Abstract
The molecular basis of bacterial cell morphogenesis remains largely an open question. Here we discover a morphogenic protein, RodZ, which is widely conserved across the bacterial kingdom. In Caulobacter crescentus, RodZ is essential for viability and is involved in all aspects of this organism's complex morphology. Depletion or over-production of RodZ results in grossly misshapen cells with stalk defects. RodZ exhibits a localization pattern during the cell cycle corresponding to sites of active peptidoglycan synthesis. The temporal transition of RodZ between patchy/helical and mid-cell localization mimics and depends on the actin-like MreB cytoskeleton. In Escherichia coli, an organism with a distinct mode of growth and MreB localization dynamics, RodZ follows MreB and retains its crucial role in cell morphogenesis, demonstrating conservation of function. Genomic analysis shows that RodZ represents an ancient function unique to bacteria. Multiple sequence alignment of 143 RodZ sequences from species across bacterial phyla identifies an N-terminal cytoplasmic domain with a helix-turn-helix motif, a transmembrane sequence, and a previously unidentified, conserved periplasmic or extracellular C-terminal domain. Both the N- and C-terminal domains are important for function, with the N-terminal domain containing localization determinants. This study uncovers a key missing player in the cytoskeleton-based growth machinery enabling heritable and defined cellular forms in bacteria.
Keywords: cell shape, MreB, cytoskeleton, helix-turn-helix, cell wall
How cell morphogenesis is achieved at the molecular level remains one of the most elusive fundamental questions of cell biology. In bacteria, some cell shape factors are limited to certain species or subsets, such as teichoic acids in gram-positive bacteria or crescentin in Caulobacter crescentus. However, there is also a core morphogenic apparatus composed of proteins with a broad, more universal function, and among them, the cytoskeleton plays a central role (1–3). This apparatus primarily dictates cell shape by topological control of cell growth. Cocci rely exclusively on the division machinery driven by the tubulin homolog FtsZ to grow as spheres, with each division producing 2 new hemispheres. Rod-shaped bacteria add an elongation phase between each division. The actin homolog MreB typically plays an essential role in this elongation by maintaining a constant cell width. When MreB function is lost, cells become progressively larger as they grow and adopt spheroid morphology over time (4–7). During elongation of Bacillus subtilis and Escherichia coli, MreB (and homologs) form helical structures that are thought to direct helical insertion of new peptidoglycan (PG) cell wall along the cell circumference (4, 8). How this occurs is not fully understood, but other proteins of the core morphogenic apparatus, such as MreC, MreD, and RodA are likely to be involved. While the exact function of these membrane-bound or periplasmic proteins remains somewhat unclear, evidence suggests that they regulate PG growth by linking MreB to cell wall enzymes or by working in concert with MreB to spatially restrict cell wall activities (9–17).
Bacteria can display many elaborations of rod morphology. For example, C. crescentus adds curvature to its rod shape and grows a thin tubular extension of the cell envelope, a stalk, at one pole. These features make C. crescentus an attractive model to study cell morphogenesis (18). In this organism, division produces a motile “swarmer cell” and a sessile “stalked cell.” In the swarmer cell, MreB has a patchy/helical localization pattern (6, 19) and cell elongation proceeds according to this pattern (15, 20). After a period of growth, the swarmer cell becomes a stalked cell by growing a polar stalk in a process that requires MreB and RodA activities (14, 15). In the early stalked cell stage, well before cell constriction is initiated, FtsZ forms a ring near mid-cell, where it recruits MreB (6, 19). From then on, cell elongation proceeds predominantly from the MreB ring location where the PG precursor enzyme, MurG, accumulates (20). The FtsZ-MreB colocalization persists for most of division until the late predivisional cell stage, when MreB readopts a patchy/helical distribution (6, 19). Such cell-cycle dynamics in MreB localization is not unusual, although the timing of ring localization appears to vary considerably among bacteria (21, 22).
No new proteins of the core morphogenic apparatus have been discovered for decades, and any missing players would represent a significant limitation to our understanding of cell morphogenesis. Here, we identify an additional core morphogenic protein that is conserved across bacterial phyla.
Results and Discussion
Identification of a Gene with an Essential Role in Cell Morphogenesis.
To find mutants with any defects in cell morphogenesis (e.g., impairment in rod shape, cell division, cell curvature, or stalk formation), we made and visually screened a transposon library (of both Tn5 and Himar1 to increase the randomness of insertions). One Himar1 mutant was impaired in virtually all aspects of C. crescentus complex morphogenesis. In addition to exhibiting slow growth [supporting information (SI) Fig. S1A], the mutant cells were fatter, straighter, often elongated, and typically stalkless (Fig. 1A and Fig. S1B). These defects were imprinted into the isolated PG sacculi (see Fig. S1B), indicating that the mutation affects PG morphogenesis.
