Abstract
Molecular chaperone heat-shock protein 90 kDa (Hsp90) is known to facilitate the conformational maturation of a diverse range of proteins involved in different signal transduction pathways during development. Recent studies have implicated Hsp90 in transcriptional regulation and an important role for Hsp90 in epigenetic processes has been proposed. Importantly, genetic and pharmacological perturbation of Hsp90 was shown to reveal heritable phenotypic variation and Hsp90 was found to play an important role in buffering genetic and epigenetic variation whose expression led to altered phenotypes. The underlying molecular mechanism remains elusive, however. Here, we show a direct molecular interaction between Hsp90 and Trithorax (Trx). Trx is a member of the TrxG chromatin proteins controlling, together with the members of the Polycomb group, the developmental fate of cells by modulating epigenetic signals. Hsp90 cooperates with Trx at chromatin for maintaining the active expression state of targets like the Hox genes. Pharmacological inhibition of Hsp90 results in degradation of Trx and a concomitant down-regulation of homeotic gene expression. A similar effect is observed with the human orthologue mixed-lineage leukemia. Connecting an epigenetic network controlling major developmental and cellular pathways with a system sensing external cues may explain the rapid fixation and epigenetic inheritance of phenotypic variation as a result of impaired Hsp90.
Keywords: chaperone, epigenetics, phenotypic variation
The heat-shock protein 90 kDa (Hsp90) is the most abundant molecular chaperone that is predominantly present in the cytosol under both normal and stressful conditions. However, Hsp90 is described as a specialized chaperone. It is distinguished from other chaperones because of its association with specific proteins that can be readily triggered to activity by growth signals. The range of Hsp90 client proteins includes steroid hormone receptors, proteins involved in signal transduction pathways, and several transcription factors (1).
There is mounting evidence for the involvement of Hsp90 in transcriptional regulation (2–4) but molecular evidence for Hsp90 playing a role in heritable gene regulation is limiting. Interaction between Hsp90 and histone proteins led to the suggestion that Hsp90 may affect chromatin structure and organization (5, 6). Importantly, involvement of Hsp90 in the disassembly of transcriptional regulatory complexes (2) revealed a potential molecular link between Hsp90 and gene regulation at the chromatin level. The proposed significance of Hsp90 in epigenetic processes is limited to evidence that a histone H3 lysine 4 methyltransferase, SMYD3, was shown to interact with Hsp90, enhancing the activity of SMYD3 in vitro (7). Analysis of the Hsp90 interaction network in yeast revealed that Hsp90 may influence global gene expression as a variety of transcription factors were identified as partner proteins (8). Recently, Hsp90 was also shown to be required for rapid nucleosome removal upon induction of GAL genes in yeast (4). Links between molecular chaperones and chromatin regulators of the Polycomb group (PcG) have been identified, which were shown to affect homeotic phenotypes in Drosophila (9, 10).
Studies in Drosophila discovered that heritable morphological abnormalities occur in almost any adult structure of Drosophila heterozygous for Hsp90 mutations or when Hsp90 function was pharmacologically impaired during development (11). The phenotypic variation in different Hsp90 mutants was ascribed to differences in genetic background, suggesting that preexisting cryptic genetic variations had become phenotypically expressed. Many of these strain-specific phenotypes could also be revealed by modest environmental changes without manipulating Hsp90 function (11). Surprisingly, Hsp90-dependent phenotypic alterations were shown to become fixed and inherited in subsequent generations, independent of the original Hsp90 perturbations, thereby providing a stage for the evolution of new traits (11, 12). On the basis of studies in Drosophila and Arabidopsis (11, 13, 14), the Hsp90 has been described as a capacitor of phenotypic variation that buffers cryptic genetic (11, 13) and epigenetic variation (14). In conjunction with its proposed evolutionary capacitance, Hsp90 has been regarded as a candidate gene for developmental canalization (11, 13, 14), which is defined as the ability of an organism to maintain a stable phenotype despite genetic variations or environmental perturbations (15, 16). In Caenorhabditis elegans, 6 highly conserved chromatin remodeling proteins were also identified as candidate genes for developmental canalization and it was proposed that they act as buffers of genetic variation in signaling processes (17).
The transgenerational inheritance of Hsp90-dependent traits was shown to have a genetic and an epigenetic basis (11, 14). Although, Hsp90-buffered genetic variation responsible for Hsp90-dependent phenotypic variation has been identified in Arabidopsis and Drosophila (18–21), no molecular link to epigenetic factors explaining transgenerational inheritance of Hsp90-dependent traits is known yet. Interestingly, among a variety of developmental alterations caused by Hsp90 mutations (11, 22), many resemble phenotypes caused by mutations in PcG and Trithorax group (TrxG) genes controlling the expression of developmental regulators at the chromatin level (23). PcG and TrxG genes encode a diverse array of transcriptional regulatory proteins, including components of complexes that function in chromatin remodeling and histone modifications that have been associated with the maintenance of epigenetic gene expression patterns (23). Although mutant alleles of Hsp90 and several TrxG genes in isogenic Drosophila strains were shown to buffer the same phenotypic variation by inducing epigenetically heritable altered chromatin states (14), the exact relationship between Hsp90 and TrxG or PcG proteins is not yet known. We therefore assessed whether epigenetic inheritance controlled by the PcG or the TrxG and the function of Hsp90 may be molecularly linked, which may explain epigenetic inheritance of Hsp90-dependent phenotypic variation. Here, we report that Drosophila mutations in Hsp90 behave similarly to Trithorax (Trx) mutations, resulting in loss of expression of target genes. The molecular characterization has revealed that pharmacological inhibition of Hsp90 function led to depletion of Trx protein and a subsequent down-regulation of the homeotic gene Deformed. Interestingly, we also observe a similar effect with the human orthologue of Trx, mixed-lineage leukemia (MLL). We show that Hsp90 interacts with the Trx protein at Polycomb response elements (PRE) controlled targets like the homeotic genes. Our results indicate that maintenance of active gene expression states of PcG and TrxG targets requires a functional interaction between Hsp90 and Trx.
