Abstract
Gram-negative bacteria use specific heme uptake systems, relying on outer membrane receptors and excreted heme-binding proteins (hemophores) to scavenge and actively transport heme. To unravel the unknown molecular details involved, we present 3 structures of the Serratia marcescens receptor HasR in complex with its hemophore HasA. The transfer of heme over a distance of 9 Å from its high-affinity site in HasA into a site of lower affinity in HasR is coupled with the exergonic complex formation of the 2 proteins. Upon docking to the receptor, 1 of the 2 axial heme coordinations of the hemophore is initially broken, but the position and orientation of the heme is preserved. Subsequently, steric displacement of heme by a receptor residue ruptures the other axial coordination, leading to heme transfer into the receptor.
Keywords: heme binding, iron uptake, membrane protein, membrane transport, protein complex
Iron is essential for most living organisms. Abundant on earth, it is nevertheless insoluble under aerobic conditions. Reduced iron, on the other hand, is soluble but highly toxic, because it promotes the formation of reactive oxygen species. As a consequence, iron solubilization and uptake are strictly regulated. To retrieve the ferric iron, most microbes excrete potent Fe3+ chelating compounds called siderophores.
In Gram-negative bacteria, ferrisiderophore uptake through the outer membrane involves their binding to specific outer membrane receptors. This is followed by active transport using energy derived from proton motive force transduced by the inner membrane TonB complex. Energy transduction by the TonB complex relies on the recognition of the “TonB box,” a short stretch of weakly conserved residues of the receptor. This stretch is exposed to the periplasm and involved in a β-strand exchange with the C-terminal periplasmic domain of TonB. Although the TonB protein is often unique and highly conserved across species (1), multiple ferrisiderophore receptors, which reflect the diversity of siderophore chemical nature and share a common 3D structure, exist within a single bacterial species.
Bacteria can also use iron sources other than ferrisiderophores such as iron carried by ferriproteins [transferrin (2) or lactoferrin (3)], heme, hemoproteins (4) (hemoglobin, hemopexin, etc.), and hemophores (small bacterial extracellular proteins that scavenge heme owing to their very high affinity) (5, 6). This raises the question of how heme/iron that is often very tightly bound to its carrier protein is transferred to the receptor.
The Serratia marcescens hemophore HasA, a 188-residue protein, has a very high affinity for heme (Kd = 18 pM), among the strongest in heme-binding proteins (7). It consists of a β-sheet layer and a layer of 4 α-helices. The heme iron is bound by axial coordination with His-32 and Tyr-75, which reside in opposing loops at the edge of each layer (8). The receptor of HasA, HasR, can internalize both free or hemophore-bound heme into the periplasm (9). It binds 1 heme molecule with weaker affinity than HasA (Kd = 0.2 μM) (10) via 2 essential histidine residues that are mostly conserved in the heme receptor family (4). HasA complexed with heme (holoHasA) as well as without (apoHasA) forms tight complexes with HasR (11) (in vivo apparent Kd = 7 nM). The high affinities of the hemophore for both heme and the receptor allow hemophore-driven heme acquisition to proceed at lower concentrations than with free heme. When holoHasA binds to HasR, heme is spontaneously transferred to the HasR heme-binding site (10). Energy derived from the proton motive force and transduced by the TonB complex is not required for this process, but for subsequent heme entry into the periplasm and apoHasA dissociation from the receptor (12). HasB, a specialized TonB paralog of the Has system, can serve as an energy transducer to the receptor (13). HasR belongs to the class of receptors subject to transcriptional autoregulation with an additional N-terminal extension. The signaling stimulus is the presence of both HasA and heme bound to the receptor (14, 15).
Precise understanding of HasA binding, heme transfer from HasA to HasR, triggering of the signaling cascade, heme uptake, and apoHasA release requires structural information. Until now we have determined crystal structures of 2 ternary complexes HasA∼HasR∼heme, with wild-type HasR plus a mutant, and of the binary complex HasA∼HasR at 2.7, 2.7, and 2.8-Å resolution, respectively.
Results
The wild-type ternary complex was solved by MAD using anomalous scattering from 8 selenomethionines and the 2 other structures by difference Fourier methods. The final models consist of 752 residues from HasR (the N-terminal extension involved in signal transduction and transcriptional autoregulation, as well as the following TonB box, are not seen in the structure), 161 from HasA and, in case of the ternary complexes, 1 additional b-type heme (see Methods).
