 |
Involved in apoptosis |
[219] |
|
TWIST |
TWIST homolog of Drosophila |
Involved in p53-mediated cell death |
[147] |
| WIF1 | WNT inhibitory factor 1 | Inhibitor of Wnt signaling | [50] |
■ Connections between DNA methylation, histone modifications & nucleosomal remodeling
Gene-silencing through DNA methylation is tightly associated with chromatin modifications mediating the packaging of DNA. Thus, knowledge of how chromatin structure is organized and maintained is key to understanding the origins of epigenetic alterations in cancer. The basic building unit of chromatin is the nucleosome, a protein–DNA complex structure composed of an octamer of four core histone proteins (H2A, H2B, H3 and H4) that is wrapped around by a 147-bp stretch of DNA [51]. Core histones are subject to a variety of covalent modifications including acetylation, methylation, phosphorylation, ubiquitylation and sumoylation [52-54], which are critically implicated in regulation of chromatin structure and gene expression. Each histone modification is a unique mark to show the status of chromatin structure (active or repressive). Acetylation of histone lysines has been known to be associated with open chromatin structure and active transcription; methylation of these residues is associated with either active or repressive states of chromatin architecture and transcription depending on the modified site [13,53]. For example, the promoters of transcription-active genes are associated with active histone marks, such as acetylation at lysine 9 (K9) of H3 as well as K5, K8, K12 and K16 of H4 and methylation at K4 of H3 (H3K4me), which are involved in a loosening of chromatin structure (euchromatic state) [6,53]. By contrast, when genes are silenced in cancer cells through hypermethylation of their promoters, these active histone marks are replaced by repressive histone marks, including mono-, di- and tri-methylation of histone H3 lysine 9 (H3K9), H3K27 and H4K20, that are implicated in initiating and maintaining closed chromatin structure (heterochromatic state) [5,6,8,13,14]. The mechanisms underlying histone-modification-mediated chromatin remodeling and control of gene transcription have been known to involve the binding of chromatin-associated proteins to these histone modified sites, including bromodomain-containing proteins that recognize acetylated lysine residues and chromodomain-containing proteins that recognize methylated lysine residues [13,55,56].
The dynamics of histone acetylation is balanced by enzymatic actions of histone acetyltransferases (HATs) and histone deacetylases (HDACs), and histone methylation by histone methyltransferases (HMTs) and histone demethylases [6,13,57-59]. Histone modifications are further characterized by the presence of mono-, di-, and tri- forms of lysine methylation, which are catalyzed by various HMTs [57,60]. Some of these histone modification enzymes have been recognized as components of nucleosomal remodeling complexes, which work together to regulate chromatin structure and gene expression [8,13,60].
In humans, 18 HDACs have been identified that are subdivided into four classes (summarized in Table 2) based on their homology to yeast HDACs, their subcellular localization, and their enzymatic activities [61-63]. Class I HDACs with homology to yeast Rpd3 (reduced potassium deficiency 3), including HDAC-1, -2, -3 and -8, are generally localized in nucleus and associate with various transcriptional repressors and cofactors to form protein complexes. It has been found that HDAC1 and HDAC2 can associate with corepressor complexes such as nucleosome remodeling and histone deacetylase (NuRD), Sin3A and CoREST [64-66]; whereas another class 1 HDAC enzyme, HDAC3, is found to associate with nuclear receptor corepressor (NCoR) and silencing mediator for retinoid and thyroid hormone receptors (SMRT) core-pressor complexes [67]. Importantly, it has been found that DNMTs can be recruited by class I HDACs and function as corepressors [68-70]. Class II HDACs are larger proteins homologous to the yeast histone deacetylase 1 (Hda1) and shuttle between the cytoplasm and the nucleus. Based on the protein structure, class II HDACs are further subdivided into class IIa with one catalytic domain (HDAC-4, -5, -7 and -9) and class IIb with two catalytic domains (HDAC-6 and -10). The HDAC proteins of class I and II contain a hydrophobic catalytic pocket domain that requires a pivotal zinc ion to stabilize its domain structure to allow the insertion of a lysine side chain. Class III HDACs are composed of homologs of yeast silencing information regulator 2 (Sir2) and their enzymatic activities require nicotinamide adenine dinucleotide (NAD+) as a cofactor. The newly identified HDAC11 is the sole enzyme categorized as the class IV HDACs. HDAC11 has no significant similarity to both class I and II HDACs except for its catalytic core domain [71]. For the role of HDACs in carcinogenesis, it has been shown that global hypoacetylation of histone H4 is a common hallmark of human cancers. Aberrant alterations in H4 acetylation, preferentially at K16, occur early in the tumorigenic process, indicating that HDACs may be engaged in abnormal post-translational modification of H4 [72]. In addition to histones, many nonhistone proteins have been identified to be the substrates of HDACs, such as proteins involved in transcription (p53, p73, E2F1, STAT1, STAT3, GATA1, YY1, HMGB1, and NF-κB), hormone response (AR, ERα, GR), nuclear transport (importin-α7), cytoskeletal structure (α-tubulin), DNA repair (Ku70), DNA architecture (WRN helicase), WNT signaling (β-catenin) and heat shock/chaperone response (HSP90) [61,63,73-79]. The broad spectrum of HDAC substrates indicates the complexity of HDAC functions to regulate the functions of substrate proteins and gene expression.
Table 2.
Classification and characteristics of histone deacetylases.
| Class type | Class I | Class IIa | Class IIb | Class III | Class IV |
|---|---|---|---|---|---|
| Human HDAC |
HDAC-1, -2, -3, -8 |
HDAC-4, -5, -7, -9 |
HDAC-6, -10 |
SIRT1−7 |
HDAC11 |
| Yeast homolog |
RPD3 |
HDA1 |
HDA1 |
SIR2 |
RPD3 |
| Subcellular localization |
Nucleus |
Nucleus/cytoplasm |
Mostly cytoplasm |
Nucleus |
Nucleus/cytoplasm |
| Tissue distribution |
Ubiquitous |
Brain, heart, skeletal muscle |
Liver, Kidney, spleen, pancreas |
Unknown |
Brain, heart, skeletal muscle, kidney |
| Substrates (partial list) |
Histones, p53, E2F1, STAT1, STAT3, NF-κB, GATA1, YY1, MyoD, SHP, RelA, MEF2D, AR, GR |
Histones, GATA1, HP-1, SMAD7, PLAG1, PLAG2, GCMa |
Histones, tubulin, HSP90, SHP, SMAD7 |
Histones, tubulin, p53 |
Unknown |
| Associated protein complexes (partial list) |
NuRD, SIN3, CoREST, NCoR, SMRT |
NCoR, SMRT |
|
PcG (PRC4) |
|
| Binding partners (partial list) |
RB, BRCA1, DNMT1, DNMT3a, DNMT3b, MBD2−3, MeCP2, ATM |
HIF-1α, BCL-6, actinin, AR, ER, MEF2 |
RUNX2 |
|
|
| Cofactor | Zn | Zn | Zn | NAD+ | Zn |
AR: Androgen receptor; ATM: Ataxia telangiectasia mutated; BCL-6: B-cell lymphoma 6; BRCA1: Breast cancer 1, early onset; CoREST: Corepressor for element-1-silencing transcription factor; DNMT1: DNA methyltransferase 1; ER: Estrogen receptor; GATA1: GATA binding protein 1; GCMa: Glial cells missing homolog α; GR: Glucocorticoid receptor; HDAC1: Histone deacetylase 1; HIF-1a: Hypoxia-inducible factor-1α; HP-1: Heterochromatin protein 1; HSP90: Heat shock protein 90; MBD2−3: Methyl-CpG binding domain 2−3; MeCP2: Methyl-CpG binding protein 2; MEF2: Myelin expression factor 2; MEF2D: Myocyte enhancer factor 2D; MyoD: Myogenic differentiation 1; NCoR: Nuclear receptor corepressor; NuRD: Nucleosome remodelling and histone deacetylase; PcG: Polycomb group; PLAG1−2: Pleiomorphic adenoma gene 1−2; RB: Retinoblastoma; RelA: v-rel reticuloendotheliosis viral oncogene homolog A; RPD3: Reduced potassium deficiency 3; RUNX2: Runt-related transcription factor 2; SHP: Small heterodimer partner; SIN3: SWI-independent 3; SIR2: Silencing information regulator 2; SMAD7: Mothers against decapentaplegic homolog 7; SMRT: Silencing mediator for retinoid and thyroid hormone receptors; STAT-1 and -3: Signal transducers and activators of transcription-1 and -3; YY1: YIN-YANG-1 transcription factor.
For histone methylation, the modifications are regulated in an even more complicated manner than histone acetylation, involving a large number of chromosomal remodeling regulatory complexes. The dimethyl- and trimethyl-H3K4 modifications (an active histone mark) have been reported to be catalyzed by the Trithorax group of histone methyltransferases, such as SET1 and MLL [13,80,81]. It has been shown recently that Chd1, a component of Spt-Ada-Gcn5-acetyltransferase (SAGA) complex with HAT activity, interacts specifically and directly with methylated H3K4 to modulate chromatin structure to the euchromatic state [82]. Trithorax group factors have long been known to be implicated in the transcriptional activation of developmental regulatory genes and their actions are balanced by the opposing effects of the Polycomb group (PcG) factors [83]. In an analogous manner, PcG proteins also participate in gene silencing in normal and cancer cells by modulating methylation modifications of H3K27 (a repressive histone mark) [13,84,85]. Increasing evidence from cancer epigenomic studies suggests a critical role for PcG factors in abnormal epigenetic silencing of tumor suppressor genes in cancer cells [5,8,9,86,87]. There are at least four different PcG complexes identified in mammalian, including the maintenance complex, PRC1, composed of RING, HPC, HPH, and BMI1, and three different initiation complexes, PRC2 through PRC4, which are formed by enhancer of zeste homolog 2 (EZH2), suppressor of zeste 12 (SUZ12), and different isoforms of embryonic ectoderm development (EED) [5,8,88,89]. In particular, PRC4 exists in embryonic, stem, progenitor and cancer cells and associates with a class III HDAC called SIRT1 [5,8]. The crucial function of PRC complexes in H3K27 methylation is mediated by EZH2, a histone lysine methyltransferase, that catalyzes this lysine methylation [13,84,85]. Methylation of H3K27 possibly stabilizes the binding of PcG complexes to this histone mark to facilitate long-term gene silencing [13,90]. Importantly, H3K27me is often present at the promoters of the DNA hypermethylated and silenced cancer genes investigated thus far [91], indicating that PcG proteins play an essential role in aberrant gene silencing in cancer cells. A recent study also showed that PcG-targeted genes in normal cells are closely associated with de novo DNA methylation in cancer cells, suggesting that PcG may preprogram its targeted genes as targets of subsequent DNA methylation in cancer cells [92,93]. In addition, several studies have shown that expression of PcG proteins such as EZH2, SUZ12 and BMI1 is aberrantly elevated in breast cancer and other cancers [94,95], suggesting deregulation of components of nucleosomal remodeling complexes can also be a mechanism resulting in gene silencing in cancer cells. In the case of another repressive histone mark, H3K9me2 (me3), this lysine methylation is catalyzed by several histone lysine methyltransferases, including SUV39H, SETDB1, G9a and GLP among others [13,96-98]. Although the defined role of H3K9 methylation in epigenetic gene silencing remains elusive, one possible mechanism is that this mark can serve as a binding site for heterochromatin protein HP1, which has an intrinsic ability to recruit DNA methyltransferases to the silenced genes [99,100].
The epigenetic mechanisms for gene silencing involve the interplay between DNA methylation, histone modifications and nucleosomal remodeling (Figure 1). The families of methyl-CpG binding proteins (MBD and Kaiso families) have been identified to play a key role in this interplay. The molecular functions of methyl-CpG binding proteins are dependent on their ability to recognize and bind methylated DNA [8,101,102]. Accumulating evidence suggests that methyl-CpG binding proteins can associate directly or indirectly with DNMTs, HDACs and HMTs and cooperate with them to modify chromatin structure and suppress initiation of gene transcription [102-106]. The associated partners of methyl-CpG binding proteins have also been found to include many nucleosomal remodeling complexes such as NuRD, CoREST, NCoR/SMRT, Sin3A, SUV39H and SWI/SNF [64,102,106-111]. The significant role of methyl-CpG binding proteins in cancer epigenetics is supported by the findings that they are localized to DNA hypermethylated and aberrantly silenced cancer genes [14,112,113]. Thus, it has been postulated that methyl-CpG binding proteins initially recognize and bind to methylated DNA, and then bring down nucleosomal remodeling complexes to modify chromatin to the repressive compact heterochromatin structure, which causes gene silencing. Inversely, the results from some other studies show that chromatin remodeling activities can further facilitate binding of methyl-CpG binding proteins to methylated DNA sites [110,114], suggesting interaction between methyl-CpG binding proteins and nucleosomal remodeling complexes results in mutual stimulation of each others’ activity. Taken together, methyl-CpG binding proteins represent an important class of chromosomal proteins that associate with multiple protein partners to modify surrounding chromatin and silence transcription, providing a functional link between DNA methylation and chromatin modification and remodeling. Needless to say, this is an emerging field and much remains to be understood regarding the components of the complex intersecting pathways that together engage in epigenetic regulation of gene expression.
