Abstract
Protein phosphorylation is central to the understanding of cellular signaling, and cellular signaling is suggested to play a major role in the regulation of human embryonic stem (ES) cell pluripotency. Here, we describe the use of conventional tandem mass spectrometry-based sequencing technology—collision-activated dissociation (CAD)—and the more recently developed method electron transfer dissociation (ETD) to characterize the human ES cell phosphoproteome. In total, these experiments resulted in the identification of 11,995 unique phosphopeptides, corresponding to 10,844 nonredundant phosphorylation sites, at a 1% false discovery rate (FDR). Among these phosphorylation sites are 5 localized to 2 pluripotency critical transcription factors—OCT4 and SOX2. From these experiments, we conclude that ETD identifies a larger number of unique phosphopeptides than CAD (8,087 to 3,868), more frequently localizes the phosphorylation site to a specific residue (49.8% compared with 29.6%), and sequences whole classes of phosphopeptides previously unobserved.
Keywords: phosphoproteomics, phosphorylation, MS/MS, PTM analysis, ion-ion chemistry
Reversible protein phosphorylation plays a critical role in cellular signaling and process regulation (1). Over the past several years, the field of phosphorylation site discovery has greatly evolved, allowing researchers to develop and test new views of the proteins that bear these marks. Evidence of this expansion can be found in multiple phosphorylation site repositories that are curated by researchers. One of these databases, PHOSIDA, contains 8,969 human phosphorylation sites, gathered from the collective literature reports (2). Two main technological advancements have enabled the creation of such lists: (i) phosphopeptide enrichment methodologies and (ii) large-scale mass spectrometry (MS)-based proteomics. Low stoichiometric levels coupled with transient regulation hindered the field for years. This problem has been largely overcome by a variety of chromatography-based enrichment techniques, e.g., strong cation exchange (SCX), immobilized metal affinity (IMAC), titanium dioxide (TiO2), and hydrophilic interaction (HILIC) (3–11). These methods rely on the unique chemical characteristics of the modification to purify phosphoryl-containing peptides from the bulk. Once purified, the captured phosphoproteome is characterized by nanoflow liquid chromatography tandem MS (nHPLC-MS/MS). MS-based protein sequencing technologies have continually progressed so that recent studies combining these technologies report the discovery of several thousand phosphorylation sites in a single large-scale experiment (4, 6, 10–15).
For all of the progress detailed above, complications still persist. In an ironic twist, the very chemical features of phosphopeptides that allow for efficient enrichment often impede the MS-based sequence identification (16). The problem occurs during MS/MS—the process of dissociating peptide cations into a collection of fragment ions (17–19). The customary method of imparting MS/MS is to collide a population of selected peptide cations with inert atoms (collision-activated dissociation, CAD). Protonated amide linkages are particularly weakened and are often cleaved upon such collisions (20). Random dissociation of the amide linkages along the peptide backbone produces fragment ions that allow for interpretation of the amino acid sequence. The presence of phosphoryl groups on Ser or Thr residues, however, can change this chemistry. In the gas phase, the phosphoryl group competes with the backbone as the preferred site of protonation and, upon collisional activation, is subject to nucleophilic attack from a neighboring amide carbonyl group. The result is that cleavage is often directed toward the phosphoryl group, rather than the peptide backbone. For this reason, many, if not most, MS/MS spectra of phosphorylated peptides have insufficient information to allow for confident sequence assignment (21).