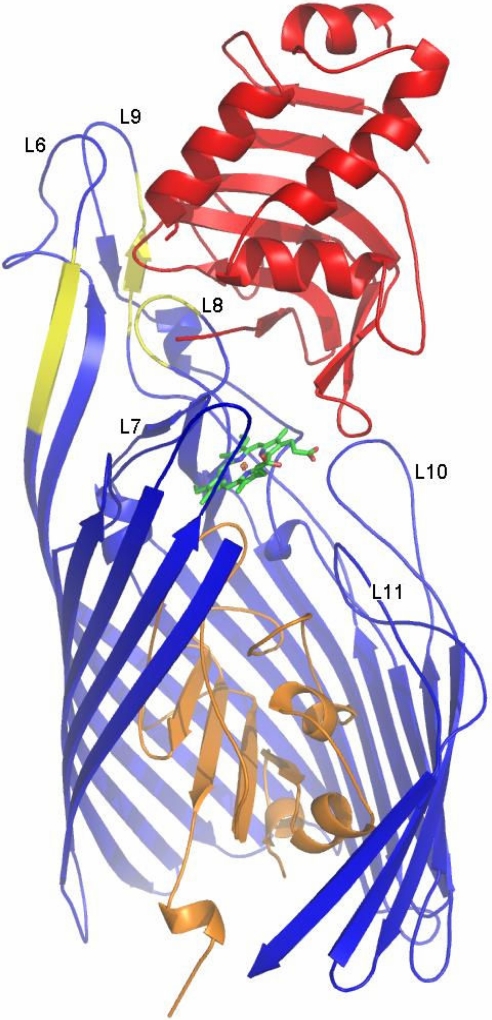
The HasR receptor complies to the overall structure of TonB dependent receptors with a C-terminal β-barrel consisting of 22 antiparallel β-strands filled partially by the N-terminal “cork” (also called “plug” or “hatch”) domain (16) (see Fig. 1). A structural alignment of HasR with known structures of TonB-dependent siderophore receptors and the cobalamin receptor BtuB [see supporting information (SI) Fig. S1] shows that all receptors share the same fold, but that the extracellular loops L2, L6, and L9 of HasR are exceptionally large. All of these loops form contacts with HasA, and deletions of 6 residues in either one of the latter 2 loops and L8 have been shown to abolish HasA binding (17). In accord with these findings, the structure shows that L6 and L9 form long arms, and L8 and L9 bury a large surface with HasA (see Fig. 1 and Table 1). L6 and L9 each contact 1 HasA β-strand that was previously shown to contribute independently to HasR binding (18).
Fig. 1.
Structure of the ternary complex HasA∼HasR∼heme. HasA (red) and HasR (cork domain beginning with residue Asn-113 in orange; barrel domain beginning at residue Lys-241 in blue) are indicated as a ribbon model. The first 5 strands and the loops L1–L3 of HasR are omitted to allow a view into the barrel interior. The heme is indicated as a wire frame model (green). The extracellular loops 6–11 are labeled. Yellow parts of or near L6, L8, and L9 mark positions of 6 residue deletions that have been found to abolish HasA binding to the receptor.
Table 1.
Buried surface areas between external loops of HasR and HasA in the HasA∼HasR∼heme complex in Å2, determined by the PISA Server (www.ebi.ac.uk/msd-srv/prot_int/pistart.html) (40)
| Loop | 2 | 3 | 6 | 7 | 8 | 9 | 10 | 11 |
| Buried surface | 212 | 256 | 230 | 122 | 479 | 477 | 196 | 48 |
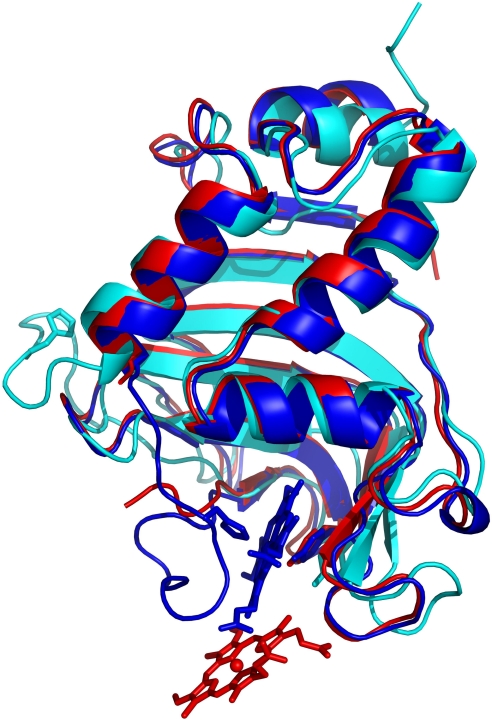
During formation of this tight complex, 1,732 Å2 of protein surface are buried from solvent, and HasA conserves most of its uncomplexed structure. It is oriented with its heme-binding loops toward the extracellular aperture of the HasR barrel, but the loop containing His-32 is turned away from its heme-binding conformation and could not be modeled because of missing electron density (residues 29–38) indicating multiple conformations or disorder (see Fig. 2).
