Abstract
A surprising example of interspecies competition is the production by certain bacteria of hydrogen peroxide at concentrations that are lethal for others. A case in point is the displacement of Staphylococcus aureus by Streptococcus pneumoniae in the nasopharynx, which is of considerable clinical significance. How it is accomplished, however, has been a great mystery, because H2O2 is a very well known disinfectant whose lethality is largely due to the production of hyperoxides through the abiological Fenton reaction. In this report, we have solved the mystery by showing that H2O2 at the concentrations typically produced by pneumococci kills lysogenic but not nonlysogenic staphylococci by inducing the SOS response. The SOS response, a stress response to DNA damage, not only invokes DNA repair mechanisms but also induces resident prophages, and the resulting lysis is responsible for H2O2 lethality. Because the vast majority of S. aureus strains are lysogenic, the production of H2O2 is a very widely effective antistaphylococcal strategy. Pneumococci, however, which are also commonly lysogenic and undergo SOS induction in response to DNA-damaging agents such as mitomycin C, are not SOS-induced on exposure to H2O2. This is apparently because they are resistant to the DNA-damaging effects of the Fenton reaction. The production of an SOS-inducing signal to activate prophages in neighboring organisms is thus a rather unique competitive strategy, which we suggest may be in widespread use for bacterial interference. However, this strategy has as a by-product the release of active phage, which can potentially spread mobile genetic elements carrying virulence genes.
Keywords: hydrogen peroxide, SOS response, Staphylococcus aureus, Streptococcus pneumoniae, bacterial interference
The interactions among bacteria living communally are highly complex and extremely interesting—illuminating, as they do, a long-ignored but nevertheless critical aspect of microbial biology. One can readily envision interactions such as direct competition for scarce nutrients, mutual cooperation for the conversion of substrates to utilizable metabolites, “borrowing” of quorum-sensing signals, DNA transfer, biofilm formation and maintenance, and interference or inhibition mediated by antibacterial products, including bacteriocins, antibiotics, and low-molecular-weight toxic compounds such as H2O2 (1). In this article, we consider a specific case of H2O2-mediated bacterial interference, that between pneumococci and Staphylococcus aureus, which, although well documented, occurs by an entirely unknown mechanism.
Several epidemiological studies have shown a negative association between carriage of Streptococcus pneumoniae and S. aureus (2, 3), raising public health concern that mass pneumococcal vaccination may cause an increase in S. aureus colonization and infection. As a case in point, it has been reported that children with recurrent otitis media vaccinated with the heptavalent pneumococcal vaccine had increased incidence of S. aureus-related acute otitis media and S. aureus colonization (3). Recent in vitro and in vivo studies have demonstrated that the interference between these 2 pathogens is related to hydrogen peroxide production by S. pneumoniae, which is bactericidal to S. aureus (4, 5). Similar observations have been reported for certain other pairs of bacteria (6). It is highly intriguing how the relatively low levels of hydrogen peroxide produced safely by some bacteria are bactericidal to others, despite the relative abundance of mechanisms protecting bacterial cells from oxidative damage, such as H2O2-inactivating enzymes and antioxidants (7) or DNA lesion repair systems (8).
Here, we shed light on the mechanism of interference between H2O2-producing bacteria and S. aureus. We present data supporting the idea that prophages may have a much greater role in bacterial ecology than has hitherto been suspected—namely, that killing of a target organism by “remote control” prophage induction may represent a major modality of directional bacterial interference. We show also that lysogenic staphylococci are much more sensitive to DNA-damaging antibiotics, such as fluoroquinolones, than nonlysogens, almost certainly for the same reason. Given the high prevalence of lysogeny, we can now predict that small, SOS-inducing molecules, produced in the environment at subinhibitory concentrations, may have strong selective value as effectors of directional interference.
Results
Hydrogen Peroxide Kills Only Lysogenic S. aureus.
