Abstract
Adipocyte differentiation is controlled by many transcription factors, but few known downstream targets of these factors are necessary for adipogenesis. Here we report that retinol saturase (RetSat), which is an enzyme implicated in the generation of dihydroretinoid metabolites, is induced during adipogenesis and is directly regulated by the transcription factor peroxisome proliferator activated receptor γ (PPARγ). Ablation of RetSat dramatically inhibited adipogenesis but, surprisingly, this block was not overcome by the putative product of RetSat enzymatic activity. On the other hand, ectopic RetSat with an intact, but not a mutated, FAD/NAD dinucleotide-binding motif increased endogenous PPARγ transcriptional activity and promoted adipogenesis. Indeed, RetSat was not required for adipogenesis when cells were provided with exogenous PPARγ ligands. In adipose tissue, RetSat is expressed in adipocytes but is unexpectedly downregulated in obesity, most likely owing to infiltration of macrophages that we demonstrate to repress RetSat expression. Thiazolidinedione treatment reversed low RetSat expression in adipose tissue of obese mice. Thus, RetSat plays an important role in the biology of adipocytes, where it favors normal differentiation, yet is reduced in the obese state. RetSat is thus a novel target for therapeutic intervention in metabolic disease.
Keywords: adipocyte, PPAR
Adipose tissue plays a fundamental role in whole-body energy homeostasis (1). Since the discovery of leptin (2, 3) and other adipokines secreted by adipocytes, our understanding of the complex interaction between adipose tissue and other organs has been tremendously increased. An excessive amount of adipose tissue, with the consequence of obesity and insulin resistance, is the main cause for the development of type 2 diabetes (4). Many transcription factors contribute to adipogenesis (5), but the nuclear receptor peroxisome proliferator activated receptor γ (PPARγ) is the master regulator of adipocyte differentiation (6, 7). Thiazolidinediones (TZDs), which have clinical utility as insulin-sensitizing drugs, are potent PPARγ ligands that promote adipogenesis (8). However, although many direct targets of PPARγ play a role in adipocyte function, few if any that do not function as transcription factors have been demonstrated to be important for adipocyte differentiation (9).
Retinoids and their related binding proteins have been increasingly recognized to contribute to metabolic regulation in adipose tissue. Adipose tissue is an important primary storage location for retinoids (10, 11), and all-trans retinoic acid (atRA), one of the active metabolites of retinol, has been described as a potent inhibitor of early stages of adipocyte differentiation by activation of the retinoic acid receptor α (RARα) and inhibition of CCAAT/enhancer-binding protein β (C/EBPβ)-mediated transcription (12, 13). In addition, the retinol oxidation product retinaldehyde has been shown to be reduced in adipose tissue of obese mice and to inhibit in vitro adipogenesis by suppressing PPARγ and retinoid X receptor α (RXRα) responses (14). Furthermore, retinol binding protein 4 (RBP4) is secreted by adipocytes and seems to modulate the development of type 2 diabetes (15).
In the search for missing pieces of adipose retinoid metabolism, we found that the oxidoreductase retinol saturase (RetSat), which has been described to catalyze the reaction of retinol to 13,14-dihydroretinol (dhretinol) (16), is induced during murine and human fat cell differentiation. RetSat expression is controlled by PPARγ through an intronic PPARγ response element (PPRE) in adipocytes. Ablation of RetSat expression in preadipocytes inhibited adipogenesis, whereas ectopic expression of RetSat enhanced differentiation. The adipogenic effect of RetSat required its catalytic activity yet, surprisingly, the differentiation block cause by loss of RetSat could not be rescued by 13,14-dhretinol, the putative product of RetSat activity. RetSat was not required for adipogenesis when cells were provided with exogenous PPARγ ligands, and RetSat increased endogenous PPARγ transcriptional activity, suggesting that the enzymatic function of RetSat is in this pathway. Furthermore, we found that RetSat is downregulated in the obese state in mice and humans, most likely owing to an increase of macrophage infiltration in the obese state, and that TZD treatment rescues RetSat expression in adipose tissue. These findings suggest an important role for RetSat in adipose tissue.
Results
RetSat Is Induced During Adipocyte Differentiation and by PPARγ Activation.