Results
Hsp90 Mutants Behave Similarly to TrxG Mutations.
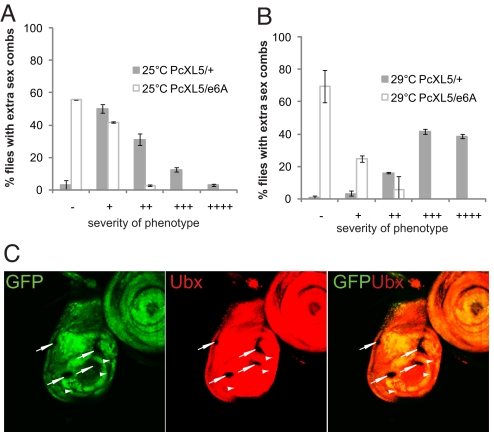
To investigate whether Hsp90 genetically interacts with the PcG or the TrxG system, Hsp90 mutant alleles (e1D, e6A, and j5C2) were crossed to 4 different alleles of Pc (1, 3, XL5, and XT109) (Table S1) at 2 different temperatures, 25°C and 29°C (Fig. 1 A and B, Fig. S1 A–I, and data not shown). Pc heterozygous mutants showed a strong extra sex comb phenotype that was more pronounced at the higher temperature (Fig. 1 A and B and Fig. S1 A–F). Importantly, all three Hsp90 alleles suppressed the extra sex comb phenotype at both temperatures (Fig. 1 A and B, Fig. S1 A–I, and data not shown). This finding correlates with previous observations by using PRE-based transgene assays, showing that TrxG activation is sensitive to high temperature (23). Our analysis of the extra sex comb phenotype supports the role for Hsp90 as a TrxG-type factor controlling homeotic phenotype.
Fig. 1.
Hsp90 mutations suppress the Polycomb phenotype. (A and B) Hsp90 allele e6A crossed with Pc (allele PcXL5 at 25°C and 29°C). The Pc allele crossed to wild-type flies, PcXL5/+, represents the control cross and strong extra sex combs are seen at 29°C (B) as compared with 25°C (A). The Hsp90 allele e6A strongly suppressed to none or few extra sex combs the Pc phenotype, PcXL5/e6A, at 25°C (A) and 29°C (B). More than 200 male flies were analyzed for each cross and mean values of 2 independent experiments are presented. Error bars represent the standard deviation. Where error bars are absent shows small standard deviation values. On the basis of strength of phenotype (number of hairs on second and third legs of males), flies were categorized as follows: −, no extra sex combs; +, 1–2 hairs on the second leg; ++, 3 or more hairs on the second leg; +++, 3 or more hairs on the second leg and 1–2 hairs on the third leg; and ++++, strong sex combs on both the second and the third pairs of legs. (C) Loss of Ubx expression in Hsp90 mutant clones. A haltere disc with clones mutant for Hsp90 (arrows) marked by absence of GFP is shown. The clones marked with white arrows show clear loss of Ubx expression in the haltere disc whereas clones marked with arrowheads indicate no effect on expression of Ubx.
Homozygous Hsp90 mutants die at early embryonic or larval stages. To analyze the consequences of Hsp90 loss on developmentally advanced stages, we generated mutant clones in larval imaginal discs by using the flp/FRT system (24). Hsp90 clones in the haltere discs show a loss of Ultrabithorax (Ubx) expression (Fig. 1C), a Hox gene controlled by the PcG and TrxG proteins (25, 26). This loss of expression reiterates similar effects observed in Trx mutant clones (25). In contrast, no deregulatory effects on Ubx expression were observed in Hsp90 mutant clones in the wing imaginal discs (data not shown), where Ubx is known to be maintained in a silent state by PcG genes (26). In summary, this genetic evidence suggests that Hsp90 interacts with TrxG proteins and as a result with their major developmental regulatory targets, thus providing possibly a molecular link through which Hsp90 could influence phenotypic variation.
Inhibition of Hsp90 Leads to Depletion of Trx Protein.