Fig. 2.
Superimposed structures of holoHasA (8) (blue), apoHasA (39) (cyan), and HasA in the HasA∼HasR∼heme complex (red) shown as ribbon models. The hemes of the holoHasA and the HasA∼HasR∼heme structures are represented as wire frame models with the ferric iron indicated as a sphere. The superposition shows the translocation of the heme from holoHasA into the HasA∼HasR∼heme complex. The distance between the ferric iron atoms is 9.2 Å.
In the HasA∼HasR∼heme complex, the heme is no longer bound to HasA but axially coordinated to 2 His residues, HasR-His-603 from L7 and HasR-His-189 from apex C of the plug (see Fig. 3A) that are conserved in hemophore receptors (10). Compared with its position in holoHasA, the heme is translated into the receptor by 9.2 Å with a concomitant rotation of the tetrapyrrole ring, leading to the exposure of one of the propionates to solvent. Of the 2 coordinating histidines in HasR, His-603 from L7 contributes less to heme supported growth as concluded from mutants (10). This correlates with significantly higher B-factors of His-603 from L7 in the HasA∼HasR∼heme structure as compared with His-189 from apex C. A double mutant of HasR, H603A-H189A, showed no detectable heme binding (10) hinting against further substantial interactions with protein residues. Moreover, typical interactions with the heme face and edge found in other b-type heme-binding proteins (19) are absent except for a van der Waals contact of HasA-Phe-78 with a pyrrole ring.
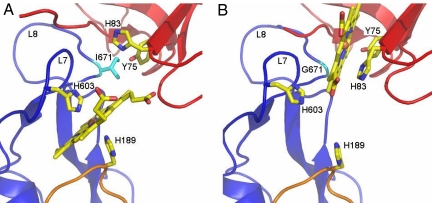
Fig. 3.
Heme-binding after HasA-docking and in a transition state of docking. (A) Heme-binding site in HasA∼HasR∼heme. Both proteins are shown in ribbon representation, HasA in red, the HasR barrel in blue, and the HasR apex C in orange. The axial heme ligands are HasR-His-603 from L7 and HasR-His-189 from apex C of the plug domain. HasR-Ile-671 is shown in cyan. The backbone of the HasA-His-32-bearing loop is seen only partially because of disorder. (B) Heme-binding site in HasA∼HasR-Ile671Gly∼heme. The axial heme ligand is HasA-Tyr-75, hydrogen bonded with HasA-His-83. HasR-Gly-671 is shown in cyan in the backbone of L8. HasA-His-83 in the side-chain conformation seen here would clash with heme in the binding site shown in A, where it is rotated toward HasA. The backbone of the HasA-His-32-bearing loop is seen only partially because of disorder.
The transfer of heme to HasR into axial coordination with the conserved histidines has been inferred from UV-vis spectrocopy, resonance Raman, isothermal calorimetry, and mutant data (10). This endergonic transfer of heme occurs spontaneously and is enabled by coupling with the complex formation between HasA and HasR, which is very exergonic and exothermic. The heat of complex formation of apoHasA and HasR is −201 kJ/mol (10), more than that of many other tight protein–protein complexes (20).