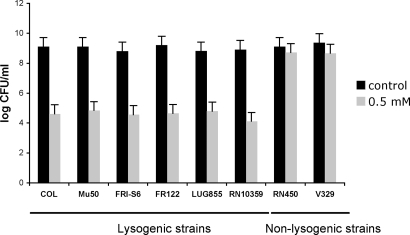
Several species of bacteria have H2O2-dependent bactericidal activity toward S. aureus (4, 9). However, the mechanism by which the relatively low levels of H2O2 produced by these organisms are bactericidal to S. aureus remains to be determined. One possibility is that H2O2 produced by one organism induces the SOS response in a competing (target) organism, lethally activating resident prophages in the latter. If so, staphylococcal lysogens but not nonlysogens should be sensitive to H2O2 and pneumococci should be insensitive, even though they are often or always lysogenic. Accordingly, we tested 8 strains of S. aureus, 6 lysogenic, 2 nonlysogenic (RN450 and V329), including a congenic pair in which one (RN10359) was an 80α lysogen of the other (RN450) and a strain producing the phage-carried PVL toxin (strain LUG855), lately implicated in serious staphylococcal infections [10; supporting information (SI) Table S1]. We used H2O2 at 0.5 mM, in the range ordinarily seen with pneumococcal cultures, and observed that all of the lysogenic strains were highly sensitive, whereas the nonlysogens were insensitive (see Fig. 1), consistent with the above prediction.
Fig. 1.
Killing of S. aureus by H2O2. Survival of lysogenic (COL, Mu50, FRI-S6, RF122, LUG855, and RN10359) or nonlysogenic (V329, RN450) S. aureus strains in media supplemented with hydrogen peroxide at a concentration of 0.5 mM, and unsupplemented medium (control). Values represent the average of 3 independent experiments. Variation was within ± 5% in all cases.
H2O2 Induces SOS in S. aureus.
It is well known that H2O2 induces the SOS response in Escherichia coli and other bacteria (8), and it is strongly predicted that it would do so in S. aureus, and that this would be responsible for H2O2 killing of lysogens, although no clear test of this has been published. Accordingly, we tested the same lysogenic strains as in Fig. 1, for H2O2 induction of prophages. Because recA-dependent prophage induction is a highly typical feature of the SOS response, we included RN1030, a lysogenic recA-defective derivative of RN451. Because cleavage of the prophage repressor is also necessary for prophage induction, we also included JP3592, an 80α lysogen with a noncleavable mutant phage repressor. As shown in Tables 1 and S2, H2O2 induced the prophages of all strains tested, in a dose-dependent manner, with the exception of RN1030, and JP3592. However, the response to H2O2 was quite variable, perhaps owing to differences in the response to oxidative stress. Phage induction by H2O2 was, as one would expect, accompanied by a corresponding decrease in viability, as also shown in Tables 1 and S2. No reduction in viability was seen with the nonlysogen or with the phage repressor mutant, except for a slight reduction in viability owing to direct H2O2 toxicity at the highest H2O2 concentration used.
Table 1.