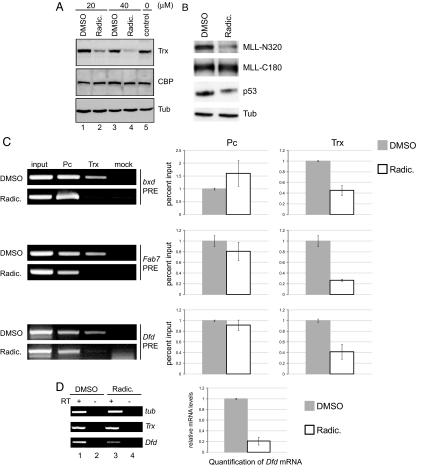
Hsp90 is a molecular chaperone that plays an essential role in the conformational maturation and stability of numerous proteins including nuclear receptors and protein kinases (1). The TrxG-like behavior of Hsp90 mutants prompted us to analyze effects of Hsp90 inhibition on the stability of TrxG proteins. We primarily focused on the TAC1 complex (27) containing the Trx protein required for histone H3 lysine 4 methylation and associated with active gene expression (23). In Drosophila cultured cells, we inhibited the Hsp90 protein with radicicol, a drug interfering with the ATP binding function (28). Drosophila Kc cells were treated with different concentrations for 4 h and the cellular amount of Trx was quantified by immunoblot (Fig. 2A and data not shown). We found that the inhibition of Hsp90, in a dose-dependent manner, resulted in depletion of Trx in Kc cells within 4 h of drug treatment (Fig. 2A). The reduction in Trx levels was specific because levels of CREB-binding protein (CBP), which together with Trx is part of the TAC1 complex (27), and the unrelated Tubulin (tub) remained unaffected in mock and drug-treated Kc cells (Fig. 2A). Notably, Hsp90 inhibition by radicicol showed no influence on transcriptional levels of Trx and tub genes used as a control (Fig. 2D).
Fig. 2.
Hsp90 is required for the stability of the Trx protein. (A) In Drosophila Kc cells, inhibition of Hsp90 with radicicol (Radic) resulted in depletion of Trx protein. After 4 h of Radic treatment with 20 μM (lane 2) and 40 μM (lane 4), Trx protein was specifically depleted. No effect on CBP and α-Tubulin was observed. Mock (DMSO vehicle, lanes 1 and 3)-treated Kc cells and untreated Kc cells (lane 5) were used as a control. The data presented are representative of 3 independent experiments. (B) HEK293 cells treated with 40 μM Radic showed specific depletion of MLL-N320 and no effect on MLL-C180 or Tub proteins in Western blot analysis. The human p53 protein reduction in drug-treated HEK293 cells was used as a positive control. (C) PCR analysis of chromatin immunoprecipitated DNA from DMSO and 40 μM Radic-treated Kc cells with primers specific for bxd, Fab7, and Dfd PREs. ChIPs performed with anti-Trx and anti-Pc antibodies were compared with no-antibody control (mock). Drug-treated Kc cells showed no effect on the association of Pc to bxd, Fab7, and Dfd PREs whereas Hsp90 inhibition specifically resulted in depletion of Trx from these PREs in drug-treated Kc cells. The Right graphs show the quantification of the ChIP data by using quantitative real-time PCR. Data presented represent mean values from 4 independent experiments. (D) Treatment of Kc cells with 40 μM drug (lane 3) showed no effect on transcript levels of Trx and tub genes. The Radic treatment (lane 3) shows down-regulation of the Dfd gene in Kc cells as compared with mock (DMSO)-treated cells (lane 1). Quantification of Dfd mRNA levels using real-time PCR revealed 70–80% reduction in drug-treated cells as compared with mock (DMSO)-treated cells. RT, reverse transcriptase; +, with RT; −, without RT.
We further examined a possible evolutionary conservation of the relationship between Hsp90 and the MLL (mammalian orthologue of Drosophila Trx) protein in mammalian cells. MLL undergoes proteolytic processing to generate ≈320-kDa (MLL-N320) and ≈180-kDa (MLL-C180) amino- and carboxyl-terminal proteins, respectively (29). Inhibiting Hsp90 with radicicol in HEK293 cells for 4 h resulted in a specific depletion of MLL-N320 with no effect on the C-terminal stub (Fig. 2B). To monitor the efficacy of different drug concentrations of radicicol, p53 protein, which is known to interact with Hsp90 (30), was used as a positive control (Fig. 2B). Our results suggest that therapeutic targeting by inhibitors of Hsp90 may be a promising approach to modulate deleterious activities of the wide spectrum of MLL-N fusion proteins that have been identified in leukemic cells (31).
Inhibition of Hsp90 Affects Trx Association with Chromatin.
Next, we monitored if inhibition of Hsp90 by radicicol also impinges on the association of Trx with chromatin and as a consequence results in a down-regulation of target genes. We performed ChIP by using anti-Trx and anti-Pc antibodies from radicicol-treated cells and compared them with DMSO vehicle-treated control Kc cells. The purified DNA from ChIP was analyzed by quantitative PCR for effects of Hsp90 inhibition on the association of Trx at bxd, Fab7, and Dfd PREs of the homeotic bithorax complex (BX-C) and Antennapedia complex (ANT-C), respectively, known to be bound by both Pc and Trx proteins (32). Hsp90 inhibition by radicicol had no significant effect on the association of Pc protein with any of the tested PREs (Fig. 2C). In contrast, inhibition of Hsp90 in Kc cells resulted in a highly specific reduction in binding of Trx from all of the PREs analyzed (Fig. 2C). Importantly, the Hox gene Deformed (Dfd), which is normally expressed in Kc cells and is controlled by the Dfd PRE (32), was down-regulated after inhibition of Hsp90 (Fig. 2D), demonstrating the specific functional consequences of depleting Trx from the PRE. The quantitative PCR revealed a 70–80% decrease in Dfd gene expression (Fig. 2D). In contrast, no effect on the silent Ubx gene, regulated by bxd PRE, was observed. Inhibition of Hsp90 in Kc cells also resulted in a global reduction of histone H3 acetylation, an epigenetic mark associated with active chromatin (data not shown). This global chromatin effect may also explain why pharmacological inhibition of histone deacetylases by Trichostatin A reduced the penetrance of some phenotypes caused by Hsp90 mutations (14).
Hsp90 Colocalizes with Trx at Puffs in Polytene Chromosomes.