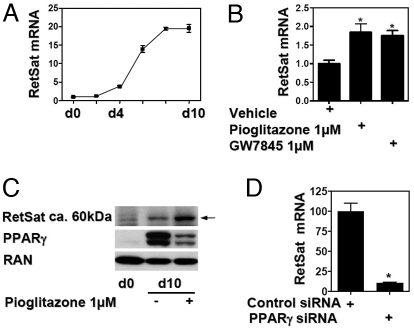
We identified RetSat as a differentially expressed gene by comparing mouse 3T3-L1 preadipocytes and adipocytes. The relevance of this finding to human adipocyte biology was confirmed in human Simpson-Golabi-Behmel syndrome (SGBS) adipocytes, where RetSat mRNA expression was markedly elevated compared with preadipocytes [supporting information (SI) Fig. S1A]. During adipocytic differentiation of 3T3-L1 cells, RetSat mRNA levels began to rise after 2–4 days and reached maximal levels in mature adipocytes at day 8 (Fig. 1A). Because PPARγ has a central role in the transcriptional regulation of adipogenesis, we investigated whether ligand activation of PPARγ can modulate RetSat expression. Incubation of mature adipocytes with pioglitazone or the non-TZD ligand GW7845 (17) increased RetSat mRNA expression (Fig. 1B). Furthermore, the time-dependent induction of RetSat by TZD treatment of mature adipocytes was similar to the activation of other known PPARγ target genes (Fig. S1B). To investigate the regulation of RetSat protein, we generated a rabbit polyclonal antibody directed against a small C-terminal peptide of RetSat. RetSat protein expression showed a robust induction in adipocytes and was modestly increased further by pioglitazone treatment (Fig. 1C). Thus, RetSat mRNA and protein expression showed a very similar pattern of regulation during fat cell differentiation and PPARγ activation and exhibits similarities with other well-characterized PPARγ target genes. The observation that 2 chemical classes of ligands induced RetSat suggested that PPARγ was critical for its expression in adipocytes. Indeed, the PPARγ antagonist PD068235 (18) prevented pioglitazone induction of RetSat (Fig. S1C), confirming the involvement of PPARγ. However, there could be other factors involved in the regulation of RetSat, considering the residual RetSat expression after antagonist treatment. Indeed, preliminary data suggest that RetSat has several intergenic C/EBP binding sides (M. Schupp, M.I.L., and M.A.L., unpublished data).
Fig. 1.
RetSat expression is induced during adipocyte differentiation and is dependent on PPARγ. (A) RetSat mRNA expression was determined during the time course of differentiation of 3T3-L1 fibroblasts. (B) 3T3-L1 adipocytes were stimulated as indicated for 24 h and mRNA expression determined. (C) Whole-cell protein from 3T3-L1 preadipocytes and adipocytes treated for 48 h with or without pioglitazone was isolated and assessed for expression of RetSat, PPARγ, and the GTPase RAN. (D) 3T3-L1 adipocytes were electroporated with PPARγ or nontargeting siRNA oligonucleotides. After 48 h, RetSat mRNA expression was determined.
PPARγ Is Required for RetSat Expression in Mature Adipocytes.
We next investigated the effect of PPARγ knockdown in adipocytes on RetSat expression. Electroporation of specific siRNA oligos into mature adipocytes efficiently reduced PPARγ protein levels 48 h later (Fig. S1D). There was no obvious phenotypic change or dedifferentiation of the adipocytes within this period as assessed by microscopy and in confirmation of previous reports (19). However, RetSat mRNA decreased by approximately 90% in the PPARγ siRNA-treated cells (Fig. 1D), underlining the importance of PPARγ in mature adipocytes to maintain RetSat gene expression.
The Activation of RetSat by PPARγ Is Mediated by an Intronic PPRE.
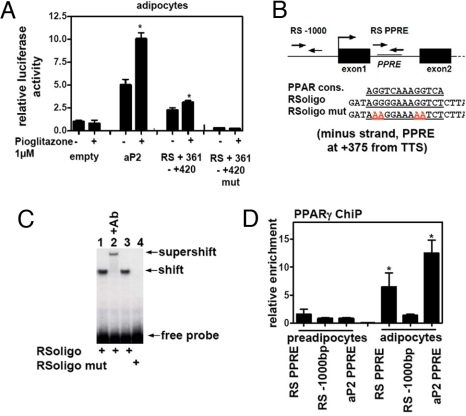
To investigate the transcriptional mechanism by which PPARγ activation induces RetSat expression, we fused genomic sequences 3 kb 5′ of the transcriptional start site upstream of a luciferase reporter vector. Electroporation of this vector into adipocytes showed some basal activity compared with the empty vector, but this was not increased by pioglitazone at a concentration (1 μM) that did induce the expression of a control PPRE-containing reporter (data not shown). Because these 3 kb already included parts of the coding region of the neighboring gene (Rbed1), we searched the transcribed region of RetSat for putative PPARγ binding sites. Inspection of the sequence of intron 1 revealed an imperfect direct repeat of the consensus nuclear receptor half-site separated by 1 bp (Fig. 2B). This site (+361 to + 420) conferred TZD-responsiveness to a luciferase reporter gene transfected into mature adipocytes, and mutation of the putative PPARγ/RXR binding site abrogated the TZD response. The aP2 enhancer region (−5533 to −5011) was used as a control and, as expected, also showed induction by TZD treatment (Fig. 2A).
Fig. 2.
An intronic PPRE regulates RetSat in adipocytes. (A) Adipocytes were electroporated with the indicated enhancers containing pGl3-promoter vectors. After stimulation with pioglitazone for 24 h, cells were assessed for luciferase activity. (B) Schematic presentation of the RetSat gene with amplicons (RS −1000 and RS PPRE) for ChIP experiments and oligonucleotide sequences used for gel shifts. (C) Gel shifts of the PPRE-containing oligonucleotides after incubation with RXRα/PPARγ (lanes 1 and 3). The complex was supershifted by a PPARγ antibody (Ab, lane 2) and binding abolished by PPRE mutation (lane 4). (D) ChIP experiments of the PPRE-containing sequence and an upstream area of the RetSat gene and an aP2 PPRE (−5642 of the aP2 transcriptional start site). Real-time PCR results are expressed as fold enrichments over amplification of a promoter fragment of the insulin gene divided by the amplification of the input material.