■ The role of estrogen signaling-regulated epigenetic processes in the development of breast cancer & drug resistance
Much of the research effort to date has concentrated on the identification of silenced genes implicated in breast tumorigenesis. However, it is also critical to understand the factors leading to epigenetic alterations in cancer. Findings in animal models and supported by epidemiological studies, have shown that prolonged exposure of undifferentiated (immature) breast cells to estrogen or estrogen-mimetic compounds during early development increases breast cancer risk in adult life. This phenomenon is called estrogen imprinting [115]. These studies can explain why, in addition to genetic factors, the risk of breast cancer is affected by pregnancy, lifestyle in terms of intake of food and drink, and environment. Although the tumorigenic mechanism underlying this phenomenon and its connection with epigenetic regulation are still largely unknown, recently published findings provide insight into this mechanism. One line of evidence is from the study of DNA methylation patterns in several subtypes of breast cells. Bloushtain-Qimron et al. found that several transcription factor genes involved in stem cell function were hypomethylated and highly expressed in breast progenitor/stem (undifferentiated) cells compared with differentiated breast epithelial cells [116], suggesting the epigenetic programs define mammary epithelial cell phenotypes. Since breast progenitor/stem cells possess self-renewal and proliferating ability and more sensitively respond to estrogenic action, this subtype of cells has been thought to be potent targets of malignant transformation [117]. The second line of evidence is from the study of the effects of estrogen exposure on breast progenitor/stem cells, using a primary culture system to decipher the phenomenon of estrogen imprinting. Cheng et al. compared DNA methylation profiles of epithelial progeny of estrogen-exposed breast progenitor cells with those of epithelial progeny of nonestrogen-exposed progenitor cells. They found that estrogen exposure caused epithelial progeny to exhibit a cancer-like methylome, leading to silencing of some tumor suppressor genes [118]. Although the dose of estradiol (E2) used in their study was higher than normal physiological levels, their findings suggest abnormal exposure to estrogen or estrogenic chemicals induces epigenetic alterations in breast progenitor cells, which have been previously implicated in breast cancer.
Although aberrant activation of estrogen signaling can lead to tumor-associated alterations in the epigenome of breast progenitor cells, approximately 30% of diagnosed breast cancer cases lack estrogen signaling due to loss or downregulation of estrogen receptor (ER)-α, also subject to epigenetic silencing [119,120]. ER-negative breast cancers exhibit more aggressive characteristics than ER-positive breast cancers and are resistant to anti-estrogen therapy. How ER-negative breast cancer cells acquire more aggressive properties after loss of estrogen signaling is a very important issue in the field of breast cancer research. The study by Leu et al. provides evidence to link loss of ER signaling to epigenetic silencing of ERα downstream target genes [121]. Their study showed that abrogation of ERα signaling by small interfering RNA-mediated knockdown of ERα expression resulted in epigenetic inactivation of ERα targets, which began from recruiting PcG repressors and HDACs to their promoters and was then progressively followed by DNA methylation of their promoters [121]. Their results suggest that epigenetic regulation on ERα target genes is required for establishing ERα-independent growth and other characteristics of ER-negative breast cancer cells.
Another important issue is whether epigenetic regulation plays a role in the development of drug resistance in breast cancer. Fan et al. performed a genome-wide analysis of gene expression and DNA methylation profiles in breast cancer cells after adapting the cells to the anti-estrogens, tamoxifen and fulvestrant [38]. Their results demonstrate that the development of anti estrogen resistance was associated with the changes in methylation profiles of multiple genes [38]. Their findings imply that aberrantly epigenetic regulation is involved in adaptation of breast cancer cells to anti-estrogens. This proposed concept is further supported by the recent finding that epigenetic silencing of cyclin-dependent kinase 10 (CDK10) is implicated in the development of tamoxifen resistance [122]. These several lines of evidence, taken together, point out the significance of epigenetic regulation in initiating breast carcinogenesis, promoting tumorigenic phenotypes, and assisting in the development of drug resistance.
Current technological approaches to breast cancer epigenomics
To facilitate understanding of the scope of epigenetic modifications that occur in normal and cancer cells, a range of gene-specific or genome-wide technological approaches have been developed. We present an overview of recent technological developments and discuss the merits and the limitations of these approaches with respect to studies on cancer cells.
■ Technologies for detection of DNA methylation
DNA methylation is the first identified epigenetic mechanism of gene regulation. The initial methods of detection of methylation were restricted to the quantitation of total 5-methylcytosine content by high-performance liquid chromatography (HPLC) or high-performance capillary electrophoresis (HPCE) [123], and the study of DNA methylation of selected sequences using restriction enzymes that can distinguish between methylated and unmethylated recognition sites in genes of interest [124]. For restriction-enzyme-based methods, incomplete restriction-enzyme digestion and the limitation of enzyme-recognition sites restricted their extensive application. These technical limitations caused delay in advances in the cancer epigenetic field in contrast to the rapid development in cancer genetics field.
The development of bisulfite-conversion technique that reproducibly changes unmethylated cytosines to uracil but leaves methylated cytosines unchanged [125] was a key development that drastically speeded up progress in the field. Several sensitive DNA methylation detection techniques (summarized in Table 3) were developed upon the basis of bisulfite conversion, including bisulfite sequencing, methylation-specific PCR (MSP), combined bisulfite restriction analysis (COBRA) and so on [125-127]. Among these technologies, MSP is the most popular and powerful method for DNA methylation detection, which needs limited amounts of DNA material [126]. Since MSP is a gel-based assay, it cannot provide quantitative information and is subjective, several real-time methylation-specific PCR methods, such as bisulfite treatment in combination with MethyLight™ [128], quantitative multiplex-MSP (QM-MSP) [129,130], or pyrosequencing [131], have been developed and used in DNA methylation studies, which have improved features to detect minimal amounts of aberrant DNA methylation in a quantitative manner.
Table 3.
Technologies for assessing and profiling DNA methylation and histone modifications.
| Technology | Description | Ref. |
|---|---|---|
| Gene-specific detection of DNA methylation | ||
| Bisulfite sequencing |
Bisulfite-converted DNA is PCR-amplified to enrich the target templates. The purified DNA templates are subjected to sequencing analysis directly or after cloning into plasmid. |
[125] |
| Methylation-specific PCR (MSP) |
This technique takes advantage of the altered sequence of bisulfite-converted unmethyated and methylated DNA for designing primers, which can amplify DNA in a methylation-state-specific manner. |
[126] |
| Combined bisulfite restriction analysis (COBRA) |
The qualitative and quantitative detection of methylated alleles is achieved by restriction enzyme-mediated digestion of PCR amplified target amplicons from bisulfite-modified DNA. |
[127] |
| MethyLight™ |
By including the fluorescent probe technology (TaqMan®), this method is able to quantitatively and sensitively detect methylated alleles. |
[128] |
| Quantitative multiplex MSP (QM-MSP) |
This technology is a modified version of fluorogenic probe-based quantitative MSP assay. This method includes the multiplex PCR step that allows amplification of multiple target alleles. The diluted multiple PCR products are subjected to quantitative MSP assay for multiple gene detection. |
[129,130] |
| Pyrosequencing | This method is based on sequencing-by-synthesis technology to quantitative detect methylation levels of individual CpG site by monitoring the real-time incorporation of nucleotides through the enzymatic conversion of released pyrophosphate into a bioluminometric signal. | [131] |
| Genome-wide profiling of DNA methylation | ||
| Restriction landmark genomic scanning (RLGS) |
In the RLGS technique, restriction-enzyme digestion, radioactive labeling and two-dimensional electrophoresis combine to quantitatively display DNA methylation levels of thousands of CpG islands. |
[132] |
| Ampliferation of intermethylated sites (AIMS) |
The AIMS method combines methylation-sensitive restriction enzyme digestion with the display of methylation fingerprint of PCR amplified DNA fragments, which can then be isolated and characterized individually by sequencing. |
[134] |
| Differential methylation hydrization (DMH) |
DMH is a promoter-sequence-microarray-basis method that combines methylation-sensitive restriction-enzyme digestion with linker-basis PCR labeling to serve as probes for array hybridization, capable of globally displaying methylated CpG islands. |
[135] |
| Hpall tiny fragment enrichment by ligation-mediated PCR assay (HELP) |
The HELP method adopts differential digestion with a methylation-sensitive restriction enzyme or its methylation-insensitive isoschizomer and ligation-mediated PCR amplification of digested templates for cohybridization to a genomic microarray, enabling the display of genome-wide methylated CpG islands. |
[136] |
| Methylated DNA immunoprecipitation (methy-DIP) |
The methyl-DIP technique uses antibodies against methyl-CpG-binding domain proteins (MBDs) to immunoprecipitate sheared genomic DNA for isolation of methylated DNA fragments. Methyl-DIP has been combined with tiling microarrays or with high-density promoter arrays to map the human methylome. |
[92,137,138] |
| Microarray-based gene expression profiling |
Gene-expression microarray has been applied to display expression-profile changes in cells treated with epigenetic inhibitors for identification of methylation-targeted genes. |
[139-141] |
| Methylation-specific digital karyotyping (MSDK) | The MSDK method is a modified serial analysis of gene expression (SAGE) assay that combines a methylation-sensitive restriction enzyme and a fragmenting restriction enzyme to generate short sequence tags for providing information on gene loci and their methylation levels. | [142] |
| Genome-wide profiling of histone modifications | ||
| Chromatin immunoprecipitation (ChIP)-on-chip |
This method combines ChIP technology with high-density microarrays for measuring and mapping histone-binding genomic loci. |
[143] |
| ChIP-SAGE |
This method adopts a combination of ChIP and SAGE technologies to globally quantify and map genomic binding sites for specifically modified histones. |
[144] |
| ChIP-Seq | This new method employs a high-throughput sequencing technique to analyze ChIP DNA for genome-wide mapping of histone-DNA binding patterns. | [145,146] |
In addition to these gene-locus specific DNA methylation detection methods mentioned above, several recently developed genome-wide techniques have been applied to the study of global DNA methylation profiles in normal and cancer cells (summarized in Table 3). Restriction landmark genomic scanning (RLGS) is one of the earliest methods to be applied to genome-wide methylation analysis [132]. RLGS has an ability to globally analyze the methylation status of approximately 1000 unselected CpG islands. Other important techniques for analy zing altered DNA methylation patterns across the genome have relied on an arbitrary primed PCR technique (e.g., AIMS) [133,134]. Further advances in this field are derived from the application of the DNA microarray technology. A widely used example is differential methylation hydridization (DMH) that uses CpG-island and promoter sequence microarrays, which enables the simultaneous analysis of the methylation levels of a large number of CpG-islands in the genome [135]. In addition, a recently developed related technique called HpaII tiny fragment enrichment by ligation-mediated PCR assay (HELP) uses a modified approach to globally analyze DNA methylation patterns [136]. Methods (e.g., Methylated DNA immunoprecipitation [methyl-DIP]) based on chromatin immunoprecipitation (ChIP) using the ChIP-on-chip technology are other seminal recent advances in the epigenomic profiling of cancer cells [92,137,138]. Finally, gene-expression profiling using microarrays is another powerful and widely used technique for assessing genome-wide DNA methylation patterns. This approach is used to compare gene expression levels from cancer cell lines before and after treatment with a demethylating drug, a HDAC inhibitor, or a combination of both drugs [139-141]. As with all microarray-based technologies, the identified candidate genes are further verified, in this case, by quantitative RT-PCR and promoter methylation analyses. In addition to microarray-based techniques, a serial analysis of gene expression (SAGE)-technology-based method, known as methylation-specific digital karyotyping (MSDK), has been developed recently for the genome-wide analysis of methylation profiles [142]. The advantage of this new technique is that there is no need of prior sequence information for analysis and the obtained data can be used to map the methylated gene loci and to determine their methylation levels.
