Abstract
Through the agr quorum-sensing system, staphylococci secrete unique autoinducing peptides (AIPs) and detect their concentration via the AgrC transmembrane receptor, coordinating local bacterial population density with global changes in gene expression. Unique AIP and AgrC variants exist within and between species, and although autologous interactions lead to agr activation, heterologous interactions usually lead to cross-inhibition, resulting in natural quorum-sensing interference. To gain insight into the mechanisms responsible for these phenomena at the level of the receptor, we used random mutagenesis to isolate variants of Staphylococcus aureus AgrC-I with constitutive activity. Constitutive mutations in the sensor domain of the receptor were localized to the last transmembrane helix, whereas those in the histidine kinase domain were mostly clustered to a region near the phosphorylation site histidine. Analysis of these mutants with a range of noncognate AIPs revealed that inhibition is manifested by inverse agonism in certain heterologous pairings and by neutral antagonism in others. In addition, we isolated and characterized an AgrC sensor domain mutant with dramatically broadened activation specificity and reduced sensitivity to inhibition, identifying a single amino acid as a critical determinant of ligand-mediated inhibition. These results suggest that certain noncognate AIPs stabilize an inhibitory receptor conformation that may be a critical feature of the ligand–receptor interaction not initially appreciated in previous analyses of agr inhibition.
Keywords: constitutive mutant, inverse agonism, staphylococci, quorum sensing
Through the secretion and sensing of a diverse array of low-molecular-weight signaling molecules known as autoinducers, bacteria communicate by using a shared chemical language, enabling local populations to rapidly modify gene expression and engage in group behavior. In Gram-positive bacteria, most autoinducers are oligopeptides, which interact with a polytopic receptor-histidine protein kinase (HPK) at the cell surface (1). At suprathreshold concentrations, the autoinducer peptides activate their cognate HPKs and set in motion a two-component system phosphorelay, leading to modulation of downstream gene expression and induction of a cell density-dependent phenotype.
Staphylococcus aureus is a prototypical facultative pathogen in which autoinducer signaling, or quorum sensing, plays a central biological role. The agr quorum-sensing system allows S. aureus to navigate its host and strategically switch, in response to bacterial density, between attachment and aggressive lifestyles during the course of infection. The agr autoinducing peptide (AIP) is 7–9 residues in length and contains a 5-membered ring in which the C terminus forms a thiolactone bond with a conserved, central cysteine, a structure that is critical for activity (2–4). Binding of the AIP, via a conserved C-terminal hydrophobic patch (5) and additional specific contacts (6, 7), to the hexahelical transmembrane (TM) sensor domain of its cognate receptor, AgrC, results in trans-autophosphorylation of the receptor via its cytoplasmic histidine kinase (HK) domain. This is followed by phosphotransfer to the response regulator, AgrA, which binds and activates the bidirectional agr promoters, P2 and P3. Because the protein units of the agr system are cotranscribed from P2, an autocatalytic circuit is induced, whereas P3 activation unleashes the effector molecule, RNAIII, which mediates the multitude of changes in virulence gene expression defining the agr phenotypic switch.
S. aureus is not alone in its use of agr quorum sensing. Most if not all staphylococci possess an agr locus (8), as do many other bacterial genera (refs. 9 and 10; K. Winzer, personal communication), so that certain bacterial ecosystems may be permeated by a heterogeneous mix of diverse AIPs. agr diversity is based on sequence variation in the AIP, its processor AgrB, and the AgrC receptor, which form a specific functional unit and thus have evolved in concert (11). This variation has resulted in 4 agr specificity groups in S. aureus (3, 12) and 2 or more in each of several other staphylococcal species (8). Generally, only the cognate ligand–receptor interaction causes activation, whereas most heterologous interactions inhibit the receptor, resulting in a form of bacterial interference directly affecting accessory gene expression. Inhibition by heterologous AIPs is reversible (13), is equivalent in strength to activation (2), and is highly tolerant of AIP sequence variation (2, 6, 14). agr cross-inhibition may drive evolutionary diversification among the staphylococci (11) and has important implications for host infections, because treatment with S. aureus AIP-II was shown to prevent the formation of an experimental murine abscess by agr-I cells (2, 15).
In previous studies we and others have identified molecular determinants of ligand-mediated activation in various S. aureus AIPs (2, 4, 6) and in AgrC-I and AgrC-IV, in which critical residues in the second extracellular loop determine group specificity (7). To begin to elucidate the mechanism by which the activation signal is transmitted within the receptor, we used random mutagenesis to isolate AgrC mutants with constitutive activity, as well as those with altered specificity for divergent AIPs. In addition to identifying critical residues involved in activation, this approach led to new insights on the mechanism of inhibition by noncognate peptides. In the interpretation of these results we considered the “two-state” model, in which the receptor exists in an equilibrium between its resting (R) and active (R*) states, the latter of which is favored by an activating ligand.
