Abstract
In response to a DNA double-strand break (DSB), chromatin is rapidly modified by the damage dependent checkpoint kinases. Also, disassembly of chromatin occurs at the break site. The damage-induced modification of chromatin structure is involved in the maintenance of the checkpoint. However, it has not been determined how chromatin is restored to its undamaged state when DSB repair is complete. Here, we show the involvement of two chromatin assembly factors (CAFs), Asf1 and CAF-1, in turning off the DNA damage checkpoint in budding yeast. DSB repair or formation of γ-H2AX does not depend on either the CAF-1 protein, Cac1, or Asf1. Absence of these proteins does not impair the ability of cells to resume cell cycle progression in the presence of an unrepaired DSB (adaptation). However, recovery from cell cycle checkpoint arrest when the DSB is repaired by gene conversion is substantially defective in the absence of both CAF-1 and Asf1, whereas deleting CAC1 or ASF1 individually had little effect. We suggest that CAF-1 and Asf1 function redundantly to deactivate the checkpoint by restoring chromatin structure on the completion of DSB repair.
To maintain genomic integrity, cells have to perform accurate and efficient DNA repair in response to different kinds of DNA damage. In eukaryotic cells, DNA repair is intimately connected to chromatin structure. Originally, chromatin structure was considered merely as a physical obstacle to the access of DNA repair machinery (1). However, a number of studies provide evidence that modifications of chromatin structure have active roles in the DNA damage-dependent response (2). Modification of chromatin structure by the DNA damage checkpoint kinases at the early stages of DNA damage response has been extensively studied (3–7). Also, the disassembly of chromatin at DNA double-strand breaks (DSBs) has been detected (8–10), implying that histones are removed before or during DSB repair. However, it is still unknown how the modified and disassembled chromatin is restored to its original status when DSB repair is complete.
In budding yeast, the induction of even a single DSB activates the DNA damage checkpoint, leading to the cell cycle arrest until the repair of the break is complete (11). By arresting the cell cycle at the G2/M phase, the checkpoint provides enough time for cells to repair DSBs before cells enter into mitosis. After the formation of a DSB, Mec1 and Tel1 (yeast orthologs of mammalian ATR and ATM, respectively) are initially activated. Mec1 is more critical for the activation of the DNA damage checkpoint, whereas Tel1 has a less important role (12). Both Mec1 and Tel1 can phosphorylate histone H2A on serine 129 (3), whereas Mec1 principally phosphorylates the checkpoint signal transducer kinases, Chk1 and Rad53 (yeast's homolog of mammalian Chk2), as well as the checkpoint mediator protein, Rad9 (13, 14). Budding yeast histone H2A contains the same SQE motif at its C′-terminus that is found in mammalian histone H2AX and is a consensus target sequence of Mec1 and Tel1 kinases. Hereafter, we will refer to phosphorylated yeast H2A-S129 as γ-H2AX. The Mec1/Tel1-dependent phosphorylation of Rad53 is required for autophosphorylation of Rad53 (11). Hyperphosphorylated Rad53 is then able to modulate downstream effectors of cell cycle progression.
Hyperphosphorylated Rad53 is maintained during the persistence of the checkpoint (11). As DNA damage is repaired, cells deactivate the checkpoint to reenter the cell cycle; this process is called recovery. Alternatively, cells can resume the cell cycle even in the presence of an unrepaired DNA break, a process termed adaptation (11). In both processes, a prominent phenomenon is the disappearance of hyperphosphorylated Rad53, which indicates inactivation of the checkpoint (11, 15). Deletions of DNA metabolism proteins such as yKu70, yKu80, and Tid1 (Rdh54), as well as the cdc5-ad mutation, are adaptation defective; deletions of the PP2C phosphatases Ptc2 and Ptc3, casein kinase II, and the helicase Srs2 are defective both in adaptation and recovery (12, 15–18).
Chromatin assembly factor (CAF)-1 and Asf1 are cooperatively involved in replication-coupled nucleosome assembly via the interaction with histone H3/H4 (19–21). Also, studies in mammalian cells have shown that CAF-1 is linked to nucleotide excision repair (NER) as well as DNA replication (22, 23). In an in vitro assay, CAF-1 mediates the assembly of nucleosomes onto naked DNA templates in a PCNA-dependent manner (24, 25). Recently, by visualizing transiently expressed tagged histone H3.1 with a specific antibody in human cells, Polo et al. (26) demonstrated that CAF-1 is involved in the deposition of histones onto repaired UV lesions in vivo. In budding yeast, deletions of the genes encoding the subunits of CAF-1, CAC1, CAC2, and CAC3 result in cellular sensitivity to UV (27). Deletion of Asf1, a histone H3/H4 chaperone, also leads to the UV sensitivity of mutant cells (28). Also, asf1 mutants are defective in completion of DNA replication when exposed to hydroxyurea (29). Asf1 has other important roles in regulating gene silencing (30) and in facilitating the acetylation of lysine 56 residue on nascent histone H3, which is synthesized mostly during S phase (31, 32). Also, the fact that mutant cells lacking either a subunit of CAF-1 or Asf1 are sensitive to DSB-inducing agents (33, 34) suggests that CAF-1 and Asf1 are involved in the DSB-mediated response. Yeast cells lacking Asf1 accumulate in G2/M phase because the DNA damage checkpoint is activated by spontaneous DNA damage during replication, although DNA DSB repair is competent in the cells (35). Double deletion of Asf1 and 1 of the subunits of CAF-1, Cac1, Cac2, or Cac3, enhances the G2/M accumulation of the cells (36); however, DNA replication itself is proficient in the mutant cells, and the transient checkpoint arrest phenotype of these mutants is rescued by deletion of checkpoint proteins such as Mec1 and Rad53 (35, 36). These results raise the possibility that the CAFs are related to the DNA damage-activated checkpoint.
