Abstract
Many bacteria can rapidly traverse surfaces from which they are extracting nutrient for growth. They generate flat, spreading colonies, called swarms because they resemble swarms of insects. We seek to understand how members of any dense swarm spread efficiently while being able to perceive and interfere minimally with the motion of others. To this end, we investigate swarms of the myxobacterium, Myxococcus xanthus. Individual M. xanthus cells are elongated; they always move in the direction of their long axis; and they are in constant motion, repeatedly touching each other. Remarkably, they regularly reverse their gliding directions. We have constructed a detailed cell- and behavior-based computational model of M. xanthus swarming that allows the organization of cells to be computed. By using the model, we are able to show that reversals of gliding direction are essential for swarming and that reversals increase the outflow of cells across the edge of the swarm. Cells at the swarm edge gain maximum exposure to nutrient and oxygen. We also find that the reversal period predicted to maximize the outflow of cells is the same (within the errors of measurement) as the period observed in experiments with normal M. xanthus cells. This coincidence suggests that the circuit regulating reversals evolved to its current sensitivity under selection for growth achieved by swarming. Finally, we observe that, with time, reversals increase the cell alignment, and generate clusters of parallel cells.
Keywords: gliding motility, stochastic model, pattern formation, cell alignment, oscillate
When fish school, birds flock, insects and bacteria swarm, a population of similar individuals cruises along similar, sometimes cyclic, paths. How do the members of a swarm anticipate the movement of others to coordinate movement with them? And how do members, when each is moving rapidly near to others, avoid impeding their neighbor's motion? These general issues, which have yet to be resolved for any schooling, flocking, or swarming organism, can be investigated in swarming bacteria (1). Such investigation might have practical value because swarming enables pathogens like Proteus (2), for example, to invade the urinary tract and to spread along the surface of catheters placed in the urethra (3). The behavior of swarming individuals might also suggest ways to speed high density automobile or pedestrian traffic and ways to avoid traffic jams (4). Bacterial sensory systems and behaviors are more amenable to analysis than those of animals. Because they are relatively small; several thousand bacteria can be followed in a time lapse movie of a swarm. Among bacteria that swarm are the hyperflagellated bacteria (5), all of the Myxobacteria (6), which lack flagella, and the Bacteriodetes that include Cytophaga, Flavobacteria, and Bacteriodes (7). Millions of Myxococcus xanthus cells are found to swarm outward for more than a week at a steady rate of ≈0.1 mm/hr, a rate close to half the speed of individual cell movement (8, 9). Swarming gives them a significant growth advantage: Whereas cells in the swarm center are competing with each other for nutrient and oxygen, cells at the swarm edge have practically unfettered access to both (10). In all of the movies made for Myxobacteria (11, 12, 13) and some of the Bacteriodetes (7), we see the cells reverse their gliding direction, but none of these studies considered why. Individual M. xanthus cells are seen to reverse their gliding direction every few minutes (14, 15). This observation prompts the question: what benefit does M. xanthus gain when it expends an estimated 5% of the cell's ATP production (16, 17) only to move back and forth? We suggest that the oscillatory motion is actually necessary for the net migration of cells from the center of the swarm, where most are born, out to the swarm edge. By escaping from the central region of high cell density to the low density region at the swarm edge, M. xanthus moves to an unoccupied area where nutrient and oxygen are abundant and there is little contest for either (10)—a much greener pasture, to use Purcell and Berg's term (18).
Cell movement in M. xanthus is propelled by polar S engines (pulling type IV pili) at the leading end of the cell, and polar A gliding engines (for secretion of slime) at the trailing end of the cell. Evidence for sites of lateral cohesion of cells to the substrate has been presented in a context of challenging a strictly polar location of the A engines (19). However, we believe that the evidence presented fails to support the injection of a propulsive force at the sites of lateral cohesion as claimed, but instead support the view that lateral cohesions are necessary for propulsion by polar slime secretion. Arguments for these beliefs are summarized in the supporting information (SI) because they have been detailed elsewhere (20). Many of the proteins that constitute the polar A and the polar S engines have been characterized (20), and many single gene motility mutants have been examined (21, 22, 23). Social interactions, such as attachment by the S engines (pili) to fibrils on other cells and the following of slime trails left by the A engines, facilitate swarming (20). As long as fresh surface is available to them, the disk-shaped swarms of M. xanthus expand at a uniform rate, for weeks (8, 9). Uniform expansion implies that the flow of cells from the center of the swarm to the edge is steady. By using A−S− mutants, it has also been shown that 90% of the swarm expansion rate results from cell movement; only 10% of the rate results from cell growth (9). Nevertheless, M. xanthus must be growing to swarm: When growth stops [even though a low level of nutrient may still be available, as in clone-fruiting medium (24)] the swarm stops expanding (10). Although the steady flow of cells to the edge is driven by growth, the spatial distribution of cells at the edge is the consequence of cell movement, which depends on the mechanics of the A and S gliding engines and on their social interactions. Because the cell flow to the swarm edge is constant over long periods of time, relative to the generation time of ≈3.5 h, time-lapse movies of several hours offer large samples of the patterns of cell movement to be found in the steady state.