Results
Library Construction and Mutant Selection.
To isolate constitutively active receptor variants, we generated mutagenized clone libraries representing each functional domain in agrC-I via error-prone PCR. The first library contained PCR products covering the C-terminal half of the sensor domain and the entire HK domain, whereas the second library represented the sensor domain through most of the last TM helix (TM6). The library DNA was introduced into an S. aureus strain containing agrA plus a fusion of the dosage-dependent resistance gene tetK (16) to the agrP3 promoter, but lacking the genes responsible for AIP synthesis. Mutants were plated on agar containing a selective concentration of tetracycline. The agrC-expressing plasmids in the tetracycline-resistant clones were outcrossed to confirm linkage and constitutivity, and the entire agrC gene was sequenced. The outcross recipient contained an agrP3-blaZ fusion (7), whose activity depends on AgrA activation by AgrC. This strain was then used to measure the activity of the mutants in the presence and absence of various AIPs.
Constitutive Mutations in the HK and Sensor Domains.
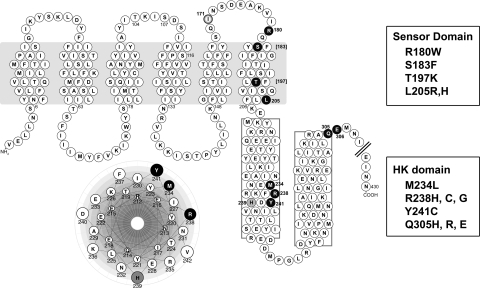
Among the constitutive mutants obtained from the first library, 4 sites within the cytoplasmic domain were hit repeatedly: M234, R238, Y241, and Q305. The first 3 of these residues are clustered within the dimerization histidine phosphotransfer (DHp) subdomain (Fig. 1) which, based on sequence comparison and secondary structure prediction, is defined by a helix-turn-helix motif forming a 4-helix bundle in the dimeric receptor. Notably, helical wheel prediction places these residues on the same helical face, on the opposite side of that containing the phosphorylation site histidine (Fig. 1 Inset). The fourth residue is located at the predicted junction between the DHp subdomain and the catalytic ATP-binding (CA) subdomain, which provides the autokinase function. In addition to these cytoplasmic domain sites, 1 mutation site, L205, is located in TM6 at the predicted interface between the sensor and HK domains. With the second library, there were 2 unique mutations, S183F and T197K, located near the predicted extracellular and intracellular ends of TM6, respectively (Fig. 1). The residue changes associated with these mutations are listed in Fig. 1.
Fig. 1.
AgrC-I topology, domain prediction, and mutagenized residues. AgrC-I is a 430-aa protein with a polytopic TM sensor domain (7, 28) and a cytoplasmic HK domain, which is further divided into the DHp and CA subdomains. Two cytoplasmic regions with high α-helical propensity predicted to form the DHp domain are boxed. The active site histidine (H239) is shown in gray. Residues analyzed in this study in which mutation (listed at right) resulted in constitutive activity are shown in black. Residue I171, in which mutation resulted in broadened specificity, is shown in spherical relief. (Inset) Helical wheel analysis of a region within the first predicted DHp α-helix.
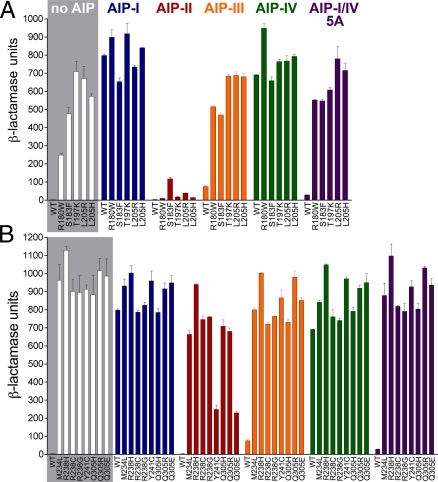
The activity of each mutant receptor was assayed with the β-lactamase reporter strain. In cells expressing the sensor domain mutants, the ligand-independent activities were lower than that of the induced wild type (WT) and were increased by the agonist ligand, AIP-I (Fig. 2A). In contrast, the HK domain mutants were fully constitutive—that is, had levels of ligand-independent activation roughly equivalent to that seen with the AIP-I-induced WT AgrC-I (about 200-fold induction; Fig. 2B); in addition, they were unaffected by treatment with AIP-I.
Fig. 2.
AgrC-I constitutively active mutants. β-Lactamase reporter cells expressing WT AgrC-I, constitutive AgrC-I sensor domain mutants (A), or constitutive AgrC-I HK domain mutants (B) were incubated without or with the indicated AIP at 1 μM. Data are presented as BLU ± SEM.