■ Technologies for detection of histone modifications
Characterization of post-translational histone modifications is a greater challenge than analysis of DNA methylation and needs special technology. Currently, the gold standard for accurately assessing global levels of histone modifications is mass spectrometry [6]. Since antibodies specifically recognizing the amino acid modifications of histone proteins are available, a simple Western blot analysis is also used for detecting histone modifications.
In addition to determining the types and relative levels of histone modification in cells, characterization of distribution of each type of histone modification on chromosomes also provides very important information. The current techniques for genome-wide analysis all adopt the ChIP technology with antibodies against specifically modified histones (summarized in Table 3). The first-developed and widely used technique is ChIP-on-chip [143]. In contrast to the ChIP-on-chip method, another new technique for profiling histone modifications at a genomic scale is ChIP-SAGE, which combines ChIP experiments with the SAGE technology [144]. The merit of ChIP-SAGE is that, unlike ChIP-on-chip that requires sequence information of preselected genomic regions to manufacture genomic microarrays, no prior genomic sequence information is required in this assay. However, the cost for ChIP-SAGE is higher than ChIP-on-chip due to the use of the more expensive traditional sequencing methods. Besides, the fact that not every region of chromosomes contains restriction-enzyme recognition sites used to cleave the ChIP DNA limits the capacity of ChIP-SAGE to study the entire genome. More recently, a new technique, chromatin immunoprecipitation combined with high-throughput sequencing techniques (ChIP-Seq), has been developed for analyzing ChIP DNA using a high-throughput massively parallel signature sequencing-like technique developed by Solexa [145,146]; this technique is more powerful and cost-effective than the ChIP-SAGE technique. There are several advantages in this new technique, including the use of less PCR amplification of ChIP DNA after ligation to adaptors and a highly efficient sequencing procedure (sequencing-by-synthesis). In contrast to ChIP-on-chip, more quantitative data of histone modification levels at different chromosomal regions can be obtained in ChIP-Seq experiments. In addition to mapping the genome-wide histone-DNA binding patterns, it can be envisioned that this new technique has great potential for globally defining the methylome of a particular cell type. With rapid and striking technological advancements, global analysis of DNA methylation and histone modification mapping on chromosomes has become eminently practical.
Translational application of epigenomics in breast cancer
■ Epigenomic profiles as breast cancer biomarkers for diagnosis & prognosis
In addition to cancer-specific genetic and gene-expression signatures, epigenetic signatures have emerged as potential biomarkers for cancer detection. The sensitivity and specificity of this type of diagnostic assay relies on the use of those DNA-methylation markers that are frequently hypermethylated in cancer cells, but are always unmethylated in normal cells. For breast cancer, as for other cancers, early detection is the most efficient way to decrease mortality. The current widely used method for early diagnostic screening is mammography. However, while mammography is highly sensitive, its lack of specificity has created the need for other highly sensitive diagnostic methods for early detection of breast cancer. As a result of this need, cancer-specific hypermethylated genes have been considered as potential and promising biomarkers for early detection of breast cancer.
The use of a panel of cancer-specific methylation markers in conjunction with MSP, MethyLight or QM-MSP has proven to be promising for breast cancer detection. For example, Evron et al. successfully used a three-gene panel (Cyclin D2, RARβ and TWIST) to detect malignant breast cancer cells in ductal fluid from routine operative breast endoscopy (ROBE) and ductal lavage [147]. Fackler et al. improved this method and tested a four-gene panel (RASSF1A, TWIST, HIN1 and Cyclin D2) using the QM-MSP assay to examine clinical tissue samples [129]. The cumulative methylation of these four genes is commonly observed to be higher in primary invasive breast cancers compared with reduction mammoplasty specimens from healthy women [129]. Fackler et al. further used the same technique but adopted a nine-gene panel (RASSF1A, TWIST, HIN1, Cyclin D2, RARβ, APC, BRCA1, BRCA2 and p16) to examine ductal lavage samples from women with or without breast cancer. This trial demonstrated that methylation-marker detection was twice as sensitive as cytological diagnosis of ductal lavage cells [148]. In addition to biopsied tissue sections and ductal fluid, methylated DNA is also detected in blood since the blood of patients with manifest breast cancer contains detectable amounts of circulating methylated DNA [149]. The blood detection of tumor-specific methylated DNA has been pursued for its potential for prognostic prediction and monitoring relapse of breast cancer after therapy [149-151].
In addition to cancer detection, it has been reported that the DNA methylation assay might be used for risk assessment and prognosis of breast cancer. Lewis et al. studied five frequently methylated genes, including RASSF1A, APC, H-cadherin, RARβ, and cyclin D2, and found a higher methylation frequency of both RASSF1A and APC genes in unaffected women at high risk for breast cancer compared with those at low or intermediate risk based on the Gail model analysis. This suggests that promoter hypermethylation of these genes is associated with epidemiologic markers of increased breast cancer risk [152]. This finding needs confirmation that such alterations do indeed occur earlier than abnormal histological findings, and by follow-up studies to examine whether these changes are associated with subsequent development of breast cancer. The prognostic significance of aberrant DNA methylation has been investigated by Muller et al. After screening 39 genes in DNA from serum of normal control patients and patients with primary or metastatic breast cancer, they identified two genes, RASSF1A and APC, whose methylation has a statistically significant association with poor outcome [151]. Other methylated genes, such as GSTP1, SFRP1, have also been identified to be associated with poor prognosis [153,154].
■ Pharmacoepigenomics as a predictor of response to chemotherapy of breast cancer
When the functions of genes are involved in mediating the therapeutic effects of certain chemotherapeutic agents, silencing of this class of genes through epigenetic mechanisms results in abrogating or blunting the drug's effects. Therefore, they can be used as a predictor to evaluate the response of patients to chemotherapy. The prototypic case is that of the expression of ERα gene, which is recognized as an important predictor of response to endocrine therapy of breast cancer. As mentioned above, approximately 30% of breast tumors lack expression of ERα and are resistant to endocrine therapy. Previous studies have shown that loss of ERα expression is often associated with promoter hypermethylation of ERα gene [119,120]. Therefore, in addition to the immunohistochemical detection of ERα protein expression, analysis of ERα methylation status presents an alternative to determine whether endocrine therapy will be effective in breast cancer patients [155]. More recently, through an RNAi screen, Iorns et al. identified downregulation of cyclin-dependent kinase 10 (CDK10) expression leading to tamoxifen resistance [122], which has been addressed previously. Their study showed that patients with ERα-positive breast tumors that expressed low levels of CDK10 relapsed early under tamoxifen treatment. More importantly, downregulated CDK10 expression is associated with methylation of CDK10 promoter [122], suggesting that CDK10 gene methylation can serve as a novel marker for determining response to endocrine therapy.
Besides ERα, inactivation of retinoic acid receptor β (RARβ) by epigenetic mechanisms is another important predictor for determining the response to retinoic acid-based chemo prevention and therapy of breast cancer as well as other types of cancer [3]. Binding of retinoids to RARβ triggers receptor dimerization and transactivation of retinoid-responsive genes, which in turn induce cellular differentiation and apop tosis [156]. Therefore, the expression and functional status of RARβ can critically affect the anti cancer activity of retinoids. Loss or down regulation of RARβ has been known to occur in many kinds of cancers, including breast cancer, and correlate with the malignant phenotype [157,158]. Therefore, RARβ promoter hypermethylation in cancer is an important mechanism causing abrogation of the RARβ2 signaling. The epigenetic mark that shows the silencing of RARβ2, therefore, appears to be a potential determinant of retinoic acid response.
■ DNA methylation & histone deacetylation as therapeutic targets in breast cancer
The frequent occurrence of epigenetic alterations in the pathogenesis of cancer has created targets for the development of novel anticancer therapeutics. Unlike genetic alterations that are irreversible processes, epigenetic alterations in cancer are potentially reversible. This feature of epigenetics has promoted the development of pharmacologic inhibitors of DNA methylation and histone deacetylation [61-63,159]. These inhibitor compounds can induce DNA demethyl ation and inhibit histone deacetylation to reverse epigenetic silencing of tumor suppressor genes, leading to re-expression of these genes in cancer cells and reactivation of pivotal cellular tumor-suppression pathways.
DNA methyltransferase inhibitors
Since DNMTs are the core enzymes that catalyze DNA methylation, inhibitors of DNMTs are promising anticancer agents. 5-azacytidine (Vidaza™) and 5-aza-2′-deoxycytidine (Decitabine) are the most extensively studied DNMT inhibitors. The pharmacological action of these two compounds is mediated by their incorporation into DNA in the place of the natural base, cytosine, during DNA replication, leading to covalent trapping of DNMTs [160]. This causes the depletion of active DNMT enzymes and demethylation of genomic DNA. The inhibitory effect of 5-aza-2′-deoxycytidine on DNMTs is stronger than 5-azacytidine since 5-aza-2′-deoxycytidine incorporates only into DNA, unlike 5-azacytidine that incorporates into both DNA and RNA [159]. Both compounds have been approved by the US FDA as elective therapeutic agents for treatment of a pre-leukemic disease, myelodysplastic syndrome. A major disadvantage of these two compounds is their instability in neutral aqueous solution. This has resulted in a search for more stable cytosine analog-based demethylating agents, such as zebularine. Zebularine is a novel DNMT inhibitor with several merits, such as very stable chemical property suitable for oral administration, low toxicity and a high selectivity for tumor cells. [161]. However, the requirement of high levels of drug to achieve efficacy of zebularine may negatively affect its potential in clinical application. In addition, non-nucleotide DNMT inhibitors have been developed recently for avoiding the toxicity inherent in nucleotide analogs. These small-molecule demethylating agents include antiarrhythmic agents such as procainamide and procaine, the antihypertensive agent such as hydralazine, epigallocathechin-3--gallate derived from green tea, and the novel compound RG108 [162-165]. They all have the ability to bind to the active sites of all DNMTs and perturb inter actions between the enzymes and their target sites. The therapeutic agents that specifically target one type of DNMTs have also been developed, such as MG98, an antisense oligonucleotide that specifically inhibits DNMT1 function [166]. It has been demonstrated that the major anticancer mechanism of demethylating agents is attributable to reactivation of silenced tumor suppressor genes in cancer cells.
Histone deacetylase inhibitors
Abnormal recruitment of HDACs to the promoters of tumor suppressor genes, which leads to cancer, has provided a strong mechanistic rationale for the use of HDAC inhibitors (HDACIs) in cancer therapy. It has been demonstrated that transcriptional repression of multiple genes implicated in cell cycle progression, differentiation and apoptosis could be reversed by inhibition of HDAC function [6,61,63]. Furthermore, HDACIs have proven potent anti-tumor activities in human cancer cells and xenograft models [61,63,167]. More importantly, transformed cancer cells have been shown to be at least ten-times more sensitive to growth inhibition by HDACIs compared with nontransformed cells, demonstrating that HDACIs have a tumor-selective feature [168]. The tumor-selective characteristics of HDACIs may be attributable to the fundamental epigenetic differences between normal and tumor cells. Taken together, these findings support the notion that HDACIs can be used as potential anticancer agents. Currently, a large number of structurally diverse HDACIs have been synthetically developed or purified from natural sources. According to their chemical characteristics and pharmacological mechanism for HDAC inhibition, HDACIs are classified into the following groups: hydroxamates, short-chain fatty acids, cyclic peptide, benzamides, and anilides [61,63,159,169] (summarized in Table 4). Several of them have progressed to clinical trials and suberoylanilide hydroxamic acid (SAHA) has been approved by FDA for treatment of cutaneous T-cell lymphoma (CTCL) [169].
Table 4.