DNA mobility shift assays demonstrated that the putative RetSat PPRE was capable of binding the PPARγ/RXRα heterodimers (Fig. 2C, lanes 1 and 3). Incubation with a specific PPARγ antibody supershifted the complex (lane 2) and mutation of the PPRE completely abolished binding (lane 4). In addition, the site was not bound by either PPARγ or RXRα alone, and binding of the heterodimer was effectively competed away by increasing concentrations of oligonucleotides containing the consensus or RetSat PPRE (Fig. S2A). In intact adipocytes, ChIP demonstrated that PPARγ bound in the region of the intronic PPRE but not further upstream (Fig. 2D). As expected, the region of the aP2 promoter containing a PPRE was also bound by PPARγ, and little PPARγ association with either gene was observed in preadipocytes, consistent with low expression of PPARγ in these cells (Fig. 2D). Acetylation of histone H3 at lysine 9, which marks sites of transcriptional activation (20–22), was accordingly increased at these genomic sites in adipocytes (Fig. S2B).
RetSat Is Required for Normal Adipocyte Differentiation Downstream of C/EBPβ Induction Without Generating 13,14-Dhretinol.
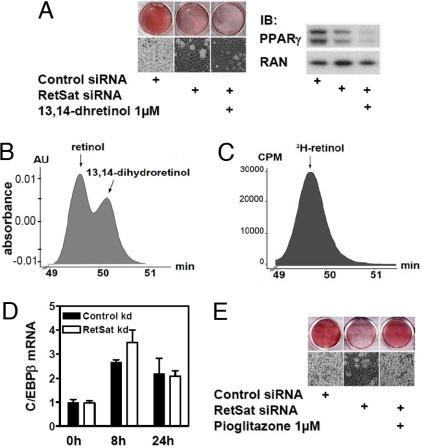
We next investigated whether RetSat induction was important for adipogenesis. We electroporated nontargeting control or RetSat siRNA oligos into preadipocytes and assayed for RetSat expression 2 days after reaching confluency. RetSat mRNA and protein were reduced by approximately 90% in these cells (Fig. S3A), and the effective knockdown was maintained for at least 7 days (data not shown). Remarkably, cells depleted of RetSat exhibited impaired adipocyte differentiation, as determined by Oil Red O staining of intracellular lipids and phase contrast microscopy (Fig. 3A). An alternative RetSat siRNA oligo gave a similar reduction in adipocyte conversion (Fig. S3B). Consistently, the expression of PPARγ was strongly reduced in cells treated with RetSat siRNA (Fig. 3A), as were the expression of C/EBPα and aP2, as well as the secretion of adiponectin (Fig. S3C). To further validate the effect of RetSat on adipocyte differentiation, we measured mRNA expression of Gata2, Pref-1, and Wnt5a, factors that are known to be expressed in preadipocytes and downregulated during differentiation (5). All of these factors showed a stronger decrease in the control siRNA-treated cells after 8 days of differentiation, underlining the inhibitory effect of RetSat ablation on the adipogenic program (Fig. S3D).
Fig. 3.
RetSat ablation inhibits differentiation. (A, D, and E) RetSat or nontargeting oligo electroporated 3T3-L1 cells were stimulated to differentiate into adipocytes. Cells were supplemented with either 13,14-dhretinol (A) or pioglitazone (E) for the first 4 days of differentiation, as indicated. After 8 days, adipocyte differentiation was assessed by Oil Red O staining and microscopy (Left) and protein expression for PPARγ and RAN (A, Right). (B) Absorbance of coinjected retinol and 13,14-dhretinol standards at 325 nm. (C) Analysis of a representative adipocyte cell extract overexpressing RetSat incubated for 24 h with 3H-retinol at retention times for standard retinol/13,14-dhretinol. (D) mRNA was isolated at indicated time points and assayed by real-time PCR for C/EBPβ expression.
We next investigated the possibility that RetSat ablation prevented triglyceride accumulation rather than adipocyte differentiation per se by knocking down RetSat in mature adipocytes. In contrast to the effect of RetSat knockdown early in adipogenesis, expression of PPARγ (Fig. S4A) and genes involved in lipid metabolism (Fig. S4B), as well as triglyceride accumulation (Fig. S4C), were not reduced. Furthermore, gene expression profiles of the RetSat knockdown cells did not reveal enrichment of lipid metabolism pathways (Fig. S4D). Thus, the impaired adipocyte conversion after RetSat ablation in preadipocytes is likely due to an adipogenic defect rather than a failure to store triglycerides.
RetSat has been shown to catalyze the conversion of retinol to 13,14-dhretinol (16), and thus we hypothesized that this retinoid would rescue the effect of RetSat ablation during differentiation. Surprisingly, however, 13,14-dhretinol did not compensate for the effect of RetSat ablation (Fig. 3A). On the contrary, cells supplemented with 13,14-dhretinol showed the tendency to differentiate less well. We also tested synthetic (13R)-13,14-dhretinol, the actual enantiomer produced by RetSat, in adipocyte differentiation but failed to see any promoting effect on adipogenesis (23).