Divergent AIPs Demonstrate Inverse Agonism.
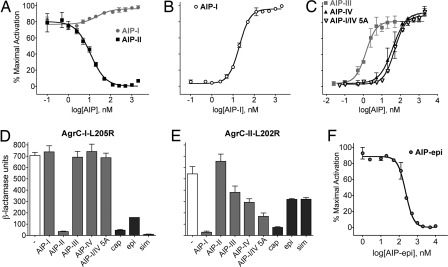
We next examined the effects of noncognate AIPs on the activities of the constitutive AgrC-I mutants. Remarkably, treatment with AIP-II, an inhibitor of WT AgrC-I, resulted in a dramatic decrease in the activity of several, but not all, of the constitutive receptors (Fig. 2). All of the sensor domain mutants were affected, whereas most of the HK domain mutants were resistant to this effect. Incubation with the heterologous peptides AIP-III and AIP-I/IV 5A, the latter of which is a synthetic variant that inhibits the AgrCs of all 4 S. aureus specificity groups (6), did not result in inhibition of constitutive activity in any mutants tested, nor did addition of the weak agonist AIP-IV (Fig. 2). In dose–response tests with AgrC-I-L205R, increasing concentrations of AIP-II progressively reduced its constitutive activity (EC50 = 11 ± 3 nM) (all EC50 and IC50 values are presented with 95% confidence interval), to a baseline level similar to that observed with the resting WT receptor (Fig. 3A, black curve). The reduction in activity was reversed by AIP-I (IC50 = 20 ± 2 nM; Fig. 3B) up to the maximal receptor activation level of this agonist peptide (Fig. 3A, gray curve). By definition, an inhibitor of a receptor that also inhibits a constitutive mutant of that receptor is an inverse agonist. AIP-II is thus an inverse agonist for AgrC-I, stabilizing an AgrC-I signaling state associated with reversal of constitutive activity. This state would correspond functionally to the resting state of the receptor.
Fig. 3.
Inverse agonism by staphylococcal AIPs. (A–C and F) β-Lactamase dose–response assays of AgrC-I-L205R. Reporter cells expressing AgrC-I-L205R were incubated with increasing concentrations of the indicated AIP alone (A and F) or in the presence of a constant dose (100 nM) of AIP-II (B and C). Data are presented as percent maximal activation ± SEM. (D and E) Assay of AgrC-I-L205R (D) or AgrC-II-L202R (E) in the absence or presence of the indicated AIP (at 1 μM) or the indicated non-aureus culture supernatant. cap, S. caprae; epi, S. epidermidis; sim, S. simulans. Data are presented as BLU ± SEM.
The effects of other peptides on the AIP-II-mediated inverse agonism of AgrC-I-L205R were next tested. AIP-III, AIP-IV, and AIP-I/IV 5A each reversed the inhibition by AIP-II in a dose-dependent manner (IC50 = 1.4 ± 0.5 nM, 39 ± 18 nM, and 49 ± 8 nM, respectively; Fig. 3C), with AIP-III causing a particularly strong degree of antagonism. Thus, although these AIPs do not affect constitutive activation, they must clearly bind to the constitutively active form of AgrC-I in addition to binding the native receptor. An inhibitor that binds to both the native and constitutively active forms of a given receptor but has no intrinsic inverse agonist activity is defined as a neutral antagonist. AIP-I/IV 5A and AIP-III therefore behave as neutral antagonists for AgrC-I, manifesting their inhibition of the native receptor only by competition with an activating ligand.
