Abstract
Although expanded polyglutamine (polyQ) repeats are inherently toxic, causing at least nine neurodegenerative diseases, the protein context determines which neurons are affected. The polyQ expansion that causes Huntington's disease (HD) is in the first exon (HDx-1) of huntingtin (Htt). However, other parts of the protein, including the 17 N-terminal amino acids and two proline (polyP) repeat domains, regulate the toxicity of mutant Htt. The role of the P-rich domain that is flanked by the polyP domains has not been explored. Using highly specific intracellular antibodies (intrabodies), we tested various epitopes for their roles in HDx-1 toxicity, aggregation, localization, and turnover. Three domains in the P-rich region (PRR) of HDx-1 are defined by intrabodies: MW7 binds the two polyP domains, and Happ1 and Happ3, two new intrabodies, bind the unique, P-rich epitope located between the two polyP epitopes. We find that the PRR-binding intrabodies, as well as VL12.3, which binds the N-terminal 17 aa, decrease the toxicity and aggregation of HDx-1, but they do so by different mechanisms. The PRR-binding intrabodies have no effect on Htt localization, but they cause a significant increase in the turnover rate of mutant Htt, which VL12.3 does not change. In contrast, expression of VL12.3 increases nuclear Htt. We propose that the PRR of mutant Htt regulates its stability, and that compromising this pathogenic epitope by intrabody binding represents a novel therapeutic strategy for treating HD. We also note that intrabody binding represents a powerful tool for determining the function of protein epitopes in living cells.
Keywords: Huntington, neurodegeneration, gene therapy, immunotherapy, intrabody, polyproline
Introduction
Huntington's disease (HD) is an autosomal dominant, progressive, neurodegenerative disorder that results from the expansion of a polyglutamine (polyQ) tract in the first exon (HDx-1) of huntingtin (Htt) (The Huntington's Disease Collaborative Research Group, 1993). At least nine other neurodegenerative diseases are caused by the expansion of a polyQ tract, including several types of spinocerebellar ataxia (Orr et al., 1993; Kawaguchi et al., 1994; Imbert et al., 1996; David et al., 1997), dentatorubral pallidoluysian atrophy (Koide et al., 1994), and spinobulbar muscular atrophy (La Spada et al., 1991). In each case, the polyQ expansion is in a different protein, and although the mutant protein is expressed widely, only a specific subset of neurons unique to each disease die. Although expression of pure polyQ is sufficient to cause toxicity (Marsh et al., 2000; Yang et al., 2002), it is the protein context surrounding the polyQ expansion that makes particular neurons susceptible in each disease. In HD, the mutant protein exhibits toxic gain of function, which includes aggregation, sequestering of important cellular proteins such as transcription factors, and aberrant protein–protein interactions, including disruption of the ubiquitin proteasome (Duyao et al., 1995; Ross, 1997; Wanker, 2000; Jana et al., 2001; Ramaswamy et al., 2007). This leads to chorea, dementia, and loss of medium spiny striatal as well as some cortical neurons (Reddy et al., 1999; Zoghbi and Orr, 2000; Nakamura and Aminoff, 2007). HDx-1 consists of 17 N-terminal amino acids followed by the polyQ tract, the P-rich region (PRR), which consists of two polyP stretches that are separated by a P-rich domain, and 13 additional amino acids (Fig. 1 A). The non-polyQ domains in HDx-1 are known to modulate the toxicity of the mutant protein, although the mechanisms by which this occurs are not well understood (Duennwald et al., 2006). Understanding how these non-polyQ domains contribute to the toxicity and specificity of mutant Htt (mHtt) could lead to new therapeutic strategies.
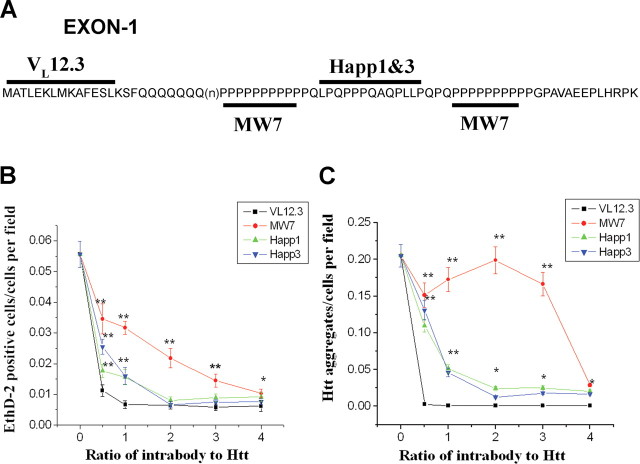
Figure 1.
The anti-Htt intrabodies reduce mHDx-1-induced toxicity and aggregation in cell culture. A, The epitopes in HDx-1 for the various intrabodies are depicted. B, To quantify mHtt toxicity, HEK293 cells were cotransfected with mHDx-1-GFP and intrabody at various intrabody/Htt ratios and incubated for 48 h. Cells were stained with EthD-2 to identify dead cell nuclei, fixed, and stained with DAPI to identify all cell nuclei. The number of dead cells was normalized to total cell number. All of the intrabodies reduce mHDx-1-induced cell death in a saturable, dose-dependent manner, with maximal effects at different intrabody/Htt ratios (1:1 for VL12.3, 2:1 for Happ1 and Happ3, and 4:1 for MW7). C, Aggregation was determined by counting GFP foci and normalizing to total cell number. *Different from VL12.3 at p < 0.05; **p < 0.01. The point labeled as 0 on the intrabody:Htt axis corresponds to the value for HDx-1 + CVL.
Classically, the function of a protein domain would be studied by removal of that domain followed by functional testing. Although a great deal of knowledge has been acquired through such methods, the deletion of a domain may cause altered folding of the remaining protein or otherwise generate effects not related directly to the function of the missing domain. Perturbation of a protein domain by intrabody binding is a more specific method for exploring function. Intrabodies are intracellular, recombinant, single-chain antibody fragments (scFv) that contain the heavy and light antigen-binding domains (VH and VL) connected by a linker. Alternatively, single-domain antibody fragments consist of either VH or VL. Intrabodies are highly specific reagents that can be targeted to subcellular compartments, distinct protein conformations, posttranscriptional modifications, and nonprotein targets such as oligosaccharides (Biocca and Cattaneo, 1995; Stocks, 2005; Messer and McLear, 2006; Lo et al., 2008). Intrabodies thus have great potential to increase our understanding of the functions of individual protein domains in living cells.
We sought to use intrabodies to better understand the role of the polyP and P-rich domains (the PRR) of Htt in HD pathology. The PRR is known to be important for mHtt toxic gain of function (Passani et al., 2000; Steffan et al., 2000; Modregger et al., 2002; Khoshnan et al., 2004; Qin et al., 2004), and although a number of binding partners, including WW domain-containing proteins, vesicle-associated proteins, P53, and IKKγ, have been identified, the mechanism of the modulation of mHtt toxicity by these domains remains unclear. The role of the P-rich domain is not known. To investigate this aspect of PRR function, we used MW7, an scFv intrabody that binds polyP. MW7 reduces mHtt-induced aggregation and promotes cell survival in culture (Khoshnan et al., 2002). It also inhibits mHtt-induced neurodegeneration in a Drosophila HD model (Jackson et al., 2004). However, the specificity of this intrabody for pure polyP could allow binding to other cellular proteins containing a polyP domain, although there is no evidence of the latter binding to date. To characterize the role of the PRR, we produced novel intrabodies (Happs) against the P-rich domain of Htt. Happ1 and Happ3 are single-domain, light chain intrabodies (VLs) that bind mHtt in a PRR-dependent manner. We then tested the Happs, MW7 and VL12.3, a single-domain light chain intrabody that binds the 17 N-terminal amino acids of Htt (Colby et al., 2004b), for efficacy in blocking mHDx-1 aggregation and toxicity, as well as their effects on subcellular localization and mHDx-1 protein levels. The most striking findings are that both the anti-polyP and anti-P-rich intrabodies reduce toxicity by increasing mHtt turnover and lowering the mHtt levels, whereas the anti-N-terminal intrabody appears to reduce mHtt toxicity by a different mechanism.