These unexpected findings led us to analyze the concentrations of endogenous dihydroretinoids in 3T3-L1 cells. Synthetic 13,14-dhretinol served as a standard, and we optimized our system to detect and distinguish between the nonreduced/reduced retinol by HPLC. 13,14-dhretinol had the described maximum absorbance at approximately 290 nm (16) and eluted slightly later than retinol, showing a characteristic double peak when both forms were present and analyzed at 325 nm, the maximum absorbance for retinol (data not shown). Retinoid content was measured at 275 and 325 nm in the cells and media during adipogenesis with this methodology. Preadipocytes contained low levels of retinyl esters, which increased during adipogenesis, whereas retinol was detected only in the cell media, or after saponification of endogenous retinyl esters in the cell extracts (data not shown). When adipocytes were incubated with 1 μM retinol for 24 h, cellular content of retinol and retinyl esters increased. However, under no conditions did we detected any 13,14-dhretinol (data not shown). To further increase the detection sensitivity, we decreased the HPLC flow rate to longer retention times to further resolve the overlapping peaks and incubated adipocytes with 3H-labeled retinol. As in the previous conditions, we detected a characteristic double peak when both retinol and 13,14-dhretinol standards were present (Fig. 3B). Furthermore, we infected adipocytes with an adenovirus expressing RetSat, to facilitate our detection of dihydroretinoids by increasing the abundance of the enzyme. Similar to the results with nonlabeled retinol, we did not detect a species corresponding to 3H-13,14-dhretinol in extracts of adipocyte or adipocytes overexpressing RetSat (Fig. 3C). These results suggest that expression of RetSat during differentiation or in adipocytes does not lead to the formation of detectable 13,14-dhretinol. They also confirm the dissociation between the effect of RetSat on differentiation and the impact of 13,14-dhretinol on this process.
To gain insight into the mechanism by which RetSat ablation blocks differentiation, we next investigated the stage of adipogenesis that requires RetSat. The bZip transcription factors C/EBPβ and C/EBPδ, which are induced early in adipogenesis and are important for induction of PPARγ (24, 25), were induced normally within the first 24 h in cells lacking RetSat (Fig. 3D and data not shown). It has been described that C/EBPβ then induces a peak in the transcriptional activity of PPARγ which involves the production of an endogenous ligand between 24 and 48 h after initiating differentiation (26), followed by a concomitant increase of PPARγ expression. The observation that RetSat knockdown blocked differentiation upstream of the full induction of PPARγ led us try to rescue the differentiation defect by supplementing with the TZD ligand pioglitazone. Indeed, pioglitazone completely overcame the defect in differentiation by RetSat ablation, as shown by Oil Red O staining (Fig. 3E). This suggested that RetSat could be involved in the regulation of the transcriptional activity of PPARγ.
RetSat Increases PPARγ Transcriptional Activity and Adipocyte Differentiation, Depending on Its Integrity of the Dinucleotide-Binding Site.
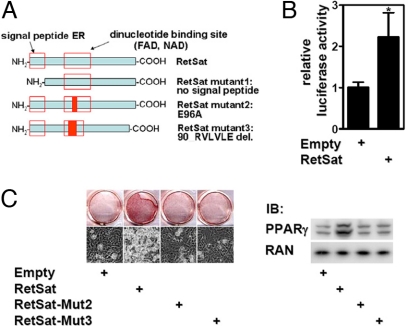
Considering the increase of RetSat expression during adipogenesis, we hypothesized that ectopic expression of RetSat in preadipocytes might promote differentiation. The oxidoreductase RetSat has been described as containing an N-terminal localization peptide that targets it to the endoplasmic reticulum (ER) and a dinucleotide-binding motif for binding of FAD or NAD/NADP (16, 27). We therefore generated mutants of RetSat lacking the signal peptide (RetSat-Mut1) or with 2 mutations within a highly conserved region of the dinucleotide-binding motif. One of these (E34A; corresponding to RetSat-Mut2: E96A) has been shown to reduce the FAD binding and enzyme activity of another oxidoreductase, the human monoamine oxidase B, by more than 95% (28). Mut3 has a deletion of 5 AS following E96 (RetSat-Mut3). The RetSat mutants are depicted in Fig. 4A. Expression of the WT and RetSat-Mut1 GFP fusion proteins in 293T cells confirmed the previously reported ER localization of RetSat by colocalization with the ER marker protein disulfide isomerase (PDI; Fig. S5A Top). In contrast, deletion of the signal peptide in RetSat-Mut1 led to cytoplasmic expression and loss of colocalization with PDI (Fig. S5A Bottom). We compared mRNA and protein expression of RetSat and Mut1–3 in transiently transfected 293T cells (data not shown) and retrovirally infected preconfluent 3T3-L1 preadipocytes. Although mRNA levels were comparable, we could detect only low amounts of RetSat-Mut1 protein (Fig. S5B). This suggests that ER localization and/or an intact signal peptide is important for RetSat protein stability in our expressed RetSat proteins void of the GFP fusion.
Fig. 4.