In the last decade, alternative peptide dissociation methods based on electron capture, and later transfer, rather than collisions (ECD and ETD, respectively) have been developed (22–24). Electron-based methods impart cleavage of the backbone N–Cα bond via free radical chemistry. Posttranslational modifications (PTMs) that are labile upon CAD (e.g., phosphoryl, glycosyl, sulfonyl, etc.) are preserved during ECD/ETD (12, 13, 25–30). Recently, we modified a hybrid linear ion trap–orbitrap mass spectrometer to perform ETD and examined the probability of either method (i.e., CAD or ETD), resulting in a successful sequencing outcome for both phosphorylated and nonphosphorylated peptides (30, 31). In that work, we discovered 8,359 phosphorylation sites on proteins harvested from human embryonic stem (ES) cells (31). By performing further analysis, we have extended this dataset to include 10,844 phosphorylation sites—the largest collection ever reported for human ES cells. In this report we present these collective sites and use the dataset to quantify the relative performance of either fragmentation method for phosphopeptide sequence identification and phosphorylation site localization.
Results
Phosphopeptide Identification.
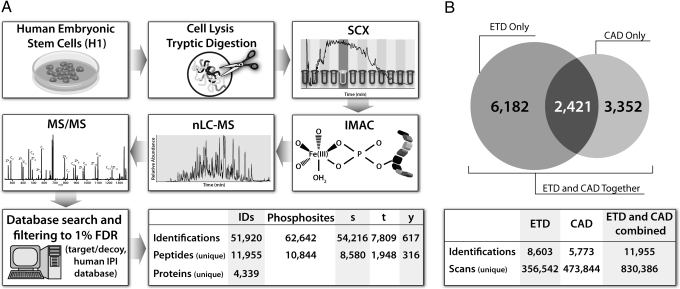
Fig. 1A presents an overview of the large-scale phosphorylation site identification strategy used here. The process began with the harvest of 10 mg of protein from federally registered human ES cells (line H1), followed by brief digestion with trypsin (45 min). Strong cation exchange chromatography (SCX) was then applied to separate the mixture into 12 fractions, each of which was subjected to phosphopeptide enrichment via immobilized metal affinity chromatography (IMAC). After IMAC enrichment, the eluate of each fraction was analyzed via nanoflow reversed-phase LC-MS/MS on a hybrid linear ion trap–orbitrap mass spectrometer that had been modified to perform ETD (30). High mass accuracy and resolution MS1 analysis (30,000) was performed in the orbitrap mass analyzer, followed by data-dependent MS/MS acquisition in the ion trap mass analyzer. The type of dissociation used for MS/MS analysis was either CAD or ETD. All MS/MS spectra were searched against a concatenated version of the forward and reversed human IPI database by using the open mass spectrometry search algorithm (OMSSA) and filtered to a 1% false discovery rate (FDR) (32, 33). A total of 62,642 phosphorylation sites on 51,920 peptides were identified after analyzing each fraction up to 8 times (Fig. 1A).
Fig. 1.
Method and results for analysis of human ES cell phosphoproteome. (A) Cells were lysed and digested with trypsin to produce peptides that were separated via strong cation exchange chromatography. Each fraction was then enriched for phosphopeptides via IMAC and analyzed via nano-RPLC-MS/MS on an ETD-enabled orbitrap mass spectrometer. All raw data were searched using OMSSA against a target/decoy version of the human IPI database and filtered to a 1% FDR, resulting in the total phosphopeptide identifications and phosphorylation sites shown above. (B) Overlap of the phosphopeptides identified via ETD and CAD. The number of overlapping peptides is shown in the diagram, whereas the total unique identifications are shown below.
Next, using in-house written software, we analyzed the MS/MS spectra to determine the most probable phosphopeptide positional isomer for each identification. All of the theoretical fragment ions for each positional isomer were compared with the ions present in the raw MS/MS spectrum. The most probable positional isomer was that with the greatest number of matches to the MS/MS spectrum. All CAD phosphopeptide identifications were then reduced to a list of unique peptides, resulting in a total of 5,773 unique identifications (Fig. 1B). This was also performed for all phosphopeptides identified by ETD, resulting in 8,603 unique identifications. Of the 14,376 sequences, 2,421 were identified by both dissociation methods (Fig. 1B); however, only the best scoring of these was retained in the unique peptide list. When combined, the 2 approaches resulted in the detection of 11,955 unique phosphopeptides (3,868 via CAD and 8,087 via ETD), corresponding to 14,954 total phosphorylation sites, of which 10,844 were nonoverlapping (8,850 serine, 1,948 threonine, and 316 tyrosine). In total, 113,315 unique amino acids spanning 4,339 proteins within the human IPI database were identified. These results make this the largest collection of human ES cell phosphorylation sites identified to date and exemplify the utility of ETD for phosphopeptide analysis.