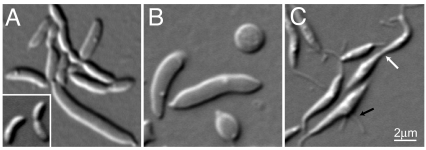
Fig. 1.
Identification of a cell morphogenic gene. (A) DIC micrograph of CB15N rodZ::Himar1 mutant cells. Inset shows wild-type cells for comparison. (B) Depletion of RodZ in CJW2747 cells (CB15N rodZ::Ω xylX::pXrodZ) was initiated by substituting xylose (inducer) in PYE with glucose to halt rodZ expression. The DIC image was taken after 28 h of depletion. (C) Overproduction of RodZ in CJW2158 cells (CB15N/pJS14PxylrodZ) was initiated by addition of xylose. The DIC image was taken after 20 h of over-production. White and black arrows show thin cellular connections between cells and a medial bulge with 2 stalks, respectively.
The transposon was inserted in open reading frame (ORF) cc_0850 (Fig. S1C). Sequence analysis of the presumed gene product revealed no identifiable domains or homologs. We were unable to obtain transcriptional or translation fusions to cc_0850, suggesting that this putative gene was not expressed. Examination of this DNA region revealed an ORF on the strand opposite cc_0850. This previously unidentified ORF, now renamed rodZ, is predicted to encode a 354-residue protein with a transmembrane segment (TM) separating an N-terminal cytoplasmic domain with a helix-turn-helix (HTH) motif of the XRE family (23) from a C-terminal periplasmic domain (see Fig. S1C). The Himar1 insertion resulted in a 152-residue truncation of the RodZ periplasmic domain, leaving the rest of the protein intact (see Fig. S1C). A low-copy plasmid bearing rodZ complemented all of the Himar1 insertion mutant phenotypes (see Fig. S1 A and D).
While rodZ::Himar1 was viable (although severely impaired), chromosomal deletion of rodZ was only possible with rodZ in trans, indicating that rodZ is essential for viability (Table S1). To deplete RodZ (strain CJW2747), rodZ was placed under the control of the xylose promoter (Pxyl) on the chromosome. The cells grew normally with xylose, whereas depletion of RodZ (by substituting xylose with glucose) slowed growth over time (not shown) and caused progressive cell widening along with cell division and stalk formation defects. In late stages of depletion, cells formed large spheroids or gigantic elongated masses (Fig. 1B). There were also many ghost cells and debris in the culture, indicating cell lysis (not shown).
Over-production of RodZ (from Pxyl on a medium-copy plasmid pJS14) resulted in misshapen and straighter cells, typically with a bulge along the long axis (Fig. 1C). These cells often had multiple stalks at the same pole, opposite poles, or along the cell body (see Fig. 1C and Fig. S1E). Over-production of RodZ also impaired division in general, and cell separation in particular, generating long, thin connections of cellular material between daughter cells (see Fig. 1C, white arrow). Thus, RodZ plays a major role in all aspects of the complex morphogenesis of C. crescentus.
Accumulation of RodZ at Sites of PG Growth.
To examine the localization of RodZ, we N-terminally tagged RodZ with CFP or GFP. When chromosomally expressed as the only copy of rodZ under its own promoter (strain CJW2745) or Pxyl (strain CJW2908), CFP-RodZ appeared stable based on Western blotting (Fig. S2A) and fully functional based on growth rate (see Fig. S1A) and cell morphology (Fig. S2B). Microscopy of asynchronous cell populations showed that, in addition to a membrane/patchy distribution (most obvious under native expression), RodZ accumulated at the stalked pole and near the mid-cell region (see Fig. S2B).
Covisualization of RodZ and FtsZ in synchronized cell populations revealed a reproducible cell cycle pattern (Fig. 2A). In swarmer cells, CFP-RodZ accumulated at the old pole, identified by the FtsZ-YFP focus at the opposite pole (24). CFP-RodZ remained at the pole throughout the cell cycle, although it started diminishing in intensity and became more transient when CFP-RodZ formed a medial band (characteristic of a ring) with FtsZ-YFP early in the stalked cell stage. CFP-RodZ remained colocalized with the FtsZ ring during division, but dispersed before cell separation. Thus, RodZ accumulates where PG synthesis is known to occur: at the old pole where a stalk forms and elongates, and at the mid-cell region during preseptal elongation and division (15, 20). In fact, the accumulation of RodZ at the old pole precedes the formation of the stalk, suggesting that RodZ may mark future sites of PG growth.