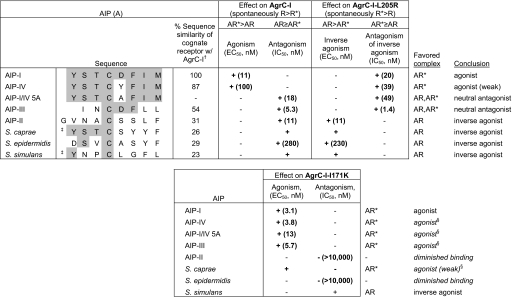
agr-II and agr-I are the two most distantly related S. aureus agr specificity groups, and their respective AIPs share only the central cysteine residue. We thus hypothesized that the behavior of an AIP ligand toward a given AgrC depends on the relatedness between the associated AIPs—the more distant the relation, the more likely the outcome will be inverse agonism as opposed to neutral antagonism. To address this and to determine whether other native peptides could act as inverse agonists of AgrC-I, we tested AIPs of other staphylococcal species, including Staphylococcus caprae, Staphylococcus epidermidis, and Staphylococcus simulans (see Table 1 for sequences) for inverse agonism with AgrC-I-L205R. Most of these had to be tested as culture supernatants because their N termini have not been identified. In one case, that of an S. epidermidis agr-I strain (17), we were able to compare the synthetic AIP with the culture supernatant. This synthetic AIP or postexponential-phase AIP-containing culture supernatants were added to reporter cells expressing AgrC-I WT or L205R. Each of these inhibited AgrC-I activation by AIP-I [supporting information (SI) Fig. S1A] and acted as inverse agonists for AgrC-I-L205R (Fig. 3 D and F). The reduction in constitutive activity was commensurate, in each case, with the degree of inhibition of AIP-I-mediated activation of WT. With the synthetic S. epidermidis AIP, increasing concentrations had a dose-dependent inverse agonist effect on AgrC-I-L205R (EC50 = 230 ± 30 nM; Fig. 3F), consistent with the result observed by using the corresponding culture supernatant. Of note, the supernatants had no effect on AgrC-I-R238H (Fig. S2), a mutant that was also resistant to inverse agonism by AIP-II (Fig. 2), arguing against nonspecific inhibition by these supernatants. These interspecies experiments thus indicate that 3 distantly related staphylococcal AIPs are inverse agonists for AgrC-I. The above hypothesis, equating evolutionary distance with inverse agonism was not, however, supported by these results, because S. caprae AIP shares up to 4 residues with AIP-I (the neutral antagonist AIP-III shares 3). This suggests that other features of AIP structure are responsible for inverse agonism.
Table 1.
Summary of effects of various AIPs on wild-type and mutant AgrC-I
†Sensor domains compared.
‡Predicted N terminus.
§Broadened specificity.
We next asked whether peptides that behaved as agonists or neutral antagonists for AgrC-I would act as inverse agonists in other contexts, such as with AgrC-II, whose sensor domain shares only 31% sequence identity with that of AgrC-I. Accordingly, we mutated AgrC-II residue L202, which corresponds to AgrC-I L205, to arginine, and we observed partial constitutivity and a response to AIP-II similar to the response of AgrC-I-L205R to AIP-I (Fig. 3E). This implies that although AgrC-I and AgrC-II possess divergent sensor domains, they likely share a similar mechanism of activation. With AgrC-II-L202R, however, the various noncognate AIPs all inhibited constitutive receptor activity, but to differing degrees. AIP-I caused a full reduction, demonstrating a reciprocity between agr groups I and II in heterologous ligand-induced inverse agonism. S. caprae supernatant also caused a nearly full reduction, whereas AIP-III, AIP-IV, AIP-I/IV 5A, and the S. epidermidis and S. simulans supernatants all had intermediate inhibitory effects (Fig. 3E). As with AgrC-I, the non-aureus supernatants inhibited WT AgrC-II, but the degree of inhibition was not entirely commensurate with their inverse agonist effect on AgrC-II-L202R (Fig. S1B). No correlation between AIP sequence divergence and strength of inverse agonism was observed. These data in total reveal that variations in autologous and heterologous AIP–AgrC interactions result in a spectrum of possible outcomes reflecting positive, negative, and neutral ligand functionalities.
Role of Charged Residues in AgrC Activation.
Of all of the AgrC-I mutants isolated from our clone library, the most frequently represented were those involving residue R238, which directly precedes the phosphorylation site H239. Mutation of R238 to H, as well as to the unrelated C or G, led to constitutivity (Fig. 2B), suggesting that loss of the arginine side chain, instead of the gain of a new side chain characteristic, was responsible for receptor activation. Notably, all members of the HPK10 family of peptide-sensing polytopic receptors (18), to which AgrC belongs, contain an R or a K at the position directly preceding the phosphorylation site histidine, as does the Bacillus subtilis phosphotransferase Spo0B, in which the crystal structure shows the arginine to be part of a salt bridge with a distal glutamate (19). These observations highlight the importance of a positively charged side chain at this position.
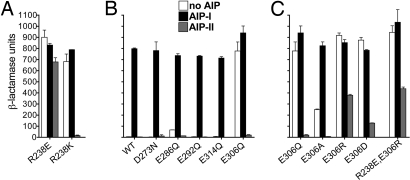
To probe the role of AgrC residue R238 further, we constructed additional mutations at this site, substituting for arginine the negatively charged glutamic acid and the positively charged lysine. The R238E mutation caused constitutive activation that was relatively unaffected by AIP addition (Fig. 4A), similar to that seen with previous mutations at that site (Fig. 2B) and therefore independent of a negative charge at that position. The R238K mutation also resulted in constitutive activity, but the activity was lower than that seen with other mutations at this site and was both increased by AIP-I and greatly decreased by AIP-II (Fig. 4A). Thus, unlike the other mutations at this position, which do not show inverse agonism, the positively charged lysine, conferring partial constitutivity, does so. The fully regulated WT receptor, however, appears to require the arginine guanidinium group at this site, with its characteristic delocalized positive charge and hydrogen-bonding features.