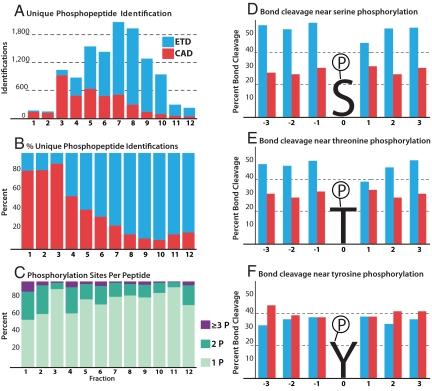
The percentage of unique phosphopeptide identifications per fraction via either CAD or ETD is shown in Fig. 2 A–C. CAD was most effective for the earlier fractions (1–3), those that contain peptides of lower net charge state. These results support previous reports that have used a shallow SCX gradient to extend this phosphopeptide enriched region to increase the number of identifications via CAD. Although CAD soundly outperformed ETD in this region, contributing >80% of the phosphopeptide identifications, these fractions only represent 11% of the total unique phosphopeptides discovered (Fig. 2B). Conversely, in the remaining SCX fractions, 4–12, the majority of phosphopeptides were identified via ETD; an anticipated result as the later fractions contained phosphopeptides of higher net charge. On average 996 unique human ES cell phosphopeptides were identified in each strong cation exchange fraction.
Fig. 2.
Summary of phosphopeptide identifications and assessment of MS/MS spectral quality. ETD identifications are shown in blue and CAD in red. (A) The number of unique identifications in each fraction and the fragmentation method by which it was identified or best identified if discovered by both. (B) The percentage of peptides identified by either CAD or ETD in each fraction. (C) The number of phosphorylation sites per peptide across all fractions. (D–F) MS/MS spectral quality can be assessed by examining the percentage of backbone bonds cleaved via either ETD (blue) or CAD (red) for 3 amino acids up- or downstream of a phosphorylated serine (D), threonine (E), or tyrosine (F) residue.
Phosphorylation Site Localization.
Phosphopeptide identification and site localization depends highly on the tandem mass spectral quality. To examine the relative qualities of the spectra resulting from either collisional or chemical activation (i.e., CAD or ETD), we next calculated fragmentation efficiencies for identified phosphopeptide spectra from all unique sequences for either method. Here, we define fragmentation efficiency as the number of observed fragments divided by the number of possible fragments present in a tandem mass spectrum. The average fragmentation efficiency was 74.8% for ETD and 55.3%, for CAD. These fragments corresponded to an average sequence coverage—defined as the detection of at least 1 fragment representing each cleavage of each backbone bond—of 86.8% for ETD and 74.0% for CAD. The greater fragmentation efficiency and sequence coverage of peptides identified via ETD allowed for site-specific phosphorylation localization of 49.8% of all of the peptides identified, whereas 29.6% were localized via CAD.