In this study, we provide evidence that CAF-1 and Asf1 have redundant roles in regulating deactivation of the damage-activated checkpoint after DSB repair is complete. When the DNA damage checkpoint is robustly activated in response to the induction of a DSB, asf1Δ cac1Δ double mutants fail to recover from the checkpoint-mediated arrest. However, the absence of these factors affects neither repair of a DSB nor the removal of γ-H2AX from regions surrounding the repaired locus. These results indicate that the failure of the checkpoint deactivation is not caused by persistent DNA damage or modified histones around the site or by defects in γ-H2AX disassembly. However, adaptation, which occurs in the absence of repair, is not defective in asf1Δ cac1Δ cells. Therefore, we suggest that when DSBs are repaired, CAF-1 and Asf1 cooperatively restore chromatin at the repair sites and mediate efficient deactivation of the checkpoint to terminate DSB response.
Results
The Absence of Both Functional CAF-1 and Asf1 Leads to a Defect in the Viability of the Cells Experiencing a Repairable DSB.
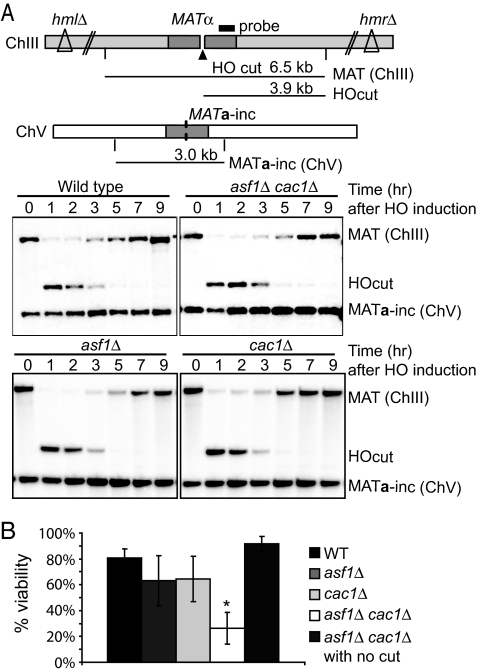
To determine the involvement of CAFs in the DSB-response process, we first examined whether the absence of CAFs affects the viability of cells in response to an HO endonuclease-induced DSB. In strain YJK17 (37), HO endonuclease induced by addition of galactose to the medium generates a DSB at the MATα locus on chromosome III. Because the cells lack both HML and HMR, which contain sequences homologous to MAT, the DSB cannot be repaired by intrachromosomal homologous recombination (HR), which normally leads to mating type switching (38). Instead, the strain contains MATa-inc sequences on chromosome V, so that the DSB at MATα can be repaired by ectopic recombination by using the MATa-inc sequence as a homologous donor (Fig. 1A). The MATa-inc sequence carries a mutated HO recognition site (a-inc) that cannot be cleaved by HO endonuclease (39). Therefore, once the HO-induced DSB at the MAT locus is repaired by ectopic HR, cells can proliferate without enduring additional HO-induced DSBs in the medium containing galactose; ≈80% wild-type cells are viable on galactose plates (Fig. 1B), whereas isogenic cells lacking either Asf1 or Cac1 generated a small but significant reduction in viability, 63% (P = 0.038) and 64% (P = 0.047), respectively. Deletion of both ASF1 and CAC1 led to only ≈26% of viability, a result statistically significantly different from the viability of wild-type cells (P < 0.001), as well as from either cac1Δ or asf1Δcells (P = 0.001 and P < 0.001, respectively). Because CAFs could affect gene expression and prevent cells from growing on galactose-containing medium, we tested whether isogenic cells lacking an HO cleavage site (a cell that carried MATa-inc instead of MATα) were unable to proliferate on galactose medium. However, there was no reduction in the viability of the cells in the absence of Asf1 and Cac1 (Fig. 1B). Therefore, we concluded that only on induction of a DSB, do the cells lacking both Asf1 and Cac1 have a defect in viability. We note that both asf1Δ and asf1Δ cac1Δ cells, which were derived from the strain YJK17 and used for this study, exhibit the expected sensitivity to different DNA damaging agents as previously reported [supporting information (SI) Fig. S1] (28), suggesting that strain differences do not affect the phenotype of cells lacking Asf1 and Cac1 in our assay.
Fig. 1.