Fig. 4.
Probing charged residues in the DHp domain. Reporter cells expressing AgrC-I with mutation to R238 (A) or a DHp residue (B and C) were incubated without or with the indicated AIP at 1 μM. Data are presented as BLU ± SEM.
In an attempt to identify the putative negatively charged salt bridge partner for R238, we mutated the 5 most likely candidates located in or near the predicted second α-helix of the DHp domain (D273, E286, E292, E306, and E314) to the corresponding amide. One mutation (E306Q) resulted in constitutive activity, whereas the others did not and generally preserved normal receptor function (Fig. 4B). Additional mutation of E306 to R, A, and D resulted similarly in constitutive activation (Fig. 4C). Unlike the mutations affecting R238, those affecting E306 were all associated with inverse agonism by AIP-II, and the conservative replacement E306D was no less active than the other variants. Finally, an R238E, E306R double mutant was fully constitutive (Fig. 4C); a return to normal receptor function was expected if the exchange of charge occurred between true salt bridge partners. Therefore, although E306 is important for AgrC activity, it is unlikely to be the putative salt bridge partner for R238. Assuming, on the basis of the conserved positive charge at this site and on the Spo0B structure, that the R238 salt bridge is a general feature of the class 10 HPKs, it is highly likely that a different negatively charged residue, elsewhere in the receptor, interacts with R238.
Isolation and Characterization of an AgrC-I Mutant with Altered Specificity.
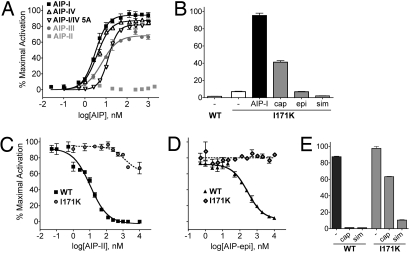
As an additional application of the mutagenized AgrC clone library, we sought to identify mutations that would illuminate the sensor domain determinants of the inhibitory effect of a given noncognate peptide. To this end, we selected for variants of AgrC-I that could be activated by the antagonist AIP-III, by growing the S. aureus sensor domain mutant library on tetracycline plates containing this peptide. Using this method we isolated one positive, AIP-III-dependent clone, which contained 2 replacements, L62I and I171K; the latter of these (located in the predicted third extracellular loop; Fig. 1) proved to be responsible for the response to AIP-III. In the absence of any AIPs, the mutant receptor AgrC-I-I171K had a basal activity level slightly higher than that seen with WT (Fig. 5B, white bars). Interestingly, this mutant was not only activated by AIP-III (EC50 = 5.7 ± 1.5 nM; Fig. 5A), but it was also activated strongly by the original cognate peptide AIP-I (EC50 = 3.1 ± 0.5 nM), the weak agonist AIP-IV (EC50 = 3.8 ± 0.6 nM), and the antagonist AIP-I/IV 5A (EC50 = 13 ± 1 nM). A partial but reproducible response was observed with the S. caprae supernatant (Fig. 5B) in which, as noted earlier, the AIP tail sequence closely resembles that of AIP-I. In total, these results demonstrate that a single amino acid change in AgrC-I broadens ligand specificity to include some, but not all, heterologous peptides.
Fig. 5.
Tests of the altered specificity variant AgrC-I-I171K with various AIPs. (A) β-Lactamase reporter cells expressing AgrC-I-I171K were incubated with increasing doses of the indicated AIP. (B) Cells expressing AgrC-I WT or I171K were assayed without or with AIP-I (1 μM) or the indicated supernatant. (C–E) Inhibition assays. AgrC-I WT or I171K was assayed with increasing concentrations of AIP-II (C) or S. epidermidis AIP (D) in the presence of a constant dose (50 nM) of AIP-I. (E) The indicated supernatant was added in the presence of AIP-I at 25 nM. Data are presented as percent maximal activation ± SEM.
AgrC-I-I171K was not activated by AIP-II or AIPs from S. epidermidis or S. simulans. To determine whether AgrC-I-I171K binds these peptides, we evaluated their ability to inhibit activation of AgrC-I-I171K. Starting with AIP-II, at concentrations sufficient to inhibit activation of WT AgrC-I by AIP-I, we observed little if any significant inhibition of AgrC-I-I171K (Fig. 5C); this was true regardless of the AIP used for activation. An analogous result was observed with S. epidermidis AIP (Fig. 5D). Inhibition of AIP-I signaling by S. caprae supernatant was also much weaker with AgrC-I-I171K than with the WT, whereas inhibition by S. simulans supernatant was only slightly reduced (Fig. 5E). These results suggest that the broadened activation specificity characterizing this receptor variant is also associated with a decreased capacity to bind certain divergent, inverse agonist peptides, or that binding per se is not sufficient for inhibition. In either case, I171 has a critical role in the inhibition of AgrC-I by these noncognate AIPs.