These data demonstrate that ETD spectra are more often correlated to the correct phosphopeptide sequence and that the site of phosphorylation is more often located to a single Ser, Thr, or Tyr residue. To further investigate the probability of site localization for each method, we examined the frequency of cleavage of the 3 bonds on either side of a localized phosphorylation site for each of these residues (Fig. 2 D–F). We conclude that ETD spectra are almost twice as likely to produce cleavage of the backbone in the vicinity of phosphorylated serine residues as compared with CAD (Fig. 2D). Fig. 2E displays a similar but somewhat lesser edge for ETD when the phosphorylated residue is threonine. These data provide direct evidence for 2 long-standing observations: (i) CAD-driven fragmentation of phosphoserine- and -threonine-containing peptides often results in the formation of spectra devoid of the necessary backbone fragments for phosphosite localization and (ii) ETD is more or less indifferent to the presence of CAD-labile PTMs. Note that phosphotyrosine residues are less labile during CAD fragmentation; our data confirms this, there is virtually no difference in the probabilities of generating backbone cleavages in proximity of phosphotyrosine sites for either CAD or ETD (Fig. 2F). Another interesting trend is the reduced cleavage efficiency for the bond in the +1 position of serine and threonine phosphorylation sites observed for ETD spectra. A common phosphorylation motif places a Pro residue on the C-terminal side of phosphorylated Ser or Thr residues—that is, the +1 position. Because ETD fragmentation is directed at the N–Cα bond, such peptides do not show cleavage of the +1 position because the cyclical structure of proline tethers the newly formed fragments. Recall CAD targets the amide linkage and therefore is not subject to that phenomenon.
Amino Acid Frequency.
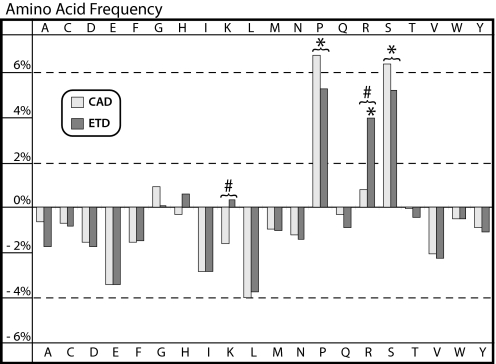
With the 11,955 unique phosphopeptide sequences in hand, we next sought to address 2 targeted questions: (i) what are the relative frequencies of all amino acids found in the vicinity of a phosphorylation site and (ii) does the applied dissociation method alter these distributions? To do this, we plotted the frequency of every amino acid detected in this phosphopeptide dataset relative to that observed from another large-scale analysis of human ES cell peptides that were not phosphorylated (Fig. 3) (31). This dataset contained 11,919 peptide sequences that were the result of endoproteinases Lys-C digestion, SCX fractionation, and both ETD and CAD MS/MS interrogation. Pro, Ser (unphosphorylated), and Arg are significantly overrepresented in phosphorylated peptides (P < 0.05, n = 20), whereas Ile and Leu were all found to be marginally underrepresented (P < 0.18, n = 20) in the phosphopeptide dataset. Note, the residue that was phosphorylated was removed from peptide sequences before amino acid frequency analysis to eliminate artificial inflation of the frequency of those amino acids. The average frequency of any amino acid was 4.6%. As noted above, Pro is a player in a common phosphorylation motif, so its enrichment is expected; however, to our knowledge, the enrichment of Ser and Arg residues surrounding phosphorylation sites has neither been previously reported nor quantified. By increasing the number of potential positional isomers, the frequent presence of Ser in phosphopeptides presents increased difficulty in identifying the exact site of modification. Surprisingly, the differences in amino acid frequency between CAD and ETD were minimal, but for 2 exceptions—Arg and Lys. The largest discrepancy between the 2 activation methods was the significantly greater (P < 0.03, n = 20) frequency of Arg (3.1%) and Lys (2.0%) residues within sequences identified via ETD. From these data we conclude that the complementary nature of CAD and ETD will be essential to uncovering the entire phosphoproteome and to reveal the dynamics that predispose certain residues/regions to modification.
Fig. 3.
The percentage difference between the frequency of every amino acid within phosphorylated peptide sequences with respect to the frequency within nonphosphorylated human ES cell peptide sequences. *, values that are statistically different from the nonphosphorylated human ES cell peptides; #, statistically significant differences between CAD and ETD frequencies (P < 0.05, n = 20).
Motif Analysis.