Phage titer and survival of H2O2-induced lysogenic staphylococcal strains
| Donor strain | [H2O2], mM | CFU* | Phage titer† |
|---|---|---|---|
| RN450 | NI | 8.4 × 108 | – |
| 0.05 | 7.1 × 108 | – | |
| 0.1 | 2.9 × 108 | – | |
| 0.5 | 1.8 × 108 | – | |
| 1 | 6.3 × 107 | – | |
| RN10359 | NI | 5.9 × 108 | 8.8 × 105 |
| (RN450 lysogenic for 80α) | 0.05 | 1.2 × 109 | 1.9 × 106 |
| 0.1 | 2.9 × 107 | 9.0 × 107 | |
| 0.5 | 1.8 × 104 | 5.3 × 109 | |
| 1 | 5.6 × 103 | 2.0 × 108 | |
| JP3592 | NI | 6.7 × 108 | <10 |
| (RN10359 cI G130E) | 0.05 | 1.8 × 108 | <10 |
| 0.1 | 1.9 × 108 | <10 | |
| 0.5 | 1.8 × 108 | <10 | |
| 1 | 0.8 × 108 | <10 | |
| RN451 | NI | 5.9 × 108 | 2.8 × 104 |
| (RN450 lysogenic for φ11) | 0.05 | 4.2 × 108 | 4.9 × 104 |
| 0.1 | 4.9 × 107 | 3.1 × 105 | |
| 0.5 | 3.8 × 105 | 2.8 × 106 | |
| 1 | 5.6 × 104 | 8.9 × 105 | |
| RN1030 | NI | ND | <10 |
| (RN451 recA-mutant) | 0.05 | ND | <10 |
| 0.1 | ND | <10 | |
| 0.5 | ND | <10 | |
| 1 | ND | <10 |
The means of results from 3 independent experiments are presented. Variation was within ± 5% in all cases. NI, not induced; ND, not determined.
*Number of cells recovered after 4 h of incubation.
†Number of plaque-forming phages per milliliter of induced culture, using RN4220 as indicator strain.
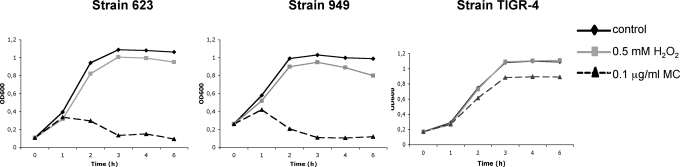
As would be expected for the producer of an antibacterial substance, S. pneumoniae is entirely resistant to H2O2 at the concentrations present in its cultures, and it has been shown that this is because it is insensitive to the toxic products of the Fenton reaction (11). Because these products cause DNA damage, which is responsible for SOS induction, we hypothesized that H2O2 would not induce the SOS response in S. pneumoniae and therefore that even lysogenic S. pneumoniae would be able to use H2O2 to interfere with competing species. To test this hypothesis, we compared lysogenic and nonlysogenic S. pneumoniae for sensitivity to H2O2 and mitomycin C (MC). As shown in Fig. 2, neither was sensitive to H2O2; however, the lysogens (strains 623 and 949), but not the nonlysogen (TIGR-4) were sensitive to SOS induction by MC, leading to lysis and, presumably, to the release of active phage, although we have not confirmed this in the present study. Nevertheless, it is clear that neither pneumococcal lysogens nor nonlysogens are sensitive to H2O2 at concentrations normally present in pneumococcal cultures and that H2O2 does not induce the SOS response in pneumococci, which accounts for the ability of this organism to produce H2O2 for use as a weapon of mass destruction with impunity.
Fig. 2.
Lysis of S. pneumoniae induced by mitomycin C or H2O2. Cultures of lysogenic (623 and 949) or nonlysogenic (TIGR4) strains received mitomycin C (0.1 μg/mL) or H2O2 (0.5 mM) at time 0, and the OD of cultures was monitored at 600 nm.
Effect of H2O2 Production by S. pneumoniae.
To confirm that H2O2 is responsible for the killing of S. aureus by S. pneumoniae, we cultured a lysogenic, H2O2-sensitive, S. aureus strain (RN10395) with 2 S. pneumoniae strains, TIGR4 and Pn-20. It has reported that these strains naturally produce H2O2 (4), with full-grown cultures (≈107 cfu/mL) containing ≈1 mM hydrogen peroxide (4). As shown in Table 2, H2O2-producing S. pneumoniae had the expected effects in coculture with the 80α WT lysogen—phage induction and loss of viability. We also tested mutants of TIGR4 and Pn-20 with deletions of the pyruvate oxidase gene, spxB, required for H2O2 production (4). Neither of the ΔspxB mutants had any effect on viability or prophage induction with lysogenic S. aureus. Further evidence was provided by a test with the phage 80α mutant defective in the ability to cleave its prophage repressor, and therefore, not SOS inducible. This mutant was as insensitive to H2O2 as the nonlysogens tested.