Histone deacetylase inhibitors.
| Class | Compound | HDAC targets (potency) | Effects on cancer cells | Stage of development (company/sponsor) |
|---|---|---|---|---|
| Hydroxamate | Suberoylanilide hydroxamic acid (SAHA, Vorinostat) | Class I and II (μM) | Differentiation, growth arrest, apoptosis, mitotic failure, senescence, polyploidy | US FDA approval for CTCL (Merck) |
| PXD101 (Bellinostat) | Class I and II (μM) | Growth arrest, apoptosis | Phase I, II (CuraGen, TopoTarget) | |
| LAQ824 | Class I and II (nM) | Growth arrest, apoptosis | Phase I (Novartis) | |
| LBH589 (Panobinostat) | Class I and II (nM) | Growth arrest, apoptosis | Phase I (Novartis) | |
| PCI-24781 | Class I and II (NA) | Apoptosis | Phase I (Pharmacyclics) | |
| Pyroxamide | Class I and II (μM) | Differentiation, growth arrest, apoptosis | Phase I (NCI) | |
| ITF2357 | Class I and II (nM) | Growth arrest, apoptosis | Phase I (Italfarmaco) | |
| SK-7041 | HDACs 1 and 2 (nM) | Growth arrest, apoptosis | NA | |
| SK-7068 | HDACs 1 and 2 (nM) | Growth arrest, apoptosis | NA | |
| TSA | Class I and II (nM) | Differentiation, growth arrest, apoptosis | NA | |
| Tubacin | Class IIb (μM) | Inhibition of HDAC6-mediated cell motility | NA | |
| CBHA | NA (μM) | Apoptosis | NA (Merck) | |
| Oxamflatin | NA (μM) | Differentiation, growth arrest | NA | |
| |
Scriptaid |
NA (μM) |
NA |
NA |
| Short-chain fatty acid | Phenylbutyrate | Class I and IIa (mM) | Differentiation, growth arrest, apoptosis | Phase I, II |
| Valproic acid (VPA) | Class I and IIa (mM) | Differentiation, growth arrest, apoptosis, senescence | Phase I, II (Abbot) | |
| AN-9 (prodrug) | NA (μM) | Differentiation, growth arrest, apoptosis | Phase I, II (Titan Pharmaceuticals) | |
| Baceca | Class I (NA) | NA | Phase I, II (Topotarget) | |
| |
Savicol |
NA (NA) |
NA |
Phase I, II (Topotarget) |
| Cyclic peptide | Depsipeptide (Romidepsin, FK228) | Class I (nM) | Differentiation, growth arrest, apoptosis, mitotic failure | Phase IIb for CTCL and PTCL (Gloucester Pharmaceuticals) |
| Trapoxin A | Class I and IIa (nM) | Differentiation, growth arrest | NA | |
| Apicidin | HDACs 1 and 3, not HDAC8 (nM) | Differentiation, growth arrest | NA | |
| |
CHAPs |
Class I (nM) |
NA |
NA |
| Benzamide | MS-275 | Class I (μM) | Differentiation, growth arrest, apoptosis | Phase I, II (Schering AG) |
| |
CI-994 (Tacedinaline) |
NA (μM) |
Growth arrest, apoptosis |
Phase I, II, III (Warner-Lambert) |
| Anilide | MGCD0103 | Class I (NA) | Differentiation, growth arrest, apoptosis | Phase I, II (Methylgene) |
CBHA: m-carboxycinnamic acid bis-hydroxamide; CHAPs: cyclic-hydroxamic-acid-containing peptides; CTCL: Cutaneous T-cell lymphoma; HDAC: Histone deacetylase; NA: Not available; NCI: National Cancer Institute; PTCL: Peripheral T-cell lymphoma; TSA: Trichostatin A.
Multiple anticancer activities of HDACIs involve cell cycle arrest, promotion of apoptosis, induction of differentiation and suppression of angiogenesis [61-63,159]. Although the mechanisms underlying the pleiotropic cellular effects of HDACIs are still not completely deciphered, a growing body of evidence has shown that HDACI-mediated anti-tumor effects involve both transcriptional and nontranscriptional mechanisms [61,63]. Treatment with HDACIs causes hyperacetylation of histones and nonhistone transcription factors such as p53, p73, E2F1, STAT1, STAT3 and NF-κB, which lead to transcriptional activation or repression of their target genes [61,170-173]. Many HDACI-induced genes have been reported (summarized in Table 5), including cell cycle regulatory genes, tumor suppressor genes, apoptotic genes, differentiation regulatory genes and immune response genes. [61,63]. For example, HDACIs can induce p21WAF1/CIP1 expression through hyperacetylated histone-mediated transactivation in a p53-dependent or p53-independent manner, leading to cell cycle arrest [174]. HDACIs can also upregulate many apoptotic genes (e.g., CD95, TRAIL, DR4, DR5, Bax, Bak, Bim, Bmf and Apaf1) involved in the extrinsic death-receptor and intrinsic mitochondrial death pathways [61]. In addition, treatment with HDACIs has been found to repress the expression of genes required for cell cycle progression (cyclin D1, cyclin A), antiapoptosis (Bcl-2) and angiogenesis (VEGF, HIF-1α) [63].
Table 5.
Genes whose mRNA or protein expression is altered by treatment with histone deacetylase inhibitors.
| Class | Gene list |
|---|---|
| Histone deacetylase inhibitor-upregulated gene expression | |
| Cell cycle inhibitory gene |
p21WAF1, p16INK4A, p27KIP1, GADD45 |
| Tumor suppressor gene |
p53, VHL, p107, gelsolin, IGFBP3 |
| Proapoptotic gene |
CD95, TRAIL, DR4, DR5, Bax, Bak, Bim, Bmf, Apaf1 |
| Differentiation gene |
RARβ2, TGFβ1 |
| Antimetastatic gene |
TIMP1, TIMP2, RECK |
| Immune response gene | MHC-I, MHC-II, CD40, CD80, CD86, ICAM1 |
| Histone deacetylase inhibitor-downregulated gene expression | |
| Cell cycle regulatory gene |
Cyclin D1, Cyclin A, thymidylate synthetase, CTP synthase |
| Antiapoptotic gene |
Bcl-2, Bcl-XL, Bcl-w, MCL1, XIAP |
| DNA repair gene |
Ku70, Ku86, DNA-PKcs |
| Hormone receptor gene |
EGFR, ERα, HER2/neu |
| Metastatic gene |
MMP2, MMP9 |
| Signal transduction gene |
Akt, Flt-3, Raf-1, Abl |
| Angiogenic factor gene | HIF1α, VEGF, IL2, IL10, bEGF, TIE2, eNOS, CXCR4 |
In addition to transcriptional mechanisms for anticancer effects of HDACIs, many lines of evidence point to nontranscriptional mechanisms as mediators of the anti-tumor effect of HDACIs. [61-63]. For example, it has been reported that HDACIs could induce defective mitoses in tumor cells and in turn trigger cell death. This effect may be attributed to changes in chromatin conformation caused by hyperacetylation of centromeric histones, resulting in abnormal chromosomal segregation or impaired mitotic progression [175]. For the apoptotic effect, it has been found that acetylation of K70 in response to HDACIs results in the release of its binding partner Bax, causing translocation of Bax to the mitochondrial outer membrane to stimulate apoptosis [78,176]. Besides these examples, HDACIs deplete protein levels of many oncoproteins whose stability is regulated by chaperonic heat-shock proteins (HSPs) [61,63]. HSP90 has been found to be hyperacetylated after treatment with HDACIs, which leads to the proteasomal degradation of HSP90 client proteins such as HER2/neu, Akt, c-Raf, ERα [61,63,77,177]. Taken together, these examples demonstrate that the interplay between transcriptional effects on the regulation of gene expression and nontranscriptional effects on mitosis and nonhistone targets contributes to anticancer activity of HDACIs. These attractive combinational anti-tumor activities enable HDACIs to be potentially very effective anticancer agents.
Clinical application of epigenetic drugs in breast cancer therapy
As single-agent anticancer drugs, epigenetic modulators such as demethylating agents and HDACIs have shown the promising efficacy against multiple types of cancers in the laboratory studies and clinical trials [61,63,159]. Few of these agents (e.g., Vidaza, Decitabine and SAHA) have been approved by FDA and applied to hematological malignancies. For breast cancer, epigenetic drugs still remain in the stage of preclinical and clinical trials and await more promising clinical data for application to patients. Here we provide a review of advances in the preclinical and clinical testing of epigenetic therapeutic agents in breast cancer therapy.
Since abnormal epigenetic gene silencing mediated by aberrant DNA methylation and chromosomal remodeling (including histone modifications) is one of the major mechanisms leading to breast tumorigenesis, it is reasonable to argue that the combination of both demethylating agents and HDACIs can synergistically transactivate a set of epigenetically silenced tumor-suppressor genes and more effectively eliminate cancer cells than treatment with a single agent [178]. This rationale has been tested in ER-negative breast cancer cells, MDA-MB-231, which are resistant to traditional anti-estrogen therapy. Yang et al. have demonstrated that cotreatment of these cells with DNA methyltransferase and HDAC inhibitors led to synergistic demethylation of ERα promoter and re-expression of functional ERα [179]. Subsequent studies by Sharma et al. showed that restoration of expression and function of ERα resensitized ER-negative breast cancer cells to tamoxifen treatment [180]. Their findings, taken together, suggest that the approach of combining demethylating agents and HDACIs is a very promising regimen for treatment of tamoxifen-resistant breast cancer patients. In addition to ERα restoration, this two-drug-combination regimen has also been demonstrated to induce RARβ2 re-expression in breast cancer cells, which caused growth inhibition of breast cancer cells regardless of ER status by cotreatment with retinoic acids [158,181]. A very recent study also introduces a new strategy that the therapeutic regimen of combining two chromatin remodeling drugs (5-aza-2′-deoxycytidine and SAHA) can be used to synergize with artificial transcription factors for the reactivation of the tumor suppressor gene maspin in breast cancer cells [182]. This novel therapeutic approach that takes advantages of both epigenetic and genetic strategies to reactivate silenced tumor suppressor genes will be an effective regimen if the expression constructs of artificial transcription factors can be designed to be specifically delivered to breast tumors.
Given that HDACIs have been clinically tested with minimal toxicity to normal cells and can lower the apoptotic threshold in cancer cells due to their abilities to stimulate extrinsic and intrinsic cell death pathways as described above, the effect from cotreatment with HDACIs and conventional chemotherapeutic drugs has been examined. Recent studies that used this cotreatment approach have shown that HDACIs have synergistic effects with other chemotherapeutic agents. For example, it has been demonstrated that treatment with HDACIs such as SAHA and LAQ824 can further sensitize breast cancer cells with amplification of HER2/neu gene to trastuzumab (Herceptin®) and other chemotherapeutic agents such as gemcitabine (antimetabolite), docetaxel and epothilone B (antimicrotubule) [77,183,184]. This synergistic anti-tumor effect might result from HDACI-induced hyperacetylation of HSP90 that further causes degradation of its client HER2/neu protein [61,63,77,177], implying that inhibition of the growth-promoting and survival HER2/neu signaling pathway lowers the apoptotic threshold and renders tumor cells more vulnerable to chemotherapeutic drugs. Moreover, it has been recently reported that treatment of EGFR-overexpressing, ER-negative breast cancer cells with SAHA could decrease the EGFR levels through destabilizing EGFR mRNA [185], suggesting that combining anti-EGFR agents and HDACIs may provide a promising strategy for dual targeted therapy. Since ERα has also been found to be one of the HSP90 clients, several lines of evidence demonstrate that HDACIs can trigger depletion of ERα protein in the ERα-positive breast cancer cells through ubiquitin-mediated degradation [177,186,187]. These findings are clinically significant based on the further supportive studies that HDACIs can sensitize ERα-positive breast cancer cells to anti-estrogenic agents [177]. Taken together, these preclinical findings support the development of HDACIs in combination with conventional chemotherapeutic drugs to treat both ER-positive and -negative breast tumors. A recent clinical study of a small group of patients with locally advanced breast cancer has shown that the therapeutic regimen with a combination of demethylating hydralazine and HDAC inhibitor magnesium valproate could cause a statistically significant decrease in global 5-methylcytosine content and HDAC activity, and this cotreatment in combination with neoadjuvant doxorubicin and cyclophosphamide appeared to increase the chemotherapeutic efficacy [188]. Although the result from this trial still needs to be further extensively evaluated, it is a first proof-of-concept study to support the efficacy of epigenetic therapy for breast cancer.