During the docking of holoHasA to HasR, heme is not lost to the medium (10) indicating that a late stage of docking creates an event that triggers dissociation of heme coordination to HasA and heme transfer. A superposition of holoHasA with HasA in HasA∼HasR∼heme showed that HasR-Ile-671 in L8 is the only HasR residue whose side chain sterically clashes with heme in holoHasA. We hypothesized that this clash might trigger heme transfer from HasA to HasR and constructed a site-directed mutant, HasR-Ile671Gly, alleviating this clash. As the wild-type receptor, the HasRIle671Gly mutant is able to transport free heme. However, in contrast to the wild-type receptor, it is unable to acquire and transport HasA-bound heme (P.D. and J. Raimon, unpublished observation). We solved the structure of the ternary complex with HasR-Ile671Gly. The structure shows heme coordinated by HasA-Tyr-75 and arranged with HasA as in holoHasA (see Fig. 2), but the HasA-His-32 loop pivoted away as in HasA∼HasR∼heme (see Fig. 3B).
Based on this structure, which displays the heme-loaded complex before heme transfer but is itself inhibited with respect to transfer, a likely scenario is the following: In the initial phase of docking of holoHasA to HasR, L7 and L8 of HasR [the so-called “β-cantilever” (16)] displace the HasR-His-32 bearing loop, causing rupture of the axial coordination between HasA-His-32 and heme. The stronger axial coordination between HasA-Tyr-75 and heme (7), however, persists. It is stabilized by deprotonation of the phenol group that acts as hydrogen bond donor to the Nε of HasA-His-83 (21, 22). At the end of this phase of docking the arrangement of heme relative to HasA is still preserved, despite the approaching loops L7 and L8. In the next phase, Nδ of His-83 may get transiently protonated (8), which will weaken the coordination of the heme iron. Finally, Ile-671 sterically displaces heme from HasA.
After heme translocation, the rotation of the His-83 side chain (as observed in the native HasR-HasA-heme complex) could impair sliding back of the heme to the HasA heme-binding site. This hypothesis is in accordance with growth experiments undertaken by using HasA and HasR mutants of heme-coordinating ligands (10). HasR-H603A supports growth only with HasA mutants devoid of the Tyr-75-His-83 association. Furthermore, a structural comparison of holoHasA with HasA∼HasR shows that significant conformational changes in HasA upon complex formation are restricted to the interface area with HasR, arguing against a major exergonic structural change of HasA upon docking, which might contribute to heme transfer.
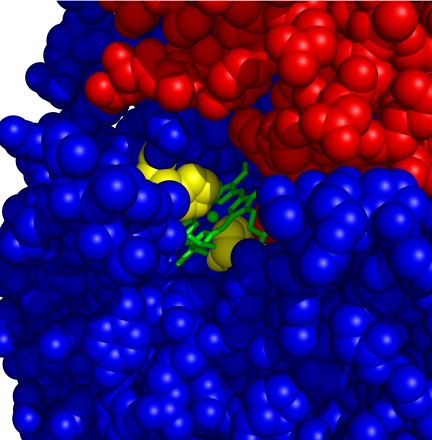
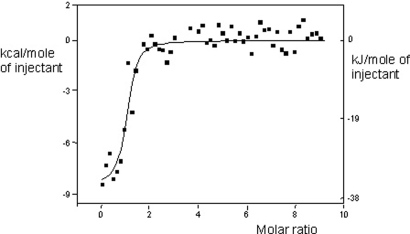
Interestingly, in vivo heme can also access its binding site on HasR when apoHasA is bound to HasR. To understand the structural basis of this, we have determined the structure of the HasA∼HasR complex without heme (see Methods), which, apart from absence of heme, shows no changes compared with that of HasA∼HasR∼heme. The structure of this complex shows a channel from the heme-binding site opening to the external medium between L3 and the bent loop L4 (see Fig. 4). This could allow access of heme into the HasA∼HasR complex in accordance with isothermal titration calorimetry (ITC) measurements that showed that heme affinity of HasA∼HasR is smaller than that of HasA (Kd = 0.06 μM versus 18 pM) (see Fig. 5), close to that of HasR [0.2 μM (10)]. The UV-vis absorption spectrum of this complex is identical to that of HasR∼heme and HasA∼HasR∼heme, showing that heme is in the same environment.
Fig. 4.
Detail of the HasA∼HasR∼heme complex showing a putative heme access channel extending from the external medium between L3 and the bent loop L4 to the receptor heme-binding site. HasA and HasR are indicated as a CPK model in red and blue, respectively, except for the heme-coordinating HasR histidines that are colored yellow. The heme is indicated as a wire frame model in green and the Fe3+ atom as a red sphere.