Materials and Methods
Cell culture.
HEK293 (American Type Culture Collection) or ST14A striatal precursor (Cattaneo, 1998) cells were grown in DMEM (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum, 2 mm glutamine, 1 mm streptomycin, and 100 U of penicillin (Invitrogen). Cells were maintained in 37°C (HEK293) or 33°C (ST14A) incubators with 5% CO2 unless otherwise stated. Transfections were performed using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's protocol.
Immunoblotting.
Protein concentration was determined using a BCA assay (Pierce). Seventy-five micrograms of total protein per sample in a volume of 30 μl was combined with 6 μl of 6× protein loading buffer (Ausubel et al., 1993) and boiled for 5 min. Samples were separated by SDS-PAGE using 4–20% criterion precast gels (Biorad) and Precision Plus Protein Kaleidoscope molecular weight standard (Bio-Rad). Samples were then transferred overnight to nitrocellulose membranes for immunoblotting. Appropriate primary and horseradish peroxidase (HRP)-conjugated secondary antibodies were then applied as described by Ausubel et al. (1993). SuperSignal West Dura (Pierce) substrate was applied to membranes according to the manufacturer's protocol. Chemiluminescence was detected and densitometry was performed using a FluorChem 8900 (Alpha Innotech) gel documentation system.
Selection of phage display library for binding to P-rich epitope of Htt.
Intrabodies were selected from the Griffin.1 human recombinant, scFv phage display library (Griffiths et al., 1994). One well of a six-well plate was coated with a synthetic peptide (200 μg/ml) derived from the P-rich epitope of Htt (PQLPQPPPQAQP) located between the two polyP stretches by incubating at 4°C overnight. The coated well was then used to select phage-expressing intrabodies specific for this epitope according to the provider's instructions. After the fourth round of selection, the P-rich enriched phage library was purified by polyethylene glycol/NaCl precipitation and suspended in 2 ml of PBS.
Generation of bait peptide for isolation of Happ intrabodies.
A plasmid encoding PQ50-glutathione S-transferase (GST) (Scherzinger et al., 1997) was transformed into XL-10 gold ultracompetent bacteria (Stratagene) according to the manufacturer's protocol. Cells were grown to an OD of 0.6 at 600 nm and induced with 1 mm isopropyl β-d-1-thiogalactopyranoside for 4 h. Bacteria were collected by centrifugation, and GST fusion proteins were isolated in 1 ml of 50% Glutathione Sepharose bead slurries containing bound peptide (Ausubel et al., 1993). Twenty-five microliters of each bead slurry were added to 10 μl of protein loading buffer, boiled for 5 min, and separated by SDS-PAGE. Peptide expression was verified by Coomassie staining of PAGE gels and comparison to a protein molecular weight marker (data not shown).
Selection of Happ intrabodies from P-rich-specific phage display library.
One milliliter of the preselected P-rich-specific phage was selected with PQ50-GST as described in the library provider's instructions. Briefly, GST-fusion bait peptide bound to glutathione agarose beads was incubated with replication-deficient phages displaying preselected, P-rich scFvs and then washed in PBS with 0.1% Triton X-100 to remove any unbound phage particles. Bound phages were allowed to infect log phase bacteria. To repeat selection, M13 helper phages, which do not display scFvs but enable the replication of scFv-displaying phages from preinfected bacteria, were used to recover selected phages. This selection was repeated an additional two times. After the final round of selection, individual clones were selected and screened for inserts by the PCR(Griffiths et al., 1994). Six clones with inserts were identified (Happ1–Happ6). Inserts were sequenced and analyzed for open reading frames (ORFs). Three clones were found to contain ORFs, two of which were redundant. The two unique ORFs (Happ1 and Happ3) were amplified by PCR using primers designed to add both appropriate restriction sites and a C-terminal hemagglutinin (HA) epitope tag, and cloned into the adeno-associated virus genome plasmid (AAV) (Stratagene) mammalian expression vector for characterization in cell culture, and into the pGEX-6p1 GST fusion (GE Healthcare) bacterial expression vector for protein purification. A control intrabody that does not bind HDx-1 (CVL) was also isolated from the library and cloned into these vectors. Cloning was performed according to the Invitrogen One Shot Top10 competent cell protocol.
Htt aggregation and toxicity assays.
HEK293 cells were cotransfected with HDx-1-green fluorescent protein (GFP) and intrabody in poly-d-lysine-coated 24-well plates at ∼60% confluency. Each well received 0.2 μg of PQ103 DNA in pcDNA3.1 vector and intrabody (VL12.3, MW7, Happ1, Happ3, or CVL) DNA in AAV vector at various ratios to HDx-1 (0.5:1, 1:1, 2:1, 3:1, and 4:1). DNA levels were normalized to 1 μg per well using CVL in AAV vector. Nontransfected wells were used as a negative control, and each condition was performed in triplicate. Cultures were moved to a 33°C incubator 8 h after transfection to slow cell division and maintain a monolayer. At 40 h after transfection, cells were incubated in medium containing 1 mm ethidium homodimer-2 (EthD-2) (Invitrogen) for 15 min at 33°C for detection of dead cell nuclei. Cells were then fixed in 4% PFA at 4°C for 30 min and permeabilized with PBS containing 0.1% Triton for 15 min. For detection of all nuclei, cells were treated with PBS containing 0.5 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI). Fluorescence microscopy was used to visualize dead cells (red channel), large Htt aggregates (green channel), and total cell number (blue channel). Three representative microscope fields were analyzed for each well (nine per condition). Dead cells and aggregates were counted for each field and normalized to the total cell number. p values were computed using a Student's t test.
Brain slice neurodegeneration assay.