Here we describe the rich dynamics of cell movement visible at the swarm edge observed in time-lapse movies. To find characteristic movement patterns in the images, we used a cell-based, stochastic computational model that was recently described (25). Because the model incorporates the mechanics of A and S motility as well as their social interactions, it accurately translates the macroscopic and readily measurable swarm expansion rates into microscopic cell fluxes that expose the average behavior of many individual cells.
Results
Dynamic Patterns at the Swarm Edge.
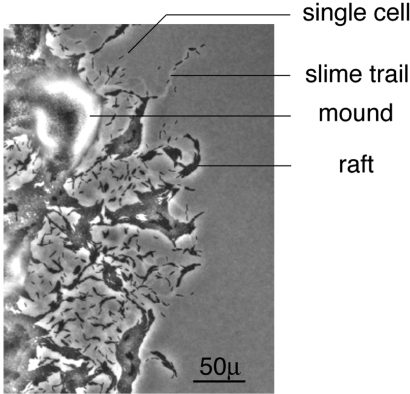
A typical pattern of cells at the swarm edge is shown in Fig. 1. In every frame of the movie (see Movie S1 in the SI), there are many single, roughly 5 μ × 0.5 μ cells. The single cells are always found on slime trails, because they are secreting propulsive slime from their poles (20). Two kinds of multicellular structures are also evident in Fig. 1 (see labels): Elongated rafts and mounds. Elongated rafts resemble two dimensional nematic liquid crystals and are composed of several files of cells, with the long axes of the files adjacent and parallel to each other but the individual cell ends in adjacent files out of register with each other. Lack of registration is most evident at the ends of a raft where cells protrude. Mounds have several layers of cells stacked on top of one another. Each layer of a mound is roughly circular and concentric with the others, contrasting with the rafts that are rectangular and have fewer cells. The layering, evident in all mounds and on some rafts (indicated in Fig. 1) shows that cells readily glide over the surface of other cells. Single cells, small clusters of 2–3 cells, rafts and mounds are evident in every frame of the movie (see Movie S1 in the SI). Both multicellular structures exchange various numbers of cells with neighboring structures by branching or converging flows. In mounds and rafts, cells frequently move between adjacent layers. Because the movie was photographed at 2 frames/min for 3 h, the apparent smoothness of flow indicates that cell birth and death are negligible for 3 h. Finding smooth flow and the constant swarm expansion rate data (9) give firm support to the steady state assumption of the model. Individual cells are seen to glide in the direction of their long axis, consistent with a polar location for both the A and the S engines. Although cells can flex (26) and U-turns are very infrequent (27), cells never seem to move sideways, also consistent with a strictly polar engine location. Cells appear to be oriented in many directions; consequently the swarm disk would be expected to expand in radial directions; this justifies the assumption of a random initial cell orientation around the radius (25).
Fig. 1.
Distribution of cells and multicellular structures at the swarm edge. In addition to some individual cells and slime trails, multicellular rafts and multicellular mounds are labeled. The swarm is expanding in the radial direction, which is to the right in this image. (Scale bar, 50μ.) This is the first frame of a 3 h time lapse movie (see Movie S1 in the SI).
Solitary cells are observed to reverse their gliding direction in the movie with remarkable regularity: they reverse at 8.8 ± 2.1 min intervals (reversal data collection and analysis are in the SI). As noted elsewhere (23), reversing cells simply stop, pause for about a minute, then lead off with their formerly lagging pole. It has been proposed that reversal is effected by an MglA G protein switch (15). The proposal explains in detail the reversal behavior of RomR mutants (20), as described by ref. 28. Because mglA mutants lack the switch, they move abnormally, never reverse, and fail to swarm (23). Moreover, mglA mutant colonies are heaped up and sharp-edged, like colonies of a nonswarming E. coli strain (29). Switching the MglA G protein from its GDP to its GTP-state (30) is proposed to follow a pulse of FrzE∼P from the frizzilator, a two-component system with negative feedback that causes it to oscillate with a definite period (31). As shown in Table 1 and the SI, none of the Frz deletion mutants have a sharp-edged, Mgl-like phenotype; all Frz mutants retain some capacity to reverse and to swarm. The data of Table 1 show that the rate of Frz mutant swarm expansion is correlated with a nonoptimal reversal period. Despite intensive genetic screens, no mutant that is still motile but has lost the ability to reverse, has ever been isolated. But, such a mutant can be simulated by deleting reversals from the computational model, and any value of the reversal period can be prescribed.
Table 1.
Swarm expansion rates of wild-type and frizzy mutants
| Strain | Rate of swarm expansion, mm/hr* | Gene deleted | Published reversal period† |
|---|---|---|---|
| DZ 2 | 0.155, r = 0.99 | None | 7.21 min |
| DZ 4480 | 0.028, r = 0.99 | Δ6–393 Frz CD | 34.1 min |
| DZ 4481 | 0.03, r = 0.95 | Frz E | 34.1 min |
| DZ 4482 | 0.123, r = 0.99 | Frz G | 4.15 min |
| DZ 4483 | 0.033, r = 0.97 | Frz F | 34.1 min |
| DZ 4486 | 0.055, r = 0.98 | Δ6–152 Frz CD | 1.41 min |
Reversals Facilitate Swarming.