Table 2.
Effects of coculture of S. aureus with H2O2-producing S. pneumoniae
| Donor strain | Inducer* | CFU† | Phage titer‡ |
|---|---|---|---|
| RN10359 | H2O2 | 7.0 × 105 | 5.0 × 109 |
| (RN450 lysogenic for 80α) | Pn-20 | 4.6 × 105 | 8.7 × 107 |
| Pn-20 ΔspxB | 1.4 × 109 | 3.3 × 105 | |
| TIGR4 | 4.2 × 106 | 5.9 × 107 | |
| TIGR4 ΔspxB | 1.2 × 109 | 5.3 × 105 | |
| NI | 1.9 × 109 | 4.6 × 105 | |
| RN450 | H2O2 | 7.3 × 108 | – |
| Pn-20 | 7.8 × 108 | – | |
| Pn-20 ΔspxB | 1.1 × 109 | – | |
| TIGR4 | 1.2 × 109 | – | |
| TIGR4 ΔspxB | 1.0 × 109 | – | |
| NI | 1.1 × 109 | – | |
| JP3592 | H2O2 | 5.3 × 108 | <10 |
| (RN10359 cI G130E) | Pn-20 | 8.8 × 108 | <10 |
| Pn-20 ΔspxB | 2.1 × 108 | <10 | |
| TIGR4 | 1.2 × 108 | <10 | |
| TIGR4 ΔspxB | 9.7 × 108 | <10 | |
| NI | 1.0 × 109 | <10 |
The means of results from 3 independent experiments are presented. Variation was within ± 5% in all cases.
*H2O2 (0.5 mM). NI, not induced.
†Number of cells recovered after 4 h of incubation.
‡Number of plaque-forming phages per milliliter of induced culture, using RN4220 as indicator strain.
Role of Catalase in Hydrogen Peroxide SOS Induction.
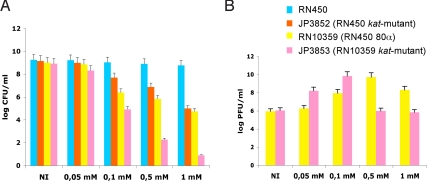
Because S. aureus produces a potent catalase as a defense against H2O2 toxicity, it seemed paradoxical that it would nevertheless be susceptible to SOS induction of prophages by such low concentrations of H2O2. To evaluate the role of catalase in SOS induction by H2O2, we constructed katA mutants of RN450 (JP3852) and RN10359 (JP3853) by transducing the katA mutation from strain KS100 (7). The 2 katA mutants and their katA+ parents were incubated for 6 h in TSB broth with various concentrations of H2O2. As shown in Fig. 3, KatA strongly protected the nonlysogenic RN450 at the higher H2O2 concentrations, but had a much less dramatic effect on the congenic lysogen, RN10359, both with respect to viability and to phage production, suggesting that KatA is much more strongly protective of general viability in the presence of H2O2 than of susceptibility to SOS induction. This may be because peroxide lethality involves a wide range of oxidative damages, whereas SOS induction involves only the RecA-LexA pathway, which is induced by only minimal DNA damage, much less than that necessary to kill the cell. Perhaps catalase does not act rapidly enough or efficiently enough at these lower concentrations to prevent minimal DNA damage. This suggests that general toxicity and SOS induction by H2O2 may target different processes, and that KatA can protect a nonlysogen but not a lysogen against H2O2 killing at relevant concentrations. Additionally, the lower phage production by the kat-mutant at high concentrations of H2O2 is probably the result of decreased capacity of the cells to support phage growth.
Fig. 3.