RetSat overexpression enhances differentiation depending on the integrity of the dinucleotide-binding site and increases PPARγ activity. (A) Schematic display of RetSat with known domains for subcellular localization and dinucleotide-binding and -generated mutations. (B and C) Subconfluent 3T3-L1 preadipocytes were infected with empty, WT-, or mutant RetSat-containing retroviruses and stimulated to differentiate under suboptimal conditions. (B) At day 2 of differentiation, cells were transiently transfected with a PPRE-containing luciferase construct and 24 h later assayed for luciferase activity. (C) After 8 days, adipocyte differentiation was assessed by Oil Red O staining and microscopy (Left) and protein expression for PPARγ and RAN (Right).
Mutations of the dinucleotide-binding motif had no influence on protein stability (Fig. S5B). We confirmed the lack of enzymatic activity of RetSat-Mut 2 and 3 by adenoviral overexpression in 293T cells and monitoring the generation of 13,14-dhretinol (Fig. 4C). We then treated infected 3T3-L1 preadipocytes with a suboptimal differentiation mix, which led to a low degree of differentiation in the control cells. Interestingly, at day 2 of differentiation the transcriptional activity of a PPRE-containing reporter gene that is responsive to PPARγ activation/antagonism under these conditions (data not shown) was significantly increased by ectopic expression of RetSat (Fig. 4B). In accordance with this finding, RetSat strongly increased differentiation under these conditions, as shown by Oil Red O staining and phase microscopy and the expression of PPARγ (Fig. 4C). Unlike WT RetSat, which enhanced adipogenesis, ectopic expression of the enzymatic mutants did not increase differentiation (Fig. 4C). We thus conclude that integrity of the dinucleotide-binding motif is essential for enhanced differentiation with ectopic RetSat expression during adipogenesis.
RetSat Gene Expression Is Downregulated in the Obese State and Induced by PPARγ Activation In Vivo.
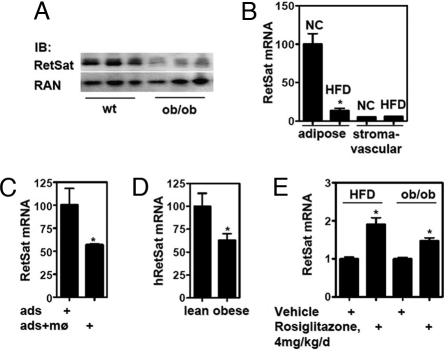
Increased adiposity in obese individuals results from an increase in both adipocyte number and size of individual fat cells (9). Given the important role of RetSat during adipocyte differentiation, we determined adipose RetSat expression in murine and human obesity. RetSat is highly expressed in mouse adipose tissue, at a level approximately 10%–25% of that in liver (Fig. S6A), where RetSat expression has been reported to be highest (16). We compared adipose tissue expression of RetSat in lean vs. obese and diabetic ob/ob mice (weight WT 32.7 ± 2.94 g and ob/ob 77.4 ± 6.75 g). Given the positive role of RetSat in adipogenesis, we predicted that RetSat expression would be higher in the obese mice. Surprisingly, however, RetSat protein expression in epididymal fat pads of obese mice was markedly reduced (Fig. 5A). To test whether the counterintuitive reduction in RetSat expression in ob/ob adipose tissue is an idiosyncratic effect of leptin deficiency in this monogenic obesity model, we next measured RetSat expression in a model of diet-induced obesity [weight on normal chow 29.7 ± 1.61 g vs. 47.9 ± 2.83 g on the high-fat diet]. Consistent with the expression of RetSat in the 3T3-L1 adipocytes, RetSat mRNA was expressed almost exclusively in the adipocyte fraction of epididymal adipose tissue (Figs. 5B and Fig. S6B). In comparison with animals receiving the control chow, mice with diet-induced obesity showed an approximately 80% reduction of RetSat expression in their adipocyte fraction (Fig. 5B).
Fig. 5.
RetSat gene expression is reduced in obese mice and humans, downregulated in adipocytes by coculture with primary macrophages in vitro, and TZD responsive in vivo. (A) Proteins from epididymal fat pads from wild-type (wt) and ob/ob mice were isolated and RetSat expression determined. (B) Adipose tissue from mice receiving normal chow (NC) or a high-fat diet (HFD) was fractionated and RetSat mRNA expression determined. (C) 3T3-L1 adipocytes were cocultured with or without primary macrophages for 48 h and adipocyte RetSat mRNA expression determined. (D) Abdominal s.c. adipose tissue samples from postmenopausal women were processed for RetSat mRNA expression. (E) Mice fed HFD or ob/ob mice were gavaged with either vehicle or rosiglitazone for 5 or 10 days and epididymal fat pad mRNA expression of RetSat determined.
A major difference between 3T3-L1 adipocytes and adipose tissue is that the latter contains multiple cell types. Adipose tissue from obese mice has been shown to contain an increase in macrophage infiltration (29, 30). In line with these observations, both our mice models showed a strong increase in the expression of inflammatory markers in the epididymal fat pad in the obese state (Fig. S6C). We hypothesized that macrophage infiltration could regulate adipocyte RetSat expression and tested this in vitro by coculturing adipocytes and macrophages. Indeed, the presence of primary macrophages reduced adipocyte RetSat mRNA expression by approximately 50% (Fig. 5C). Therefore, the decrease in RetSat expression in obesity could be related to the impact of inflammatory cells on adipocytes. Finally, we tested whether these findings regarding RetSat expression were pertinent to human obesity. RetSat gene expression was reduced by approximately 40% in adipose tissue from postmenopausal obese women compared with lean controls (body mass index lean 22.4 ± 1.77 and obese 40.8 ± 2.23) (Fig. 5D).