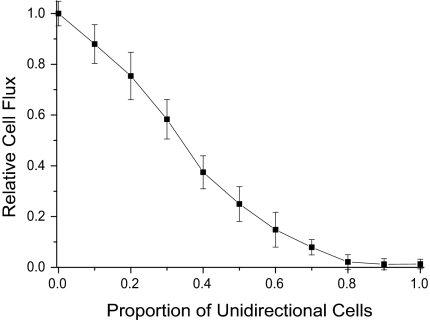
In the steady state, the number of cells that flow across the edge of the swarm per unit time (the cell flux), should be proportional to the rate at which the swarm expands on the macroscopic scale. The swarm expansion rate can be measured readily, and its value is used to set the model parameters. As shown in Fig. 2, the cell flux decreases gradually as the proportion of unidirectional (nonreversing) cells in a mixed population is increased in simulations. Such decreases are expected for wild-type cells (A+S+ in Fig. 2) and for an A+S− mutant (see SI). A−S+ mutant swarms have yet to be modeled, but they are expected to depend on reversals as well because their swarms expand at almost the same rate as the A+S− (9) and do have the same reversal period as A+S+ (23). The net fluxes diminish to zero in Fig. 2 when 100% of cells are unidirectional. Even though cells are moving, the swarms do not expand at all because unidirectional cells are easily jammed.
Fig. 2.
Dependence of the flux on the proportion of unidirectional, wild-type cells. The average flux of 10 independent runs is plotted against the proportion of unidirectional cells in the mixture. Unidirectional cells were randomly distributed within the initial domain of cells, as described in Methods. A reversal period of 8 min was used for the reversing cells.
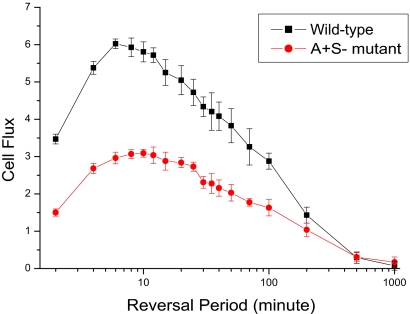
Seeking to understand why moving cells should need to reverse to swarm, we used the model to test how the flux would be expected to change as the length of the reversal period was changed. As shown in Fig. 3, both the A+S+ and the A+S− mutant reach their maximal fluxes at reversal periods between 5 and 12 min. The flux is significantly less on both sides of the 5 to 12 min range, and the wild-type maximum is narrower than that of the mutant. Because the flux of wild-type is twice that of the A+S− mutant in Fig. 3, the two engines are making equal contributions to the flux, just as they make roughly equal contributions to the maximum swarm expansion rates (9). Moreover, the wild-type (A+S+) and the A+S− mutant appear to have the same optimal reversal period within measurement error. For future reference, we note that the optimal value obtained by simulation corresponds to the observed reversal period of strains that have wild-type Frz systems (14, 23, 32).
Fig. 3.
The average cell flux of 10 independent runs is shown with error bars that represent the standard deviation. Flux is plotted against the reversal periods for A+S+ bacteria and for an A+S− mutant. The x-axis is scaled by log10.
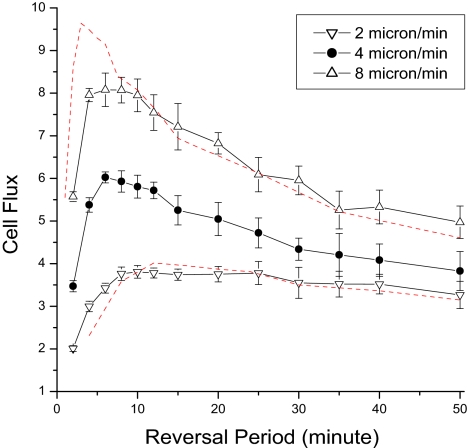
Next, we investigated whether and how the optimum reversal period depended on the average speed at which the cells move. The speed of any cell is observed to vary from moment to moment as well as from cell to cell (9), possibly because cells differ in the number of pilus fibers, in the numbers of polar nozzles that happen to be secreting slime, and in whether they have stalled after colliding with another cell. The optimal reversal period was not found to be affected by noise in the distribution of reversal periods (data in the SI). Despite fluctuations around their mean, the average speed is found to be reproducible from one experiment to another (9, 33, 34). For these reasons and for simplicity, we assumed that all cells moved at the observed average speed of 4 μ/min. To the first approximation, the speed would have been expected to fix the time scale for the simulation. The scaling expectations are shown by the dashed red lines in Fig. 4 for an 8 μm/min speed and a 2 μm/min speed. These speeds were chosen to lie on either side of the experimentally observed average cell speed of 4 μm/min (Fig. 4). Simulations are shown for both A+S+ (Fig. 4) and A+S− (SI). For both strains the 8 μm/min optimum (upright triangles) is shifted to a shorter reversal period, whereas the 2 μm/min optimum (inverted triangles) is shifted toward a longer reversal period. Whether deviations of both sets of points from the red line are significant (in which case they could reflect the model's nonlinear social interactions) or not, they clearly show that the optimal reversal period changes with the cell speed. Consistent with these data, we suggest that Myxococcus evolved an approximately 8 min reversal period because the cells had adopted an average speed of 4 μm/min.