Discussion
In previous AIP structure-function analyses, agr cross-inhibition was determined to require the AIP ring structure, but it was relatively tolerant of amino acid replacements (6, 14), as well as replacement of the thiolactone bond with a lactam (4, 13, 17). These results suggested that autologous and heterologous AIPs interact with AgrC via unique yet overlapping binding sites, consistent with a process of competitive inhibition. In this work constitutively active AgrC variants were used as substrates to illuminate additional aspects of staphylococcal cross-inhibition, revealing that many heterologous interactions are defined by a mechanism more complex than simple competitive antagonism. We demonstrate that certain noncognate, inhibitory AIPs reverse constitutive activity, affecting receptor function in the absence of any activating ligand. In other words, these act as inverse agonists. Other inhibitory AIPs have no effect on constitutive receptor mutants and act on the WT receptor only by competitively inhibiting an activating ligand. In other words, these act as neutral antagonists. In addition, we localized a molecular determinant of inhibition in AgrC, residue I171, mutation of which almost completely abolished the sensitivity of the receptor to inhibition by noncognate ligands. The results presented here highlight both the malleability of AgrC and the remarkably ambidextrous nature of native AIPs, which can function actively as either agonists or inverse agonists depending on the specific AgrC with which they interact. This is a demonstration of natural bacterial inverse agonists, and it is an intriguing possibility that the different outcomes for heterologous AIP–AgrC interactions (inverse agonism, neutral antagonism, and partial agonism) within and between species could have been evolutionarily selected. The mutants isolated in this work also have interesting clinical implications, which are discussed separately in the SI Text.
This study was initiated with a search for constitutively active AgrC variants through random mutagenesis. Most of the residues targeted in the HK domain (M234, R238, Y241) are near the active site histidine on the opposite helical face, and thus may be involved in intersubunit contacts, because H239 must be solvent exposed. Such contacts could involve another DHp helix or the CA domain, possibly sequestering its kinase function from H239 in the resting state. Consistent with this idea, the crystal structure of the cytoplasmic domain of a Thermotoga maritima HPK revealed hydrophobic interactions between the DHp and CA subdomains that are critical for regulation of autokinase activity (20). By analogy with Spo0B (19), we have assumed that R238 is involved in a regulatory salt bridge; however, we have been unable to identify genetically the putative negatively charged partner residue. A possible explanation is that the putative partner may participate in an essential secondary function not tolerant of mutation (for example, a CA subdomain residue necessary for subsequent kinase activity). Single mutations in the sensor domain resulting in constitutivity all clustered to TM6, even though the entire domain was mutagenized and clones were selected to saturation, underlining the importance of this helix in signaling. Interestingly, the replacements on the external pole of TM6 (R180W and S183F) were polar-to-hydrophobic mutations, whereas the converse changes occurred on the cytoplasmic pole (T197K and L205R/H). A possible interpretation of these results is that ligand-mediated activation involves an inward displacement or rearrangement of this helix relative to the membrane, as has been shown for the aspartate chemoreceptor (21). Because the sensor domain mutants were uniformly sensitive to inverse agonism, this sort of conformational change is presumably reversed by inverse agonist binding.
The constitutive receptor mutants can be placed into 2 classes based on their sensitivity to inverse agonism. Variants involving certain mutations in the cytoplasmic domain were resistant to inverse agonism (irreversible constitutive); these residue replacements have presumably engendered a “locked” activated state, indifferent to input from the sensor domain. This is exemplified by the isolated R238 mutants, in which constitutive activation may be due to the loss of a critical regulatory contact. In contrast, other mutants, including all those involving the sensor domain, were sensitive to inverse agonism (reversible constitutive); that is, their resting configuration could be restored by inverse agonist binding. This differentiation provided insight into the role of R238, in which mutation to lysine was the only replacement at that position associated with inverse agonism.
A spectrum of activities, including weak to full agonism, neutral antagonism, and weak to full inverse agonism, was observed in the various interactions analyzed in this work. We initially hypothesized that where a peptide's effect with a given receptor fell within this range depended on the evolutionary distance between the cognate and noncognate AIPs; however, this proved not to be strictly the case, because S. caprae AIP, although relatively similar in sequence to AIP-I, is an inverse agonist of AgrC-I. Other aspects of AIP structure must determine its behavior, and clues may be found in the sequences of the cognate sensor domains; although the AIPs are related in sequence, the S. caprae AgrC sensor domain shares only 26% identity with AgrC-I, as opposed to that of S. aureus AgrC-III, which shares 54% identity. Indeed, sensor domain divergence from AgrC-I broadly correlates with the behavior of the corresponding cognate AIP with AgrC-I (Table 1). Consistent with this trend, the cognate receptors of the AIPs acting as inverse agonists with AgrC-II-L202R were each less than 40% similar to the AgrC-II sensor domain. Structure–function analyses using additional natural and variant AIPs will likely elucidate the precise AIP determinants of ligand behavior in each case.