On the basis of the above findings, we hypothesized that TZD treatment could rescue RetSat expression in vivo by at least 2 different mechanisms: by direct activation of the RetSat PPRE and by decreasing macrophage infiltration/activation in adipose tissue. Indeed, rosiglitazone treatment of mice receiving a high-fat diet or ob/ob mice resulted in a significant increase in adipose RetSat mRNA expression after rosiglitazone treatment (Fig. 5E). As expected, rosiglitazone treatment of these mice also reduced the expression of TNFα in adipose tissue (Fig. S6D).
Discussion
We have discovered that RetSat is regulated by the nuclear receptor PPARγ in adipose tissue and that it is required for adipocyte differentiation. We show that the effect of RetSat ablation on adipocyte differentiation is unlikely to be due to effects on triglyceride storage or to its known enzymatic activity and that ectopic RetSat is able to enhance differentiation. Thus, RetSat has a novel function that interferes with adipocyte differentiation. Furthermore, this function is lost by mutating the dinucleotide-binding motif, suggesting that RetSat works as a FAD- or NAD/NADP-dependent oxidoreductase with a yet-unidentified substrate in this cellular context.
RetSat fulfills all of the criteria for a classic adipocyte PPARγ target gene: it is induced by PPARγ ligands in a manner that is blocked by antagonist; PPARγ is required for its expression in adipocytes; the RetSat gene contains a functional PPRE that binds recombinant PPARγ in vitro and endogenous PPARγ in living adipocytes; and RetSat is induced by TZDs in vivo. However, it is to our knowledge the only nontranscription factor and direct PPARγ target gene with an important role in differentiation. It could therefore be part of the signaling pathway and amplification loop that early on lead to activation and expression of PPARγ. Another oxidoreductase, xanthine oxidase, has been described as promoting adipocyte differentiation by playing a role in the generation of an endogenous but as-yet unidentified species that activates PPARγ (31). RetSat could be epistatic with this pathway, because provision of a synthetic PPARγ ligand overcame the effect of RetSat ablation. We have noted that knockdown of RetSat increased xanthine oxidase expression 2-fold in adipocytes (unpublished data), which could be a compensatory effect. Interestingly, xanthine oxidase has been shown to oxidize retinol (32) in addition to its known substrate, xanthine.
Szatmati et al. (33) have suggested that cross-talk between PPARγ and RARα signaling is a critical feature of human dendritic cells, where activation of PPARγ leads to an increased expression of enzymes involved in the formation of atRA and consequently higher cellular levels of atRA. We have also found that PPARγ ligands induce retinoid-metabolizing enzymes in adipocytes. Therefore, it is possible that RetSat plays a role in a novel retinoid biosynthetic pathway in adipocytes, such that its PPARγ-dependent induction leads to an unidentified retinoid(s) that promotes adipocyte differentiation.
While this article was in preparation, a study implicated RetSat in the cellular response to oxidative stress (34). This finding, which was not further related to retinoid metabolism, raises the possibility of the involvement of RetSat in the regulation of oxidative species. Whether these observations are connected to RetSat's role during adipogenesis has to be elucidated.
Considering the substantial expression of RetSat in adipocytes and its effect on differentiation, it is possible that RetSat plays an important functional role in mature adipose tissue. Although obesity is a cause of insulin resistance, in some settings expansion of adipose tissue is actually associated with insulin sensitization; these include administration of antidiabetic TZDs and transgenic expression of adiponectin (35). Conversely, ablation of PPARγ2 in ob/ob mice resulted in severe insulin resistance, despite a reduction in adipose tissue (36). Furthermore, several studies showed that adipocyte- or differentiation-related genes are downregulated in adipose tissue of obese or type 2 diabetic individuals (37–39) and that hypertrophic, older adipocytes are more insulin resistant than newly formed adipocytes (40), suggesting that adipocyte dysfunction contributes to these conditions. A potential cause for the loss of differentiation potential could be the increased infiltration of macrophages into adipose tissue in the obese state, supported by our experiments showing the effect of primary macrophages on an adipogenic gene in our coculture model. Our finding that RetSat expression is reduced in obese mice and obese humans supports this hypothesis and underlines its critical role in adipose tissue plasticity.
Materials and Methods
Mouse and Human Studies.
Sample preparation and human patient characteristics are described elsewhere (41). For comparing RetSat expression in lean and obese women, the number of samples was reduced to 10 per group to obtain extreme body mass index phenotypes. Specifics of murine samples are described in the SI Materials and Methods.
Cell Culture and Differentiation.
293A, BOSC23, and murine 3T3-L1 cells were maintained in DMEM supplemented with 10% FBS (U.S. Bio-Technologies), 100 U/mL penicillin, and 100 μg/mL streptomycin. 3T3-L1 cells were treated, differentiated, and cocultured with macrophages as described in the SI Materials and Methods.
Plasmid Cloning, Transfections and Luciferase Assay.
The RetSat promoter constructs and enhancer regions were cloned by using standard methods. Detailed procedures for cloning, transfections, and cell electroporations can be found in the SI Materials and Methods.
Chromatin Immunoprecipitation.