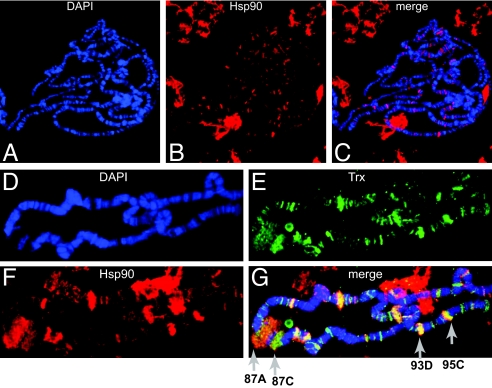
To identify the subcellular site of action of Hsp90 with Trx, we generated a Drosophila transgene expressing, under its own promoter, the Hsp90 coding sequence fused with green fluorescent protein (GFP). The Western blot analysis of Hsp90GFP fly extracts revealed the expected size of the fusion protein, however, expressed at a lower level than endogenous Hsp90 (Fig. S2 A and B). Nevertheless, Hsp90GFP rescued the lethality of the Hsp90 transheterozygote allele combination e1D/e6A (33), suggesting that the fusion is fully functional (data not shown). In Hsp90GFP flies, the analysis of unfixed salivary glands containing giant polytene chromosomes revealed that Hsp90GFP was abundant in the cytoplasm. However, it was also weakly distributed throughout the nucleus (Fig. S3 A–F), which is consistent with the previously described immunoelectron microscopic localization of Hsp90 in the perichromatin region in Drosophila cells (34). Immunostaining of polytene chromosomes from third instar larvae with anti-Hsp90 antibody revealed Hsp90 associated with chromosomes at interband regions (Fig. 3 A–C and Fig. S4 A–D). The specificity of this pattern was further confirmed by demonstrating a complete overlap of signals on chromosomes of transgenic lines carrying the Hsp90GFP fusion protein with anti-GFP and anti-Hsp90 antibodies (Fig. S4 A–D). We next investigated whether Hsp90 colocalizes with Trx protein at polytene chromosomes and found only a limited overlap (Fig. S5 A–D). This picture changed substantially when polytene chromosomes from heat-shocked larvae were analyzed (Fig. S5 E–I). A colocalization of Hsp90 and Trx at many sites, especially at heat-shock puffs, was observed, suggesting that Trx requires Hsp90 at active gene expression sites (Fig. 3 D–G and Fig. S5). Indeed, Trx has been shown to be required for modulation of heat-shock gene expression (35). However, Trx is also bound at genes that are kept silent by PcG proteins (32, 36). This might explain the relatively modest overlap of Hsp90 and Trx in chromosomes of non-heat-shocked salivary glands (Fig. S5 A–D), where many PcG/TrxG targets are repressed (37).
Fig. 3.
Hsp90 binds to chromatin at active Trx target genes. (A–C) Immunostaining of polytene chromosomes from third instar larvae with anti-Hsp90 antibody shows Hsp90 bound to chromatin at interband regions. (D–G) Immunostaining of polytene chromosomes from heat-shocked larvae stained with anti-Hsp90 and Trx antibodies shows a clear overlap at many sites, including the major heat-shock puffs marked as 87A, 87C, 93D, and 95D.
Hsp90 Associates with Trx at the Active Abd-B Gene in SF4 Cells.
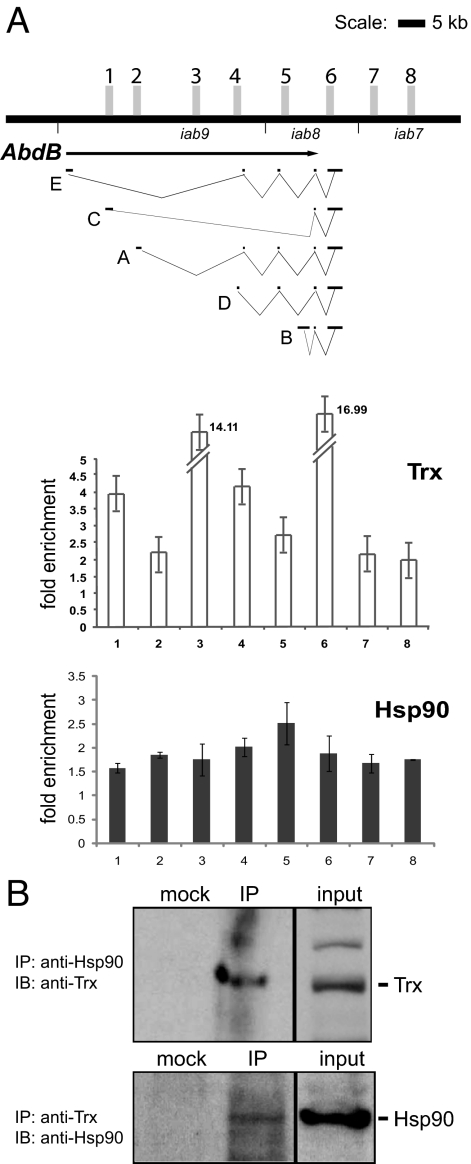
To search for an interaction of Hsp90 and Trx proteins at a molecular resolution, we performed ChIP by using Drosophila SF4 cells. On the basis of the finding that Hsp90 and Trx appear to colocalize over the entire transcribed region in puffs (Fig. 3G), we applied quantitative PCR ChIP to analyze the presence of Hsp90 at 8 different well-characterized Trx binding sites in the Abd-B gene (32), which spreads over a region of 84 kb in the BX-C (Fig. 4A). These 8 Trx binding sites were mapped to transcription start sites for different Abd-B transcripts (E, C, A, D, and B; Fig. 4A) and boundary elements (iab7, iab8, and iab9) in SF4 cells where the Abd-B gene is active (32). ChIP with Hsp90-specific antibody revealed association of Hsp90 with all of the 8 Trx binding sites analyzed across the Abd-B gene (Fig. 4A). In contrast, analysis of ChIP DNA with Dfd, bxd, and Ubx gene-specific primers revealed no association of Hsp90, although Trx was found to be bound (data not shown). This supports the notion that Hsp90 associates with Trx only at active genes because both Dfd and Ubx genes are silent in the SF4 cell line. Hsp90 was also not found to be associated at transcription start sites or coding regions of tubulin (tub), ImpL1, and white (w) genes, which were chosen as non-Trx targets (data not shown). In addition, the absence of Hsp90 from the tub gene, a highly expressed housekeeping gene, suggests that this chaperone is not required for general gene activation. We found that endogenous Hsp90 and Trx proteins could be coimmunoprecipitated from Drosophila embryonic nuclear extracts, indicating a possible direct physical interaction (Fig. 4B). In contrast, no coimmunoprecipitation with CBP, which together with Trx is part of the TAC1 complex (27), could be observed (data not shown).