Fig. 2.
RodZ localizes to areas of peptidoglycan growth and depends on FtsZ for its medial localization. (A) CJW2867 cells (CB15N xylX::pXCFPN-1rodZ vanA::pMT400) producing CFP-RodZ under Pxyl and FtsZ-YFP under Pvan were grown in the presence of vanillic acid and xylose for 2 h before synchronization. Synchronized swarmer cells were resuspended in liquid medium containing vanillate and xylose, and samples were taken at 15-min intervals for microscopy. Under these conditions, the cell cycle takes about 90 min. Time is annotated in cell cycle units; 0 immediately follows synchronization and 1 represents cell separation. Arrows show stalks. (Scale bar, 2 μm.) (B) CJW2907 cells (CB15N rodZ::pHL23Pxylgfp-rodZ ftsZ::pVMCS-6FtsZ5′) grown in vanillate-containing medium were washed and resuspended in vanillate-free medium containing xylose to induce GFP-RodZ synthesis while depleting FtsZ. Cells were imaged 3 h later.
Dependence of RodZ Localization on MreB.
Colocalization between RodZ and the FtsZ ring near mid-cell suggested FtsZ dependence. Consistent with this notion, CFP-RodZ failed to form bands (rings) in FtsZ-depleted cells; instead it displayed a patchy distribution along the cell filaments (Fig. 2B).
Kymographs of time-lapse recordings of GFP-RodZ and FtsZ-mCherry during the normal cell cycle (Fig. S2C) showed that RodZ is recruited to the FtsZ ring shortly after the FtsZ ring forms during cell elongation (note that RodZ polar localization disappears over time because of photobleaching). Before the end of constriction, RodZ leaves the FtsZ ring and redisperses within the late predivisional cell.
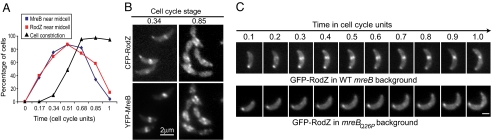
This behavior was reminiscent of that of MreB during the cell cycle (6, 19, 20). This was confirmed by covisualizing CFP-RodZ and YFP-MreB (Fig. S3) and quantifying their localization over the course of the cell cycle (Fig. 3A). The recruitment of CFP-RodZ and YFP-MreB near mid-cell (i.e., FtsZ ring location) coincided in time and space. When the YFP-MreB localization changed back to a patchy (helix-like) pattern at the late predivisional stage, CFP-RodZ did likewise (see Fig. 3A and Fig. S3), and YFP-MreB and CFP-RodZ patchy localization showed a similar pattern (see Fig. 3B and Fig. S3), suggestive of a possible dependence between these proteins.
Fig. 3.
RodZ localization depends on MreB. (A) Quantitative analysis of cells with medial localization of YFP-MreB (blue), with medial localization of CFP-RodZ (red) and with discernible cell constrictions (black) during the course of the cell cycle. Results obtained from 2 separate time-course experiments in which over 100 cells were considered at each time point. Strain CJW2866 (CB15N rodZ::cfp-rodZ xylX::pXYFP-mreB) was grown with xylose for 2 h before synchronization to induce the expression of yfp-mreB. Synchronized swarmer cells were resuspended in PYE with xylose and samples were taken at regular time intervals converted to cell-cycle units to examine the localization of YFP-MreB and CFP-RodZ over the cell cycle. (B) Representative images of YFP-MreB and CFP-RodZ localizations from the time-course experiments described in (A). See Fig. S3 for the whole time-course sequence. (C) Time-lapse microscopy of GFP-RodZ in an mreB wild-type background (CJW2748; CB15N xylX::pXGFPN-2rodZ) or in an mreBQ26P mutant background (CJW2767; CB15N mreBQ26P xylX::pXGFPN-2rodZ) over the cell cycle. GFP-RodZ synthesis was induced with xylose for 2 h before microscopy. The agarose-padded slide contained xylose. (Scale bar, 1 μm.)