All animal experiments were performed in accordance with the Institutional Animal Care and Use Committee and Duke University Medical Center Animal Guidelines. Brain slice preparation and biolistic transfection were performed as previously described (Lo et al., 1994; Khoshnan et al., 2004). Briefly, brain tissue was dissected from CD Sprague Dawley rats killed on postnatal day 10 (P10) (Charles River Laboratory) and placed in ice-cold culture medium containing 15% heat-inactivated horse serum, 10 mm KCl, 10 mm HEPES, 100 U/ml penicillin/streptomycin, 1 mm sodium pyruvate, and 1 mm l-glutamine in Neurobasal A (Invitrogen). Brain tissue was cut into 250-μm-thick coronal slices using a Vibratome and incubated for 1 h at 37°C under 5.0% CO2 before biolistic transfection. Gold particles (1.6 μm gold microcarriers; Bio-Rad) were coated with the appropriate DNAs (see below) as per the manufacturer's instructions and loaded into Tefzel tubing (McMaster-Carr) for use with the Helios biolistic device (Bio-Rad), which was used at a delivery pressure of 95 psi. Gold particles were coated with expression constructs encoding yellow fluorescent protein (YFP) as a morphometric marker, cyan fluorescent protein (CFP)-tagged mHDx-1 Q-73, and the relevant intrabody; for control transfections, particles were coated with YFP + CVL, YFP + mHDx-1Q73 and vector backbone DNA, or YFP + mHDx-1Q73 + CVL. For each condition, transfections were done on 12 brain slices, and numbers of healthy medium spiny neurons (MSNs) expressing the YFP reporter were assessed 4–5 d after brain slice preparation and transfection using fluorescence microscopy. MSNs with normal-sized cell bodies, even and continuous expression of YFP in the cell body and dendrites, and having more than two discernable primary dendrites more than two cell bodies long were scored as healthy. p values were computed using a Student's t test.
Immunohistochemical HDx-1 localization.
ST14A cells were grown in six-well plates containing coverslips and cotransfected with HDx-1-GFP and intrabody in six-well plates at ∼60% confluency. Each well received 1 μg of PQ103 and intrabody DNA at optimal ratios. Nontransfected wells were used as a negative control. At 48 h after transfection, cells were fixed and permeabilized as described above. Intrabodies were then labeled using M2 anti-Flag for MW7 and 3F10 anti-HA for VL12.3, Happ1, and Happ3. Secondary antibodies were conjugated to Alexa Fluor 568 (Invitrogen) (Hockfield et al., 1993). Cells were processed for microscopy as above. Mean fluorescence intensity for whole-cell and nuclear HDx-1 (green channel) and intrabody (red channel) was measured in three microscope fields per well. The ratio of nuclear HDx-1 or intrabody to cellular HDx-1 or intrabody was determined by (mean intensity of nucleus/mean intensity of whole cell). p values were computed using a Student's t test.
HDx-1 immunoblot assay.
HEK293 cells were cotransfected with HDx-1-GFP and intrabody in 10 cm dishes at ∼80% confluency. Each dish received 4 μg of PQ103 or PQ25 DNA in pcDNA3.1 vector and intrabody DNA in AAV vector at the optimal ratio for each intrabody (4 μg of VL12.3, 16 μg of MW7, 8 μg of Happ1 and Happ3). A nontransfected dish was used as a negative control. Cells were dislodged by mechanical dissociation and pipetting 48 h after transfection, harvested by centrifugation, washed with PBS, and lysed by sonication in 500 μl of lysis buffer (25 mm HEPES, 50 mm NaCl, 1 mm MgCl2, and 0.5% Triton) containing 1 Complete, Mini, EDTA-free Protease Inhibitor Cocktail tablet (Roche) per 7 ml of buffer. The soluble protein fraction was collected by centrifugation for 20 min at 4°C at 20,000 × g. The insoluble pellet was sonicated in 150 μl of 6 m urea and incubated for 20 min at room temperature. Immunoblots were then performed using rabbit anti-GFP (1:1000; Invitrogen) as primary antibody and HRP-conjugated, goat anti-rabbit (1:10,000; Santa Cruz Biotechnology) as secondary antibody to detect HDx-1-GFP. For a loading control, membranes were stripped using Restore Western blot stripping buffer (Pierce) and reprobed with mouse anti-β-tubulin (1:1000; Sigma) as primary and HRP-conjugated, goat anti-mouse (1:10,000; Santa Cruz Biotechnology) as secondary antibody. Densities of HDx-1 and β-tubulin bands were determined. Each HDx-1 band was normalized to the level of the β-tubulin band for that sample. The ratio of HDx-1 level in the presence of intrabody to HDx-1 level alone was determined by (density of intrabody plus HDx-1/density of intrabody plus HDx-1 tubulin)/(density of HDx-1 alone/density of HDx-1 alone tubulin). The experiment was repeated three additional times, giving an n of 4. p values were computed using a Student's t test.
HDx-1 turnover assay.
ST14A cells were grown in six-well plates containing coverslips and cotransfected with HDx-1-SNAP and intrabody at ∼60% confluency. Each well received 1 μg of either PQ97-SNAP (97Q-HDx-1 fused to the SNAP tag) or PQ25-SNAP (25Q-HDx-1 fused to the SNAP tag) DNA in pSEMXT-26m vector (Covalys Witterswil) and intrabody DNA in AAV vector at optimal ratios (1 μg of VL12.3, 4 μg of MW7, 2 μg of Happ1 and Happ3). Nontransfected wells were used as a negative control, and each condition was performed twice. To covalently label HDx-1 present at 24 h after transfection, cells were treated with decay-accelerating factor (DAF) green fluorescent SNAP-substrate (Covalys Witterswil) according to the manufacturer's protocol. After labeling, cells were handled in low light conditions to avoid photobleaching the DAF substrate. One well of each condition was then fixed and permeabilized as described above. For detection of all nuclei, cells were treated with blocking solution (3% BSA w/v, 10% NGS, and 0.1% Triton X-100 in PBS) containing 1:2000 Toto-3 iodide (Invitrogen). Coverslips were then mounted with Prolong gold anti-fade reagent (Invitrogen). The remaining well of each condition was incubated for an additional 24 h (48 h after transfection) to allow turnover of labeled HDx-1, and processed for microscopy as above. Mean fluorescence intensity of individual cells was observed in three microscope fields per well using LCS software (Leica). Mean cellular fluorescence intensities were computed for both 24 and 48 h conditions. The percentage of labeled HDx-1 at 24 h and remaining at 48 h was determined by [(mean intensity at 48 h/mean intensity at 24 h) × 100]. The experiment was repeated three additional times, giving an n of 4. p values were computed using a Student's t test.
Results
Isolation of Happ intrabodies
Novel intrabodies against the PRR domain were selected in a two-stage protocol. First, a nonimmune, human recombinant scFv phage library (Griffin.1) (Griffiths et al., 1994) was used to select clones that bind a unique, P-rich sequence between the two polyP domains in mHDx-1. The second stage involved three rounds of selection using PQ50 (HDx-1 containing 50 Q and the PRR) (Scherzinger et al., 1997). After the second stage, individual clones were analyzed for inserts containing ORFs. Although the Griffin.1 library consists of full scFv fragments, the two clones selected had only the VL ORFs. A control VL that does not bind Htt (CVL) was also isolated from the library. These three VLs (Happ1, Happ3, and CVL) were then inserted into a mammalian expression vector for cell culture and brain slice studies. To verify the specificity of these intrabodies, they were expressed as GST fusion proteins and used as primary antibodies to stain membranes containing the lysates of HEK293 cells transfected with HDx-1 or HDx-1ΔPRR. The lysates of nontransfected cells were used to test for binding non-Htt cellular proteins (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). As expected, MW7 and the Happs bind only to HDx-1 containing the PRR, whereas VL12.3 binds both forms of HDx-1. None of the intrabodies bind the nontransfected lysates. These results confirm that the Happs require the Htt PRR epitope for binding.