An HO endonuclease-induced DSB leads to the reduced viability of asf1Δ cac1Δ. (A, Upper) diagram of MATα in chromosome III (ChIII) and MATa-inc in ChV in strain YJK17. Vertical lines indicate EcoRI cleavage sites. The black bar denotes the probe specific to the MAT distal region (for PCR primers, see Table S1). Prior to HO-induced DSB formation, a 6.5 kb EcoRI fragment [MAT (ChIII)] is detected by the probe. After HO cleavage, a 3.9-kb band (HOcut) is generated. In addition, a 3.0-kb fragment surrounding MATa-inc [MATa-inc (ChV)] is also detected. (A, Lower) Exponentially growing cells were collected at different time points after HO induction. DNA from each time point was digested with EcoRI, and then run on a 0.8% agarose gel for the Southern analysis using the probe described above. The HO-induced DSB is repaired by interchromosomal homologous recombination by using MATa-inc on ChV as a homologous donorm resulting in the replacement of MATα on ChIII is replaced by MATa-inc sequences. (B) Percentage viability of a wild type (YJK17) and its derivatives lacking Asf1, Cac1, or both of them on galactose plates. Wild-type and mutant cells were grown in YEP/Lactate media to the exponential phase and then ≈100–200 cells were plated either on YEP/Dextrose plates or YEP/Galactose plates. After 5 days, colonies were counted. Percent viability was calculated by dividing colony numbers on galactose with that on dextrose plates. “asf1Δcac1Δ with no cut” indicates asf1Δcac1Δ containing MATa-inc sequence at MAT on chromosome III prior to HO induction. The graph shows the average of 5 experiments with error bar representing SDs. * indicates the value of asf1Δcac1Δ, which is significantly different wild type as well as either asf1Δ or cac1Δ.
DSB Repair in asf1Δ cac1Δ Is As Efficient As that in Wild-Type Cells.
After induction of a DSB, cells could fail to proliferate for 2 different reasons: defects in repairing DNA damage or defects in resuming cell cycle by failing to turn off the damage-mediated checkpoint. We first monitored the HR-mediated DSB repair in mutants lacking Asf1 and/or Cac1. By using a probe specific to the MAT distal region (Fig. 1A), formation of an HO-induced DSB and the repair of the break were detected; 1 h after the induction of HO endonuclease, all of the mutant as well as wild-type cells exhibited efficient HO cleavage at the MAT locus on chromosome III (Fig. 1A). In wild-type cells, the appearance of repair product was seen as early as 3 h after DSB induction and by 9 h, ≈80% of the wild-type cells completed the repair. DSB repair in all of the mutant cells were as efficient as that in wild-type cells (Fig. 1A). Therefore, it is unlikely that the reduced viability of asf1Δ cac1Δ cells after the induction of a DSB is caused by a defect in DSB repair.
The asf1Δ cac1Δ Cells Do Not Resume Cell Cycle As Efficiently As Wild-Type Cells Due to a Defect in Turning Off the Damage Checkpoint.
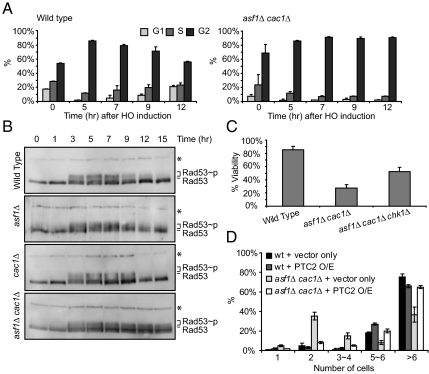
Next, we determined whether the DNA damage checkpoint activated in response to a DSB is involved in the viability of asf1Δ cac1Δ cells. We monitored cell cycle progression microscopically; 5 h after HO induction in cycling cells, ≈80% wild-type cells were arrested at the G2/M phase and, subsequently, the population of G2/M declined (Fig. 2A). In contrast, the majority of asf1Δ cac1Δ cells (>80%) persisted at the G2/M phase as late as 12 h after DSB induction. Although asf1Δ cac1Δ cells tend to be constitutively delayed in the G2/M stage than wild-type cells (Fig. 2A, 0 hr time point), the induction of a DSB was definitely required for the complete arrest of the asf1Δ cac1Δ cells, which remained as late as 12 h after the break induction. These results suggest that the DNA damage checkpoint is not efficiently deactivated in the absence of Asf1 and Cac1.
Fig. 2.
asf1Δcac1Δ cells are deficient in recovering from the DNA damage checkpoint-mediated arrest. (A) Wild-type and asf1Δ cac1Δ MATα cells lacking HML and HMR were grown in YEP/Lactate medium to the exponential phase and then 2% galactose was added to induce HO endonuclease. Cells were observed microscopically. Unbudded cells (G1), small budded cells (S), and large budded cells (G2/M) were counted. Three different experiments counting 100–200 cells were performed to obtain the average shown in this figure. Error bars indicate SDs. (B) Western analysis with anti-Rad53 antibody determined the extent of phosphorylated Rad53 in whole cell extracts of wild-type and mutant cells during DSB repair. Phosphorylated Rad53 (Rad53∼p) migrates slowly compared to unphosphorylated Rad53 (Rad53). * indicates a nonspecific band detected by cross-reactivity of the Rad53 antibody, which was used as a loading control. (C) Percent viabilities were measured as described in Fig. 1. (D) Wild-type (YJK17) and its derivatives lacking Asf1 and/or Cac1 were grown in YEP/Lactate media to the exponential phase and then the unbudded cells (G1) were micromanipulated on galactose plates. Overexpression (O/E) of Ptc2 was driven by galactose-inducible promoter. One day after plating, the number of cells derived from a single cell was counted under the microscope. The graph shows the average of 3 different experiments with error bars representing SDs.