Examining localization of RodZ in MreB-depleted cells or vice versa was not particularly informative (not shown) because of the poor health and the severe cell shape defect of these cells, conditions which affect FtsZ ring formation and likely cause other secondary effects. The drug A22 rapidly disrupts MreB localization (7), but the disruption is incomplete (20, 25). Therefore, the maintenance of some GFP-RodZ medial localization in A22-treated cells (not shown) was inconclusive. To obtain conclusive results, we used MreBQ26P, an MreB point mutant unable to consistently form a band at the FtsZ ring location (20). In an mreBQ26P background (CJW2767), medial localization of RodZ was severely disrupted (Fig. 3C). This was particularly striking, as other FtsZ ring-dependent proteins, such as the PG precursor enzyme MurG (20), PBP3, and other cell-wall proteins (unpublished data from our laboratory) retain a medial localization in the mreBQ26P mutant, implying that the localization of these proteins (including MurG) is independent of the localization of RodZ. The severe abrogation of medial RodZ accumulation in cells unable to form an MreB ring strongly supports the notion that MreB affects RodZ localization.
Analysis of RodZ Functional Domains.
Next, we examined whether localization defects accompanied the morphogenetic defects caused by the rodZ::Himar1 mutation and rodZ over-expression (see Fig. 1 A and C). Not surprisingly, over-production of CFP-RodZ resulted in a largely diffuse membrane distribution (Fig. S4).
To examine the behavior of the Himar1-generated RodZ truncation, we fused GFP to the first 201 residues of RodZ, which include the cytoplasmic domain, the TM, and about 52 residues of the periplasmic domain. In a wild-type rodZ background, GFP-RodZ1–201 (produced from Pxyl from the xylX locus) exhibited partial polar and medial localizations, but a fraction of the fluorescence signal appeared diffuse (Fig. 4A). Western blot analysis showed the presence of degradation products and varying the length of the linker between GFP and RodZ1–201 did not improve the stability of the fusion (see Fig. S2A). Thus, RodZ1–201 appears more unstable than the full-length protein. As expected, GFP-RodZ1–201 supported viability in a ΔrodZ background (see Table S1), and recapitulated the cell morphological defects of the rodZ::Himar1 mutant (not shown).
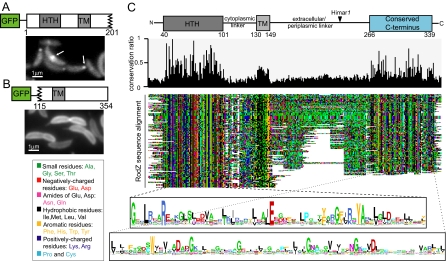
Fig. 4.
Mutagenesis and multiple sequence alignment identify regions and residues of RodZ important for its structure, function, and localization. (A) Fluorescence micrograph of CJW2936 cells (CB15N xylX::pXGFPN-2rodZ1–201) preinduced with xylose for 2 h to synthesize GFP-RodZ1–201 (in a wild-type rodZ background). White arrows indicate polar and medial localizations. (B) Fluorescence micrograph of CJW2869 cells (CB15N xylX::pXGFPN-2rodZ115–354) preinduced with xylose for 2 h to synthesize GFP-RodZ115–354 (in a wild-type rodZ background). (C) Colored dotplot showing alignment of 143 RodZ sequences and identifying conserved regions. (Top) Conservation histogram ranging from 0 (no conservation) to 1 (100% conservation). (Bottom) logos showing consensus sequences for the HTH and C-terminal domains; letter height is proportional to the degree of conservation.
A GFP-RodZ115–354 fusion containing the entire periplasmic domain, the TM, and the last 15 residues of the cytoplasmic domain was stable (see Fig. S2A) but was unable to accumulate at the pole or to form a band near mid-cell in wild-type rodZ background. Instead, it displayed a smooth distribution around the membrane (Fig. 4B). This C-terminal fragment, whether fused to GFP or not, failed to support viability as indicated by our inability to transduce the ΔrodZ::Ω knockout (see Table S1). Collectively, these data indicate that the N-terminal cytoplasmic domain is essential for viability (and thus function) and contains localization determinants, whereas the C-terminal periplasmic domain is dispensable for viability and localization but remains important for overall protein stability and function.