The varied effects observed in this work have prompted a consideration of AgrC in the context of the 2-state receptor model, which has been widely used to describe the influence of agonists, inverse agonists, and neutral antagonists on GPCR signaling in eukaryotes (22). This model has also been invoked to describe the function of chemoreceptors (23) and the LuxN HPK (24) in Gram-negatives in the presence and absence of specific agonists. According to the 2-state model, receptors exist in an equilibrium between a resting state (R) and an activated state (R*). Agonists and inverse agonists act by stabilizing/enriching R* and R, respectively, shifting the equilibrium in the respective direction; a neutral antagonist, in contrast, has no intrinsic stabilizing activity and binds R and R* with equal affinity, imposing no effect on the preexisting equilibrium (22, 25). The observed AIP–AgrC-I interactions can be evaluated by using the 2-state model, with WT AgrC-I representing an equilibrium spontaneously favoring the R state, and the constitutively active AgrC-I-L205R representing one spontaneously favoring R* (Table 1). For example, the inhibitory effect of AIP-II and the non-aureus AIPs would be to bias the receptor equilibrium toward inactivity through stabilization of the R state. In this model the “irreversible constitutive” mutants may represent extremely biased preexisting equilibria not easily influenced by ligand binding.
In searching for AgrC-I variants that could respond to AIP-III, we isolated a single mutant, I171K, that demonstrated not only nonspecific activation by several noncognate AIPs, but also dramatically reduced inhibition by inverse agonists. Generally, partial agonists and neutral antagonists behaved as full agonists, and inverse agonist efficacy was diminished (Table 1). It is apparent, in keeping with the 2-state model, that the AIP-R complex was strongly disfavored. I171 thus has a critical role in heterologous inhibition, representing a specific determinant of the inhibitory conformational state. I171 could directly interact with peptides, or it could indirectly influence the ligand-binding pocket and its overall sensitivity. This may be related to its interaction with the membrane, because a change to lysine from the highly hydrophobic isoleucine would have the potential to significantly reposition this residue relative to the lipid bilayer. It is noted that position 171 is a constant isoleucine in AgrC-I, III, and IV, whereas in AgrC-II the equivalent residue is an asparagine. It appears that the loss of a putative inhibition-determining contact, as with the I171K mutation, results in a receptor that is primed for activation upon binding one of many possible noncognate peptides; the previously described intergroup sensor domain chimeras involving AgrC-III, which also displayed greatly broadened specificity (5), represent a similar case. Indeed, the observation that a single residue change can lead to constitutive AgrC activation or to broadened specificity is a powerful argument that the resting receptor is sensitively primed for activation, with critical residues representing hair-triggers that hold AgrC in a controlled but highly and specifically activatable state. A speculative biological function for inverse agonist peptides in heterologous staphylococcal interactions would hence be to increase the stability of the resting, inactive AgrC signaling state.
Materials and Methods
The strains and plasmids used in this study are listed in Table S1, and primers are listed in Table S2. Construction of mutant library and plasmids and synthesis of AIPs are described in the SI Text.
For assay of receptor activity, S. aureus reporter cells containing plasmid-borne agrC were grown to log phase in CYGP broth (26) without antibiotics, normalized for cell density, and transferred to microtiter plates. Synthetic peptides at various concentrations or culture supernatants at 1:10 dilution were added to cells, followed by incubation with shaking at 37 °C for 60 min (or 105 min for experiments with constitutive mutants) in a ThermoMax microplate reader (Molecular Devices) with monitoring of cell density at 650 nm. The β-lactamase activity was assayed by the nitrocefin method (27). Data were normalized to β-lactamase units (BLU; defined as Vmax/OD650) or to percent maximal activation and were plotted as initial β-lactamase reaction velocity versus log AIP concentration. Individual dose-response curves were fitted as described in ref. 7.
Supplementary Material
Acknowledgments.
We are grateful to Elizabeth George Cisar for producing synthetic AIPs. This work was supported by National Institutes of Health Grant R01-AI42783 (to R.P.N.) and Medical Scientist Training Program Grant 5T32 GM07308 from the National Institute of General Medical Sciences (to E.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807760106/DCSupplemental.