Fig. 5.
ITC analysis of the interaction of heme with HasA-HasR. The binding isotherm derived from the signal is shown. The molar ratio heme/HasA-HasR is indicated. Thermodynamic parameters deduced from the fit are: Kd = 0.06 μM; ΔH = −36 kJ·mol−1; ΔG = −41 kJ·mol−1; TΔS = +5 kJ·mol−1.
Discussion
Experiments with radiolabeled HasA in living cells showed that TonB/HasB-dependent uptake of heme is accompanied by release of apoHasA from HasR to the external medium (12), and this occurs only in the presence of bound heme, i.e., with HasA∼HasR∼heme complexes.
This raises the question of the structural signal that triggers TonB-dependent chemiosmotic energy transduction to the receptors and what the results of the transduction are. Because TonB forms functional complexes with several receptors in a given cell, one would expect a common mechanism. Indeed, HasR can be well-superimposed with the known structures of the TonB-dependent receptor family except for the extracellular loops and shares a set of conserved motifs (16) with them (see Fig. S1). In all of the receptors, combinations of residues from 3 apices (or ligand-binding loops) of the plug domain, A, B, and C, contribute to ligand binding (23–27), but individual receptors use different combinations to bind their respective ligands.
In FhuA and FecA (23–25), residues from all apices of the plug contribute to ligand binding accompanied by an induced fit and significant allosteric changes on the periplasmic surface of the plug. In BtuB, only residues from apices C and A contribute to binding, causing a less conspicuous allosteric effect (26). In FptA, residues from only apex A (27) contribute to ligand binding; allosteric changes are unknown because the ligand-free structure has not been determined. In HasR, only residues from apex C contribute to heme binding. Because the structure of HasA∼HasR is virtually identical to that of HasA∼HasR∼heme, we can rule out large allosteric changes due to heme binding. Because it was observed that TonB/HasB-dependent release of apoHasA requires presence of heme (12) and because the binding of heme and HasA to HasR triggers the signaling cascade (14, 15), HasR in an intermediate stage of its engagement with TonB should have a different conformation when complexed with heme as compared with uncomplexed. However, within the accuracy given by their resolution, our structures do not provide evidence for such a conformational change.
Despite striking structural differences, the course of events from heme binding by HasA to translocation through HasR reminds us of binding-protein dependent bacterial ABC transport that performs translocation of cargo molecules from the periplasmic-binding protein through an inner-membrane protein complex (28). In both cases, translocation starts with a spontaneous reaction in which a binding protein with its cargo molecule binds on the cis side to its membrane bound receptor and the cargo molecule is transferred from the former to the latter. Subsequently, energy derived either from proton motive force or ATP hydrolysis is used to translocate the cargo to the trans compartment and to detach the binding protein from the cis side.
The mechanism of ligand transfer from one protein with high ligand affinity, the “donor,” to another with lower ligand affinity, the “acceptor” is of general importance. Our results show that ligand dissociation and transfer is controlled by events occurring successively in the course of docking of the 2 proteins. Initially the donor with bound ligand approaches the acceptor, and a part of the binding energy to the donor is consumed for displacement of a donor loop involved in ligand binding. This preserves the arrangement of the ligand relative to the donor. Ligand movement is triggered only in a later phase of docking when the donor–acceptor complex is almost complete by a localized steric clash involving a small relocation. We think that this ensures close to 100% efficiency of transfer from a high- to a lower-affinity binding site. Bacteria acquiring iron or heme bound very tightly to host proteins, such as lactoferrin or hemopexin, are likely to use mechanisms similar to those of the Has system so that the subsequent reactions of hemophore docking, heme uptake, and complex dissociation described by us, could be of paradigmatic value.
Methods
Protein Expression and Purification.