We exploited the widespread distribution of RodZ homologs across bacterial phyla to identify functionally and structurally important regions based on sequence conservation over long evolutionary time. Multiple sequence alignment analysis of 143 RodZ homologs revealed a domain organization consisting of a well-conserved N-terminal HTH domain, a conserved TM (rich in hydrophobic residues), and a conserved C-terminal domain that had not been identified by STRING, Pfam, or SMART, thereby identifying a previously unidentified domain (Fig. 4C). The conserved domains of RodZ are capped by N- and C-terminal extensions and are separated by cytoplasmic and periplasmic/extracellular linkers of variable length (35 ± 7 and 99 ± 40 residues, respectively). The linker separating the conserved C-terminal domain from the TM is enriched in prolines and small residues, such as glycine and alanine (see Fig. 4C). The Himar1 insertion disrupts this linker and effectively deletes the conserved periplasmic domain of C. crescentus RodZ. The conservation of this domain is consistent with an important function. To examine whether this previously unidentified domain is used by proteins besides RodZ, we built a hidden Markov model (HMM) of the domain and scanned all protein sequences in the STRING 7.1 database for its presence. All 121 proteins identified by the HMM contained the N-terminal HTH domain and were RodZ homologs, indicating that this C-terminal domain plays a role unique to RodZ. Interestingly, this C-terminal domain is dispensable in some species, as 16 RodZ sequences consist solely of a membrane-anchored HTH domain with a C terminus of fewer than 10 residues (Fig. S5). These RodZ sequences were typically found in obligate intracellular pathogens with reduced genomes (such as Rickettsia spp., Xanthomonas campestris, and Chlamydophila spp.) and in some round-shaped bacteria, such as Staphylococcus (see Fig. S5).
Conservation of RodZ Function and MreB-Driven Localization in E. coli.
The conservation of the RodZ protein in general, and particularly its cytoplasmic domain, suggests that its function and MreB-dependent localization might also be well conserved. We tested this in E. coli, which differs from C. crescentus with respect to cell morphology and cell-cycle pattern of MreB localization. Notably, E. coli does not form a stalk and it elongates as a straight rod predominantly via helical/patchy insertion of PG along the long cell axis (26). This growth pattern is presumably driven by the helix-like distribution of MreB (16, 27).
Conveniently, a deletion of E. coli rodZ (originally annotated yfgA) was already available from the Keio deletion mutant library (28). Because rodZ is essential for viability in C. crescentus (at least under the conditions tested), we attempted to move, by P1 transduction, the rodZEc deletion (ΔyfgA::Kan) into a clean E. coli MC1000 background. At room temperature, transduction was only successful in the presence of a plasmid-borne rodZEc, suggesting an essential role for rodZEc in sustaining life under these growth conditions. However, at 37 °C, a similar number of transductants were obtained whether a plasmid-encoded rodZEc copy was present or not (Table S2). It is possible that viability is sustained thanks to suppressive mutations, which may arise more easily at faster growth rates. In any case, even at 37 °C, the deletion strain had severe cell-shape defects ranging from very large and grossly misshapen in log-phase cultures to largely spheroid in stationary phase cultures (Fig. 5A).
Fig. 5.
The cell shape-determining function and the MreB-dependent localization of RodZ is conserved in E. coli. (A) DIC images of rodZ knockout cells (CJW2910; MC1000 ΔrodZEc::Kan) from log-phase (Left) and stationary phase (Right) populations. The insets show wild-type E. coli MC1000 cells for comparison. (B) Fluorescent and DIC images of GFP-RodZEc-overproducing cells (CJW2912; MC1000/pBAD18gfprodZEc) after 10 h of growth in arabinose. (C) Fluorescence image of cells producing GFP-RodZEc (CJW2871; MC1000/pBAD33gfprodZEc). After a 10-min arabinose induction, glucose was added for another 20 min before microscopy. (D) CJW2911 cells (MC1000/pBAD33gfprodZEc/pLE7) were treated with isopropyl-beta-D-thiogalactopyranoside (IPTG) for 2 h (to induce YFP-MreB synthesis) and cephalexin for 1.5 h before microscopy. Induction of GFP-RodZEc was achieved as in (C). (E) GFP-RodZEc localization in ΔmreB cells (CJW2913; PB103mreB<>frt/pFB112/pBAD33gfprodZEc). Induction of GFP-RodZEc was achieved as in (C). (F) YFP-MreB localization in a rodZ knockout background (CJW2946; MC1000ΔrodZEc::Kan/pLE7) after 2 h of growth in the presence of IPTG to induce YFP-MreB synthesis.
Over-production of RodZEc or GFP-RodZEc in E. coli also caused severe cell enlargement and shape defects (Fig. 5B). Localization of the over-produced membrane-bound GFP-RodZEc revealed membrane invaginations and vesicles in the misshapen cells, strikingly reminiscent of the intracytoplasmic vesicles and membrane involutions observed in ΔmreB cells (29). Both deletion and over-expression phenotypes show that cell morphogenesis in E. coli, as in C. crescentus, is severely impaired when RodZ function is altered.