To examine the persistence of the DNA damage checkpoint more directly, we also monitored phosphorylation of Rad53. In wild-type cells, phosphorylated Rad53 appears ≤3 h after DSB induction and then disappears at the same time that repair of the DSB is complete, 9–12 h after HO induction (Fig. 2B). In asf1Δ or cac1Δ cells, the appearance of phosphorylated Rad53 occurred almost at the same time as wild-type cells, whereas its disappearance occurred either earlier or at the same time as in wild type (Fig. 2B). By contrast, in asf1Δ cac1Δ cells, phosphorylated Rad53 appears by 3 h, but persists as late as 15 h after the formation of an HO-induced DSB (Fig. 2B), even though DSB repair is almost complete by 9 h (Fig. 1A). These results indicate that deactivation of the DNA damage-mediated checkpoint is strikingly defective in cells lacking both functional CAF-1 and Asf1.
Consistent with the idea that the CAFs engage in cross-talk with the DNA damage checkpoint, we found that deletion of the checkpoint kinase, Chk1, which is required for maintenance of checkpoint-mediated arrest beyond ≈8 h (40), partially suppressed the failure of asf1Δ cac1Δ cells to resume growth after ectopic recombination in strain YJK17 (Fig. 2C). PP2C protein phosphatases Ptc2 and Ptc3 are involved in deactivation of the damage checkpoint (16, 41). Although cells lacking Ptc2 and Ptc3 are defective both in recovery and adaptation and fail to dephosphorylate Rad53 (16, 41), overexpression of Ptc2 rescues the adaptation defect by dephosphorylating Rad53 (16). Recovery from the DNA damage-activated checkpoint can be monitored by observing division of individual cells on galactose plates (15). When we overexpressed Ptc2 in asf1Δ cac1Δ cells on galactose plates, by 24 h, 70% the cells generated >6 cells, which were as many as their wild-type counterpart. In contrast, ≈60% asf1Δ cac1Δ cells containing empty vector were still arrested either at G2/M phase or at 4 cell stage (Fig. 2D). These results indicate that the defect in the recovery from the damage-activated checkpoint in asf1Δ cac1Δ cells is suppressed by turning off the damage checkpoint. We note that overexpressed Ptc2 results in the lethality of even wild-type cells, so we could not determine the effect of overexpressed Ptc2 on the viability of cells.
Although Asf1 and Cac1 Are Not Involved in Removing γ-H2AX from Chromatin, the Nucleosome Assembly Function of Asf1 Is Required for the Viability of asf1Δ cac1Δ Cells After DSB Repair.
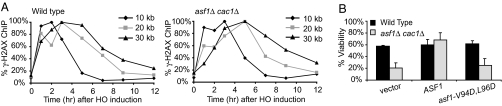
Coincident with time that the DSB is repaired in strain YJK17, γ-H2AX begins to be removed from the 50-kb region around the DSB (37). We compared the kinetics of γ-H2AX appearance and disappearance in wild-type and mutant cells by using a ChIP assay (3); 1 h after HO induction, γ-H2AX was enriched 10, 20, and 30 kb away from the DSB in asf1Δ cac1Δ as well as in wild-type cells and reached its maximum between 2 and 3 h (Fig. 3A and Fig. S2). In wild-type cells, the ChIP signal then decreased when the break is repaired. The decrease of γ-H2AX occurred progressively from the break site (Fig. 3A). However, there was no substantial difference in the disappearance of γ-H2AX between wild-type and asf1Δ cac1Δ cells, or in the 2 single mutants (data not shown). These results suggest that removal of γ-H2AX from chromatin during DSB repair does not depend on either Asf1 or CAF-1.
Fig. 3.
Although appearance and disappearance of γ-H2AX is not affected by the absence of both Asf1 and Cac1, the nucleosome assembly function of Asf1 is required for the viability of asf1Δ cac1Δ cells after DSB repair. (A) γ-H2AX ChIP values at 10 kb (black bars), 20 kb (gray bars) or 30 kb (whites bars) to the right of the DSB at MAT in wild-type and asf1Δ cac1Δ cells were measured several time points (1, 2, 3, 5, 7, 9, and 12 hrs) after HO induction (see Fig. S2) and the percent maximum value of γ-H2AX ChIP is shown. (C) Percent viabilities of wild-type (black bars) and asf1Δcac1Δ (gray bars) cells with an empty plasmid, a plasmid with ASF1, or a plasmid with asf1-V94D, L96D. Cells grown in SC-Ura/Raffinose (synthetic complete medium containing raffinose instead of dextrose but lacking uracil) were plated either on YEP/Dextrose or on YEP/Galactose plates. Viability was measured as described in Fig. 1.