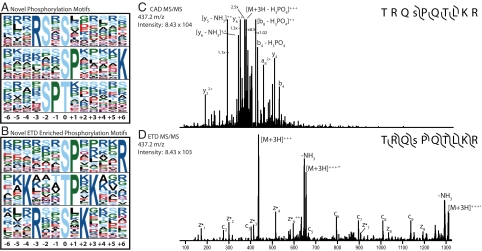
Given the enrichment of Arg and Lys in the ETD phosphopeptide dataset, we reasoned that this sizeable collection—the largest reported to date—might contain evidence for previously unidentified phosphorylation motifs. To test this hypothesis, all unique phosphorylation sites were subjected to motif analysis using the Motif-X algorithm (34). Sequences were centered on every phosphorylation site and extended to 6 residues on either side, by using the corresponding protein sequence from the human IPI database. The frequency of all motifs was compared with the human IPI database in its entirety. We first sought to identify motifs for phosphorylated Ser, Thr, and Tyr from our entire dataset (i.e., CAD and ETD sequences). This yielded 20 Ser and 11 Thr motifs with high significance (P < 10−6, [supporting information (SI) Table S1]. Ten of these motifs had been previously been identified in human, and corresponded to known substrates such as PKA and CDK1. Five additional motifs have been previously identified in large-scale phosphorylation analyses of other organisms (10, 15). The remaining 16 motifs were neither found in the human PHOSIDA database nor any other large-scale phosphorylation experiments. A selection of these motifs are shown in Fig. 4A—note the complete list can be found in Table S1. Several of the motifs we have identified add clarity and detail to known motifs. For example, the motif xxxRSxsxxxxxx we report here contains the previously described motif of xxxRxxsxxxxxx, where “s” indicates the site and residue of phosphorylation (10, 15).
Fig. 4.
Analysis of the phosphopeptide sequences identified here resulted in the identification of several previously unreported phosphorylation motifs. (A) Motifs identified within the entire dataset (CAD and ETD) when using the human IPI database as the background (note the residue at position 0 is the site of phosphorylation). (B) Motifs enriched in the ETD dataset when using CAD-identified peptides from PHOSIDA as a background. A list of all identified motifs can be found in Table S1. (C and D) MS/MS spectra from either CAD (C) or ETD (D) of a peptide having the sequence TRQsPQTLKR (“s” is phosphorylated), which represents an ETD-enriched motif (xxxxxxsPxxxxR). The CAD spectrum (C) lacked sufficient backbone cleavage to make a successful identification; however, ETD results in near-complete backbone cleavage.
To test our theory that ETD contributed largely to the discovery of these motifs, we examined whether any of the patterns were enriched in either the ETD or CAD analyses. We compared all unique phosphopeptides identified via ETD (those in this report only) to the human subset of the PHOSIDA database, which comprises 18,869 phosphopeptides identified from CAD analyses by multiple researchers (2). Not surprisingly, all of the phosphoserine and threonine motifs we discovered contained 1 or more basic residues (18 of 28 total motifs, P < 10−6, Table S2). A selection of these motifs is displayed in Fig. 4B). In contrast, several recent large-scale experiments using CAD have reported the percentage of phosphopeptides fitting a basic motif to be less than that of those fitting Pro-directed or acidic motifs—the subset that is most readily identified via CAD (10, 15). Here, we show that insights can be made simply by combining complementary methods of phosphopeptide dissociation. Fig. 4 C and D display CAD and ETD tandem mass spectra, respectively, for a phosphopeptide having the sequence TRQsPQTLKR (“s” indicates phosphoserine). The CAD tandem mass spectrum of this phosphopeptide could not be matched to the correct sequence upon database searching. Manual inspection reveals only 6 of 18 possible backbone cleavages are present, with the major peaks resulting from neutral loss of phosphoric acid. Application of ETD to this precursor, however, resulted in the observation of 14 of 18 possible backbone cleavages and a high-confidence match to the correct sequence. This peptide represents an example of an ETD-enriched motif, xxxxxsPxxxxR, that is evidently present but particularly CAD-unfriendly. We conclude that for broad, expansive identification of protein phosphorylation, both collisional and electron-based peptide dissociation technologies should be applied.