Besides chemotherapy, radiation therapy is one of therapeutic modalities for breast cancer treatment, especially for advanced metastatic disease. Although radiation has well-documented palliative effects on metastatic disease of brain and meninges, the survival rate may vary from a few months to a couple of years. A further improvement in the standard radiotherapy, therefore, might greatly benefit many metastatic patients [189]. Since HDACIs can alter chromatin structure by promoting hyperacetylation of histones, it is possible that changes in chromosomal remodeling can modulate DNA damage response. Indeed, it has recently been shown that treatment with trichostatin A could activate ATM-p53 DNA damage signaling pathway and had the synergistic effect on enhancing ionizing-radiation-induced ATM activation [190]. This raises the possibility that HDACIs are able to augment the effects of DNA damaging agents. Furthermore, HDACIs have been found to radiosensitize human melanoma cells by inhibiting DNA repair activity, concomitantly correlating with HDACI-induced decreased expression of DNA repair-related genes such as Ku70, Ku86 and DNA protein kinase catalytic subunit (DNA-PKcs) [191]. These data suggest that HDACIs may override the DNA damage defense response and facilitate radiation-induced mitotic cell death. Recently, Nome et al. have tried a similar strategy using a breast carcinoma cell line with an ability to metastasize to the brain and found that following pretreatment with the HDACI (trichostatin A), the clonogenic regrowth of breast cancer cells after ionizing radiation was significantly reduced [192]. Their finding is in accordance with the notion that the disruption of chromatin structure increases the probability of mitotic cell death. These studies support the possibility that HDACIs may have the therapeutic potential to serve as radiosensitizers.
Conclusion
Dramatic advances in powerful technologies (e.g., ChIP-seq) for epigenomic studies have enabled us to move forward in big steps towards understanding the role of epigenetics in normal and cancerous cells. The comprehension of the interplay between DNA methylation, histone modifications and nucleosomal remodeling, and epigenetic mechanisms regulating chromatin structure, gene transcription and diverse cellular responses has provided a strong basis for the development of epigenetics-based therapeutic biomarkers and drugs for cancer diagnosis and treatment. In breast cancer, the discovery of cancer-specific methylated genes has led to the development of potential DNA methylation markers for early detection, risk assessment, prognosis and prediction of drug response. Discovery and development of epigenetic drugs (e.g., demethylating agents and HDACIs) have also shed new light on the development of new therapeutic regimens for the treatment of breast cancer.
Future perspective
Genome-wide profiling of mutated genes in breast and colorectal tumors has been recently catalogued [193]. This advance in the human cancer genetic field has urged epigenetic researchers to undertake national and international human epigenome projects, with the ultimate aim to map all epigenetic modifications in both normal and diseased cells [194]. It is plausible that the implementation of these projects will enable us to more completely define the progressive changes in epigenomic patterns during the development of a particular cancer type such as breast cancer and elucidate the pathological roles of these epigenetic alterations. The outcome of the human epigenome projects is essential for the mature development of epigenetic markers and therapeutics, which will be very important for application of these advances to diagnosis, prognosis and therapy of breast cancer patients. Given that the roles of epigenetic alterations in development of resistance to anticancer drugs during breast cancer therapy remain elusive, exploration of drug-resistance mechanisms by the genome-wide epigenetic study is also very important. Furthermore, although diagnostic screening using epigenetic markers has been demonstrated to be a promising prospect, several issues still need to be solved, including development of specific and sensitive screening panels, and the necessity of improving and optimizing assay protocols for screening and final testing in large prospective clinical studies. The clinical significance of epigenetic markers also needs to be evaluated by comparing the efficacy of these panels with classic screening methods and other developing screening procedures derived from proteomics, mRNA expression, or microRNA arrays.
Although the current development of epigenetic therapeutics is promising for the treatment of breast cancer, there are many issues that need to be addressed before moving forward. The more detailed comprehension of the molecular mechanisms underlying the anticancer activities of demethylating agents and HDACIs is essential. Further translational studies of epigenetic drugs are also required to find the correlation between reversion of epigenetic changes, gene re-expression and therapeutic response of breast tumor. For clinical use of epigenetic agents in breast cancer
Executive summary.
The epigenome in breast cancer
Epigenetic alterations are one of main driving mechanisms leading to breast cancer. Epigenetic alterations are heritable and reversible, and do not involve any change in the primary DNA sequence itself.
Epigenetics involves three molecular events including DNA methylation, histone modifications and chromatin remodeling. A sophisticated interplay between these three epigenetic events modulates chromatin conformation and gene expression.
DNA methylation is mediated by the catalytic activity of DNA methyltransferases. During tumorigenesis, alterations in DNA methylation involve global hypomethylation and locus-specific hypermethylation, resulting in genomic instability and rearrangement, activation of oncogenes and inactivation of tumor suppressor genes.
Histone modifications mediated by various histone tail-modifying enzymes collaborate with or without DNA methylation to communicate with chromatin remodeling factors, which adapt chromatin structure to the open euchromatin (active transcription) or the closed heterochromatin (repressive transcription) states.
Epigenetic regulation controls the expression program of stem-cell-function genes and defines mammary cell phenotypes.
In vitro cultured breast progenitor-cell model shows that abnormal exposure to estrogen causes epigenetic alterations in epithelial progeny, leading to silencing of tumor suppressor genes.
Epigenetically regulatory mechanisms are implicated in the development of estrogen receptor (ER)-negative breast cancer phenotypes and drug resistance.
Current technological approaches to breast cancer epigenomics
The development of bisulfite-conversion-based techniques facilitated the detection of methylated genes and made a revolutionary advance in the epigenetic field; a combination with real-time PCR made quantitation of gene methylation possible.
The development of chromatin immunoprecipitation (ChIP)-based techniques promoted progress in profiling of genome-wide methylome and histone-modification maps on chromosomes from normal and diseased cells.
Translational application of epigenomics in breast cancer
Epigenetic signatures specific to breast cancer have emerged as innovative biomarkers for early detection, risk assessment, prognostic prediction and the evaluation of drug response.
The reversibility of epigenetic modifications provides the rationale for developing epigenetic agents to reverse the epigenetic silencing of tumor suppressor genes.
To reduce side effects, maximize selective anticancer effects and prevent drug resistance, the developed epigenetic agents, such as demethylating agents and histone deacetylase inhibitors, can be administered in combination with other epigenetic agents, conventional chemotherapeutic agents, differentiation agents or radiation.
Conclusion
The advances in comprehension of epigenetic mechanisms implicated in modulating chromatin conformation, gene transcription and diverse cellular signaling pathways have provided the essential basis for the current development of epigenetics-based biomarkers and drugs for the diagnosis and treatment of breast cancer.
therapy, it will be important to determine their optimal therapeutic doses, timing and mode of administration. To obtain synergistic therapeutic efficacy and lower toxicity for breast cancer patients, the epigenetic anti-tumor drugs should be tested and analyzed in preferential combination with other currently used anti-breast-cancer drugs such as anti-estrogen agents, conventional chemotherapeutic agents, kinase inhibitors and differentiation agents. Another way to reduce the side effects of epigenetic drugs is to identify specific epigenetic modulators (e.g., some specific HDACs) involved in breast tumori genesis and to develop the specific drugs to target these molecules. Since the entire picture of the upstream regulatory pathways and downstream events of epigenetic modifications that are specific to breast cancer are not yet completely deciphered, the continuation of efforts to define these molecular events is still very important for the development of the next new generation of specific anti-tumor agents against breast cancer. In addition, relapse of breast cancer has been assumed to involve the failure of cancer stem cells to respond to current cancer therapy. Given that it has been shown that epigenetic modulation is required for the maintenance of cancer stem-cell features, whether epigenetic agents have effects against breast cancer stem-cells is worthy of investigation.
From the tremendous progress we have witnessed in both basic and clinical studies with regard to epigenetics, we have clearly entered a remarkable era of the epigenome. A greater understanding of the molecular events for the initiation and maintenance of epigenetic gene silencing during the process of breast tumorigenesis could promote the development of promising epigenetic-marker strategies for early detection, risk assessment, prognosis and drug-response prediction, and could also speed up the progress in developing potential clinical strategies for breast cancer prevention and therapy.
Acknowledgements
We thank the 2007 AVON Breast Cancer Prevention Initiative and the NIH SPORE grant, P50 CA888431 for support.
Financial & competing interests disclosure
Saraswati Sukumar was a consultant for, and has received research funding from Oncomethylome Sciences Inc. for performing research mentioned in this article. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat. Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 3■.Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002;21:5462–5482. doi: 10.1038/sj.onc.1205606. [of interestProvides an organized review of functional roles for identified methylated genes in breast tumorigenesis.] [DOI] [PubMed] [Google Scholar]
- 4.Polyak K. Breast cancer: origins and evolution. J. Clin. Invest. 2007;117:3155–3163. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5■■.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer – a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [of considerable interestExcellent overview of how epigenetic changes might addict cancer cells to altered signal-transduction pathways during the early stages of tumor development.] [DOI] [PubMed] [Google Scholar]
- 6■■.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [of considerable interestExcellent overview of the current advances in epigenomic studies from DNA methylomes to histone-modification profiles, technological development and translational application to diagnosis and therapy.] [DOI] [PubMed] [Google Scholar]
- 7■.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [of interestHighlights how aberrant alterations in patterns of DNA methylation and chromatin structure cause abnormal changes in gene expression in cancer.] [DOI] [PubMed] [Google Scholar]
- 8.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome – components and functional correlates. Genes Dev. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 9.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richardson BC. Role of DNA methylation in the regulation of cell function: autoimmunity, aging and cancer. J. Nutr. 2002;132:S2401–S2405. doi: 10.1093/jn/132.8.2401S. [DOI] [PubMed] [Google Scholar]
- 11■■.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [of considerable interestPioneering report demonstrates that hypomethylation can drive genomic instability and some types of tumorigenesis using animal model experiments.] [DOI] [PubMed] [Google Scholar]
- 12.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 13■■.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell. Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [of considerable interestExcellent review that summarizes recent advances in the current understanding of molecular events implicated in histone lysine methylation and the biological functions of these lysine modifications.] [DOI] [PubMed] [Google Scholar]