To relate the defect in checkpoint deactivation after DSB repair in asf1Δ cac1Δ directly to the cooperative nucleosome assembly functions of Asf1 and CAF-1, we tested whether introduction of a plasmid-borne asf1-V94D,L96D mutant that is defective in histone binding (42) would affect the viability of asf1Δ cac1Δ cells. Unlike the wild-type counterpart, the mutant asf1-V94D,L96D did not suppress the viability defect in asf1Δ cac1Δ cells (Fig. 3B). This result suggests that the histone binding and presumably the chromatin assembly activity of Asf1 is required for the cells experiencing an HO-induced DSB repair to deactivate the damage-activated checkpoint on the completion of DSB repair.
Asf1 and CAF-1 Do Not Affect Recovery When DSB Repair Occurs Without Strongly Activating the DNA Damage Checkpoint.
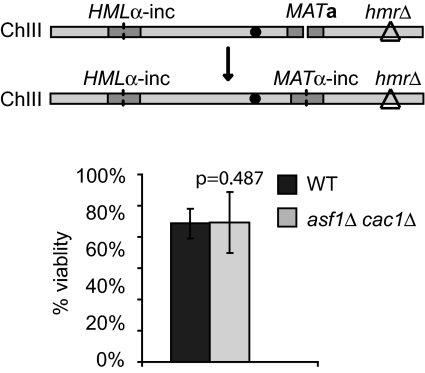
When there is a homologous donor either at HML or at HMR, an HO-induced DSB at the MAT locus is rapidly repaired by intrachromosomal HR. In this circumstance, the repair of the HO-induced DSB is completed as early as 2 h, and the Rad53 checkpoint kinase does not become hyperphosphorylated; however, Mec1 and/or Tel1 carry out the early checkpoint responses and phosphorylate γ-H2AX around the MAT locus (Fig. S3) (11). Consistent with previous reports (37, 43), more γ-H2AX is detected at a region 10 kb away than 5 kb away from the break. To monitor the viability of the cell experiencing intrachromosomal HR on galactose plates, we constructed MATa strain YJK139, deleted for HMR and carrying HMLα-inc sequences. Thus, cells switch from MATa to MATα-inc, and cannot be again cleaved by HO on galactose medium. In this background, deletion of both ASF1 and CAC1 did not reduce viability of the cells (Fig. 4), even though ≥700 bp of new DNA must be synthesized to replace the MAT Ya sequence with Yα-inc copied from HMLα-inc (and packaged into nucleosomes). Therefore, we conclude that only when the DNA damage checkpoint is strongly activated, which is indicated by hyperphosphorylation of Rad53, is the cooperative function of Asf1 and CAF-1 required for cell proliferation after DSB repair.
Fig. 4.
Asf1 and CAF-1 do not affect recovery when DSB repair occurs without strongly activating the DNA damage checkpoint. Percentage viabilities of YJK139 wild type and its derivatives lacking both Asf1 and Cac1 were measured as described in Fig. 1A. The diagram shows the HO-induced DSB at the MAT locus on ChIII and the consequence of the DSB repair by intrachromosomal HR by using the HMLα-inc donor located at the left arm of chromosome III. The CEN3 locus is marked as a black dot.
Asf1 and CAF-1 Are Not Required for Adaptation in the Absence of DSB Repair.
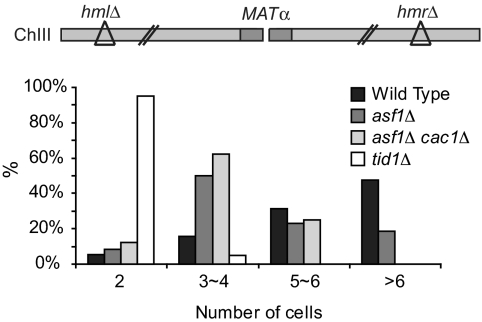
To determine whether repair per se was required before the loss of Asf1 and Cac1 displayed an arrest phenotype, we examined the ability of cells to resume cell cycle progression in the presence of an unrepairable DSB, a process termed adaptation (12). A DSB was induced in strains carrying MATα but deleted for both HML and HMR donors; 8 h after DSB induction, wild-type cells and an isogenic derivative lacking Asf1 only or both Asf1 and Cac1 arrest as large dumbbell-shaped cells characteristic of G2/M arrest, and by 48 h, > 80% of both wild-type and mutant cells had adapted by dividing 1 or more times (Fig. 5). On the contrary, 95% tid1Δ cells were persistent as a dumbbell-shaped cell as late as 48 h, which is a characteristic of adaptation-defective (17). These results show that the CAFs are not required to silence the checkpoint during adaptation, but only when the DSB has been repaired, presumably, because in repaired cells, chromatin must be reestablished at the repaired, intact DNA. We note that, although ≈50% of wild-type cells reach >6 cells by 48 h after a DSB induction, only 20% asf1Δ and none of asf1Δ cac1Δ cells have >6 cells by then (Fig. 5). By observing growth of wild-type and asf1Δ cac1Δ cells, both of which lack the HO cut site (inc), on galactose plates, we found that asf1Δ cac1Δ cells grow approximately twice as slowly as wild-type cells (Fig. S4). This result indicates that the smaller number of asf1Δ cac1Δ cells reached >6 cells by 48 h because of the slow division of the cells, compared with wild-type cells. In the adaptation assay we use, cells eventually die because the broken chromosome III is not repaired (44); because degradation of the DSB ends continues even in cells arrested in G2/M (11), they will reach a point of inviability with fewer cell divisions when the cells grow substantially slower. Given that most cells progress beyond the 2-cell arrest seen for tid1Δ, we conclude that asf1Δ and asf1Δ cac1Δ cells are competent for the checkpoint adaptation.