Pathway Analysis.
Gene symbols corresponding to all identified phosphoproteins were submitted to the KEGG PATHWAY Database (www.genome.jp/kegg/) (35). A list of several pathways and the number of genes identified in each pathway is listed in Table S3. One pathway identified, and suggested to be critical to the self-renewal of human ES cells, was the TGFβ signaling pathway (36). The activation of this pathway maintains the expression of genes associated with pluripotency such as OCT4, SOX2, and NANOG (36–39). In this study, we have characterized a total of 17 nonredundant phosphorylation sites on proteins within the TGFβ signaling pathway, as displayed in Fig. S2.
Biological Significance.
Human ES cells hold great promise for their ability to self-renew indefinitely and to differentiate into any type of cell in the adult human body. Ethical concerns of using human embryos, however, have made the study of such cells controversial. Recently, adult human cell lines were reprogrammed to an ES cell state (induced pluripotent stem cells, iPS cells) (40, 41). These cells possess the therapeutically desired characteristics of ES cells, namely indefinite self-renewal and pluripotency, without the requirement of human embryo destruction. Of the collection of 4 transcription factors used to induce pluripotency, OCT4 and SOX2 have been identified as vital in every report. Thus, these 2 factors play a critical role in cellular reprogramming, and greater characterization of the posttranslational modifications, such as phosphorylation, affecting the function of OCT 4 and SOX2 may facilitate future reprogramming efforts.
OCT4, a homeodomain protein, is a component of a transcriptional network including SOX2 and NANOG, which works in concert to regulate the human ES cell state. High levels of OCT4 are a marker for pluripotency, whereas diminishing levels are indicative of differentiation (42). Our large-scale phosphoproteomic study identified a phosphorylation site on OCT4 at Ser 236. This site is contained within the DNA-binding homeodomain, which spans amino acids 230–289, and hints at the potential of this phosphorylation site to influence binding to DNA and, consequently, transcription. To our knowledge, no sites of phosphorylation have previously been described on OCT4, although the homologous site of serine phosphorylation on Oct1 has previously been described (43). We also identified 4 previously unreported sites of phosphorylation within the SOX2 protein (Ser 246, 249, 250, and 251). SOX2 is believed to work closely with OCT4 to maintain human ES cell pluripotency, as half of the genes bound by OCT4 are also bound by SOX2 (44). Because of the serine-rich nature of the peptide and the presence of overlapping b- and y- or c- and z·-type ions, we located the exact sites of phosphorylation on 5 of the 8 possible isoforms. The peptides identified on OCT4 and SOX2 are shown in Table S4. Note, only those peptides with localized phosphorylation sites are displayed. Of these 6 peptides, 2 were identified via ETD and 4 via CAD, once again demonstrating the merit of using complementary methods of dissociation.
Discussion
The field of phosphoproteomics has rapidly evolved over the past decade. Phosphopeptide enrichment strategies, coupled with MS-based proteomics, have greatly catalyzed this growth. Proof of the impact of this methodology can be found in the numerous recent literature reports cataloging several thousands of phosphorylation sites across multiple organisms. From these data, researchers have launched large-scale data-mining efforts to elucidate common amino acid motifs that are targeted for phosphorylation. That said, the peptide fragmentation that often results from the conventional MS/MS strategy, CAD, often restricts phosphorylation site or even sequence identification.