A GFP-RodZEc fusion in wild-type cells exhibited a patchy/banded pattern of localization (Fig. 5C). This GFP fusion was functional based on P1 transduction analysis of the rodZ deletion and on the normal cell morphology of the resulting strain (not shown). The banded or helix-like distribution of GFP-RodZEc was best seen in filamentous cells (treated with the cell division inhibitor cephalexin). Covisualization of RodZ and MreB in filamentous cephalexin-treated cells revealed a remarkable colocalization between the two proteins (Fig. 5D), which was also observed in normal-sized cells (not shown). Deletion of MreB [in the presence of FtsZ over-production to support viability (16, 29, 30)] resulted in dispersion of GFP-RodZEc around the membrane (Fig. 5E). We obtained the same localization result in MreB-depleted cells in the absence of FtsZ over-production (not shown). Thus, the difference in RodZ localization pattern between C. crescentus and E. coli is a direct result of its dependence on MreB. Collectively, our data show that RodZ is a key element of the bacterial cell morphogenic machinery, and that its localization is spatially regulated by MreB.
Interestingly, MreB localization appeared less helical and more diffuse in the rodZ deletion strain (Fig. 5F). We could not, however, confidently say whether this defect was directly linked to the absence of RodZ or to the cell shape defects.
Comparative Genomics of RodZ Distribution.
We found that RodZ is absent from eukaryotic and archaeal genomes, but is widely conserved among bacteria (Fig. 6), indicating an ancient function specific to bacteria. To avoid bias because of over-representation of species with multiple sequenced strains, we filtered the 312 bacterial strains of the STRING 7.1 database down to 229 species, keeping only 1 strain from each species. We found 143 RodZ sequences in 141 species, with 2 cyanobacterial genomes having two RodZ copies (see Fig. S5).
Fig. 6.
Phlyogenetic tree of bacterial species showing the presence of RodZ (red), MreB (blue), MreC (cyan), MreD (purple), adjacency between rodZ and gcpE (green), and adjacency between rodZ and pgsA (black). Subtrees indicate phyla, except for the proteobacterial phylum, which is further subdivided by class. See Fig. S5 for a more detailed tree.
We also examined the distribution of other protein sequences, including cell shape-determining MreB, MreC, and MreD (see Fig. 6 and Fig. S5 for species names, cell shape information, and other details). RodA was excluded because its high sequence similarity to the cell division protein FtsW prevented delineation of a set of purely RodA homologs. RodZ sequences were present in almost all β, γ, and δ-proteobacteria but were less represented among α-proteobacteria. The absence of RodZ often correlated with the absence of MreB. Among the exceptions were the ε-proteobacteria, which lack RodZ but contain MreB (see Fig. 6). Interestingly, in the ε-proteobacterium Helicobacter pylori, deletion of MreB does not affect cell width or helical shape, indicating that these bacteria use a different, MreB-independent, mode of growth (B. Waidner and P. Graumann, personal communication), which explains the absence of RodZ. While evidence clearly indicates that RodZ is important for rod shape-determination, its role in cell morphogenesis is more general given its polar localization and essential role in stalk formation in C. crescentus. A larger role in cell morphogenesis is supported by the presence of RodZ sequences in several cocci (such as Streptococcus spp., Staphylococcus spp, Lactococcus lactis, and Enterococcus faecalis) where MreB is absent.
The conservation of the HTH domain among RodZ sequences from distantly related bacteria argues that this domain is important for function. This may suggest DNA-binding activity, although HTH domains can also mediate protein-protein interactions (23). The cytoplasmic domain, which is required for MreB-dependent localization, typically connects via a TM and a flexible linker to a conserved periplasmic (or extracellular) C-terminal domain that may provide a crucial link to the cell wall.
It is also interesting to note that rodZ is adjacent to, or in an operon with, the isoprenoid biosynthesis gene gcpE (ispG) in 88% (58/66) of proteobacterial genomes, whereas it is next to the phospholipid biosynthesis gene pgsA in 76% (25/33) of Gram-positive bacterial genomes that have RodZ (see Fig. 6 and Fig. S5). This striking correlated evolution suggests a functional link. Isoprenoids are ubiquitous compounds involved in tRNA modifications, production of respiratory quinones, and formation of bactoprenols. The latter are required for cell wall synthesis and not surprisingly, disruption of isoprenoid biosynthesis results in severe cell shape defects and ultimately cell death (31, 32). On the other hand, a connection between RodZ and the phospholipid pathway in Firmicutes is attractive, given the restricted localization of phospholipids and phospholipid synthases (such as PgsA) in Bacillus subtilis (33).