Fig. 5.
Asf1 and CAF-1 are not required for the adaptation in the absence of DSB repair. The diagram shows ChIII containing MATα but lacking both HML and HMR. Wild-type, asf1Δ, asf1Δ cac1Δ, and tid1Δ cells were synchronized to G1 by growth to saturation and unbudded cells were spread onto galactose plates. The number of buds or cells was counted at 48 h after spreading (HO induction), and the number of each population with different bud/cell numbers was converted to a percentage value.
Discussion
As DSB repair is complete, cells must extinguish the checkpoint signal to resume cell division. Previous studies have shown that turning off the checkpoint requires the PP2C phosphatases, Ptc2 and Ptc3, as well as casein kinase II; these factors appear to be involved in the dephosphorylation of Rad53 and perhaps Chk1 (16, 18, 40, 45). Srs2 DNA helicase is also required, possibly to remove a checkpoint signaling protein from the repaired region (15). Also, the presence of γ-H2AX delays efficient termination of the damage-activated checkpoint (37). We have previously shown that γ-H2AX is only dephosphorylated after it is removed from the chromatin surrounding the repaired site (37). It is not known how the modified histones are removed, or whether chromatin is restored to its original undamaged state or to a novel configuration. Specifically for repair of a DSB during ectopic recombination, our previous studies showed that 5′ to 3′ resection of DSB ends, which is required for efficient Rad51-mediated repair, removes γ-H2AX from the sites near DSBs (37, 43); however, its displacement occurs in a resection-independent manner (37). We initially hypothesized that Asf1 might be involved in the disassembly of γ-H2AX from the chromatin surrounding DSBs, given the fact that Asf1 is involved in the histone eviction for transcriptional activation (46). However, we found that both Asf1 and CAF-1 are dispensable for the removal of γ-H2AX during the repair of an HO-induced DSB. Also, although a previous report argued that asf1 cac1 double mutants were defective in DSB repair pathways (33), the absence of either Asf1 or Cac1, a subunit of CAF-1 did not affect DSB repair itself in our assay system. Also, in our hands, γ-H2AX removal also does not depend on Ino80 or Swr1 chromatin remodeling complexes or on the histone chaperone Nap1, even in multiple mutant combinations (data not shown).
However, when the damage dependent checkpoint is robustly activated, the asf1Δ cac1Δ cells are defective in the deactivation of the checkpoint that is coupled to DSB repair. Neither asf1Δ nor cac1Δ alone exhibited this defect. These results suggest that Asf1 and CAF-1 have cooperative roles in the regulation of the damage checkpoint. Importantly, we did not detect a recovery defect in asf1Δ cac1Δ cells when a DSB is repaired by intrachromosomal gene conversion that does not cause Rad53-mediated cell-cycle arrest. This result suggests the involvement of the CAFs in the DNA damage checkpoint pathway only when it is robustly activated. When the checkpoint is not maintained, in a chk1Δ mutant that releases cells from arrest after ≈8 h, the defect in asf1Δ cac1Δ cells is partially suppressed; the fact that some cells remain arrested could imply that the constitutive problems of the double mutant to traverse G2/M augment the intensity of the checkpoint signal and is not fully suppressed by chk1Δ. Asf1 and CAF-1 also do not affect the ability of cells that fail to repair a DSB from recovering by adaptation. The result is important, because it suggests that cells in which a DSB cannot be repaired (and in which there is no reestablishment of chromatin) turn off the checkpoint in a way that is fundamentally different from what happens when there is repair.