The intrabodies reduce mHDx-1 aggregation and toxicity
Each of the intrabodies was tested at various ratios to mHDx-1 (0.5:1, 1:1, 2:1, 3:1, and 4:1) for effects on mHDx-1 toxicity by counting EthD-2-positive dead cell nuclei (Fig. 1 B), and aggregation by counting green foci of the HDx-1-GFP fusion protein (Fig. 1 C). Whereas VL12.3, Happ1, and Happ3 demonstrate dose-dependent, saturable beneficial effects on aggregation, MW7 demonstrates a threshold effect requiring a 4:1 ratio for benefit. This may be the result of its specificity for pure polyP. Because there are two polyP stretches that can each accommodate binding of two intrabody molecules, reduction of aggregation by polyP binding may require complete blockade of these epitopes. Interestingly, this does not appear to be the case for the toxicity induced by mHDx-1. The VL12.3 intrabody is the most effective in reducing toxicity, with an optimal ratio to mHDx-1 of 1:1. MW7 is optimal at a ratio of 4:1, whereas Happ1 and Happ3 each show an optimal ratio of 2:1, with significant beneficial effects at 1:1. Similar effects on mHDx-1-induced toxicity were seen when measuring lactate dehydrogenase activity (data not shown). These results confirm previous findings with the N1-17 aa epitope and further demonstrate that the PRR also regulates HDx-1 toxicity. Because CVL shows no dose-dependent effects (data not shown), the median ratio of 2:1 was used as a baseline level of mHDx-1-induced toxicity or aggregation (Fig. 1 B, zero point).
The intrabodies reduce mHDx-1-induced neurodegeneration in a corticostriatal brain slice model of HD
Rat brain slices, which preserve much of the intrinsic neural circuitry, were biolistically cotransfected with YFP, mHDx-1-CFP, and an intrabody. The number of morphologically healthy, transfected MSNs in the striatum of each slice was then assessed using YFP fluorescence as an independent reporter of cell type and vitality 4–5 d after slice preparation and transfection (Fig. 2). The number of healthy MSNs per brain slice was compared between a positive control (brain slices transfected with YFP + CVL), a negative control (transfected with YFP + mHDx-1 + vector backbone DNA), and the test condition (transfected with YFP + mHDx-1 + anti-Htt intrabody). Cotransfection of mHDx-1 with CVL results in significantly reduced numbers of healthy MSNs, as is seen in slices transfected with mHDx-1 and vector. In contrast, cotransfection of mHDx-1 with VL12.3 or Happ1 results in numbers of healthy MSNs that are similar to slices transfected with YFP + CVL. Cotransfection of slices with mHDx-1 + MW7 yields intermediate results, with significantly greater numbers of healthy MSNs than with mHDx-1 + CVL, but fewer than with YFP + CVL. These results extend the findings from HEK293 cells to MSNs in a semi-intact milieu.
Figure 2.
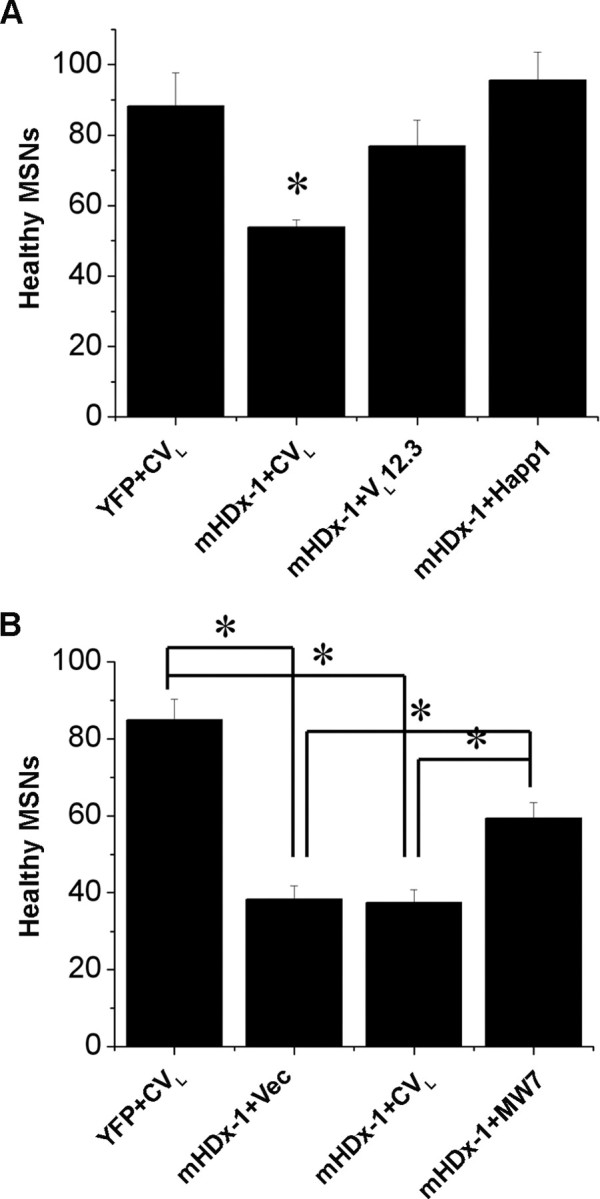
Protective effects of anti-Htt intrabodies against mHDx-1-induced neurodegeneration in corticostriatal brain slice explants. Corticostriatal brain slices were biolistically transfected with plasmid expression constructs encoding YFP, mHDx-1 (N1–66 with 73 Q), and the indicated intrabody. The number of healthy medium spiny neurons in the striatal region of each slice was scored visually 4–5 d after slice preparation and transfection. A, Slices were transfected with YFP + CVL; YFP + CVL + mHDx-1; YFP + mHDx-1 + VL12.3; or YFP + mHDx-1 + Happ1. *Different from YFP + CVL at p < 0.01. B, Slices were transfected with YFP + CVL; YFP + vector (Vec) + mHDx-1; YFP + CVL + mHDx-1; or YFP + mHDx-1 + MW7. *p < 0.01. The data in A and B are from independent experiments.
VL12.3 alters cytoplasmic versus nuclear trafficking of mHDx-1
To evaluate the effect of the intrabodies on HDx-1 intracellular trafficking, ST14A striatal neuronal precursor cells were cotransfected with mHDx-1-GFP and intrabody and incubated for 48 h. Cells were fixed, stained for both intrabody and nuclei, and then mounted for analysis by confocal microscopy. GFP fluorescence intensity was used to compare levels of mHDx-1 in the whole cell versus the nucleus (Fig. 3). The PRR-binding intrabodies do not alter the cytoplasmic/nuclear mHDx-1 ratio, whereas VL12.3 causes a dramatic increase of nuclear Htt. In terms of localization of the intrabodies themselves, VL12.3, Happ1, and Happ3 display a slight preference for the nucleus, whereas MW7 is slightly more cytoplasmic (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). This could be the result of the larger size of the MW7 scFv compared with the single-domain intrabodies. No significant differences are seen between VL12.3, Happ1, and Happ3, and the slight preference of VL12.3 for the nucleus is too small to account for the increased nuclear HDx-1 in the presence of VL12.3, indicating that this change in localization is not the result of intrabody localization. Thus, intrabody binding to the N terminus of Htt disrupts cytoplasmic versus nuclear trafficking of Htt, which may influence its nuclear functions. Because the amount of nuclear mHtt correlates with toxicity (Truant et al., 2007), this result suggests that VL12.3 may not be ideal as a therapeutic intrabody despite its clear effects on blocking mHtt toxicity.