Fig. 4.
Dependence of the optimal reversal period on the cell speed. Simulation results for different speeds are presented for A+S+ cells. The red curves are reconstructed from the data of normal speed (4 μm/min), by using the functions Y = C1 * f(2x) for 8 μm/min and Y = C2 *f(x/2) for 2 μm/min respectively to show the effect of scale change. Here x is the reversal period, f(x) is the fitted function for the original curve of normal speed, and C1 and C2 are constant factors dependent on the average speed.
Reversal Facilitates Alignment of Cells.
Fig. 1 shows the complex pattern of cells, rafts, and mounds found at the swarm edge when the 3 h movie starts. Over the movie's course, the field of the microscope starts with roughly 5,000 cells and expands outward by ≈20%. During expansion, the overall arrangement of cells at the swarm edge remains the same: an open network of rafts and mounds. Moreover, the average surface density of cells over the network remains constant, as the sizes and positions of the rafts, the cellular flows between rafts and mounds, and the number of layers in each mound changes constantly. An organizational constraint appears to be imposed on the steady state flow of cells. To reveal that constraint, we employ the 2-dimensional, discrete form of the orientation correlation function C(r) used by ref. 35 to measure the amount of cell alignment. It is represented as follows:
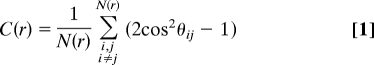 |
Here θij denotes the angle between the orientations of the i-th and j-th cells; r is the distance between a pair of cells and N(r) is the number of cell pairs separated by a distance r. C(r) measures cell alignment as a function of distance, r. Because the cells are rod-like, C(r) is large at short distances where the moving cells are aligning with their neighbors. C(r) then falls rapidly to a constant value, called the background as shown in Fig. 5. When cells do not reverse, C(r) falls rather abruptly to the background value. When cells do reverse, the reversal period shows itself in a more gentle curvature of the bend of the C(r) curve. That curvature can be described by a characteristic correlation distance, l, which is defined as the distance at which C(r) reaches its background level and no longer changes significantly with r. Thus, the correlation distance, l, depends on the reversal period as well as on any social interactions between cells. The way that l can be calculated from the correlation data are described in the SI.
Fig. 5.
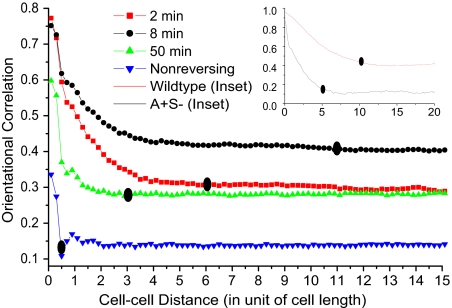
The orientational correlation functions for nonreversing cells and for reversal periods of 2, 8, and 50 min are plotted against the cell-cell distance for an A+S− mutant after 400 min of simulation. The two curves in the inset are the orientation correlation functions calculated for wild-type and A+S− cells outside the initial domain having an 8 min reversal period. The background level for the A+S− (Inset) is smaller than the main figure because cells outside the initial domain have a lower density than cells at the very edge in the steady state. The correlation distances are shown by solid ovals. The cell–cell distance is given in units of cell length, where 1 cell length = 5 μm.
Inasmuch as pilus retraction tends to pull the rod-shaped M. xanthus cells together into flat side-by-side rafts of different compactions, pilus retraction is expected to contribute substantially to C(r). To see the effects of reversals on local alignment clearly, we worked with the A+S− mutant, which lacks S motility and pilus retraction. As shown in Fig. 5, reversing A+S− swarms are indeed locally aligned, whereas a colony (not a swarm) of nonreversing cells has relatively little alignment and small C(r) values. They would be a disorderly heap of cells. A+S− swarms with reversal periods in the optimal range of 5–12 min have higher background levels of C(r) and greater correlation distances than either hypo- (50 min period) or hyper- (2 min period) reversing swarms. Fig. 5 provides values of l for each reversal period under model conditions. For 2 min reversals, l ≈ 6 cell lengths (30 μm); for 8 min reversals, l ≈ 11 cell lengths (55 μm); for 50 min reversals, l ≈ 3 cell lengths (14 μm). For nonreversing cells the correlation distance is l ≈ 0.5 cell length (2.5 μm). Thus, the reversal period with the greatest swarming efficiency (8 min) also presents the greatest correlation distance, l = 11 cell lengths (55 μm). The correlation distance l also defines the clustering diameter of cells at the swarm edge. In simulations, we find this correlation distance to be ≈10 cell lengths (50 μm) for wild-type, and, as shown in Fig. 5 inset, the clustering diameter of A+S− mutant is half that, ≈5 cell lengths (26 μm).