How could Asf1 and CAF-1 mediate the termination of the damage checkpoint? Asf1 and CAF-1 were originally identified as replication-dependent CAFs (19, 47). Also, when UV-damage is repaired by NER, human Asf1 and CAF-1 synergistically reassemble chromatin in NER-dependent manner (24), during which ≈30–50 nt of new DNA is synthesized (48). When a DSB is repaired by HR in the system we study, ≥700 base pairs must be synthesized to repair the break (49). We speculate that the newly synthesized DNA by HR-mediated repair must undergo chromatin assembly to establish and reorganize intact chromatin. If this idea is correct, our study suggests that Asf1 and CAF-1 can be strong candidates to connect the repair-mediated chromatin assembly with DSB repair and the checkpoint. Although the removal of γ-H2AX occurs even without Asf1 and CAF-1, the repaired DNA may not have a full complement of nucleosomes in the absence of these CAFs, so that proteins bound to naked DNA might maintain the damage checkpoint, which is indicated by the persistence of phosphorylated Rad53. Our result that the ectopic mutant Asf1 lacking the histone H3/H4 binding ability fails to suppress the viability defect in asf1Δ cac1Δ cells supports the idea that chromatin assembly function is required for the termination of the damage checkpoint. Alternatively, a specific histone mark might be incorporated into the chromatin at repair sites in an Asf1 and CAF-1 dependent manner. This repair mark might trigger the signal to deactivate the checkpoint. In either case, the cells without the restored chromatin could fail to turn off the checkpoint efficiently. Interestingly, Asf1 physically interacts with the checkpoint kinase, Rad53 (50). This association is weakened when Rad53 is hyperphosphorylated by the activation of the DNA damage checkpoint (50, 51). Consequently, the Asf1 that completes its chromatin assembly function might facilitate dephosphorylation of Rad53, leading to the reassociation of Asf1 with Rad53.
While we were preparing this manuscript, J. Tyler and coworkers (52) reported the involvement of Asf1 in the checkpoint recovery. However, our results (many using the same strains created in our laboratory) are different in many ways. In their study, Asf1 alone is defective in cell viability after repair of a DSB. However, the viability defect in asf1Δ cells by our ectopic HR assay is moderate, whereas the defect in asf1Δ cac1Δ cells is more striking. Also, we did not detect a viability defect of any mutant cells lacking Asf1 and/or Cac1 after intrachromosomal HR. Also, although Chen et al. (52) showed that chromatin assembly is defective during intrachromosomal MAT switching, we did not see a defect in viability of either an asf1Δ or an asf1Δ cac1Δ strain undergoing MAT switching. Last, Chen et al. (52) interpreted that asf1Δ is adaptation-defective. However, we present the evidence that both the asf1Δ and the asf1Δ cac1Δ double mutant are not adaptation defective. Some of our differences could be explained by the duration or intensity of the checkpoint signal in different repair assays. For example, we found that ptc2Δ ptc3Δ cells undergoing the same ectopic recombination event we have studied here were not permanently arrested, whereas cells undergoing a long-delayed single-strand annealing event (strain YMV002 used by Tyler's group) were permanently arrested (16). It is possible that the different duration or intensity of the checkpoint signal, generated by different repair assays, determines the degree of Asf1 dependency for turning off the checkpoint. Chen et al. (52) proposed that the Asf1- and Rtt109-dependent incorporation of H3K56Ac is involved in the recovery from the checkpoint. Whether Rtt109 affects CAF-1 activity is unknown. Further study should reveal the mechanism by which Asf1 and CAF-1 regulate the DSB repair-mediated chromatin assembly to signal checkpoint termination.
Materials and Methods
Yeast Strains.
The strains used in this study were derived either from JKM179 (MATα hoΔ hmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2-3,112 trp::hisG lys5 ura3-52 ade3::GAL::HO) (53), or YJK17 (MATα hoΔ hmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2-3,112 trp::hisG lys5 ura3-52 ade3::GAL::HO arg5,6::MATa-inc::HPH1) (37). ASF1 and CAC1 were deleted by KanMX and NatMX, respectively. YXW2 (MATa hoΔ HMLα HMRa leu2-3,112 his3-11,15 ade2-1 can1-100 ura3-1 trp1-1 bar1::hisG ade3::GAL::HO) is used to determine γ-H2AX enrichment around the MAT locus during normal mating type switching.
Western Blot Analysis.
Cell extracts were prepared by trichloroacetic acid (TCA) extraction and then subjected to the Western blot analysis. Rad53 was detected by polyclonal anti-Rad53 antibody (Santa Cruz, 1:200).
ChIP of γ-H2AX.
ChIP of γ-H2AX was carried out as previously described (3, 43), by using an antibody against yeast γ-H2AX, generously provided by C. Redon and W. Bonner (National Institutes of Health). The primers used for ChIP analysis are listed in Table S1.
Supplementary Material
Acknowledgments.
Both the plasmid containing wild-type ASF1 with its own promoter and the plasmid containing mutant asf1-V94D,L96D were generously provided by P. Kaufman (University of Massachusetts Medical School, Worcester, MA). This work was funded by National Institutes of Health Grant GM61766.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812578106/DCSupplemental.