Conclusions
Our study identifies a previously unidentified, widely conserved morphogenic protein in bacteria. Its broad conservation across bacterial species indicates an important and ancient function in cell shape determination, which we experimentally demonstrate in species as diverse as C. crescentus, E. coli, and even Bacillus subtilis (see SI Text, Fig. S6). Our studies in C. crescentus and E. coli indicate that RodZ acts with MreB to control topological growth, introducing a new key player in cytoskeleton-based cell morphogenesis.
Materials and Methods
Bacterial Strains, Plasmids, and Growth.
Table S3 contains the strains and plasmids used in this work. Their modes of construction are in the SI Text. E. coli strains were grown in LB or in M9 supplemented with 0.2% glucose, 1-μg/ml thiamine, and 0.05% casamino acids for fluorescence microscopy. C. crescentus cells were grown, synchronized, and genetically manipulated as previously described (34–36). Log-phase cultures were used unless indicated. Induction of gene expression under Pxyl, Pvan, Para, and Plac was achieved with 0.3% xylose, 0.5-mM vanillic acid, 0.2% arabinose, and 20-μm IPTG, respectively. For cell growth measurements, 96-well plates were inoculated with 1:40 of overnight cultures and 660-nm absorbance were measured every 2 min using a microplate reader at 30 °C with shaking (BioTek Instruments).
Microscopy.
For light microscopy, cells were spotted on a 1% agarose-padded slide containing appropriate antibiotics or inducers and were observed using a Nikon E1000 microscope with DIC 100× or phase-contrast objectives and a Hamamatsu Orca-ER LCD camera. Image acquisition and processing were done with Metamorph software. Electron microscopy of cells and sacculi was performed as previously described (20).
Immunoblot Analysis.
Samples were normalized based on optical density measurements, boiled, resolved by 10% SDS/PAGE, and transferred to plyvinylidene fluoride probed with anti-GFP antibodies (1:1000; Clontech).
Bioinformatic Analyses.
Detailed description of our bioinformatic analyses is provided in the SI Text.
Supplementary Material
Acknowledgments.
We thank P. de Boer and H. Niki for communicating their independent discovery of RodZ and for agreeing on common gene and protein names; N. Grindley for helpful suggestions; P. de Boer (Case Western Reserve University, Cleveland, OH), Z. Gitai (Princeton University, Princeton, NJ), L. Shapiro (Stanford University, Palo Alto, CA), L. Rothfield (University of Connecticut, Farmington, CT), and M. Thanbichler (Max Planck Institute, Marburg, Germany) for strains; B. Waidner and P. Graumann for communicating unpublished information; P. Montero-Llopis and M. Cabeen (Yale University) for reagents; M. Cabeen for manuscript editing; A. Jackson for critical reading of the manuscript; M. Jacobs-Wagner for graphic design; and the Jacobs-Wagner laboratory for valuable discussions. This work was funded in part by National Institutes of Health Grants GM065835 and GM076698 (to C.J-W.), the Pew Charitable Trusts (to C.J.-W.), and the “Fundação para a Ciência e Tecnologia” Grant POCTI/BIA-BCM/60855/2004 (to A.O.H.) and SFRH/BPD/26232/2006 (to T.C.). C.J.-W. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810794106/DCSupplemental.