Figure 3.
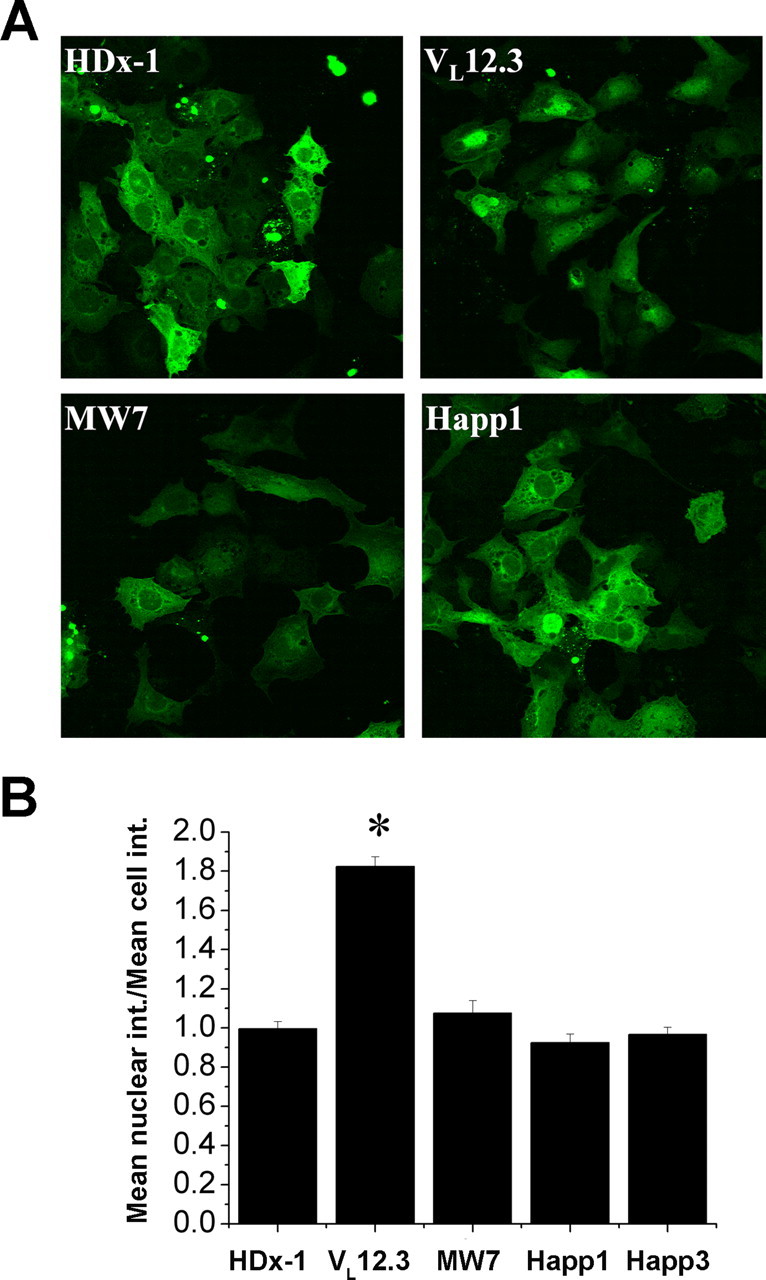
VL12.3 increases the level of nuclear HDx-1. ST14A cells were cotransfected with mHDx-1-GFP with 103Q and intrabody at optimal ratios. A, At 48 h after transfection, cells were fixed, stained for the appropriate intrabody and cell nuclei, and analyzed by confocal microscopy. B, Mean whole-cell fluorescence intensity (int.) and mean nuclear fluorescence intensity of HDx-1 were compared. Whereas MW7, Happ1, and Happ3 have no effect on HDx-1 localization, VL12.3 significantly increases nuclear HDx-1. *Different from HDx-1 at p < 0.01.
The intrabodies differentially alter the level of soluble mHDx-1
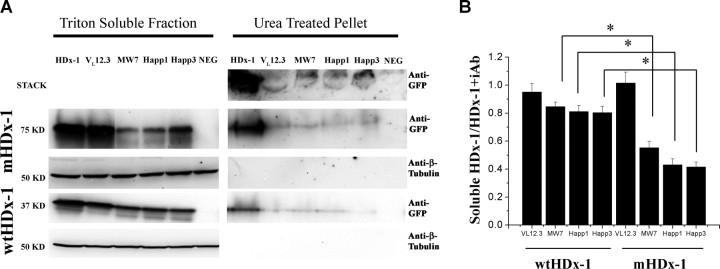
To determine the effects of the intrabodies on mHDx-1 levels, HEK293 cells were cotransfected with intrabody and either wild-type HDx-1 (wtHDx-1) or mHDx-1, using each intrabody at its optimal ratio to HDx-1, and incubated for 48 h. Soluble and insoluble cell fractions were then assayed for HDx-1 by immunoblotting and densitometry (Fig. 4). Each of the intrabodies dramatically reduces the level of insoluble mHDx-1. However, the three PRR-binding intrabodies (MW7, Happ1, and Happ3) also significantly reduce the level of soluble mHDx-1, whereas VL12.3 has no significant effect on soluble mHDx-1 or wtHDx-1 levels. From a therapeutic standpoint, it is important that only a slight reduction of wtHDx-1 protein is seen, indicating that anti-PRR intrabodies are selective for the mutant form. Although these intrabodies bind wtHDx-1, their preference for the mutant form is not unexpected as the interaction of endogenous Htt PRR-binding partners with Htt is known to increase with increasing polyQ repeat length (Passani et al., 2000; Holbert et al., 2001).
Figure 4.
All of the anti-Htt intrabodies reduce insoluble HDx-1, whereas only the anti-PRR intrabodies also reduce soluble HDx-1. HEK293 cells were cotransfected with intrabody and mHDx-1 (76 kDa) or wtHDx-1 (40 kDa) at the optimal ratio for each intrabody. A, At 48 h after transfection, cells were lysed with detergent. The soluble protein fraction was recovered, and the insoluble fraction was treated with urea. Samples were then separated by SDS-PAGE and blotted for HDx-1. Nontransfected cells were used as a negative control (NEG). B, Quantification of bands shows that reduction of HDx-1 by PRR-binding intrabodies is significantly greater for the mutant form of Htt. Chemiluminescence densitometry was used to compare the levels of soluble mHDx-1 and wtHDx-1. Each band was normalized to the level of β-tubulin (54 kDa) in that sample. Bands for HDx-1 + intrabody (iAb) were then normalized to the level of soluble HDx-1 for that blot. n = 4 independent experiments, and values for each blot were used to compile a mean. *p < 0.01.