Role of catalase in SOS induction by H2O2. (A) Survival of S. aureus strains in media supplemented with hydrogen peroxide at different concentrations. Values represent the average of 3 independent experiments. Variation was within ±5% in all cases. (B) Phage titer obtained from the lysogenic strains analyzed in A.
Hydrogen Peroxide Induces Phage-Mediated SaPI Transfer.
A by-product of H2O2 killing of lysogens by SOS induction is, of course, the release of viable phage into the environment. This could promote the spread of phage-encoded toxins, such as PVL, and of phage-related pathogenicity islands such as the SaPIs, which encode TSST-1 and other superantigen toxins (12). We have presented data on H2O2 induction of a PVL-encoding phage (Table S2); induction of SaPI particle production by coculture with S. pneumoniae is presented in Table S3.
Discussion
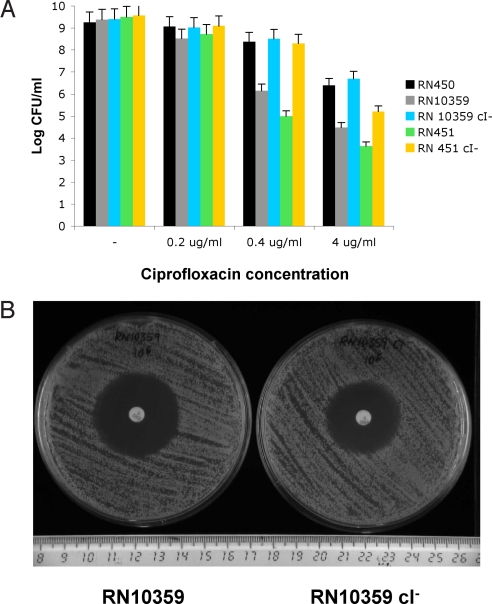
In this report, we have examined the widespread strategy of bacterial interference by the production of H2O2, demonstrating, in the case of S. pneumoniae vs. S. aureus that it acts through the lethal induction of resident prophages in the target organism. We note that our results play deeply into the overall universe of bacteria and phages. Lysogeny is extremely common among bacteria, representing a major symbiotic strategy in which the prophage, residing stably in the host's chromosome, provides protection against attack by other phages, not only through coimmunity, but also by encoding restriction-modification capabilities or other more general protective mechanisms. And prophages may also enhance the virulence of pathogens. A consequence of lysogeny is, of course, inherent in the definition, and spontaneous lysis, a universal feature of lysogens, probably has an important role in phage-mediated lateral gene transfer. Nevertheless, lysogeny is, after all, an Achilles heel for the organism as the prophage state causes vulnerability to environmental influences such as radiation and certain chemicals that cause (sublethal) DNA damage, invoking the SOS response, a stress response whose objective is to repair damaged DNA. Prophage repressors are generally inactivated during the SOS response, setting in motion the phage lytic cycle; although the biological purpose of this is not entirely obvious, it would provide a wonderful target for aggression by competing organisms, if only they knew how to cause DNA damage and thus to induce prophages. As we have shown here, the production of H2O2 at sublethal concentrations serves this purpose admirably. Biochemically, H2O2 is easily produced by the oxidation of pyruvate, generates DNA-damaging hyperoxides through the Fenton reaction (13), and thereby induces the SOS response in many bacteria. As is the case with all antibacterial substances, the producing organism is immune. With antibiotics, the immune mechanism is usually obvious. With H2O2, the mechanism of immunity is not so obvious because it involves resistance to the toxic products of the Fenton reaction (11), including, as we show here, resistance to induction of the SOS response. Pneumococci do not produce catalase, which inactivates H2O2 and could compromise the use of H2O2 as a weapon; perhaps this is precisely why the pneumococci lack catalase. And we would predict that other H2O2-producing organisms also lack catalase for the same reason. Given the occurrence of H2O2-mediated bacterial interference based on lethal prophage induction in the target organism, we note that DNA-damaging antibiotics have recently been shown to induce prophages and thereby to promote the spread of phage-coded toxins and of phage-related pathogenicity islands (14–16). One would therefore predict that lysogenic bacteria would be more sensitive to the bactericidal effects of DNA-damaging antibiotics than nonlysogens, owing to lysis caused by SOS-induced prophages in the former, as we have demonstrated above for H2O2. As shown in Fig. 4, two different lysogenic derivatives of S. aureus strain RN450, RN451 and RN10359, lysogenic for φ11 and 80α, respectively, were considerably more sensitive to ciprofloxacin (Fig. 4A) than their nonlysogenic parent and this sensitivity was eliminated in both cases by the phage repressor cleavage defect. Similarly, the noncleavable repressor mutation significantly reduced sensitivity to enrofloxacin (Fig. 4B). In a more general sense, we would predict that lysogenic bacteria would be more sensitive than nonlysogens to interference by antibiotic producers in the environment, where the antibiotic concentrations would surely be very low. This remains to be tested.