References
- 1.Lyon GJ, Novick RP. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides. 2004;25:1389–1403. doi: 10.1016/j.peptides.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 2.Mayville P, et al. Structure–activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc Natl Acad Sci USA. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 4.McDowell P, et al. Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus. Mol Microbiol. 2001;41:503–512. doi: 10.1046/j.1365-2958.2001.02539.x. [DOI] [PubMed] [Google Scholar]
- 5.Wright JS, 3rd, Lyon GJ, George EA, Muir TW, Novick RP. Hydrophobic interactions drive ligand-receptor recognition for activation and inhibition of staphylococcal quorum sensing. Proc Natl Acad Sci USA. 2004;101:16168–16173. doi: 10.1073/pnas.0404039101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyon GJ, Wright JS, Muir TW, Novick RP. Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry. 2002;41:10095–11104. doi: 10.1021/bi026049u. [DOI] [PubMed] [Google Scholar]
- 7.Geisinger E, George EA, Muir TW, Novick RP. Identification of ligand specificity determinants in AgrC, the Staphylococcus aureus quorum-sensing receptor. J Biol Chem. 2008;283:8930–8938. doi: 10.1074/jbc.M710227200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dufour P, et al. High genetic variability of the agr locus in Staphylococcus species. J Bacteriol. 2002;184:1180–1186. doi: 10.1128/jb.184.4.1180-1186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Autret N, Raynaud C, Dubail I, Berche P, Charbit A. Identification of the agr locus of Listeria monocytogenes: Role in bacterial virulence. Infect Immun. 2003;71:4463–4471. doi: 10.1128/IAI.71.8.4463-4471.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sturme MH, et al. An agr-like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J Bacteriol. 2005;187:5224–5235. doi: 10.1128/JB.187.15.5224-5235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright JS, III, et al. The agr radiation: An early event in the evolution of staphylococci. J Bacteriol. 2005;187:5585–5594. doi: 10.1128/JB.187.16.5585-5594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarraud S, et al. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J Bacteriol. 2000;182:6517–6522. doi: 10.1128/jb.182.22.6517-6522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyon GJ, Wright JS, Christopoulos A, Novick RP, Muir TW. Reversible and specific extracellular antagonism of receptor-histidine kinase signaling. J Biol Chem. 2002;277:6247–6253. doi: 10.1074/jbc.M109989200. [DOI] [PubMed] [Google Scholar]
- 14.George EA, Novick RP, Muir TW. Cyclic peptide inhibitors of staphylococcal virulence prepared by Fmoc-based thiolactone peptide synthesis. J Am Chem Soc. 2008;130:4914–4924. doi: 10.1021/ja711126e. [DOI] [PubMed] [Google Scholar]
- 15.Wright JS, III, Jin R, Novick RP. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc Natl Acad Sci USA. 2005;102:1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Projan SJ, Novick RP. Reciprocal intrapool variation in plasmid copy numbers: A characteristic of segregational incompatibility. Plasmid. 1984;12:52–60. doi: 10.1016/0147-619x(84)90066-0. [DOI] [PubMed] [Google Scholar]
- 17.Otto M, Sussmuth R, Vuong C, Jung G, Gotz F. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 1999;450:257–262. doi: 10.1016/s0014-5793(99)00514-1. [DOI] [PubMed] [Google Scholar]
- 18.Grebe TW, Stock JB. The histidine protein kinase superfamily. Adv Microb Physiol. 1999;41:139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- 19.Varughese KI, Madhusudan, Zhou XZ, Whiteley JM, Hoch JA. Formation of a novel four-helix bundle and molecular recognition sites by dimerization of a response regulator phosphotransferase. Mol Cell. 1998;2:485–493. doi: 10.1016/s1097-2765(00)80148-3. [DOI] [PubMed] [Google Scholar]
- 20.Marina A, Waldburger CD, Hendrickson WA. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 2005;24:4247–4259. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milligan G. Constitutive activity and inverse agonists of G protein-coupled receptors: A current perspective. Mol Pharmacol. 2003;64:1271–1276. doi: 10.1124/mol.64.6.1271. [DOI] [PubMed] [Google Scholar]
- 23.Keymer JE, Endres RG, Skoge M, Meir Y, Wingreen NS. Chemosensing in Escherichia coli: Two regimes of two-state receptors. Proc Natl Acad Sci USA. 2006;103:1786–1791. doi: 10.1073/pnas.0507438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swem LR, Swem DL, Wingreen NS, Bassler BL. Deducing receptor signaling parameters from in vivo analysis: LuxN/AI-1 quorum sensing in Vibrio harveyi. Cell. 2008;134:461–473. doi: 10.1016/j.cell.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leff P. The two-state model of receptor activation. Trends Pharmacol Sci. 1995;16:89–97. doi: 10.1016/s0165-6147(00)88989-0. [DOI] [PubMed] [Google Scholar]
- 26.Novick RP. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 27.Ji G, Beavis RC, Novick RP. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lina G, et al. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol Microbiol. 1998;28:655–662. doi: 10.1046/j.1365-2958.1998.00830.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.