Here, we performed a large-scale analysis of phosphorylation in human ES cells using both CAD and ETD MS/MS dissociation methods. From these experiments we conclude that ETD identifies a larger number of unique phosphopeptides than CAD (8,087 to 3,868), more frequently localizes the phosphorylation site to a specific residue (49.8% compared with 29.6%), and sequences whole classes of phosphopeptides previously unobserved. The latter conclusion is drawn from the observation of 16 previously unreported phosphorylation motifs found only the ETD-sequenced dataset. In general, these motifs can be classified as basic and will provide researchers a broadened view of kinase targets. Although ETD analysis was superior to CAD in many aspects, i.e., fragmentation efficiency, sequence coverage, phosphorylation site localization, and peptide identification rates, each method has its strengths and weaknesses. Factors such as peptide precursor charge state (z), mass-to-charge (m/z), and the identity of the phosphorylated residue, can strongly influence the probability of a sequence assignment with either method. For this reason, we envision that the routine application of both CAD and ETD, in a decision tree-driven fashion, will yield the broadest, most expansive view of the highly dynamic phosphoproteome.
Methods
Cell Culture, Protein Harvesting, Digestion, Peptide Fractionation, and Phosphopeptide Enrichment.
Cells were lysed by sonication and protein extracted. An aliquot of 10 mg of human ES cell protein was digested with trypsin and desalted, and the peptide mixture was separated by means of SCX. Each SCX fraction was enriched for phosphopeptides by means of IMAC. Further details are provided in SI Text.
nHPLC, MS, Database Searching, Phosphosite Localization, and Motif Extraction.
All experiments were performed on a hybrid linear ion trap–orbitrap mass spectrometer (Thermo Fisher Scientific) that had been modified to allow for ETD (30). All tandem MS spectra were searched against the human IPI database by using OMSSA (Open Mass Spectrometry Search Algorithm) and phosphosites localized by using a program written in-house (33). Additional details are provided in SI Text.
Supplementary Material
Acknowledgments.
We thank Judit Villen and Steven P. Gygi, from Harvard Medical School, for their invaluable assistance in performing the SCX fractionation and Travis Berggren for helping initiate this project. We gratefully acknowledge Jarrod Marto and Scott Ficarro for advice with the Waters nUPLC. This work was supported by the University of Wisconsin, the Beckman Foundation, and National Institutes of Health (NIH) Grants R01GM080148 (to J.J.C.) and P01GM081629 (to J.A.T. and J.J.C.). D.L.S. acknowledges support from an NIH predoctoral traineeship—the Genomic Sciences Training Program, NIH 5T32HG002760.
Footnotes
Conflict of interest statement: In the interest of full disclosure, J.A.T. is a cofounder and shareholder of Cellular Dynamics International, a company commercializing pluripotent stem cell-derived cells for drug screening and discovery, the activities of which are not directly related to this manuscript.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811964106/DCSupplemental.
References
- 1.Pawson T, Scott JD. Protein phosphorylation in signaling–50 years and counting. Trends Biochem Sci. 2005;30:286–290. doi: 10.1016/j.tibs.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 2.Gnad F, et al. PHOSIDA (phosphorylation site database): Management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albuquerque CP, et al. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beausoleil SA, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat Methods. 2007;4:231–237. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]
- 6.Cantin GT, et al. Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J Proteome Res. 2008;7:1346–1351. doi: 10.1021/pr0705441. [DOI] [PubMed] [Google Scholar]
- 7.Ficarro SB, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 8.Kweon HK, Hakansson K. Selective zirconium dioxide-based enrichment of phosphorylated peptides for mass spectrometric analysis. Anal Chem. 2006;78:1743–1749. doi: 10.1021/ac0522355. [DOI] [PubMed] [Google Scholar]