Fig. 4.
Killing of lysogenic S. aureus cells by antibiotics. (A) Survival S. aureus strains in media supplemented with different concentrations of the antibiotic ciprofloxacin. RN450, nonlysogenic; RN10359, RN450 lysogenic for phage 80α; RN10359 cI−, derivative of RN10359 carrying a non-SOS-inducible phage 80α; RN451, RN450 lysogenic for phage 11; RN451 cI−, derivative of RN451 carrying a non-SOS-inducible phage 11. Values represent the average of 3 independent experiments. Variation was within ± 5% in all cases. (B) Antibiogram showing the different susceptibility between strains RN10359 (lysogenic for phage 80α) and RN10359 cI− (carrying a non-SOS-inducible phage 80α) to enrofloxacin discs. Average zone of inhibition: RN10359, 36 mm; RN10359 cI−, 30 mm.
Materials and Methods
Bacterial Strains and Growth Conditions.
Bacterial strains used in these studies are listed in Table S1. Bacteria were grown at 37 °C overnight on TSA agar medium (S. aureus) or BHI with 1.5% agar (S. pneumoniae), supplemented with antibiotics as appropriate. Broth cultures were grown at 37 °C in TSB (S. aureus) or Todd Hewitt (S. pneumoniae).
Induction of Prophages.
Procedures for preparation and analysis of phage lysates, transduction, and transformation in S. aureus were performed essentially as described in refs. 17 and 18. In general, bacteria were grown in TSB to OD540 = 0.15 and induced by the addition of H2O2 at various concentrations and cultures were continued at 32 °C with slow shaking (80 rpm). Presence of the lysis was evaluated within 3 h by using previously applied criteria (17).
Coculture Assay.
Interference between S. aureus and S. pneumoniae strains was measured in coculture assay as described in ref. 4. In brief, 1 mL of the S. pneumoniae cultures (OD540 0.4–0.5) were mixed with 1 mL of the S. aureus cultures (OD540 0.3–0.4), and incubated 4 h at 37 °C. To quantify bactericidal activity, the cocultured bacterial strains were plated on selective medium (TSA supplemented with 5 μg/mL optochin), where only S. aureus can grow.
DNA Methods.
General DNA manipulations were performed by standard procedures (19, 20). Strain JP3592 was obtained by using plasmid pMAD, as described in ref. 15. The oligonucleotides pairs used were phi11–1cB (5′-cgcggatccAGTGTTAATGTGTATATGCTC-3′)/phi11–3c (5′-TAATTCTTCTCCTATCTCAGCACCAGTTGCACC-3) and phi80alpha-cI-9mE (5′- CCGGAATTCCACAGATTCGTTTTATTTCCC-3′)/phi11–4m (5′-GGTGCAACTGGTGCTGAGATAGGAGAAGAATTA-3′).
Supplementary Material
Acknowledgments.