The PRR-binding intrabodies increase mHDx-1 turnover
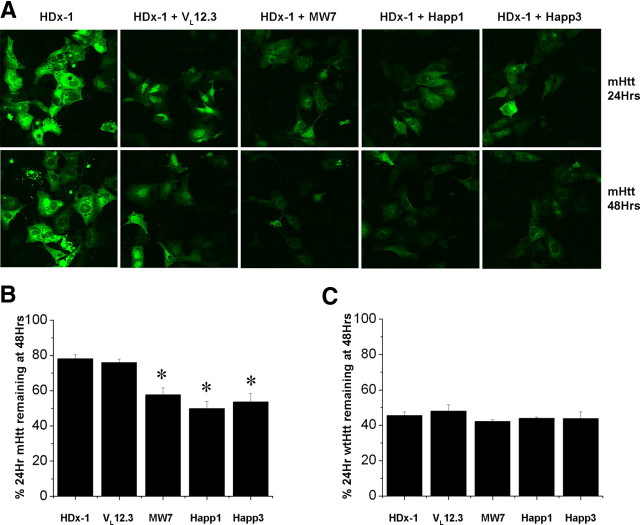
To further investigate the reduction of soluble mHDx-1, a SNAP tag fusion labeling experiment was performed (Jansen et al., 2007). A traditional pulse chase experiment was not used because mHDx-1 is known to affect transcriptional regulation. This property of mHDx-1 could conceivably be altered by intrabody binding, leading to variable transcription rates of HDx-1 in the presence of the various intrabodies. Traditional pulse-chase experiments require equal transcription and translation of the target protein in all conditions within the labeling period. The SNAP tag fusion system allows labeling of all preexisting HDx-1. By measuring the amount of Htt at the time of labeling and again at a later time point, we are able to measure a rate of turnover independent of transcription or translation rate. This system also offers greater specificity, because only the SNAP tag fusion protein is labeled as opposed to all cellular proteins translated during the labeling period as with traditional pulse-chase experiments.
To investigate HDx-1 turnover using the SNAP tag fusion system, HEK293 cells were cotransfected with intrabody and HDx-1 fused to the SNAP tag. Twenty-four hours after transfection, HDx-1 was labeled using a fluorescent, cell-permeable SNAP substrate. This substrate undergoes a covalent binding reaction with the SNAP tag and remains fluorescent until the SNAP-tag fusion protein is broken down. Some cultures were immediately examined for HDx-1 levels, whereas others were incubated for 48 h after transfection to allow turnover of labeled HDx-1. Fluorescence intensity of HDx-1-SNAP was used to determine the percentage of HDx-1 labeled at 24 h that is still intact at 48 h (Fig. 5). Cells transfected with HDx-1-SNAP alone were used to determine a baseline level of turnover. Although the percentage of mHDx-1 remaining in the presence of VL12.3 is equivalent to that in the control, this percentage is significantly reduced in the presence of MW7, Happ1, or Happ3, indicating an increase in the rate of mHDx-1 turnover specifically in the presence of the PRR-binding intrabodies (Fig. 5 B). The lack of effect of VL12.3 provides a convenient control for nonspecific effects of intrabody binding to mHDx-1. Although the mechanism by which this increase in mHtt turnover occurs is not yet clear, the levels of intrabody protein are increased in the presence of mHtt (data not shown), suggesting that mHtt is not broken down as a part of a complex with intrabody. This novel ability of PRR-binding intrabodies to increase turnover of mHtt suggests that this region of the protein is important for stability. Further evidence of the specificity of the intrabody effects is shown by the fact that none of the anti-Htt intrabodies significantly changes the rate of wtHDx-1 turnover (Fig. 5 C).
Figure 5.
Anti-PRR intrabodies increase mHDx-1 turnover. A–C, ST14A cells were transfected with mHDx-1-SNAPtag 97Q (A, B) or wtHDx-1-SNAPtag 25Q (C) and intrabody at the optimal ratio for each intrabody. DAF green fluorescent SNAP substrate was added to cultures at 24 h after transfection. Some cultures were then fixed and stained with Toto-3 iodide nuclear marker, whereas others were incubated an additional 24 h to allow turnover of labeled HDx-1. Mean fluorescence intensity of cells at 24 h was compared with intensity at 48 h to determine the percentage of labeled HDx-1 remaining. VL12.3 has no effect on mHDx-1 or wtHDx-1 turnover, whereas MW7, Happ1, and Happ3 significantly increase the rate of mHDx-1 turnover. n = 4; *p < 0.01.
Discussion
Although anti-N-terminal and anti-PRR intrabodies ameliorate the negative effects of mHtt in cell culture and brain slice models of HD, they do so with different efficacy and by different mechanisms. These different mechanisms offer clues to the specific functions of their target domains.
The VL12.3 intrabody was isolated from a yeast surface display library and initially required a 5:1 ratio to mHtt to reduce aggregation (Colby et al., 2004a). It was then reengineered, including removal of the disulfide bonds, which do not form in the reducing environment of the mammalian cytoplasm [and can cause misfolding of intrabodies (Biocca et al., 1995)], and mutated for greater binding affinity to Htt (Colby et al., 2004b). In addition to inhibiting mHtt-induced toxicity and aggregation, we find that VL12.3 also alters cytoplasmic versus nuclear trafficking of HDx-1.
Modulation of Htt intracellular targeting by the N terminus has been recently characterized. Removal of this amphipathic α helix causes an increase in the level of nuclear Htt, indicating that it functions as a cytoplasmic retention signal (Rockabrand et al., 2007). Mutation of hydrophobic residues, or the introduction of a helix-breaking proline residue in the N-terminal domain results in increased nuclear Htt, suggesting that cytoplasmic retention by the N terminus is the result of association with organelle and vesicle membranes (Atwal et al., 2007). Although the N terminus is not a dimerization domain, disruption of the helical structure also prevents the aggregation of mHtt, which is accompanied by an increase in the toxicity of the protein. Thus, the N terminus of Htt is required for cytoplasmic localization and the formation of aggregates. The effect on toxicity seen in these experiments may be related to the prevention of aggregation, because mHtt-expressing neurons without aggregates exhibit more toxicity than those with aggregates (Arrasate et al., 2004). Toxicity related to the N terminus may also involve altered Htt localization, because the addition of a nuclear localization signal to mHtt increases its toxicity in both cell culture and mouse models of HD (Peters et al., 1999; Schilling et al., 2004). Interestingly, whereas removal or mutation of the N terminus results in increased toxicity, VL12.3 binding results in reduced toxicity, suggesting that VL12.3 may inhibit formation of a toxic conformation or an oligomerization seed molecule. Thus, this intrabody may ameliorate toxicity regardless of mHtt localization or aggregation state.
The polyP and P-rich domains of mHtt are implicated in a number of aberrant protein interactions. These domains are required for mHtt binding to, and sequestering of, several SH3 domain-containing proteins, including proteins associated with vesicle function (Modregger et al., 2002; Qin et al., 2004). The PRR of Htt is required for interaction with WW domain-containing proteins (Staub and Rotin, 1996; Faber et al., 1998). These include transcription factors, and these interactions are enhanced with increased polyQ repeat length (Passani et al., 2000; Holbert et al., 2001). These domains are the site of interaction with IKKγ, a regulatory subunit of the IκB kinase complex. Activation of this complex is known to promote aggregation and nuclear localization of mHtt (Khoshnan et al., 2004). The PRR of Htt is also the site of P53 interaction and is required for transcriptional repression of P53-regulated genes (Steffan et al., 2000). Again, this interaction is enhanced by increased polyQ repeat length.