- 14.Nguyen CT, Gonzales FA, Jones PA. Altered chromatin structure associated with methylation-induced gene silencing in cancer cells: correlation of accessibility, methylation, MeCP2 binding and acetylation. Nucleic Acids Res. 2001;29:4598–4606. doi: 10.1093/nar/29.22.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esteller M, Almouzni G. How epigenetics integrates nuclear functions. Workshop on epigenetics and chromatin: transcriptional regulation and beyond. EMBO Rep. 2005;6:624–628. doi: 10.1038/sj.embor.7400456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferguson-Smith AC, Surani MA. Imprinting and the epigenetic asymmetry between parental genomes. Science. 2001;293:1086–1089. doi: 10.1126/science.1064020. [DOI] [PubMed] [Google Scholar]
- 17.Reik W, Lewis A. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nat. Rev. Genet. 2005;6:403–410. doi: 10.1038/nrg1602. [DOI] [PubMed] [Google Scholar]
- 18■.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [of interestImportant paper that reports the global and systematic analysis of chromosomes 21 and 22 for defining the properties of CpG islands.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brenner C, Fuks F. DNA methyltransferases: facts, clues, mysteries. Curr. Top. Microbiol. Immunol. 2006;301:45–66. doi: 10.1007/3-540-31390-7_3. [DOI] [PubMed] [Google Scholar]
- 20.Tajima S, Suetake I. Regulation and function of DNA methylation in vertebrates. J. Biochem. 1998;123:993–999. doi: 10.1093/oxfordjournals.jbchem.a022066. [DOI] [PubMed] [Google Scholar]
- 21.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 22.Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl Acad. Sci. USA. 2002;99:16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aapola U, Kawasaki K, Scott HS, et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65:293–298. doi: 10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- 24.Dong A, Yoder JA, Zhang X, Zhou L, Bestor TH, Cheng X. Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res. 2001;29:439–448. doi: 10.1093/nar/29.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25■.Sinkkonen L, Hugenschmidt T, Berninger P, et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [of interestOne of the first reports to demonstrate that Dicer-mediated microRNA biogenesis is implicated in regulation of DNA methylation.] [DOI] [PubMed] [Google Scholar]
- 26■.Benetti R, Gonzalo S, Jaco I, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat. Struct. Mol. Biol. 2008;15:268–279. doi: 10.1038/nsmb.1399. [of interestOne of the first reports to demonstrate that Dicer-mediated microRNA biogenesis is implicated in regulation of DNA methylation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 28.Girault I, Tozlu S, Lidereau R, Bieche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin. Cancer Res. 2003;9:4415–4422. [PubMed] [Google Scholar]
- 29.Roll JD, Rivenbark AG, Jones WD, Coleman WB. DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines. Mol. Cancer. 2008;7:15. doi: 10.1186/1476-4598-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ehrlich M. DNA hypomethylation, cancer, the immunodeficiency, centromeric region instability, facial anomalies syndrome and chromosomal rearrangements. J. Nutr. 2002;132:S2424–S2429. doi: 10.1093/jn/132.8.2424S. [DOI] [PubMed] [Google Scholar]
- 31.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 32.Narayan A, Ji W, Zhang XY, et al. Hypomethylation of pericentromeric DNA in breast adenocarcinomas. Int. J. Cancer. 1998;77:833–838. doi: 10.1002/(sici)1097-0215(19980911)77:6<833::aid-ijc6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 33.Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 34.Kaneda A, Feinberg AP. Loss of imprinting of IGF2: a common epigenetic modifier of intestinal tumor risk. Cancer Res. 2005;65:11236–11240. doi: 10.1158/0008-5472.CAN-05-2959. [DOI] [PubMed] [Google Scholar]
- 35.Soares J, Pinto AE, Cunha CV, et al. Global DNA hypomethylation in breast carcinoma: correlation with prognostic factors and tumor progression. Cancer. 1999;85:112–118. [PubMed] [Google Scholar]
- 36.Gupta A, Godwin AK, Vanderveer L, Lu A, Liu J. Hypomethylation of the synuclein γ gene CpG island promotes its aberrant expression in breast carcinoma and ovarian carcinoma. Cancer Res. 2003;63:664–673. [PubMed] [Google Scholar]
- 37.Pakneshan P, Szyf M, Farias-Eisner R, Rabbani SA. Reversal of the hypomethylation status of urokinase (uPA) promoter blocks breast cancer growth and metastasis. J. Biol. Chem. 2004;279:31735–31744. doi: 10.1074/jbc.M401669200. [DOI] [PubMed] [Google Scholar]
- 38■■.Fan M, Yan PS, Hartman-Frey C, et al. Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogens tamoxifen and fulvestrant. Cancer Res. 2006;66:11954–11966. doi: 10.1158/0008-5472.CAN-06-1666. [of interestPioneering paper provides the genome-wide analysis of gene-expression and DNA-methylation profiles for an in-depth understanding of the molecular changes specific to acquired resistance to clinically important anti-estrogens.] [DOI] [PubMed] [Google Scholar]
- 39.Knudson AG. Chasing the cancer demon. Annu. Rev. Genet. 2000;34:1–19. doi: 10.1146/annurev.genet.34.1.1. [DOI] [PubMed] [Google Scholar]
- 40.Silva J, Silva JM, Dominguez G, et al. Concomitant expression of p16INK4a and p14ARF in primary breast cancer and analysis of inactivation mechanisms. J. Pathol. 2003;199:289–297. doi: 10.1002/path.1297. [DOI] [PubMed] [Google Scholar]
- 41.Jin Z, Tamura G, Tsuchiya T, et al. Adenomatous polyposis coli (APC) gene promoter hypermethylation in primary breast cancers. Br. J. Cancer. 2001;85:69–73. doi: 10.1054/bjoc.2001.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Birgisdottir V, Stefansson OA, Bodvarsdottir SK, Hilmarsdottir H, Jonasson JG, Eyfjord JE. Epigenetic silencing and deletion of the BRCA1 gene in sporadic breast cancer. Breast Cancer Res. 2006;8:R38. doi: 10.1186/bcr1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehmann U, Hasemeier B, Christgen M, et al. Epigenetic inactivation of microRNA gene hsa-mir-9−1 in human breast cancer. J. Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- 44.Tlsty TD, Crawford YG, Holst CR, et al. Genetic and epigenetic changes in mammary epithelial cells may mimic early events in carcinogenesis. J. Mammary Gland Biol. Neoplasia. 2004;9:263–274. doi: 10.1023/B:JOMG.0000048773.95897.5f. [DOI] [PubMed] [Google Scholar]
- 45.Janzen V, Forkert R, Fleming HE, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 46.Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 47.Reynolds PA, Sigaroudinia M, Zardo G, et al. Tumor suppressor p16INK4A regulates polycomb-mediated DNA hypermethylation in human mammary epithelial cells. J. Biol. Chem. 2006;281:24790–24802. doi: 10.1074/jbc.M604175200. [DOI] [PubMed] [Google Scholar]
- 48.Esteller M. Epigenetic lesions causing genetic lesions in human cancer: promoter hypermethylation of DNA repair genes. Eur. J. Cancer. 2000;36:2294–2300. doi: 10.1016/s0959-8049(00)00303-8. [DOI] [PubMed] [Google Scholar]
- 49.Lo PK, Mehrotra J, D'Costa A, et al. Epigenetic suppression of secreted frizzled related protein 1 (SFRP1) expression in human breast cancer. Cancer Biol. Ther. 2006;5:281–286. doi: 10.4161/cbt.5.3.2384. [DOI] [PubMed] [Google Scholar]
- 50.Ai L, Tao Q, Zhong S, et al. Inactivation of Wnt inhibitory factor-1 (WIF1) expression by epigenetic silencing is a common event in breast cancer. Carcinogenesis. 2006;27:1341–1348. doi: 10.1093/carcin/bgi379. [DOI] [PubMed] [Google Scholar]
- 51.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 52.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc. Natl Acad. Sci. USA. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schubeler D, MacAlpine DM, Scalzo D, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 55.Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 56.Daniel JA, Pray-Grant MG, Grant PA. Effector proteins for methylated histones: an expanding family. Cell Cycle. 2005;4:919–926. doi: 10.4161/cc.4.7.1824. [DOI] [PubMed] [Google Scholar]
- 57.Rice JC, Briggs SD, Ueberheide B, et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 58.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 59.Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 60.Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- 61■■.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [of considerable interestAn excellent review paper provides a detailed overview of recent advances in the understanding of the molecular mechanisms underlying the anticancer effects of histone deacetylases (HDAC) inhibitors and discusses how to use such information for optimizing the development and application of these agents in the clinic.] [DOI] [PubMed] [Google Scholar]
- 62.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 63.Kim TY, Bang YJ, Robertson KD. Histone deacetylase inhibitors for cancer therapy. Epigenetics. 2006;1:14–23. doi: 10.4161/epi.1.1.2644. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc. Natl Acad. Sci. USA. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- 67.Li J, Wang J, Wang J, et al. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 69.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 70.Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J. Biol. Chem. 2002;277:25748–25755. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- 72■■.Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005;37:391–400. doi: 10.1038/ng1531. [of considerable interestFirst study to define the global changes in histone H4 modifications in cancer.] [DOI] [PubMed] [Google Scholar]
- 73.Bannister AJ, Miska EA, Gorlich D, Kouzarides T. Acetylation of importin-α nuclear import factors by CBP/p300. Curr. Biol. 2000;10:467–470. doi: 10.1016/s0960-9822(00)00445-0. [DOI] [PubMed] [Google Scholar]
- 74.Blander G, Zalle N, Daniely Y, Taplick J, Gray MD, Oren M. DNA damage-induced translocation of the Werner helicase is regulated by acetylation. J. Biol. Chem. 2002;277:50934–50940. doi: 10.1074/jbc.M210479200. [DOI] [PubMed] [Google Scholar]
- 75.Hubbert C, Guardiola A, Shao R, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 76.Wolf D, Rodova M, Miska EA, Calvet JP, Kouzarides T. Acetylation of β-catenin by CREB-binding protein (CBP). J. Biol. Chem. 2002;277:25562–25567. doi: 10.1074/jbc.M201196200. [DOI] [PubMed] [Google Scholar]
- 77.Fuino L, Bali P, Wittmann S, et al. Histone deacetylase inhibitor LAQ824 down-regulates Her-2 and sensitizes human breast cancer cells to trastuzumab, taxotere, gemcitabine, and epothilone B. Mol. Cancer Ther. 2003;2:971–984. [PubMed] [Google Scholar]
- 78.Cohen HY, Lavu S, Bitterman KJ, et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol. Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 79.Bali P, Pranpat M, Bradner J, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J. Biol. Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 80.Briggs SD, Bryk M, Strahl BD, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dou Y, Milne TA, Tackett AJ, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 82■.Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, 3rd, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [of interestFirst report to demonstrate a component of a chromatin-associated complex is able to bind methyl-H3K4 and link this active histone-methylation mark to histone acetylation.] [DOI] [PubMed] [Google Scholar]
- 83.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 84.Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 85.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 86■.Lund AH, van Lohuizen M. Epigenetics and cancer. Genes Dev. 2004;18:2315–2335. doi: 10.1101/gad.1232504. [of interestHighlights key mechanisms underlying epigenetics-mediated regulation of gene expression and connections between epigenetic deregulation of gene expression and tumorigenesis.] [DOI] [PubMed] [Google Scholar]
- 87.Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 88.Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol. Cell. 2004;14:183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- 89.Kuzmichev A, Margueron R, Vaquero A, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc. Natl Acad. Sci. USA. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyllysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- 92■■.Keshet I, Schlesinger Y, Farkash S, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 2006;38:149–153. doi: 10.1038/ng1719. [of considerable interestThrough the genome-wide analysis using Methylated DNA immunoprecipitation (methyl-DIP), this paper introduces a novel hypothesis that cancer-related de novo methylation may come about through a preprogrammed mechanism.] [DOI] [PubMed] [Google Scholar]
- 93■■.Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007;39:232–236. doi: 10.1038/ng1950. [of considerable interestThrough the genome-wide analysis using methyl-DIP, this paper introduces a novel hypothesis that cancer-related de novo methylation may come about through a preprogrammed mechanism.] [DOI] [PubMed] [Google Scholar]
- 94.Dimri GP, Martinez JL, Jacobs JJ, et al. The Bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. Cancer Res. 2002;62:4736–4745. [PubMed] [Google Scholar]