Discussion
There are 3 principal findings. The first is that motile nonreversing cells are unable to swarm. This was discovered when we simulated Myxobacterial swarming by using our cell model. The mglA mutants, which also are completely unable to swarm, are practically nonmotile. No growth defects are apparent (29, 36), and their A engines secrete slime from both ends (23). In contrast to the uni-polar secretion of motile (reversing) strains, bipolar secretion prevented net movement in either direction; it was responsible for their complete lack of ability to swarm. Their colonies are sharp-edged heaps of cells, whose growth is limited by competition among the heaped-up cells for nutrient and oxygen. The failure of nonreversing cells to swarm can be explained by their inability to build the orderly cell arrangements in the central region of the swarm that would allow a steady radial flux of cells for swarm expansion (Fig. 5). The surprise was that nonreversing cells would have zero net flux across what should be the swarm edge. Flux being the microscopic equivalent of the macroscopic rate of swarm expansion, the rate of expansion would be zero.
We also show that the flux depends on the length of the reversal period—the maximum flux is obtained with periods ≈8 min (Fig. 3). Reversal periods of 3 min or of 25 min gave significantly lower cell fluxes than 8 min. It is striking that the reversal period for swarming wild-type cells is the same, within experimental error, as that reported for cells at low density that are not interacting with each other (23). It is the same for developing cells in traveling waves, 8.2 ± 0.6 min (37). These similarities suggest that the reversal period is a robust property of M. xanthus that is independent of growth rate and of cell density, remaining constant for a more than a week. The macroscopic rate of swarm expansion can be measured accurately in culture because of those qualities. The reversal period is increased during the aggregation phase of development to allow streaming (33), by altering feedback in the frizzilator with FruA∼P (20). The expansion rate is proportional, in the model, to the average velocity of cells. This agrees with measurements, although the cell velocity is much less accurately known because it varies from cell to cell and moment to moment (9). The constant of proportionality represents the fraction of the individual cells' velocity that, because of the self-organization of cells, moves cells to the edge of the swarm. According to the data (9), wild-type cells moving at a speed of 4 μm/min have a swarm expansion rate of 1.6 μm/min, and thus a swarming efficiency of 40%. Considering that swarm expansion is unidirectional while cells move periodically in either direction, one might say that 80% of the energy expended for movement has been harnessed for swarming. This high efficiency is one reason for using M. xanthus swarm cell behavior as a model system.
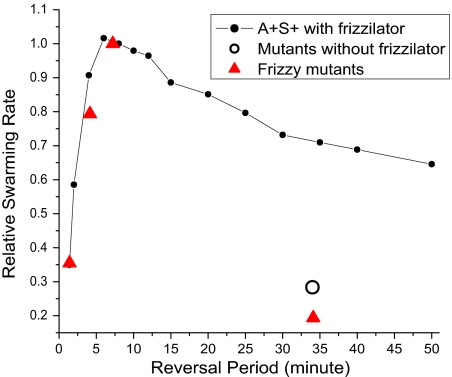
A second noteworthy finding is that the 8.8 ± 2.1 min reversal period of wild-type M. xanthus cells, which has been checked experimentally by clocking cells at the swarm edge, maximizes the cell flux that can be obtained with an average cell speed of 4μ/min. It was pointed out above that mutations in the frizzy genes are observed to change the average reversal period, significantly. Accordingly, the swarming model could be challenged with predicting the quantitative effect of changing the average reversal period on the flux, inasmuch as the reversal period of the frizzilator is an adjustable parameter of the model. For this purpose, we investigated two deletion mutants with short periods, one in Frz G and another in FrzCD, and three with long periods, comparing them all with wild-type cells. The measured changes in the reversal period (32) are presented in Table 1 along with our measurements of the corresponding swarm expansion rates, to which the flux is in direct proportion. As shown in Fig. 6, good quantitative agreement between observed and predicted rates was obtained for the whole-gene deletion mutation in FrzG and the N-terminal deletion of residues 6–152 of FrzCD that shorten the reversal period. However, the 3 whole-gene deletion mutants of FrzCD, of FrzF, and of FrzE, all with 34 min periods, fall below the value predicted by the frizzilator that is shown by the black line in Fig. 6. These three mutants are unique among the 5 tested in greatly and identically lengthening the reversal period to 34.1 min (32), see Table 1. Moreover, any one of those three mutations would have destroyed the capacity of the frizzilator to oscillate or to produce pulses of FrzE∼P. With no pulses of FrzE∼P to drive the MglAB switch, we suggest that MglAB would oscillate spontaneously between the MglA·GDP and MglA·GTP states with a period (observed) of 34 min. That period would arise from the kinetic constants of interacting MglB and MglA proteins. When the simulation is modified to approximate such an MglAB oscillator (that has no frizzilator, as detailed in the SI), a relative swarming rate of 0.28 is predicted (open circle in Fig. 6). That prediction is much closer to the Table 1 values of 0.21 observed for delFrzF, of 0.19 observed for delFrzE, and of 0.18 observed for delFrzCD.
Fig. 6.
Dependence of the swarm expansion rate on the reversal period. Observed expansion rates for WT and 3 deletion frizzy mutants (red triangles) are compared with those computed from the model (all are relative values). Measurement of the swarm expansion rate is described in the SI.