3T3-L1 preadipocytes or adipocytes were crosslinked with 1% formaldehyde for 10 min. After quenching with 2.5 M glycine, cells were harvested and resuspended in hypotonic lysis buffer (20 mM Hepes, 0.25 M sucrose, 3 mM MgCl2, 0.2% Nonidet P-40, 3 mM β-ME, and 0.2 mM PMSF and protease inhibitors) and homogenized. Nuclei lysis, immunoprecipitation, and purification are described in the SI Materials and Methods.
Retro- and Adenoviral Expression of RetSat and Mutants in 3T3-L1 Cells.
For retroviral expression of RetSat in preadipocytes, the coding sequence was cloned into a Murine Stem Cell Virus (pMSCV) vector. Adenoviral expression of RetSat and mutants in 293T cells or in mature adipocytes was carried out by using the Adeno-X Expression System 2 (Clontech). The generation of RetSat mutants and cell infection are described in the SI Materials and Methods.
Antibody Generation and Immunoblot Analysis.
A rabbit polyclonal antibody for RetSat was generated at Covance to a synthesized polypeptide corresponding to AS 470–488 of mRetSat. For immunoblot analysis, proteins were isolated and separated in 4%–20% SDS polyacrylamide gels and transferred to a polyvinylidene diflouride membrane (Invitrogen). Antibodies uses are described in the SI Materials and Methods.
Quantitative PCR.
RNA was purified with the RNeasy Mini Kit (Qiagen). cDNA was generated using the Sprint Powerscript System (Clontech; primers and probes are listed in Table S1). PCR reactions were carried out using either Taqman or Sybrgreen PCR Mastermix (Applied Biosystems) and the PRISM 7900 (Applied Biosystems) and evaluated by the standard curve method. Murine and human differentiation mRNA expression data were normalized to 36B4. Human adipose tissue biopsy mRNA expression was normalized to GAPDH.
Other Methods.
Detailed descriptions of electrophoretic mobility shift assays, cell retinoid extraction and HPLC analysis of retinoids and [3H]-retinol, generation of GFP-fusion proteins, Affymetrix gene expression arrays and analysis, and confocal microscopy are available in the SI Materials and Methods.
Statistical Analysis.
Statistical significance was determined using either the 2-tailed Student's t test or ANOVA, as appropriate, and P < 0.05 was deemed significant; experiment-specific evaluations are described in the SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Dr. M. Wabitsch (Pediatric Endocrinology, University of Ulm, Germany) for the SGBS cell line; R. Zipkin (Biomol) for the synthesis of 13,14-dhretinol, B. Norman and W. Chin (Lilly) for AGN193618, and T. Willson (GSK) for GW7845; D. Shao (Merck) for early work on RetSat; and A.R. Moise and K. Palczewski (Case Western Reserve University, Cleveland, OH) for helpful discussions. This work was supported by National Institutes of Health Grants R01 DK49780 (to M.A.L.) and R01 CA43796 (to L.J.G.). M.S. was supported by a Mentored Fellowship award from the American Diabetes Association, and J.C.C. was supported by National Institute of Diabetes and Digestive and Kidney Diseases training grants (5-F32-DK-070405 and T32-DK07314).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812065106/DCSupplemental.
References
- 1.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelleymounter MA, et al. Effects of the obese gene product on body weight regulation in ob/ob mice. Science. 1995;269:540–543. doi: 10.1126/science.7624776. [DOI] [PubMed] [Google Scholar]
- 3.Halaas JL, et al. Weight-reducing effects of the plasma protein encoded by the obese gene. Science. 1995;269:543–546. doi: 10.1126/science.7624777. [DOI] [PubMed] [Google Scholar]
- 4.Qatanani M, Lazar MA. Mechanisms of obesity-associated insulin resistance: Many choices on the menu. Genes Dev. 2007;21:1443–1455. doi: 10.1101/gad.1550907. [DOI] [PubMed] [Google Scholar]
- 5.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 6.Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- 7.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann JM, et al. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 9.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsutsumi C, et al. Retinoids and retinoid-binding protein expression in rat adipocytes. J Biol Chem. 1992;267:1805–1810. [PubMed] [Google Scholar]
- 11.O'Byrne SM, et al. Retinoid absorption and storage is impaired in mice lacking lecithin:retinol acyltransferase (LRAT) J Biol Chem. 2005;280:35647–33557. doi: 10.1074/jbc.M507924200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue JC, Schwarz EJ, Chawla A, Lazar MA. Distinct stages in adipogenesis revealed by retinoid inhibition of differentiation after induction of PPARgamma. Mol Cell Biol. 1996;16:1567–1575. doi: 10.1128/mcb.16.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarz EJ, et al. Retinoic acid blocks adipogenesis by inhibiting C/EBPbeta-mediated transcription. Mol Cell Biol. 1997;17:1552–1561. doi: 10.1128/mcb.17.3.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziouzenkova O, et al. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat Med. 2007;13:695–702. doi: 10.1038/nm1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Q, et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436:356–362. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- 16.Moise AR, Kuksa V, Imanishi Y, Palczewski K. Identification of all-trans-retinol:all-trans-13,14-dihydroretinol saturase. J Biol Chem. 2004;279:50230–50242. doi: 10.1074/jbc.M409130200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cobb JE, et al. N-(2-Benzoylphenyl)-L-tyrosine PPARgamma agonists. 3. Structure-activity relationship and optimization of the N-aryl substituent. J Med Chem. 1998;41:5055–5069. doi: 10.1021/jm980414r. [DOI] [PubMed] [Google Scholar]