Fig. 4.
Trx and Hsp90 colocalize over transcribed regions of Abdominal-B (Abd-B). (A) ChIP analysis of chromatin with anti-Trx and anti-Hsp90 antibodies showed Hsp90 associated with chromatin at Trx binding sites. (Upper) A solid line representing the part (84 kb) of BX-C that includes the Abd-B gene is shown, marked with shaded vertical bars each representing a primer pair in known Trx binding sites used to analyze the anti-Trx and anti-Hsp90 ChIPs shown below. Five different transcripts (E, C, A, D, and B) expressed from the Abd-B gene are shown below. (Lower) ChIP DNA was analyzed by quantitative PCR and percentage input was calculated for all 8 primer pairs for Trx binding sites shown above and ImpL1, white, and tubulin (non-Trx genes; data not shown). Fold enrichment for Trx and Hsp90 over ImpL1 is shown, which revealed Hsp90 association with chromatin together with Trx. Data presented represent mean values from 3 independent experiments and error bars represent standard deviations. (B) In wild-type embryonic nuclear extracts, endogenous Hsp90 and Trx were specifically coimmunoprecipitated as compared with immunoprecipitation with a mock (nonspecific antibody) control. Input represents 25% of total nuclear extract used for immunoprecipitation. IP, immunoprecipitation; IB, immunoblot.
Discussion
We have established a well-defined link between Hsp90 and the TrxG by demonstrating the genetic and molecular interaction between Hsp90 and Trx that is required for maintaining active gene expression in Drosophila. We provide evidence that genetic manipulation or pharmacological inhibition of Hsp90 strongly affects Trx target genes and results in their reduced expression because of depletion of Trx. This illustrates that Hsp90 facilitates Trx in maintaining active gene expression indicates how epigenetic gene expression maintained by the TrxG can be quickly modulated.
Ectopic activation of homeotic genes in PcG mutants relies on the TrxG, leading to morphological transformations. Appearance of additional sex combs on the first tarsal segment of the second and third leg pairs in Pc heterozygous males is one of such transformations. However, a reduced dose of TrxG proteins counteracts the reduced dose of Pc, restoring normal regulation of homeotic genes and suppressing the sex comb phenotype (23). Strong suppression of extra sex combs phenotype by Hsp90 mutants illustrates that Hsp90 interacts with the TrxG in counteracting silencing maintained by PcG genes. In corroboration, genetic or pharmacological manipulation of Hsp90 resulted in reduced Ubx and Dfd gene expression whose active expression is maintained by Trx. A molecular link between Hsp90 and Trx was validated by the evidence that pharmacological inhibition of Hsp90 specifically leads to depletion of Trx, which alleviates chromatin association of Trx and consequently affects expression of Dfd within 4 h of drug treatment. This provides strong evidence for a rapid Hsp90-dependent transcriptional modulation of active chromatin.