Maximization of the cell flux also suggests that the reversal period evolved under selection for the most rapid swarming. Before 1941, M. xanthus had evolved in the soil of Iowa, whence it was isolated (38). Since 1941, wild-type strains DZ2 and DK1622 have been separately maintained in different laboratories. Yet, these strains exhibit similar swarm morphologies and have the same reversal period within measurement error, as indicated in Table 1. The similarity suggests that laboratory cultivation has not improved the swarm expansion rate beyond Beebe's natural isolate. The following events may have occurred after both engines had achieved their current strength and the current cell speed of ≈4μ/minute. Should the average reversal period have ever been higher or lower than the current 8.8 min optimum, natural selection would have driven the period toward 8.8 min. The curves in Fig. 3 show that any change in the average reversal period (within the 2–50 min range) that brought it closer to an average of 8.8 min would exhibit a larger cell flux. Each such change, however small, would increase the cell flux and the swarm expansion rate, thereby promoting growth by limiting competition between cells for nutrients and would thus have a selective advantage. Targets for mutational refinement of the reversal period could include frz CD, frz E, frz B, frz F, mglB, and mglA—the protein components of the reversal generator (15). Among these targets, members of the protein complex that enables negative feedback are the most promising because their kinetics directly determine the reversal period. Mutations in those components strongly affect the swarm expansion rate (Table 1). The negative feedback helps to explain why a two-component system with adaptation by protein modification (methylation) serves as the reversal clock; such a system can readily be made to oscillate with the most appropriate period for cells that use the A and S engines to glide at 4μ/min. Because all these reversal period genes are conserved in the related myxobacterium, Stigmatella aurantiaca (39), they should have been present in the immediate precursor to M. xanthus and S. aurantiaca: this particular evolutionary scenario is certainly plausible. Traces of movement along this evolutionary path are evident in the fact that yeast or mammalian Ras, whose amino acid sequences differ from MglA protein, provide partial rescue to ΔmglA mutants (40).
Finally, we would like to emphasize that the flexibility and 10:1 aspect ratio of M. xanthus cells enhance their movement and that periodic reversals take advantage of those properties to organize cells. All swarming bacteria, including those that differentiate and move by many rotating flagella (5) and those that, like the Myxobacteria (11, 12, 13), use other engines, have long, thin cells for swarming. M. xanthus cells have a large aspect ratio and are unusually flexible for cells with peptidoglycan walls (25); they resemble Bdellovibrio (41) in that respect. As shown in the SI, the swarm expansion rate should be higher for long, thin cells than for other shapes because collisions, which are inevitable among cells at high density that are moving in many directions, limit the swarming efficiency. Lacking the sophisticated senses that animals use to track their neighbors, swarming bacteria try to avoid collisions by reducing their collisional cross-section. They are also able to resolve the unavoidable collisions. It is evident watching the swarm movies that M. xanthus cells stall when one collides with another. The stall ends when one cell moves away or reverses. In end to side collisions, one cell, being pushed from behind by slime secretion, bends and comes along side the second cell (23). Once side by side, both cells can start to move again (see movie for Fig. 1 in the SI).
Like the type IV pili which pull cells into parallel alignments, reversals tend to orient cells along parallel tracks, thereby decreasing the probability of collision. With a reversal period of 8 min, we find (Fig. 5) that cell orientations are correlated over 10 cell lengths, or ≈50 microns. Without reversal, the order does not extend beyond one cell length. And, when the reversal period is too long, 50 min say, the order extends for only 3 cell lengths. The same reversal period that optimizes the flux of cells across the swarm edge also maximizes the cell order that is created. As shown in Fig. 3, optimal reversal plus the social interactions inherent in both the S engine and the A engines produce multicellular order. That order is reflected in the rafts and mounds that are seen at the swarm edge in Fig. 1. We suggest that the capacity of Myxobacteria to build species-specific fruiting bodies arises from their ability to swarm.
The frz genes are parts of a reversal clock that, according to our proposal, operates the MglAB reversal switch to change the gliding direction. Because the frz chemosensory genes are differentiated orthologs of the chemotaxis genes of E. coli (42) that control the frequency of reversals of the flagellar motor, and because Frz proteins are wired up differently for the frizzilator than are the Che proteins for chemotaxis in E. coli, it appears that swarming and chemotaxis represent 2 different ways for motile bacteria to move to greener pastures (18).
Methods
The model for swarming of M. xanthus (27) has two parts describing the behavior of individual cells and the set-up for swarming (see the SI for more details).
A cell is represented by two segments connected by 3 nodes, having an observed aspect ratio of M. xanthus cells, ≈10:1. The relative positions of the nodes can change according to an energy constraint to keep a stable cell shape. The cell is led forward by the motion of its head node. The direction is determined by local rules used in the model to reflect the properties of both engines and their social interactions (20, 27). Each cell in our model has a reversal clock, whose value increases until reaching T, the reversal period. Then the cell remains motionless for one minute, resets the clock to zero, and reverses.