MW7, an intrabody recognizing pure polyP, reduces mHtt-induced aggregation and toxicity in cell culture and in Drosophila models of HD (Khoshnan et al., 2002; Jackson et al., 2004). We find that it is also effective in an acute brain slice model of HD, and that it increases the turnover of HDx-1, with greater effect on the mutant than the wild-type form. We also produced novel intrabodies, Happ1 and Happ3, which recognize the unique, P-rich epitope between the two polyP domains of Htt. The Happ intrabodies exhibit beneficial properties similar to those of MW7, such as preferential effects on the mutant form of Htt and increasing turnover without altering localization, but the Happs are effective at lower ratios to Htt than MW7. We found no evidence that the anti-PRR intrabodies bind to previously aggregated mHtt, suggesting that the observed reduction in aggregation is the indirect result of increased turnover of the soluble form of the protein, causing a shift away from the aggregated state. The increased turnover of HDx-1 in the presence of either anti-polyP or anti-P-rich intrabodies suggests that this effect is a direct result of blocking these epitopes and therefore that this domain has a role in modulating stability of the mutant protein.
Disruption of mHtt stability by Happ binding could have therapeutic potential. The success of RNA interference (RNAi) experiments show that reduction of mHtt levels is an effective therapeutic strategy (Harper et al., 2005; Rodriguez-Lebron et al., 2005; Machida et al., 2006). Unlike RNAi however, these intrabodies can distinguish between the wt and mutant forms of Htt, which is preferable, because the loss of normal Htt function can have negative effects (Dragatsis et al., 2000; Leavitt et al., 2001; Zuccato et al., 2001). The ability of the Happ intrabodies to increase turnover of mHtt may ameliorate the disruption of the ubiquitin proteasome seen in HD, although it is presently unclear if this increased turnover occurs through a ubiquitin-dependent pathway. Because the levels of intrabody protein are increased in the presence of Htt, it is likely that the intrabodies direct the breakdown of mHtt without themselves being degraded. Moreover, the Happs, although significantly more effective than the original intrabody isolated and matured to become VL12.3, have yet to undergo any reengineering and could potentially be improved by removal of disulfide bonds and mutation for greater Htt binding affinity. In addition, the present results with the Happ intrabodies highlight the importance of the unique, P-rich domain in mHtt toxicity.
Footnotes
This work was supported by grants from the High Q, Hereditary Disease, and McGrath Foundations. We thank David Colby and K. Dane Wittrup for providing us with VL12.3, the Medical Research Council Center for Protein Engineering for providing us with the Griffin.1 library, Elena Cattaneo for providing us with ST14A cells, Christian Essrich for the design and piloting of the brain slice experiments, and Jan Ko for experimental support.
References
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Atwal RS, Xia J, Pinchev D, Taylor J, Epand RM, Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. New York: Greene and Wiley; 1993. [Google Scholar]
- Biocca S, Cattaneo A. Intracellular immunization: antibody targeting to subcellular compartments. Trends Cell Biol. 1995;5:248–252. doi: 10.1016/s0962-8924(00)89019-4. [DOI] [PubMed] [Google Scholar]
- Biocca S, Ruberti F, Tafani M, Pierandrei-Amaldi P, Cattaneo A. Redox state of single chain Fv fragments targeted to the endoplasmic reticulum, cytosol and mitochondria. Biotechnology (N Y) 1995;13:1110–1115. doi: 10.1038/nbt1095-1110. [DOI] [PubMed] [Google Scholar]
- Cattaneo E, Conti L. Generation and characterization of embryonic striatal conditionally immortalized ST14A cells. J Neurosci Res. 1998;53:223–234. doi: 10.1002/(SICI)1097-4547(19980715)53:2<223::AID-JNR11>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Colby DW, Garg P, Holden T, Chao G, Webster JM, Messer A, Ingram VM, Wittrup KD. Development of a human light chain variable domain (VL) intracellular antibody specific for the amino terminus of huntingtin via yeast surface display. J Mol Biol. 2004a;342:901–912. doi: 10.1016/j.jmb.2004.07.054. [DOI] [PubMed] [Google Scholar]
- Colby DW, Chu Y, Cassady JP, Duennwald M, Zazulak H, Webster JM, Messer A, Lindquist S, Ingram VM, Wittrup KD. Potent inhibition of huntingtin aggregation and cytotoxicity by a disulfide bond-free single-domain intracellular antibody. Proc Natl Acad Sci U S A. 2004b;101:17616–17621. doi: 10.1073/pnas.0408134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G, Abbas N, Stevanin G, Dürr A, Yvert G, Cancel G, Weber C, Imbert G, Saudou F, Antoniou E, Drabkin H, Gemmill R, Giunti P, Benomar A, Wood N, Ruberg M, Agid Y, Mandel J-L, Brice A. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat Genet. 1997;17:65–70. doi: 10.1038/ng0997-65. [DOI] [PubMed] [Google Scholar]
- Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26:300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci U S A. 2006;103:11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyao MP, Auerbach AB, Ryan A, Persichetti F, Barnes GT, McNeil SM, Ge P, Vonsattel J-P, Gusella JF, Joyner AL, MacDonald ME. Inactivation of the mouse Huntington's disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, Crosby WL, Kontermann RE, Jones PT, Low NM, Allison TJ. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 1994;13:3245–3260. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. From the cover: RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci U S A. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockfield S, Carlson S, Evans C, Levitt P, Pintar J, Silberstein L. Selected methods for antibody and nucleic acid probes. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1993. [Google Scholar]
- Holbert S, Denghien I, Kiechle T, Rosenblatt A, Wellington C, Hayden MR, Margolis RL, Ross CA, Dausset J, Ferrante RJ, Néri C. The Gln-Ala repeat transcriptional activator CA150 interacts with huntingtin: neuropathologic and genetic evidence for a role in Huntington's disease pathogenesis. Proc Natl Acad Sci U S A. 2001;98:1811–1816. doi: 10.1073/pnas.041566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert G, Saudou F, Yvert G, Devys D, Trottier Y, Garnier J-M, Weber C, Mandel J-L, Cancel G, Abbas N, Dürr A, Didierjean O, Stevanin G, Agid Y, Brice A. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat Genet. 1996;14:285–291. doi: 10.1038/ng1196-285. [DOI] [PubMed] [Google Scholar]
- Jackson GR, Sang T, Khoshnan A, Ko J, Patterson PH. Inhibition of mutant huntingtin-induced neurodegeneration in vivo by expression of a polyproline-binding single chain antibody. Soc Neurosci Abstr. 2004;30:938–5. [Google Scholar]
- Jana NR, Zemskov EA, Wang G-h, Nukina N. Altered proteasomal function due to the expression of polyglutamine-expanded truncated N-terminal huntingtin induces apoptosis by caspase activation through mitochondrial cytochrome c release. Hum Mol Genet. 2001;10:1049–1059. doi: 10.1093/hmg/10.10.1049. [DOI] [PubMed] [Google Scholar]