- 95.Kleer CG, Cao Q, Varambally S, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl Acad. Sci. USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lehnertz B, Ueda Y, Derijck AA, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 98■.Tachibana M, Ueda J, Fukuda M, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [of interestReports the first identification of two methyltransferases that cooperate together to mediate the methylation of euchromatic histone H3 lysine 9 (H3K9).] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 100.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 102■■.Clouaire T, Stancheva I. Methyl-CpG binding proteins: specialized transcriptional repressors or structural components of chromatin? Cell Mol. Life Sci. 2008;65:1509–1522. doi: 10.1007/s00018-008-7324-y. [of considerable interestExcellent review that provides a detailed overview of recent advances in the roles for methyl-CpG-binding proteins in epigenetic regulation of chromatin structure.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103■■.Jones PL, Veenstra GJ, Wade PA, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [of considerable interestImportant paper that presents the first functional link between the methyl-CpG binding protein and histone deacetylase.] [DOI] [PubMed] [Google Scholar]
- 104.Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 105.Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J. Biol. Chem. 2003;278:4806–4812. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- 106.Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 107.Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 108.Fujita N, Watanabe S, Ichimura T, et al. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J. Biol. Chem. 2003;278:24132–24138. doi: 10.1074/jbc.M302283200. [DOI] [PubMed] [Google Scholar]
- 109.Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell. 2003;12:723–734. doi: 10.1016/j.molcel.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 110.Harikrishnan KN, Chow MZ, Baker EK, et al. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet. 2005;37:254–264. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- 111.Le Guezennec X, Vermeulen M, Brinkman AB, et al. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell. Biol. 2006;26:843–851. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bakker J, Lin X, Nelson WG. Methyl-CpG binding domain protein 2 represses transcription from hypermethylated pi-class glutathione S-transferase gene promoters in hepatocellular carcinoma cells. J. Biol. Chem. 2002;277:22573–22580. doi: 10.1074/jbc.M203009200. [DOI] [PubMed] [Google Scholar]
- 113.Lopez-Serra L, Ballestar E, Fraga MF, Alaminos M, Setien F, Esteller M. A profile of methyl-CpG binding domain protein occupancy of hypermethylated promoter CpG islands of tumor suppressor genes in human cancer. Cancer Res. 2006;66:8342–8346. doi: 10.1158/0008-5472.CAN-06-1932. [DOI] [PubMed] [Google Scholar]
- 114.Feng Q, Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 2001;15:827–832. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fenton SE. Endocrine-disrupting compounds and mammary gland development: early exposure and later life consequences. Endocrinology. 2006;147:S18–S24. doi: 10.1210/en.2005-1131. [DOI] [PubMed] [Google Scholar]
- 116■■.Bloushtain-Qimron N, Yao J, Snyder EL, et al. Cell type-specific DNA methylation patterns in the human breast. Proc. Natl Acad. Sci. USA. 2008;105:14076–14081. doi: 10.1073/pnas.0805206105. [of considerable interestReports important evidence to suggest epigenetically regulated transcription factors are critically implicated in controlling breast epithelial cell phenotypes, and epigenetic programs involved in defining progenitor cell characteristics exhibit the similarities to those from embryonic stem cells.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shipitsin M, Campbell LL, Argani P, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 118■■.Cheng AS, Culhane AC, Chan MW, et al. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008;68:1786–1796. doi: 10.1158/0008-5472.CAN-07-5547. [of considerable interestProvides the in vitro evidence to demonstrate exposure of breast progenitor/stem cells to estrogen can reprogram the epigenetic mechanisms to silence some critical tumor suppressor genes, an important finding that for the first time links epigenetic regulation to estrogen imprinting phenomenon.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119■■.Ottaviano YL, Issa JP, Parl FF, Smith HS, Baylin SB, Davidson NE. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res. 1994;54:2552–2555. [of considerable interestImportant paper showing that silencing of estrogen receptor (ER)α expression in human breast cancer cells is mainly attributable to ERα promoter hypermethylation.] [PubMed] [Google Scholar]
- 120■■.Lapidus RG, Nass SJ, Butash KA, et al. Mapping of ER gene CpG island methylation-specific polymerase chain reaction. Cancer Res. 1998;58:2515–2519. [of considerable interestImportant paper showing that silencing of estrogen receptor (ER)α expression in human breast cancer cells is mainly attributable to ERα promoter hypermethylation.] [PubMed] [Google Scholar]
- 121■■.Leu YW, Yan PS, Fan M, et al. Loss of estrogen receptor signaling triggers epigenetic silencing of downstream targets in breast cancer. Cancer Res. 2004;64:8184–8192. doi: 10.1158/0008-5472.CAN-04-2045. [of considerable interestProvides the first report to demonstrate abrogation of estrogen receptor signaling causes progressively epigenetic inactivation of estrogen receptor downstream targets in ER-positive breast cancer cells. This finding may explain how ER-negative breast cancer cells acquire the tumorigenic ability in the absence of ER-dependent growth-promoting signaling.] [DOI] [PubMed] [Google Scholar]
- 122.Iorns E, Turner NC, Elliott R, et al. Identification of CDK10 as an important determinant of resistance to endocrine therapy for breast cancer. Cancer Cell. 2008;13:91–104. doi: 10.1016/j.ccr.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 123.Fraga MF, Esteller M. DNA methylation: a profile of methods and applications. Biotechniques. 2002;33:632, 634, 636–649. doi: 10.2144/02333rv01. [DOI] [PubMed] [Google Scholar]
- 124.Bird AP, Southern EM. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J. Mol. Biol. 1978;118:27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- 125.Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126■.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [of interestReports an innovative technique for highly sensitive detection of methylated alleles.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128■.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [of interestReports an improved quantitative methylation-specific PCR technique for highly sensitive quantitation of methylated alleles.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fackler MJ, McVeigh M, Mehrotra J, et al. Quantitative multiplex methylation-specific PCR assay for the detection of promoter hypermethylation in multiple genes in breast cancer. Cancer Res. 2004;64:4442–4452. doi: 10.1158/0008-5472.CAN-03-3341. [DOI] [PubMed] [Google Scholar]
- 130.Swift-Scanlan T, Blackford A, Argani P, Sukumar S, Fackler MJ. Two-color quantitative multiplex methylation-specific PCR. Biotechniques. 2006;40:210–219. doi: 10.2144/000112097. [DOI] [PubMed] [Google Scholar]
- 131.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat. Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 132■.Costello JF, Fruhwald MC, Smiraglia DJ, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat. Genet. 2000;24:132–138. doi: 10.1038/72785. [of interestImportant paper that provides extensive analysis of a number of hypermethylated CpG islands in cancer cells and their distribution according to tumour type.] [DOI] [PubMed] [Google Scholar]
- 133.Toyota M, Ho C, Ahuja N, et al. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59:2307–2312. [PubMed] [Google Scholar]
- 134.Frigola J, Ribas M, Risques RA, Peinado MA. Methylome profiling of cancer cells by amplification of inter-methylated sites (AIMS). Nucleic Acids Res. 2002;30:E28. doi: 10.1093/nar/30.7.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Hum. Mol. Genet. 1999;8:459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- 136.Khulan B, Thompson RF, Ye K, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137■.Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [of interestFirst report on the use of methyl-DIP for defining the profiles of genome-wide DNA methylation patterns in normal and transformed cells.] [DOI] [PubMed] [Google Scholar]
- 138.Weber M, Hellmann I, Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 139■.Suzuki H, Gabrielson E, Chen W, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat. Genet. 2002;31:141–149. doi: 10.1038/ng892. [of interestOne of the first studies of genome-wide screening of epigenetically silenced genes in cancer cells using the pharmacologic unmasking strategy in conjunction with expression microarrays.] [DOI] [PubMed] [Google Scholar]
- 140■.Yamashita K, Upadhyay S, Osada M, et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2:485–495. doi: 10.1016/s1535-6108(02)00215-5. [of interestOne of the first studies of genome-wide screening of epigenetically silenced genes in cancer cells using the pharmacologic unmasking strategy in conjunction with expression microarrays.] [DOI] [PubMed] [Google Scholar]
- 141.Hoque MO, Kim MS, Ostrow KL, et al. Genome-wide promoter analysis uncovers portions of the cancer methylome. Cancer Res. 2008;68:2661–2670. doi: 10.1158/0008-5472.CAN-07-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hu M, Yao J, Polyak K. Methylation-specific digital karyotyping. Nat. Protoc. 2006;1:1621–1636. doi: 10.1038/nprot.2006.278. [DOI] [PubMed] [Google Scholar]
- 143.Bernstein BE, Kamal M, Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 144.Roh TY, Ngau WC, Cui K, Landsman D, Zhao K. High-resolution genome-wide mapping of histone modifications. Nat. Biotechnol. 2004;22:1013–1016. doi: 10.1038/nbt990. [DOI] [PubMed] [Google Scholar]
- 145.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 146.Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147■.Evron E, Dooley WC, Umbricht CB, et al. Detection of breast cancer cells in ductal lavage fluid by methylation-specific PCR. Lancet. 2001;357:1335–1336. doi: 10.1016/s0140-6736(00)04501-3. [of interestFirst paper to demonstrate that methylated marker genes can potentially serve as biomarkers for detection of breast cancer cells in ductal fluid specimens.] [DOI] [PubMed] [Google Scholar]
- 148.Fackler MJ, Malone K, Zhang Z, et al. Quantitative multiplex methylation-specific PCR analysis doubles detection of tumor cells in breast ductal fluid. Clin. Cancer Res. 2006;12:3306–3310. doi: 10.1158/1078-0432.CCR-05-2733. [DOI] [PubMed] [Google Scholar]
- 149.Widschwendter M, Menon U. Circulating methylated DNA: a new generation of tumor markers. Clin. Cancer Res. 2006;12:7205–7208. doi: 10.1158/1078-0432.CCR-06-2531. [DOI] [PubMed] [Google Scholar]
- 150.Silva JM, Silva J, Sanchez A, et al. Tumor DNA in plasma at diagnosis of breast cancer patients is a valuable predictor of disease-free survival. Clin. Cancer Res. 2002;8:3761–3766. [PubMed] [Google Scholar]
- 151.Muller HM, Widschwendter A, Fiegl H, et al. DNA methylation in serum of breast cancer patients: an independent prognostic marker. Cancer Res. 2003;63:7641–7645. [PubMed] [Google Scholar]
- 152.Lewis CM, Cler LR, Bu DW, et al. Promoter hypermethylation in benign breast epithelium in relation to predicted breast cancer risk. Clin. Cancer Res. 2005;11:166–172. [PubMed] [Google Scholar]
- 153.Arai T, Miyoshi Y, Kim SJ, Taguchi T, Tamaki Y, Noguchi S. Association of GSTP1 CpG islands hypermethylation with poor prognosis in human breast cancers. Breast Cancer Res. Treat. 2006;100:169–176. doi: 10.1007/s10549-006-9241-9. [DOI] [PubMed] [Google Scholar]
- 154.Veeck J, Niederacher D, An H, et al. Aberrant methylation of the Wnt antagonist SFRP1 in breast cancer is associated with unfavourable prognosis. Oncogene. 2006;25:3479–3488. doi: 10.1038/sj.onc.1209386. [DOI] [PubMed] [Google Scholar]
- 155.Widschwendter M, Siegmund KD, Muller HM, et al. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Res. 2004;64:3807–3813. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- 156.Fields AL, Soprano DR, Soprano KJ. Retinoids in biological control and cancer. J. Cell Biochem. 2007;102:886–898. doi: 10.1002/jcb.21530. [DOI] [PubMed] [Google Scholar]
- 157.Bovenzi V, Le NL, Cote S, Sinnett D, Momparler LF, Momparler RL. DNA methylation of retinoic acid receptor β in breast cancer and possible therapeutic role of 5-aza-2′-deoxycytidine. Anticancer Drugs. 1999;10:471–476. doi: 10.1097/00001813-199906000-00007. [DOI] [PubMed] [Google Scholar]
- 158.Sirchia SM, Ferguson AT, Sironi E, et al. Evidence of epigenetic changes affecting the chromatin state of the retinoic acid receptor β2 promoter in breast cancer cells. Oncogene. 2000;19:1556–1563. doi: 10.1038/sj.onc.1203456. [DOI] [PubMed] [Google Scholar]
- 159.Hellebrekers DM, Griffioen AW, van Engeland M. Dual targeting of epigenetic therapy in cancer. Biochim. Biophys. Acta. 2007;1775:76–91. doi: 10.1016/j.bbcan.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 160.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 161.Cheng JC, Yoo CB, Weisenberger DJ, et al. Preferential response of cancer cells to zebularine. Cancer Cell. 2004;6:151–158. doi: 10.1016/j.ccr.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 162.Villar-Garea A, Fraga MF, Espada J, Esteller M. Procaine is a DNA-demethylating agent with growth-inhibitory effects in human cancer cells. Cancer Res. 2003;63:4984–4989. [PubMed] [Google Scholar]