There is convincing evidence that Hsp90 regulates transcription processes (2–4) and more recently involvement of Hsp90 in epigenetic processes has been proposed (8, 14). However, molecular evidence for involvement of Hsp90 in heritable transcriptional regulation remained elusive. For example, the role of Hsp90 in steroid hormone receptor-mediated transcription activation is well characterized (38, 39). In particular, Hsp90 is known to hold glucocorticoid receptors (GR) in a conformation competent for ligand binding and activation of transcription. Importantly, Hsp90 is also proposed to interact with GR at chromatin where it facilitates the dissociation of GR from its targets and recycles the ligand-free GR back to chromatin in an Hsp90-dependent chaperone cycle (40, 41). Inhibition of Hsp90 is known to severely impair GR-mediated gene activation (38). Evidence for association of Hsp90 with Trx described in this study now strengthens a recently proposed role for Hsp90 in epigenetic processes where molecular evidence was lacking. The mechanistic link between Hsp90 and Trx may be similar to a reported interaction of Hsp90 with a histone H3 lysine 4-specific methyltransferase, SMYD3, which is up-regulated in cancer cells (7). In colorectal and hepatocellular carcinoma cells, SMYD3 was shown to interact with Hsp90, which dramatically enhanced catalytic activity of SMYD3 in vitro. Importantly, pharmacological inhibition of Hsp90 also significantly reduced SMYD3-mediated activation of a homeobox gene (7). The consequential effect of restrained Hsp90-SMYD3 association on homeobox genes appears comparable to reduced Dfd expression because of impaired Hsp90 activity described in this study. In mammals, Hsp90 was also shown to regulate the activity and stability of tumor suppressor P53 under physiological and elevated temperatures (30, 42). After heat shock, Hsp90 was found to associate with Trx at chromatin in polytene chromosomes, which may indicate that Hsp90 ensures stability and activity of Trx not only under normal conditions but also in times of stress. Because heat-shock puffs visualize active transcription, it also indicates that Hsp90 may facilitate Trx in maintaining active gene expression, which is also validated by the presence of Hsp90 at the active Abd-B gene in SF4 cells. The overlap between Hsp90 and Trx in non-heat-shocked chromosomes is limited to interband regions, which are also often active sites of transcription. The lack of overlap on some developmental puffs might be a sensitivity problem, because these are expressed at a lower level than the heat-shock genes. We cannot rule out a non-chromatin-bound role of Hsp90 in stabilizing Trx and subsequently affecting gene expression because Hsp90 was also found to be prevalent in the nucleoplasm (Fig. S3). The molecular interaction between Hsp90 and Trx could be analogous to the GR–Hsp90 partnership. Hsp90 may hold Trx in a stable, ready to be activated state before Trx receives a signal for gene activation. Further, Hsp90 may also play a crucial role in recycling Trx proteins, which may undergo a dynamic process of association and dissociation at their target sites. It is plausible to envisage that Hsp90-stabilized Trx may have a higher affinity for its interacting partners in chromatin-associated or soluble TAC1 complexes.
In this study we have uncovered how Hsp90 can directly influence an important, evolutionary highly conserved epigenetic network that supervises the appropriate cellular identities during development and homoeostasis of an organism. Because PcG and TrxG gene control has such a fundamental and evolutionary conserved role, its modulation by factors influenced by external signals could have substantial implications for the process of transcriptional memory and thus on phenotype. Interplay between Hsp90 and Trx supports the notion that chromatin regulators play an important role in developmental canalization (17) and signifies the likelihood that different canalization factors may be linked to further ensure developmental stability. Because of the reversible nature of epigenetic processes, Hsp90- and Trx-mediated switches may enable cells and organisms to adapt to environmental conditions more easily. The observed phenotypic variation in Drosophila because of genetic and environmental manipulation of Hps90 can also be explained as a consequence of reduction in Hsp90–Trx interactions affecting homeotic and other developmental genes. However, PcG and TrxG proteins not only maintain gene expression patterns heritable during mitosis but also can in certain conditions transmit epigenetically controlled states through meiosis to the next generations (43). Thus, Hsp90-dependent phenotypic variations might also become fixed and inherited through the action of the PcG/TrxG system providing an epigenetic basis for transgenerational inheritance of Hsp90-dependent traits. We support the notion that Hsp90-dependent phenotypic variations have a combined genetic and epigenetic origin and suggest that epigenetic inheritance mediated by TrxG or PcG proteins may significantly contribute in rapid fixation of heritable phenotypes.
Materials and Methods
D. melanogaster Stocks and Genetic Analysis.
Strains with independently isolated EMS-induced Hsp90 alleles (e6A, e6D, e1D, and j5C2) as described (33, 44) were obtained from the Bloomington stock center and S. Rutherford (Table S1). The Pc alleles (Table S1) were crossed with these hsp90 alleles at 25°C and at 29°C and males in the progeny were scored for extra sex combs as described (Fig. 1 A and B, Fig. S1 A–I, and data not shown).
The mutant clones for Hsp90 generated by flp-mediated recombination (24) were marked by the absence of Ubi-GFP expression. To induce recombination yw hs-flp/yw;FRT3L Hsp8308445/FRT3L Ubi-GFP larvae were heat-shocked (37°C for 2 h) 72 h after egg deposition followed by the analysis of clones in wing and haltere discs 2 days after heat shock. The Hsp90 mutant cells must have a disadvantage in growth as clones induced at earlier time points than at 72 h or analyzed later than 48 h after induction could not be observed.
DNA Construct and Generation of Transgenic Flies.
PCR-amplified Hsp90 and GFP coding sequences (CDS) were cloned in frame under the Drosophila Hsp90 promoter in the hsp83CasPer fly transformation vector as described (45). The GFP was fused C-terminally to the Hsp90 CDS and transgenic flies were generated by using standard protocol. The HspGFP flies were analyzed by Western blot for expression level of HspGFP fusion protein with anti-Hsp90 (generated during this study) (Fig. S2 A and B) and anti-GFP antibodies (Chemicon International) (Fig. S2 A and B).
Cell Culture and Pharmacological Inhibition of Hsp90.
Drosophila Kc cells were grown in Schneider's Drosophila medium (Invitrogen). A total of 3 × 106/ml cells per well in a 6-well plate were seeded 1 day before drug treatment and treated with different concentrations (3, 5, 10, 20, 40, and 80 μM) of radicicol dissolved in DMSO (Alomone labs) (Fig. 2A and data not shown) for 4 h. The Kc cells treated with equivalent amounts of DMSO vehicle were used as mock-treated cells. Finally, cells from each well were resuspended in 40 μl standard protein gel sample buffer, boiled for 5 min at 95°C, and resolved on Trizma base-acetate SDS polyacrylamide gels (Invitrogen). After transfer onto nitrocellulose membrane (Hybond-ECL, Amersham Bioscience), Trx, CBP, and αTub proteins were detected by using anti-Trx (36) (1:500 diluted), anti-CBP (27) (1:2,000 diluted), and mouse monoclonal anti-tubulin (1:5,000 diluted; Sigma) antibodies. The signal was detected by using enhanced chemiluminescence (GE Healthcare) followed by exposure to Amersham Hyperfilm ECL (GE Healthcare).