References
- 1.De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: An escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 2.Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- 3.Shroff R, et al. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morrison AJ, et al. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 6.Downs JA, et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Unal E, et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 8.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 10.Shim EY, et al. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cell Biol. 2007;27:1602–1613. doi: 10.1128/MCB.01956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- 12.Harrison JC, Haber JE. Surviving the breakup: The DNA damage checkpoint. Annu Rev Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez Y, et al. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez Y, et al. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- 15.Vaze MB, et al. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- 16.Leroy C, et al. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol Cell. 2003;11:827–835. doi: 10.1016/s1097-2765(03)00058-3. [DOI] [PubMed] [Google Scholar]
- 17.Lee SE, Pellicioli A, Malkova A, Foiani M, Haber JE. The Saccharomyces recombination protein Tid1p is required for adaptation from G2/M arrest induced by a double-strand break. Curr Biol. 2001;11:1053–1057. doi: 10.1016/s0960-9822(01)00296-2. [DOI] [PubMed] [Google Scholar]
- 18.Toczyski DP, Galgoczy DJ, Hartwell LH. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell. 1997;90:1097–1106. doi: 10.1016/s0092-8674(00)80375-x. [DOI] [PubMed] [Google Scholar]
- 19.Tyler JK, et al. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 20.Tyler JK, et al. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol Cell Biol. 2001;21:6574–6584. doi: 10.1128/MCB.21.19.6574-6584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 22.Gaillard PH, et al. Chromatin assembly coupled to DNA repair: A new role for chromatin assembly factor I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 23.Green CM, Almouzni G. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J. 2003;22:5163–5174. doi: 10.1093/emboj/cdg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mello JA, et al. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 2002;3:329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moggs JG, et al. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol Cell Biol. 2000;20:1206–1218. doi: 10.1128/mcb.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–493. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 27.Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 28.Le S, Davis C, Konopka JB, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 29.Franco AA, Lam WM, Burgers PM, Kaufman PD. Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev. 2005;19:1365–1375. doi: 10.1101/gad.1305005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharp JA, Rizki G, Kaufman PD. Regulation of histone deposition proteins Asf1/Hir1 by multiple DNA damage checkpoint kinases in Saccharomyces cerevisiae. Genetics. 2005;171:885–899. doi: 10.1534/genetics.105.044719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adkins MW, Carson JJ, English CM, Ramey CJ, Tyler JK. The histone chaperone anti-silencing function 1 stimulates the acetylation of newly synthesized histone H3 in S-phase. J Biol Chem. 2007;282:1334–1340. doi: 10.1074/jbc.M608025200. [DOI] [PubMed] [Google Scholar]
- 32.Schneider J, Bajwa P, Johnson FC, Bhaumik SR, Shilatifard A. Rtt109 is required for proper H3K56 acetylation: A chromatin mark associated with the elongating RNA polymerase II. J Biol Chem. 2006;281:37270–37274. doi: 10.1074/jbc.C600265200. [DOI] [PubMed] [Google Scholar]
- 33.Lewis LK, Karthikeyan G, Cassiano J, Resnick MA. Reduction of nucleosome assembly during new DNA synthesis impairs both major pathways of double-strand break repair. Nucleic Acids Res. 2005;33:4928–4939. doi: 10.1093/nar/gki806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linger J, Tyler JK. The yeast histone chaperone chromatin assembly factor 1 protects against double-strand DNA-damaging agents. Genetics. 2005;171:1513–1522. doi: 10.1534/genetics.105.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramey CJ, et al. Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function 1. Mol Cell Biol. 2004;24:10313–10327. doi: 10.1128/MCB.24.23.10313-10327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kats ES, Albuquerque CP, Zhou H, Kolodner RD. Checkpoint functions are required for normal S-phase progression in Saccharomyces cerevisiae RCAF- and CAF-I-defective mutants. Proc Natl Acad Sci USA. 2006;103:3710–3715. doi: 10.1073/pnas.0511102103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keogh MC, et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 38.Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 39.Weiffenbach B, et al. Deletions and single base pair changes in the yeast mating type locus that prevent homothallic mating type conversions. Proc Natl Acad Sci USA. 1983;80:3401–3405. doi: 10.1073/pnas.80.11.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dotiwala F, Haase J, Arbel-Eden A, Bloom K, Haber JE. The yeast DNA damage checkpoint proteins control a cytoplasmic response to DNA damage. Proc Natl Acad Sci USA. 2007;104:11358–11363. doi: 10.1073/pnas.0609636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guillemain G, et al. Mechanisms of checkpoint kinase Rad53 inactivation after a double-strand break in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:3378–3389. doi: 10.1128/MCB.00863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Recht J, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci USA. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee SE, et al. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 45.Marsolier MC, Roussel P, Leroy C, Mann C. Involvement of the PP2C-like phosphatase Ptc2p in the DNA checkpoint pathways of Saccharomyces cerevisiae. Genetics. 2000;154:1523–1532. doi: 10.1093/genetics/154.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schermer UJ, Korber P, Horz W. Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol Cell. 2005;19:279–285. doi: 10.1016/j.molcel.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 47.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 48.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 49.Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Trends Genet. 1992;8:446–452. doi: 10.1016/0168-9525(92)90329-3. [DOI] [PubMed] [Google Scholar]
- 50.Emili A, Schieltz DM, Yates JR, III, Hartwell LH. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol Cell. 2001;7:13–20. doi: 10.1016/s1097-2765(01)00150-2. [DOI] [PubMed] [Google Scholar]
- 51.Mousson F, Ochsenbein F, Mann C. The histone chaperone Asf1 at the crossroads of chromatin and DNA checkpoint pathways. Chromosoma. 2007;116:79–93. doi: 10.1007/s00412-006-0087-z. [DOI] [PubMed] [Google Scholar]
- 52.Chen CC, et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.