- 18.Camp HS, Chaudhry A, Leff T. A novel potent antagonist of peroxisome proliferator-activated receptor gamma blocks adipocyte differentiation but does not revert the phenotype of terminally differentiated adipocytes. Endocrinology. 2001;142:3207–3213. doi: 10.1210/endo.142.7.8254. [DOI] [PubMed] [Google Scholar]
- 19.Liao W, et al. Suppression of PPAR-gamma attenuates insulin-stimulated glucose uptake by affecting both GLUT1 and GLUT4 in 3T3–L1 adipocytes. Am J Physiol Endocrinol Metab. 2007;293:E219–E227. doi: 10.1152/ajpendo.00695.2006. [DOI] [PubMed] [Google Scholar]
- 20.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 22.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Moise AR, et al. Stereospecificity of retinol saturase: Absolute configuration, synthesis, and biological evaluation of dihydroretinoids. J Am Chem Soc. 2008;130(4):1154–1155. doi: 10.1021/ja710487q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeh WC, Cao Z, Classon M, McKnight SL. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 1995;9:168–181. doi: 10.1101/gad.9.2.168. [DOI] [PubMed] [Google Scholar]
- 25.Wu Z, Xie Y, Bucher NL, Farmer SR. Conditional ectopic expression of C/EBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev. 1995;9:2350–23563. doi: 10.1101/gad.9.19.2350. [DOI] [PubMed] [Google Scholar]
- 26.Tzameli I, et al. Regulated production of a peroxisome proliferator-activated receptor-gamma ligand during an early phase of adipocyte differentiation in 3T3–L1 adipocytes. J Biol Chem. 2004;279:36093–36102. doi: 10.1074/jbc.M405346200. [DOI] [PubMed] [Google Scholar]
- 27.Dailey TA, Dailey HA. Identification of an FAD superfamily containing protoporphyrinogen oxidases, monoamine oxidases, and phytoene desaturase. Expression and characterization of phytoene desaturase of Myxococcus xanthus. J Biol Chem. 1998;273:13658–13662. doi: 10.1074/jbc.273.22.13658. [DOI] [PubMed] [Google Scholar]
- 28.Zhou BP, Wu B, Kwan SW, Abell CW. Characterization of a highly conserved FAD-binding site in human monoamine oxidase B. J Biol Chem. 1998;273:14862–14868. doi: 10.1074/jbc.273.24.14862. [DOI] [PubMed] [Google Scholar]
- 29.Weisberg SP, et al. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu H, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheung KJ, et al. Xanthine oxidoreductase is a regulator of adipogenesis and PPARgamma activity. Cell Metab. 2007;5:115–128. doi: 10.1016/j.cmet.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Taibi G, Nicotra CM. Xanthine oxidase catalyzes the oxidation of retinol. J Enzyme Inhib Med Chem. 2007;22:471–476. doi: 10.1080/14756360701408739. [DOI] [PubMed] [Google Scholar]
- 33.Szatmari I, et al. PPARgamma controls CD1d expression by turning on retinoic acid synthesis in developing human dendritic cells. J Exp Med. 2006;203:2351–2362. doi: 10.1084/jem.20060141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagaoka-Yasuda R, et al. An RNAi-based genetic screen for oxidative stress resistance reveals retinol saturase as a mediator of stress resistance. Free Radic Biol Med. 2007;43:781–788. doi: 10.1016/j.freeradbiomed.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Kim JY, et al. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J Clin Invest. 2007;117:2621–2637. doi: 10.1172/JCI31021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medina-Gomez G, et al. PPAR gamma 2 prevents lipotoxicity by controlling adipose tissue expandability and peripheral lipid metabolism. PLoS Genet. 2007;3:e64. doi: 10.1371/journal.pgen.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dubois SG, et al. Decreased expression of adipogenic genes in obese subjects with type 2 diabetes. Obesity (Silver Spring) 2006;14:1543–1552. doi: 10.1038/oby.2006.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sewter C, et al. Human obesity and type 2 diabetes are associated with alterations in SREBP1 isoform expression that are reproduced ex vivo by tumor necrosis factor-alpha. Diabetes. 2002;51:1035–1041. doi: 10.2337/diabetes.51.4.1035. [DOI] [PubMed] [Google Scholar]
- 39.Yang X, et al. Evidence of impaired adipogenesis in insulin resistance. Biochem Biophys Res Commun. 2004;317:1045–1051. doi: 10.1016/j.bbrc.2004.03.152. [DOI] [PubMed] [Google Scholar]
- 40.Hissin PJ, et al. Mechanism of insulin-resistant glucose transport activity in the enlarged adipose cell of the aged, obese rat. J Clin Invest. 1982;70:780–790. doi: 10.1172/JCI110674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engeli S, et al. Association between adiponectin and mediators of inflammation in obese women. Diabetes. 2003;52:942–947. doi: 10.2337/diabetes.52.4.942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.