References
- 1.Shih YL, Rothfield L. The bacterial cytoskeleton. Microbiol Mol Biol Rev. 2006;70:729–754. doi: 10.1128/MMBR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pichoff S, Lutkenhaus J. Overview of cell shape: cytoskeletons shape bacterial cells. Curr Opin Microbiol. 2007;10:601–605. doi: 10.1016/j.mib.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osborn MJ, Rothfield L. Cell shape determination in Escherichia coli. Curr Opin Microbiol. 2007;10:606–610. doi: 10.1016/j.mib.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 5.Wachi M, et al. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J Bacteriol. 1987;169:4935–4940. doi: 10.1128/jb.169.11.4935-4940.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figge RM, Divakaruni AV, Gober JW. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- 7.Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Kruse T, Moller-Jensen J, Lobner-Olesen A, Gerdes K. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 2003;22:5283–5292. doi: 10.1093/emboj/cdg504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Divakaruni AV, Loo RR, Xie Y, Loo JA, Gober JW. The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus. Proc Natl Acad Sci USA. 2005;102:18602–18607. doi: 10.1073/pnas.0507937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leaver M, Errington J. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol Microbiol. 2005;57:1196–1209. doi: 10.1111/j.1365-2958.2005.04736.x. [DOI] [PubMed] [Google Scholar]
- 11.Levin PA, Margolis PS, Setlow P, Losick R, Sun D. Identification of Bacillus subtilis genes for septum placement and shape determination. J Bacteriol. 1992;174:6717–6728. doi: 10.1128/jb.174.21.6717-6728.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varley AW, Stewart GC. The divIVB region of the Bacillus subtilis chromosome encodes homologs of Escherichia coli septum placement (minCD) and cell shape (mreBCD) determinants. J Bacteriol. 1992;174:6729–6742. doi: 10.1128/jb.174.21.6729-6742.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JC, Stewart GC. Essential nature of the mreC determinant of Bacillus subtilis. J Bacteriol. 2003;185:4490–4498. doi: 10.1128/JB.185.15.4490-4498.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner JK, Galvani CD, Brun YV. Caulobacter crescentus requires RodA and MreB for stalk synthesis and prevention of ectopic pole formation. J Bacteriol. 2005;187:544–553. doi: 10.1128/JB.187.2.544-553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Divakaruni AV, Baida C, White CL, Gober JW. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol Microbiol. 2007;66:174–188. doi: 10.1111/j.1365-2958.2007.05910.x. [DOI] [PubMed] [Google Scholar]
- 16.Kruse T, Bork-Jensen J, Gerdes K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol. 2005;55:78–89. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- 17.Dye NA, Pincus Z, Theriot JA, Shapiro L, Gitai Z. Two independent spiral structures control cell shape in Caulobacter. Proc Natl Acad Sci USA. 2005;102:18608–18613. doi: 10.1073/pnas.0507708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner JK, Brun YV. Out on a limb: how the Caulobacter stalk can boost the study of bacterial cell shape. Mol Microbiol. 2007;64:28–33. doi: 10.1111/j.1365-2958.2007.05633.x. [DOI] [PubMed] [Google Scholar]
- 19.Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci USA. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aaron M, et al. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- 21.Slovak PM, Wadhams GH, Armitage JP. Localization of MreB in Rhodobacter sphaeroides under conditions causing changes in cell shape and membrane structure. J Bacteriol. 2005;187:54–64. doi: 10.1128/JB.187.1.54-64.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vats P, Rothfield L. Duplication and segregation of the actin (MreB) cytoskeleton during the prokaryotic cell cycle. Proc Natl Acad Sci USA. 2007;104:17795–17800. doi: 10.1073/pnas.0708739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 25.Karczmarek A, et al. DNA and origin region segregation are not affected by the transition from rod to sphere after inhibition of Escherichia coli MreB by A22. Mol Microbiol. 2007;65:51–63. doi: 10.1111/j.1365-2958.2007.05777.x. [DOI] [PubMed] [Google Scholar]
- 26.de Pedro MA, Quintela JC, Holtje JV, Schwarz H. Murein segregation in Escherichia coli. J Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shih YL, Le T, Rothfield L. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci USA. 2003;100:7865–7870. doi: 10.1073/pnas.1232225100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:1–11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bendezu FO, de Boer PA. Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J Bacteriol. 2008;190:1792–1811. doi: 10.1128/JB.01322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shih YL, Kawagishi I, Rothfield L. The MreB and Min cytoskeletal-like systems play independent roles in prokaryotic polar differentiation. Mol Microbiol. 2005;58:917–928. doi: 10.1111/j.1365-2958.2005.04841.x. [DOI] [PubMed] [Google Scholar]
- 31.McAteer S, Coulson A, McLennan N, Masters M. The lytB gene of Escherichia coli is essential and specifies a product needed for isoprenoid biosynthesis. J Bacteriol. 2001;183:7403–7407. doi: 10.1128/JB.183.24.7403-7407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell TL, Brown ED. Characterization of the depletion of 2-C-methyl-D-erythritol-2,4-cyclodiphosphate synthase in Escherichia coli and Bacillus subtilis. J Bacteriol. 2002;184:5609–5618. doi: 10.1128/JB.184.20.5609-5618.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishibori A, Kusaka J, Hara H, Umeda M, Matsumoto K. Phosphatidylethanolamine domains and localization of phospholipid synthases in Bacillus subtilis membranes. J Bacteriol. 2005;187:2163–2174. doi: 10.1128/JB.187.6.2163-2174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 35.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam H, Matroule JY, Jacobs-Wagner C. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev Cell. 2003;5:149–159. doi: 10.1016/s1534-5807(03)00191-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.