HEK 293 cells were grown in D-MEM medium (4.5 g/L glucose, 50 μg penicillin/ml, 50 μg/ml streptomycin, 10% FCS; Invitrogen). A total of 800,000 cells per well in a 6-well plate were seeded 1 day before radicicol treatment. After 4 h starvation of cells with serum-free medium, radicicol (or DMSO alone, as control) was added with 1 vol of medium containing 20% FCS. Cells were harvested after 4 h incubation and lysed by boiling in protein sample buffer. Finally, MLL-N320- and MLL-C180-specific antibodies (Millipore) and anti-p53 (BD Biosciences) antibodies were used.
Chromatin Immunoprecipitation and Coimmunoprecipitation.
ChIP was performed from Kc cells and SF4 cells as described (32, 46), by using anti-Trx (rabbit), anti-Pc (rabbit), and anti-Hsp90 (rabbit) antibodies. Finally, the purified DNA was resuspended in 50 μl DNase-free water and analyzed by PCR by using gene-specific primers following the PCR profile (95°C for 3 min followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with a final cycle at 72°C for 10 min). All of the ChIPs were quantified by using Light cycler 480 real-time PCR systems (Roche) and SYBR Green was used to monitor the accumulation of double-stranded DNA in PCR mixtures at the end of each cycle to produce amplification profiles. Finally, percentage inputs in each ChIP reaction were calculated as described (47). Drosophila embryonic nuclear extracts were used for coimmunoprecipitation by using a standard protocol (Millipore) with anti-Trx and anti-Hsp90 (rabbit) antibodies.
Reverse Transcription PCR Analysis.
Total RNA was isolated from radicicol- and DMSO-treated cells by using TRIzol reagent following the manufacturer's instructions (Invitrogen). The total RNA was treated with TURBO DNase (Ambion) to get rid of any contaminating DNA, following manufacturer's instructions (Ambion). Finally, 1 μg of DNase-treated total RNA was used to reverse transcribe by using an Oligo (dT) primer in a SuperScript III first-strand synthesis system (Invitrogen). Further, 2 μl of cDNA were used as a template to amplify Dfd, Trx, and tub gene products following the PCR profile (95°C for 3 min followed by 32 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s with a final cycle at 72°C for 10 min). Relative mRNA levels were quantified with Light cycler 480 real-time PCR systems (Roche)by using SYBR Green. The Dfd and Trx mRNA levels were quantified relative to the endogenous housekeeping gene ATPase cf6 (32).
Supplementary Material
Acknowledgments.
We thank the Zentrum fuer Molekulare Biologie Heidelberg, University of Heidelberg, Germany, where part of our research was performed. M.T. was supported by a European Molecular Biology Organization Long-Term Fellowship. R.P. was supported by grants from the Deutsche Forschungsgemeinschaft, the European Union-Network of Excellence Epigenome, and the Eidgenössiche Technische Hochschule Zurich.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809669106/DCSupplemental.
References
- 1.Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: Chaperones of tumorigenesis. Trends Biochem Sci. 2006;31:164–172. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Freeman BC, Yamamoto KR. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296:2232–2235. doi: 10.1126/science.1073051. [DOI] [PubMed] [Google Scholar]
- 3.Hung JJ, Wu CY, Liao PC, Chang WC. Hsp90alpha recruited by Sp1 is important for transcription of 12(S)-lipoxygenase in A431 cells. J Biol Chem. 2005;280:36283–36292. doi: 10.1074/jbc.M504904200. [DOI] [PubMed] [Google Scholar]
- 4.Floer M, Bryant GO, Ptashne M. HSP90/70 chaperones are required for rapid nucleosome removal upon induction of the GAL genes of yeast. Proc Natl Acad Sci USA. 2008;105:2975–2980. doi: 10.1073/pnas.0800053105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schnaider T, Oikarinen J, Ishiwatari-Hayasaka H, Yahara I, Csermely P. Interactions of Hsp90 with histones and related peptides. Life Sci. 1999;65:2417–2426. doi: 10.1016/s0024-3205(99)00508-1. [DOI] [PubMed] [Google Scholar]
- 6.Csermely P, Kajtar J, Hollosi M, Oikarinen J, Somogyi J. The 90 kDa heat shock protein (hsp90) induces the condensation of the chromatin structure. Biochem Biophys Res Commun. 1994;202:1657–1663. doi: 10.1006/bbrc.1994.2124. [DOI] [PubMed] [Google Scholar]
- 7.Hamamoto R, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 8.Zhao R, et al. Navigating the chaperone network: An integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 9.Mollaaghababa R, et al. Mutations in Drosophila heat shock cognate 4 are enhancers of Polycomb. Proc Natl Acad Sci USA. 2001;98:3958–3963. doi: 10.1073/pnas.061497798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang YJ, Brock HW. Polyhomeotic stably associates with molecular chaperones Hsc4 and Droj2 in Drosophila Kc1 cells. Dev Biol. 2003;262:350–360. doi: 10.1016/s0012-1606(03)00396-8. [DOI] [PubMed] [Google Scholar]
- 11.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 12.Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: Drug resistance in diverse fungi. Science. 2005;309:2185–2189. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- 13.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 14.Sollars V, et al. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat Genet. 2003;33:70–74. doi: 10.1038/ng1067. [DOI] [PubMed] [Google Scholar]
- 15.Waddington CH. Genetic assimilation of an acquired character. Evolution. 1953;7:118–126. [Google Scholar]