- 163.Segura-Pacheco B, Trejo-Becerril C, Perez-Cardenas E, et al. Reactivation of tumor suppressor genes by the cardiovascular drugs hydralazine and procainamide and their potential use in cancer therapy. Clin. Cancer Res. 2003;9:1596–1603. [PubMed] [Google Scholar]
- 164.Fang MZ, Wang Y, Ai N, et al. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–7570. [PubMed] [Google Scholar]
- 165.Brueckner B, Boy RG, Siedlecki P, et al. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305–6311. doi: 10.1158/0008-5472.CAN-04-2957. [DOI] [PubMed] [Google Scholar]
- 166.Yan L, Nass SJ, Smith D, Nelson WG, Herman JG, Davidson NE. Specific inhibition of DNMT1 by antisense oligonucleotides induces re-expression of estrogen receptor-α (ER) in ER-negative human breast cancer cell lines. Cancer Biol. Ther. 2003;2:552–556. doi: 10.4161/cbt.2.5.469. [DOI] [PubMed] [Google Scholar]
- 167.Kelly WK, Marks PA. Drug insight: Histone deacetylase inhibitors – development of the new targeted anticancer agent suberoylanilide hydroxamic acid. Nat. Clin. Pract. Oncol. 2005;2:150–157. doi: 10.1038/ncponc0106. [DOI] [PubMed] [Google Scholar]
- 168.Dokmanovic M, Marks PA. Prospects: histone deacetylase inhibitors. J. Cell Biochem. 2005;96:293–304. doi: 10.1002/jcb.20532. [DOI] [PubMed] [Google Scholar]
- 169.Garber K. HDAC inhibitors overcome first hurdle. Nat. Biotechnol. 2007;25:17–19. doi: 10.1038/nbt0107-17. [DOI] [PubMed] [Google Scholar]
- 170.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 171.Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-κB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 172.Costanzo A, Merlo P, Pediconi N, et al. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol. Cell. 2002;9:175–186. doi: 10.1016/s1097-2765(02)00431-8. [DOI] [PubMed] [Google Scholar]
- 173.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 174.Ocker M, Schneider-Stock R. Histone deacetylase inhibitors: signalling towards p21cip1/waf1. Int. J. Biochem. Cell Biol. 2007;39:1367–1374. doi: 10.1016/j.biocel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 175.Cimini D, Mattiuzzo M, Torosantucci L, Degrassi F. Histone hyperacetylation in mitosis prevents sister chromatid separation and produces chromosome segregation defects. Mol. Biol. Cell. 2003;14:3821–3833. doi: 10.1091/mbc.E03-01-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Subramanian C, Opipari AW, Jr, Bian X, Castle VP, Kwok RP. Ku70 acetylation mediates neuroblastoma cell death induced by histone deacetylase inhibitors. Proc. Natl Acad. Sci. USA. 2005;102:4842–4847. doi: 10.1073/pnas.0408351102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fiskus W, Ren Y, Mohapatra A, et al. Hydroxamic acid analogue histone deacetylase inhibitors attenuate estrogen receptor-α levels and transcriptional activity: a result of hyperacetylation and inhibition of chaperone function of heat shock protein 90. Clin. Cancer Res. 2007;13:4882–4890. doi: 10.1158/1078-0432.CCR-06-3093. [DOI] [PubMed] [Google Scholar]
- 178■■.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 1999;21:103–107. doi: 10.1038/5047. [of considerable interestDescribes the first evidence to show synergy between demethylating agents and HDAC inhibitors (HDACIs) in reactivating epigenetically silenced genes.] [DOI] [PubMed] [Google Scholar]
- 179■■.Yang X, Phillips DL, Ferguson AT, Nelson WG, Herman JG, Davidson NE. Synergistic activation of functional estrogen receptor (ER)-α by DNA methyltransferase and histone deacetylase inhibition in human ER-α-negative breast cancer cells. Cancer Res. 2001;61:7025–7029. [of considerable interestOne of two important papers to show co-treatment with demethylating agents and HDACI synergistically reverse silencing of ERα expression in ER-negative breast cancer cells and re-sensitize endocrine-resistant cancer cells to the anti-estrogen agent.] [PubMed] [Google Scholar]
- 180■■.Sharma D, Saxena NK, Davidson NE, Vertino PM. Restoration of tamoxifen sensitivity in estrogen receptor-negative breast cancer cells: tamoxifen-bound reactivated ER recruits distinctive corepressor complexes. Cancer Res. 2006;66:6370–6378. doi: 10.1158/0008-5472.CAN-06-0402. [of considerable interestOne of two important papers to show co-treatment with demethylating agents and HDACI synergistically reverse silencing of ERα expression in ER-negative breast cancer cells and re-sensitize endocrine-resistant cancer cells to the anti-estrogen agent.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mongan NP, Gudas LJ. Valproic acid, in combination with all-trans retinoic acid and 5-aza-2′-deoxycytidine, restores expression of silenced RARβ2 in breast cancer cells. Mol. Cancer Ther. 2005;4:477–486. doi: 10.1158/1535-7163.MCT-04-0079. [DOI] [PubMed] [Google Scholar]
- 182.Beltran AS, Sun X, Lizardi PM, Blancafort P. Reprogramming epigenetic silencing: artificial transcription factors synergize with chromatin remodeling drugs to reactivate the tumor suppressor mammary serine protease inhibitor. Mol. Cancer Ther. 2008;7:1080–1090. doi: 10.1158/1535-7163.MCT-07-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kim MS, Blake M, Baek JH, Kohlhagen G, Pommier Y, Carrier F. Inhibition of histone deacetylase increases cytotoxicity to anticancer drugs targeting DNA. Cancer Res. 2003;63:7291–7300. [PubMed] [Google Scholar]
- 184.Bali P, Pranpat M, Swaby R, et al. Activity of suberoylanilide hydroxamic acid against human breast cancer cells with amplification of her-2. Clin. Cancer Res. 2005;11:6382–6389. doi: 10.1158/1078-0432.CCR-05-0344. [DOI] [PubMed] [Google Scholar]
- 185.Zhou Q, Shaw PG, Davidson NE. Inhibition of histone deacetylase suppresses EGF signaling pathways by destabilizing EGFR mRNA in ER-negative human breast cancer cells. Breast Cancer Res. Treat. 2008 doi: 10.1007/s10549-008-0148-5. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 186.Margueron R, Duong V, Bonnet S, et al. Histone deacetylase inhibition and estrogen receptor α levels modulate the transcriptional activity of partial antiestrogens. J. Mol. Endocrinol. 2004;32:583–594. doi: 10.1677/jme.0.0320583. [DOI] [PubMed] [Google Scholar]
- 187.De los Santos M, Martinez-Iglesias O, Aranda A. Anti-estrogenic actions of histone deacetylase inhibitors in MCF-7 breast cancer cells. Endocr. Relat. Cancer. 2007;14:1021–1028. doi: 10.1677/ERC-07-0144. [DOI] [PubMed] [Google Scholar]
- 188.Arce C, Perez-Plasencia C, Gonzalez-Fierro A, et al. A proof-of-principle study of epigenetic therapy added to neoadjuvant doxorubicin cyclophosphamide for locally advanced breast cancer. PLoS ONE. 2006;1:E98. doi: 10.1371/journal.pone.0000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chang EL, Lo S. Diagnosis and management of central nervous system metastases from breast cancer. Oncologist. 2003;8:398–410. doi: 10.1634/theoncologist.8-5-398. [DOI] [PubMed] [Google Scholar]
- 190.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 191.Munshi A, Kurland JF, Nishikawa T, et al. Histone deacetylase inhibitors radiosensitize human melanoma cells by suppressing DNA repair activity. Clin. Cancer Res. 2005;11:4912–4922. doi: 10.1158/1078-0432.CCR-04-2088. [DOI] [PubMed] [Google Scholar]
- 192.Nome RV, Bratland A, Harman G, Fodstad O, Andersson Y, Ree AH. Cell cycle checkpoint signaling involved in histone deacetylase inhibition and radiation-induced cell death. Mol. Cancer Ther. 2005;4:1231–1238. doi: 10.1158/1535-7163.MCT-04-0304. [DOI] [PubMed] [Google Scholar]
- 193.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 194.Moving AHEAD with an international human epigenome project Nature. 2008;454:711–715. doi: 10.1038/454711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ferguson AT, Evron E, Umbricht CB, et al. High frequency of hypermethylation at the 14−3−3σ locus leads to gene silencing in breast cancer. Proc. Natl Acad. Sci. USA. 2000;97:6049–6054. doi: 10.1073/pnas.100566997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Umbricht CB, Evron E, Gabrielson E, Ferguson A, Marks J, Sukumar S. Hypermethylation of 14−3−3σ (stratifin) is an early event in breast cancer. Oncogene. 2001;20:3348–3353. doi: 10.1038/sj.onc.1204438. [DOI] [PubMed] [Google Scholar]
- 197.Virmani AK, Rathi A, Sathyanarayana UG, et al. Aberrant methylation of the adenomatous polyposis coli (APC) gene promoter 1A in breast and lung carcinomas. Clin. Cancer Res. 2001;7:1998–2004. [PubMed] [Google Scholar]
- 198.Evron E, Umbricht CB, Korz D, et al. Loss of cyclin D2 expression in the majority of breast cancers is associated with promoter hypermethylation. Cancer Res. 2001;61:2782–2787. [PubMed] [Google Scholar]
- 199.Caldeira JR, Prando EC, Quevedo FC, Neto FA, Rainho CA, Rogatto SR. CDH1 promoter hypermethylation and E-cadherin protein expression in infiltrating breast cancer. BMC Cancer. 2006;6:48. doi: 10.1186/1471-2407-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Toyooka KO, Toyooka S, Virmani AK, et al. Loss of expression and aberrant methylation of the CDH13 (H-cadherin) gene in breast and lung carcinomas. Cancer Res. 2001;61:4556–4560. [PubMed] [Google Scholar]
- 201.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 202.Zochbauer-Muller S, Fong KM, Maitra A, et al. 5′ CpG island methylation of the FHIT gene is correlated with loss of gene expression in lung and breast cancer. Cancer Res. 2001;61:3581–3585. [PubMed] [Google Scholar]
- 203.Xiang YY, Ladeda V, Filmus J. Glypican-3 expression is silenced in human breast cancer. Oncogene. 2001;20:7408–7412. doi: 10.1038/sj.onc.1204925. [DOI] [PubMed] [Google Scholar]
- 204.Esteller M, Corn PG, Urena JM, Gabrielson E, Baylin SB, Herman JG. Inactivation of glutathione S-transferase P1 gene by promoter hypermethylation in human neoplasia. Cancer Res. 1998;58:4515–4518. [PubMed] [Google Scholar]
- 205.Fujii H, Biel MA, Zhou W, Weitzman SA, Baylin SB, Gabrielson E. Methylation of the HIC-1 candidate tumor suppressor gene in human breast cancer. Oncogene. 1998;16:2159–2164. doi: 10.1038/sj.onc.1201976. [DOI] [PubMed] [Google Scholar]
- 206.Krop IE, Sgroi D, Porter DA, et al. HIN-1, a putative cytokine highly expressed in normal but not cancerous mammary epithelial cells. Proc. Natl Acad. Sci. USA. 2001;98:9796–9801. doi: 10.1073/pnas.171138398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Li B, Goyal J, Dhar S, et al. CpG methylation as a basis for breast tumor-specific loss of NES1/kallikrein 10 expression. Cancer Res. 2001;61:8014–8021. [PubMed] [Google Scholar]
- 208.Hartsough MT, Clare SE, Mair M, et al. Elevation of breast carcinoma Nm23-H1 metastasis suppressor gene expression and reduced motility by DNA methylation inhibition. Cancer Res. 2001;61:2320–2327. [PubMed] [Google Scholar]
- 209.Yu Y, Xu F, Peng H, et al. NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas. Proc. Natl Acad. Sci. USA. 1999;96:214–219. doi: 10.1073/pnas.96.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hesson L, Dallol A, Minna JD, Maher ER, Latif F. NORE1A, a homologue of RASSF1A tumour suppressor gene is inactivated in human cancers. Oncogene. 2003;22:947–954. doi: 10.1038/sj.onc.1206191. [DOI] [PubMed] [Google Scholar]
- 211.Lapidus RG, Ferguson AT, Ottaviano YL, et al. Methylation of estrogen and progesterone receptor gene 5′ CpG islands correlates with lack of estrogen and progesterone receptor gene expression in breast tumors. Clin. Cancer Res. 1996;2:805–810. [PubMed] [Google Scholar]
- 212.Dammann R, Yang G, Pfeifer GP. Hypermethylation of the cpG island of Ras association domain family 1A (RASSF1A), a putative tumor suppressor gene from the 3p21.3 locus, occurs in a large percentage of human breast cancers. Cancer Res. 2001;61:3105–3109. [PubMed] [Google Scholar]
- 213.Du Y, Carling T, Fang W, Piao Z, Sheu JC, Huang S. Hypermethylation in human cancers of the RIZ1 tumor suppressor gene, a member of a histone/protein methyltransferase superfamily. Cancer Res. 2001;61:8094–8099. [PubMed] [Google Scholar]
- 214.Lau QC, Raja E, Salto-Tellez M, et al. RUNX3 is frequently inactivated by dual mechanisms of protein mislocalization and promoter hypermethylation in breast cancer. Cancer Res. 2006;66:6512–6520. doi: 10.1158/0008-5472.CAN-06-0369. [DOI] [PubMed] [Google Scholar]
- 215.Domann FE, Rice JC, Hendrix MJ, Futscher BW. Epigenetic silencing of maspin gene expression in human breast cancers. Int. J. Cancer. 2000;85:805–810. doi: 10.1002/(sici)1097-0215(20000315)85:6<805::aid-ijc12>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 216.Xu XL, Wu LC, Du F, et al. Inactivation of human SRBC, located within the 11p15.5-p15.4 tumor suppressor region, in breast and lung cancers. Cancer Res. 2001;61:7943–7949. [PubMed] [Google Scholar]
- 217.Yuan Y, Mendez R, Sahin A, Dai JL. Hypermethylation leads to silencing of the SYK gene in human breast cancer. Cancer Res. 2001;61:5558–5561. [PubMed] [Google Scholar]
- 218.Bachman KE, Herman JG, Corn PG, et al. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- 219.Conway KE, McConnell BB, Bowring CE, Donald CD, Warren ST, Vertino PM. TMS1, a novel proapoptotic caspase recruitment domain protein, is a target of methylation-induced gene silencing in human breast cancers. Cancer Res. 2000;60:6236–6242. [PubMed] [Google Scholar]