In the beginning of each simulation, cells are distributed in an initial domain that represents a small section of the edge of a swarm. The initial cell density is set to be 0.25 cell/μm2, so that the cells occupy half of the area of the domain, simulating the dense condition of swarming. The number of cells in this domain is kept constant throughout simulations, because the cell flux is in a steady state. The ratio of the cell fluxes for wild-type and A+S− mutants is set to the ratio of their measured swarm expansion rates (27). The duration of each simulation run is 1,000 time steps, equivalent to 200 min of swarming (see SI).
We found that the optimal range of reversal periods presented in Fig. 3 were stable as varying the key parameters in the model (see SI). The effect of different aspect ratio of cells on the swarming efficiency was also studied (see SI).
Supplementary Material
Acknowledgments.
We thank Zhenyu Shi for the reversal period statistics. We thank Dr. Emilia Mauriello and Dr. David Zusman of the University of California Berkeley for providing DZ2 and frizzy mutant strains. M.S.A. and Y.W. were partially supported by National Science Foundation Grants DMS 0719895 and CCF 0622940. Y.J. was supported by the U.S. Department of Energy at Los Alamos National Laboratory under Contract No. DE-AC52-06NA25396.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811662106/DCSupplemental.
References
- 1.Harshey RM. Bees aren't the only ones: Swarming in Gram-negative bacteria. Mol Microbiol. 1994;13:389–394. doi: 10.1111/j.1365-2958.1994.tb00433.x. [DOI] [PubMed] [Google Scholar]
- 2.Belas R, Schneider R, Melch M. Characterization of Proteus mirabilis precocious swarming mutants: Identification of rsbA, encoding a regulator of swarming behavior. J Bacteriol. 1998;180:6126–6139. doi: 10.1128/jb.180.23.6126-6139.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warren JW. Catheter-associated urinary tract infection. In: Bayless T, Brain M, Cherniak R, editors. Current Therapy in Internal Medicine. Philadelphia: BC Decker Inc.; 1987. pp. 290–292. [Google Scholar]
- 4.Helbing D. Traffic and related self-driven many-particle systems. Rev Mod Phys. 2001;73:1067–1141. [Google Scholar]
- 5.McCarter LL. Dual flagellar systems enable motility under different circumstances. J Mol Microbiol Biotechnol. 2004;7:18–29. doi: 10.1159/000077866. [DOI] [PubMed] [Google Scholar]
- 6.Reichenbach H. Myxobacteria: A most peculiar group of social prokaryotes. In: Rosenberg E, editor. Myxobacteria. New York: Springer; 1984. pp. 1–50. [Google Scholar]
- 7.McBride MJ. Cytophaga-flavobacterium gliding motility. J Mol Microbiol Biotechnol. 2004;7:63–71. doi: 10.1159/000077870. [DOI] [PubMed] [Google Scholar]
- 8.Burchard RP. Studies on gliding motility in Myxococcus xanthus. Arch Microbiol. 1974;99:271–280. doi: 10.1007/BF00696242. [DOI] [PubMed] [Google Scholar]
- 9.Kaiser D, Crosby C. Cell movement and its coordination in swarms of Myxococcus xanthus. Cell Motil. 1983;3:227–245. [Google Scholar]
- 10.Kaiser D. Bacterial swarming, a re-examination of cell movement patterns. Curr Biol. 2007;17:R561–R570. doi: 10.1016/j.cub.2007.04.050. [DOI] [PubMed] [Google Scholar]
- 11.Reichenbach H. Film E777. Gottingen, Germany: Institut fur den Wissenschaftlichen Film; 1968. Archangium violaceum (Myxobacterales)—Schwarmentwicklung und Bildung von Protocysten. 16mm; black and white, 22 min at 24 frames/sec. [Google Scholar]
- 12.Reichenbach H. Film E779. Gottingen, Germany: Institut Wissenschaftlichen Film; 1974. Chondromyces apiculatus (Myxobacterales)—Schwarmentwicklung und Morphogenese. 16mm; black and white, 15 min at 24 frames/sec. [Google Scholar]
- 13.Reichenbach H. Film E778. Gottingen, Germany: Institut Wissenschaftlichen Film; 1966. Myxococcus spp (Myxobacteriales) Schwarmentwicklung und Bildung von Protocysten. 16mm; black and white, 14.5 min at 24 frames/sec. [Google Scholar]
- 14.Blackhart BD, Zusman D. “Frizzy” genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci USA. 1985;82:8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser D. In: Myxobacteria, Multicellularity, and Differentiation. Whitworth DE, editor. Washington, DC: ASM Press; 2008. pp. 93–102. [Google Scholar]
- 16.Berg H, Purcell EM. Physics of chemoreception. Biophys J. 1977;20:193–219. doi: 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merz AJ, Forest KT. Bacterial surface motility: Slime trails, grappling hooks and nozzles. Curr Biol. 2002;12:R297–R303. doi: 10.1016/s0960-9822(02)00806-0. [DOI] [PubMed] [Google Scholar]
- 18.Purcell EM. Life at low Reynolds number. Am J Phys. 1977;45:3–11. [Google Scholar]
- 19.Mignot T, Shaevitz JW, Hartzell PL, Zusman DR. Evidence that focal adhesion complexes power bacterial gliding motility. Science. 2007;315:853–856. doi: 10.1126/science.1137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaiser D. Myxococcus—from single-cell polarity to complex multicellular patterns. Ann Rev Genetics. 2008;42 doi: 10.1146/annurev.genet.42.110807.091615. 110807.091615. [DOI] [PubMed] [Google Scholar]