- 16.Waddington CH. Canalization of development and genetic assimilation of acquired characters. Nature. 1959;183:1654–1655. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 17.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 18.Milton CC, Ulane CM, Rutherford S. Control of canalization and evolvability by Hsp90. PLoS ONE. 2006;1:e75. doi: 10.1371/journal.pone.0000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carey CC, Gorman KF, Rutherford S. Modularity and intrinsic evolvability of Hsp90-buffered change. PLoS ONE. 2006;1:e76. doi: 10.1371/journal.pone.0000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sangster TA, et al. HSP90 affects the expression of genetic variation and developmental stability in quantitative traits. Proc Natl Acad Sci USA. 2008;105:2963–2968. doi: 10.1073/pnas.0712200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sangster TA, et al. HSP90-buffered genetic variation is common in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:2969–2974. doi: 10.1073/pnas.0712210105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song Y, Fee L, Lee TH, Wharton RP. The molecular chaperone Hsp90 is required for mRNA localization in Drosophila melanogaster embryos. Genetics. 2007;176:2213–2222. doi: 10.1534/genetics.107.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 24.Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 25.Klymenko T, Muller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 2004;5:373–377. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beuchle D, Struhl G, Muller J. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development. 2001;128:993–1004. doi: 10.1242/dev.128.6.993. [DOI] [PubMed] [Google Scholar]
- 27.Petruk S, et al. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science. 2001;294:1331–1334. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- 28.Sreedhar AS, Soti C, Csermely P. Inhibition of Hsp90: A new strategy for inhibiting protein kinases. Biochim Biophys Acta. 2004;1697:233–242. doi: 10.1016/j.bbapap.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 29.Yokoyama A, Kitabayashi I, Ayton PM, Cleary ML, Ohki M. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood. 2002;100:3710–3718. doi: 10.1182/blood-2002-04-1015. [DOI] [PubMed] [Google Scholar]
- 30.Walerych D, et al. Hsp90 chaperones wild-type p53 tumor suppressor protein. J Biol Chem. 2004;279:48836–48845. doi: 10.1074/jbc.M407601200. [DOI] [PubMed] [Google Scholar]
- 31.Rowley JD. The critical role of chromosome translocations in human leukemias. Annu Rev Genet. 1998;32:495–519. doi: 10.1146/annurev.genet.32.1.495. [DOI] [PubMed] [Google Scholar]
- 32.Beisel C, et al. Comparing active and repressed expression states of genes controlled by the Polycomb/Trithorax group proteins. Proc Natl Acad Sci USA. 2007;104:16615–16620. doi: 10.1073/pnas.0701538104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yue L, et al. Genetic analysis of viable Hsp90 alleles reveals a critical role in Drosophila spermatogenesis. Genetics. 1999;151:1065–1079. doi: 10.1093/genetics/151.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carbajal ME, Valet JP, Charest PM, Tanguay RM. Purification of Drosophila hsp 83 and immunoelectron microscopic localization. Eur J Cell Biol. 1990;52:147–156. [PubMed] [Google Scholar]
- 35.Smith ST, et al. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat Cell Biol. 2004;6:162–167. doi: 10.1038/ncb1088. [DOI] [PubMed] [Google Scholar]
- 36.Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petruk S, Smith ST, Sedkov Y, Mazo A. Association of trxG and PcG proteins with the bxd maintenance element depends on transcriptional activity. Development. 2008;135:2383–2390. doi: 10.1242/dev.023275. [DOI] [PubMed] [Google Scholar]
- 38.Picard D, et al. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature. 1990;348:166–168. doi: 10.1038/348166a0. [DOI] [PubMed] [Google Scholar]
- 39.Kimura Y, Yahara I, Lindquist S. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- 40.Stavreva DA, Muller WG, Hager GL, Smith CL, McNally JG. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol Cell Biol. 2004;24:2682–2697. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeFranco DB, Csermely P. Steroid receptor and molecular chaperone encounters in the nucleus. Sci STKE. 2000;2000:PE1. doi: 10.1126/stke.2000.42.pe1. [DOI] [PubMed] [Google Scholar]
- 42.Muller L, Schaupp A, Walerych D, Wegele H, Buchner J. Hsp90 regulates the activity of wild type p53 under physiological and elevated temperatures. J Biol Chem. 2004;279:48846–48854. doi: 10.1074/jbc.M407687200. [DOI] [PubMed] [Google Scholar]
- 43.Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 44.Dickson BJ, van der Straten A, Dominguez M, Hafen E. Mutations Modulating Raf signaling in Drosophila eye development. Genetics. 1996;142:163–171. doi: 10.1093/genetics/142.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhat KM, Poole SJ, Schedl P. The miti-mere and pdm1 genes collaborate during specification of the RP2/sib lineage in Drosophila neurogenesis. Mol Cell Biol. 1995;15:4052–4063. doi: 10.1128/mcb.15.8.4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orlando V, Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- 47.Dellino GI, et al. Polycomb silencing blocks transcription initiation. Mol Cell. 2004;13:887–893. doi: 10.1016/s1097-2765(04)00128-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.