We thank Simon Foster, Gabriela Bowden, and Pedro García for providing bacterial strains used in this study and Jeff Weiser, Simon Foster, Adela González de la Campa, and Alex Mira for helpful comments on the manuscript. This work was supported by Comisión Interministerial de Ciencia y Tecnología Grants BIO2005-08399-C02-02, BIO2008-05284-C02-02, and BIO2008-00642-E/C; Cardenal Herrera-CEU University Grants PRCEU-UCH25/08 and Copernicus program; and by Conselleria de Agricultura, Pesca i Alimentació (CAPiA), and from the Generalitat Valenciana (ACOMP07/258) (J.R.P.). L.S. and D.V. were supported by Cardenal Herrera-CEU University fellowships.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809600106/DCSupplemental.
References
- 1.Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Regev-Yochay G, et al. Association between carriage of Streptococcus pneumoniae and Staphylococcus aureus in Children. JAMA. 2004;292:716–720. doi: 10.1001/jama.292.6.716. [DOI] [PubMed] [Google Scholar]
- 3.Bogaert D, et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet. 2004;363:1871–1872. doi: 10.1016/S0140-6736(04)16357-5. [DOI] [PubMed] [Google Scholar]
- 4.Regev-Yochay G, Trzcinski K, Thompson CM, Malley R, Lipsitch M. Interference between Streptococcus pneumoniae and Staphylococcus aureus: In vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J Bacteriol. 2006;188:4996–5001. doi: 10.1128/JB.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park B, Nizet V, Liu GY. Role of Staphylococcus aureus catalase in niche competition against Streptococcus pneumoniae. J Bacteriol. 2008;190:2275–2278. doi: 10.1128/JB.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pericone CD, Overweg K, Hermans PW, Weiser JN. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect Immun. 2000;68:3990–3997. doi: 10.1128/iai.68.7.3990-3997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosgrove K, et al. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J Bacteriol. 2007;189:1025–1035. doi: 10.1128/JB.01524-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goerlich O, Quillardet P, Hofnung M. Induction of the SOS response by hydrogen peroxide in various Escherichia coli mutants with altered protection against oxidative DNA damage. J Bacteriol. 1989;171:6141–6147. doi: 10.1128/jb.171.11.6141-6147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strauss E. Microbiology. Fighting bacterial fire with bacterial fire. Science. 2000;290:2231–2233. doi: 10.1126/science.290.5500.2231a. [DOI] [PubMed] [Google Scholar]
- 10.Labandeira-Rey M, et al. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science. 2007;315:1130–1133. doi: 10.1126/science.1137165. [DOI] [PubMed] [Google Scholar]
- 11.Pericone CD, Park S, Imlay JA, Weiser JN. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the fenton reaction. J Bacteriol. 2003;185:6815–6825. doi: 10.1128/JB.185.23.6815-6825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novick RP, Subedi A. The SaPIs: Mobile pathogenicity islands of Staphylococcus. Chem Immunol Allergy. 2007;93:42–57. doi: 10.1159/000100857. [DOI] [PubMed] [Google Scholar]
- 13.Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, et al. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J Infect Dis. 2000;181:664–670. doi: 10.1086/315239. [DOI] [PubMed] [Google Scholar]
- 15.Maiques E, et al. {beta}-Lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J Bacteriol. 2006;188:2726–2729. doi: 10.1128/JB.188.7.2726-2729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ubeda C, et al. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol Microbiol. 2005;56:836–844. doi: 10.1111/j.1365-2958.2005.04584.x. [DOI] [PubMed] [Google Scholar]
- 17.Novick RP. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 18.Lindsay JA, Ruzin A, Ross HF, Kurepina N, Novick RP. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol Microbiol. 1998;29:527–543. doi: 10.1046/j.1365-2958.1998.00947.x. [DOI] [PubMed] [Google Scholar]
- 19.Ausubel FM, et al. Current Protocols in Molecular Biology. New York: Wiley; 1990. [Google Scholar]
- 20.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.