- 9.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJD. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci USA. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bodenmiller B, et al. PhosphoPep—A phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol Syst Biol. 2007;3:139. doi: 10.1038/msb4100182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi A, et al. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci USA. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci USA. 2007;104:2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 15.Zhai B, Villen J, Beausoleil SA, Mintseris J, Gygi SP. Phosphoproteome analysis of Drosophila melanogaster embryos. J Proteome Res. 2008;7:1675–1682. doi: 10.1021/pr700696a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schroeder MJ, Shabanowitz J, Schwartz JC, Hunt DF, Coon JJ. A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal Chem. 2004;76:3590–3598. doi: 10.1021/ac0497104. [DOI] [PubMed] [Google Scholar]
- 17.Coon JJ, Syka JEP, Shabanowitz J, Hunt DF. Tandem mass spectrometry for peptide and protein sequence analysis. BioTechniques. 2005;38:519–523. doi: 10.2144/05384TE01. [DOI] [PubMed] [Google Scholar]
- 18.Hunt DF, Yates JR, Shabanowitz J, Winston S, Hauer CR. Protein sequencing by tandem mass-spectrometry. Proc Natl Acad Sci USA. 1986;83:6233–6237. doi: 10.1073/pnas.83.17.6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLafferty FW. Tandem mass spectrometric analysis of complex biological mixtures. Int J Mass Spectrom. 2001;212:81–87. [Google Scholar]
- 20.Dongre AR, Jones JL, Somogyi A, Wysocki VH. Influence of peptide composition, gas-phase basicity, and chemical modification on fragmentation efficiency: Evidence for the mobile proton model. J Am Chem Soc. 1996;118:8365–8374. [Google Scholar]
- 21.Carr SA, Huddleston MJ, Annan RS. Selective detection and sequencing of phosphopeptides at the femtomole level by mass spectrometry. Anal Biochem. 1996;239:180–192. doi: 10.1006/abio.1996.0313. [DOI] [PubMed] [Google Scholar]
- 22.Coon JJ, Syka JEP, Schwartz JC, Shabanowitz J, Hunt DF. Anion dependence in the partitioning between proton and electron transfer in ion/ion reactions. Int J Mass Spectrom. 2004;236:33–42. [Google Scholar]
- 23.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 25.Good DM, Wirtala M, McAlister GC, Coon JJ. Performance characteristics of electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 2007;6:1942–1951. doi: 10.1074/mcp.M700073-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Kelleher NL, et al. Localization of labile posttranslational modifications by electron capture dissociation: The case of gamma-carboxyglutamic acid. Anal Chem. 1999;71:4250–4253. doi: 10.1021/ac990684x. [DOI] [PubMed] [Google Scholar]
- 27.Khidekel N, et al. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat Chem Biol. 2007;3:339–348. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- 28.Shi SDH, et al. Phosphopeptide/phosphoprotein mapping by electron capture dissociation mass spectrometry. Anal Chem. 2001;73:19–22. doi: 10.1021/ac000703z. [DOI] [PubMed] [Google Scholar]
- 29.Swaney DL, et al. Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal Chem. 2007;79:477–485. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McAlister GC, et al. A proteomics grade electron transfer dissociation-enabled hybrid linear ion trap-orbitrap mass spectrometer. J Proteome Res. 2008;7:3127–3136. doi: 10.1021/pr800264t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swaney DL, McAlister GC, Coon JJ. Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat Methods. 2008;5:959–964. doi: 10.1038/nmeth.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 33.Geer LY, et al. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 35.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan GJ, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- 37.Babaie Y, et al. Analysis of Oct4-dependent transcriptional networks regulating self-renewal and pluripotency in human embryonic stem cells. Stem Cells. 2007;25:500–510. doi: 10.1634/stemcells.2006-0426. [DOI] [PubMed] [Google Scholar]
- 38.James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGF beta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 39.Xu RH, et al. NANOG is a direct target of TGF beta/Activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 41.Yu JY, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 42.Phanstiel D, et al. Mass spectrometry identifies and quantifies 74 unique histone H4 isoforms in differentiating human embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:4093–4098. doi: 10.1073/pnas.0710515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inamoto S, Segil N, Pan ZQ, Kimura M, Roeder RG. The cyclin-dependent kinase-activating kinase (CAK) assembly factor, MAT1, targets and enhances CAK activity on the POU domains of octamer transcription factors. J Biol Chem. 1997;272:29852–29858. doi: 10.1074/jbc.272.47.29852. [DOI] [PubMed] [Google Scholar]
- 44.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.