- 21.Youderian P, Burke N, White DJ, Hartzell PL. Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol Microbiol. 2003;49:555–570. doi: 10.1046/j.1365-2958.2003.03582.x. [DOI] [PubMed] [Google Scholar]
- 22.Youderian P, Hartzell PL. Transposon insertions of magellan-4 that impair social gliding motility in Myxococcus xanthus. Genetics. 2006;172:1397–1410. doi: 10.1534/genetics.105.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu R, Kaiser D. Gliding motility and polarized slime secretion. Mol Microbiol. 2007;63:454–467. doi: 10.1111/j.1365-2958.2006.05536.x. [DOI] [PubMed] [Google Scholar]
- 24.Bretscher AP, Kaiser D. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J Bacteriol. 1978;133:763–768. doi: 10.1128/jb.133.2.763-768.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, Jiang Y, Kaiser D, Alber M. Social interactions in Myxobacterial swarming. PLoS Comput Biol. 2007;3:e253. doi: 10.1371/journal.pcbi.0030253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolgemuth CW. Force and flexibility of flailing myxobacteria. Biophys J. 2005;89:945–950. doi: 10.1529/biophysj.105.062513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spormann AM, Kaiser D. Gliding Mutants of Myxococcus xanthus with high reversal frequencies and small displacements. J Bacteriol. 1999;181:2593–2601. doi: 10.1128/jb.181.8.2593-2601.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leonardy S, Freymark G, Hebener S, Ellehauge E, Søgaard-Andersen L. Coupling of protein localization and cell movements by a dynamically localized response regulator in Myxococcus xanthus. EMBO J. 2007;26:4433–4444. doi: 10.1038/sj.emboj.7601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): Genes controlling movement of single cells. Mol Gen Genet. 1979;171:167–176. [Google Scholar]
- 30.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 31.Igoshin OA, Goldbeter A, Kaiser D, Oster G. A biochemical oscillator explains several aspects of Myxococcus xanthus behavior during development. Proc Natl Acad Sci USA. 2004;101:15760–15765. doi: 10.1073/pnas.0407111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bustamante VH, Martinez-Flores I, Vlamakis HC, Zusman DR. Analysis of the Frz signal transduction system of Myxococcus xanthus shows the importance of the conserved C-terminal region of the cytoplasmic chemoreceptor FrzCD in sensing signals. Mol Microbiol. 2004;53:1501–1513. doi: 10.1111/j.1365-2958.2004.04221.x. [DOI] [PubMed] [Google Scholar]
- 33.Jelsbak L, Søgaard-Andersen L. The cell surface-associated intercellular C-signal induces behavioral changes in individual Myxococcus xanthus cells during fruiting body morphogenesis. Proc Natl Acad Sci USA. 1999;96:5031–5036. doi: 10.1073/pnas.96.9.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jelsbak L, Søgaard-Andersen L. Pattern formation by a cell surface-associated morphogen in Myxococcus xanthus. Proc Natl Acad Sci USA. 2002;99:2032–2037. doi: 10.1073/pnas.042535699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saintillan D, Shelley MJ. Orientational order and instabilities in suspensions of self-locomoting rods. Phys Rev Lett. 2007;99 doi: 10.1103/PhysRevLett.99.058102. 058102. [DOI] [PubMed] [Google Scholar]
- 36.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): Two gene systems control movement. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- 37.Welch R, Kaiser D. Cell behavior in traveling wave patterns of myxobacteria. Proc Natl Acad Sci USA. 2001;98:14907–14912. doi: 10.1073/pnas.261574598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beebe JM. The morphology and cytology of Myxococcus xanthus, N Sp. J Bacteriol. 1941;42:193–223. doi: 10.1128/jb.42.2.193-223.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ronning CM, Nierman WC. In: Myxobacteria, Multicellularity, and Differentiation. Whitworth DE, editor. Washington, DC: ASM Press; 2008. pp. 285–298. [Google Scholar]
- 40.Hartzell PL. Complementation of sporulation and motility defects in a prokaryote by a eukaryotic GTPase. Proc Natl Acad Sci USA. 1997;94:9881–9886. doi: 10.1073/pnas.94.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borgnia MJ, Subramaniam S, Milne JL. Three-dimensional imaging of the highly bent architecture of Bdellovibrio bacteriovorus by using cryo-electron tomography. J Bacteriol. 2008;190:2588–2596. doi: 10.1128/JB.01538-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McBride MJ, Weinberg RA, Zusman DR. “Frizzy” aggregation genes of the gliding bacterium Myxococcus xanthus show sequence similarities to the chemotaxis genes of enteric bacteria. Proc Natl Acad Sci USA. 1989;86:424–428. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.