- Jansen LET, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Okamoto T, Taniwaki M, Aizawa M, Inoue M, Katayama S, Kawakami H, Nakamura S, Nishimura M, Akiguchi I, Kimura J, Narumiya S, Kakizuka A. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat Genet. 1994;8:221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- Khoshnan A, Ko J, Patterson PH. Effects of intracellular expression of anti-huntingtin antibodies of various specificities on mutant huntingtin aggregation and toxicity. Proc Natl Acad Sci U S A. 2002;99:1002–1007. doi: 10.1073/pnas.022631799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnan A, Ko J, Watkin EE, Paige LA, Reinhart PH, Patterson PH. Activation of the IκB kinase complex and nuclear factor-κB contributes to mutant huntingtin neurotoxicity. J Neurosci. 2004;24:7999–8008. doi: 10.1523/JNEUROSCI.2675-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide R, Ikeuchi T, Onodera O, Tanaka H, Igarashi S, Endo K, Takahashi H, Kondo R, Ishikawa A, Hayashi T, Saito M, Tomoda A, Miike T, Naito H, Ikuta F, Tsuji S. Unstable expansion of CAG repeat in hereditary dentatorubral-pallidoluysian atrophy (DRPLA) Nat Genet. 1994;6:9–13. doi: 10.1038/ng0194-9. [DOI] [PubMed] [Google Scholar]
- La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- Leavitt BR, Guttman JA, Hodgson JG, Kimel GH, Singaraja R, Vogl AW, Hayden MR. Wild-type huntingtin reduces the cellular toxicity of mutant huntingtin in vivo. Am J Hum Genet. 2001;68:313–324. doi: 10.1086/318207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo AS, Zhu Q, Marasco WA. Intracellular antibodies (intrabodies) and their therapeutic potential. Hand Exp Pharmacrol. 2008;181:343–373. doi: 10.1007/978-3-540-73259-4_15. [DOI] [PubMed] [Google Scholar]
- Lo DC, McAllister AK, Katz LC. Neuronal transfection in brain slices using particle-mediated gene transfer. Neuron. 1994;13:1263–1268. doi: 10.1016/0896-6273(94)90412-x. [DOI] [PubMed] [Google Scholar]
- Machida Y, Okada T, Kurosawa M, Oyama F, Ozawa K, Nukina N. rAAV-mediated shRNA ameliorated neuropathology in Huntington disease model mouse. Biochem Biophys Res Comm. 2006;343:190–197. doi: 10.1016/j.bbrc.2006.02.141. [DOI] [PubMed] [Google Scholar]
- Marsh JL, Walker H, Theisen H, Zhu Y-Z, Fielder T, Purcell J, Thompson LM. Expanded polyglutamine peptides alone are intrinsically cytotoxic and cause neurodegeneration in Drosophila . Hum Mol Genet. 2000;9:13–25. doi: 10.1093/hmg/9.1.13. [DOI] [PubMed] [Google Scholar]
- Messer A, McLear J. The therapeutic potential of intrabodies in neurologic disorders: focus on Huntington and Parkinson diseases. BioDrugs. 2006;20:327–333. doi: 10.2165/00063030-200620060-00002. [DOI] [PubMed] [Google Scholar]
- Modregger J, DiProspero NA, Charles V, Tagle DA, Plomann M. PACSIN 1 interacts with huntingtin and is absent from synaptic varicosities in presymptomatic Huntington's disease brains. Hum Mol Genet. 2002;11:2547–2558. doi: 10.1093/hmg/11.21.2547. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Aminoff MJ. Huntington's disease: clinical characteristics, pathogenesis and therapies. Drugs Today (Barc) 2007;43:97–116. doi: 10.1358/dot.2007.43.2.1050788. [DOI] [PubMed] [Google Scholar]
- Orr HT, Chung M-y, Banfi S, Kwiatkowski TJ, Jr, Servadio A, Beaudet AL, McCall AE, Duvick LA, Ranum LPW, Zoghbi HY. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- Passani LA, Bedford MT, Faber PW, McGinnis KM, Sharp AH, Gusella JF, Vonsattel J-P, MacDonald ME. Huntingtin's WW domain partners in Huntington's disease post-mortem brain fulfill genetic criteria for direct involvement in Huntington's disease pathogenesis. Hum Mol Genet. 2000;9:2175–2182. doi: 10.1093/hmg/9.14.2175. [DOI] [PubMed] [Google Scholar]
- Peters MF, Nucifora FC, Jr, Kushi J, Seaman HC, Cooper JK, Herring WJ, Dawson VL, Dawson TM, Ross CA. Nuclear targeting of mutant huntingtin increases toxicity. Mol Cell Neurosci. 1999;14:121–128. doi: 10.1006/mcne.1999.0773. [DOI] [PubMed] [Google Scholar]
- Qin Z-H, Wang Y, Sapp E, Cuiffo B, Wanker E, Hayden MR, Kegel KB, Aronin N, DiFiglia M. Huntingtin bodies sequester vesicle-associated proteins by a polyproline-dependent interaction. J Neurosci. 2004;24:269–281. doi: 10.1523/JNEUROSCI.1409-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Shannon KM, Kordower JH. Huntington's disease: pathological mechanisms and therapeutic strategies. Cell Transplant. 2007;16:301–312. doi: 10.3727/000000007783464687. [DOI] [PubMed] [Google Scholar]
- Reddy PH, Williams M, Tagle DA. Recent advances in understanding the pathogenesis of Huntington's disease. Trends Neurosci. 1999;22:248–255. doi: 10.1016/s0166-2236(99)01415-0. [DOI] [PubMed] [Google Scholar]
- Rockabrand E, Slepko N, Pantalone A, Nukala VN, Kazantsev A, Marsh JL, Sullivan PG, Steffan JS, Sensi SL, Thompson LM. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum Mol Genet. 2007;16:61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lebron E, Denovan-Wright EM, Nash K, Lewin AS, Mandel RJ. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington's disease transgenic mice. Mol Ther. 2005;12:618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA. Intranuclear neuronal inclusions: a common pathogenic mechanism for glutamine-repeat neurodegenerative diseases? Neuron. 1997;19:1147–1150. doi: 10.1016/s0896-6273(00)80405-5. [DOI] [PubMed] [Google Scholar]
- Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates GP, Davies SW, Lehrach H, Wanker EE. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- Schilling G, Savonenko AV, Klevytska A, Morton JL, Tucker SM, Poirier M, Gale A, Chan N, Gonzales V, Slunt HH, Coonfield ML, Jenkins NA, Copeland NG, Ross CA, Borchelt DR. Nuclear-targeting of mutant huntingtin fragments produces Huntington's disease-like phenotypes in transgenic mice. Hum Mol Genet. 2004;13:1599–1610. doi: 10.1093/hmg/ddh175. [DOI] [PubMed] [Google Scholar]
- Staub O, Rotin D. WW domains. Structure. 1996;4:495–499. doi: 10.1016/s0969-2126(96)00054-8. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu Y-Z, Gohler H, Wanker EE, Bates GP, Housman DE, Thompson LM. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci U S A. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks MM. Intrabodies as drug discovery tools and therapeutics. Curr Opin Chem Biol. 2005;9:359–365. doi: 10.1016/j.cbpa.2005.06.003. [DOI] [PubMed] [Google Scholar]
- The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Truant R, Atwal RS, Burtnik A. Nucleocytoplasmic trafficking and transcription effects of huntingtin in Huntington's disease. Prog Neurobiol. 2007;83:211–227. doi: 10.1016/j.pneurobio.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Wanker EE. Protein aggregation and pathogenesis of Huntington's disease: mechanisms and correlations. Biol Chem. 2000;381:937–942. doi: 10.1515/BC.2000.114. [DOI] [PubMed] [Google Scholar]
- Yang W, Dunlap JR, Andrews RB, Wetzel R. Aggregated polyglutamine peptides delivered to nuclei are toxic to mammalian cells. Hum Mol Genet. 2002;11:2905–2917. doi: 10.1093/hmg/11.23.2905. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, Timmusk T, Sipione S, Cattaneo E. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]