Cells from Escherichia coli popc4420(pFR2) (an E. coli MC4100 derivative devoid of the major outer-membrane proteins OmpF, OmpC, and LamB) and carrying the pFR2 plasmid containing the hasR gene under the control of paraBAD promoter, were grown in 300-L fermentors in M9 minimal medium supplemented with 0.2% casamino acids, 0.4% glycerol, and 60 μM iron citrate at 30 °C. At an OD600 of 0.5, arabinose was added to a final concentration of 40 mg/L to induce expression of the HasR receptor for 3 h. The whole culture was quickly chilled at 10 °C and centrifuged. The cell pellet was quickly frozen in liquid N2 and kept at −80 °C until use. Alternatively, the same medium with a 5 times higher concentration of concentrated carbon and nitrogen sources was used to increase the cell yield per liter and reduce culture size. In that case 15–20 g of cells per liter were routinely obtained. All cultures were made at the Recombinant Proteins and Antibodies Platform of the Institut Pasteur.
The purification of the HasA∼HasR∼heme complex was essentially done as described (29) with the exception that N-tetradecyl-N,N-dimethyl-3-ammonio-propanesulfonate (ZW3-14) was used at a concentration of 0.02% throughout the purification. This detergent was exchanged against octyltetraoxyethylene (C8E4) before crystallization.
Before preparing the HasA∼HasR complex, both His6HasA and HasR were purified by anion-exchange chromatography (Q-Sepharose; GE Healthcare) and His6HasA with an additional gel filtration (Bio-Gel, P-60; Bio-Rad) before forming the complex. His6HasA was either purified in the apo form or loaded with heme to obtain the holo form. The HasA∼HasR complex was purified in the same way as the HasA∼HasR∼heme complex.
Site-Directed Mutagenesis.
HasRI671G was constructed by a mutagenic PCR using pFR2 as template and the 2 mutagenic oligonucleotides 5′-TGGTGCAGCCGAGCGGCGGCAATGCCGCTT-3′ and 5′-AAGCGGCATTGCCGCCGCTCGGCTGCACCA-3′ and the quick-change kit of Stratagene. The full characterization of the mutant will be described elsewhere.
Crystallization.
Initial crystallization screenings were performed with Nextal kits (Qiagen) in sitting drops. Fine screenings were performed in hanging drops mixing 2 μL of protein solution [20 mg/ml in 50 mM Tris (pH 7.5), 0.6% C8E4] with equal volumes of reservoir solutions and equilibrating against 1 mL of reservoir.
In the final crystallization condition, the reservoir solution was 100 mM Tris (pH 8.0), 2 M NaCl. Crystals grew within 10–14 days at 18 °C to a final size of 0.2 × 0.2 × 0.05 mm and were soaked in artificial mother liquor containing 20% glycerol before freezing in liquid nitrogen.
Native datasets for HasA∼HasR∼heme, HasA∼HasR and HasA∼HasR-I671G∼heme were collected at the European Synchrotron Radiation Facility (ESRF) beamline ID 29 to a resolution of 2.6 Å and at the Swiss Light Source (SLS) beamline X06SA to a resolution of 3.0 and 2.8 Å, respectively.
Phases were determined by using a selenomethionine-labeled HasR preparation. A complete MAD dataset was collected at the SLS beamline X06SA to a resolution of 3.1 Å (see Table 2).
Table 2.
X-ray datasets used for structure determination (beamlines are X06A at the SLS and ID29 at the ESRF)
| Data collection | Remote high | Peak | Inflection | Native | Native | Native |
|---|---|---|---|---|---|---|
| HasA∼HasR∼heme | HasA∼HasR∼heme | HasA∼HasR∼heme | HasA∼HasR∼heme | HasA∼HasR | HasA∼HasR-I671G∼heme | |
| Beamline | X06SA | X06SA | X06SA | ID29 | X06SA | X06SA |
| Wavelength, Å | 0.97152 | 0.97897 | 0.97921 | 1.0 | 0.9792 | 0.9792 |
| Resolution, Å | 50–3.12 (3.31–3.12) | 50–3.16 (3.35–3.16) | 50–3.19 (3.38–3.19) | 50–2.57 (2.7–2.57) | 50–3.0 (3.18–3.0) | 40–2.8 (2.96–2.80) |
| I/σ | 10.27 (3.02) | 8.03 (2.93) | 5.97 (2.28) | 10.21 (1.59) | 12.35 (1.92) | 14.03 (0.89) |
| Completeness, % | 95.1 (80.6) | 97.1 (83.0) | 97.3 (83.8) | 91.0 (63.5) | 99.1 (97.6) | 98.0 (88.3) |
| Observations | 345,884 | 448,208 | 446,611 | 673,585 | 501,683 | 616,190 |
| Unique reflections | 124,640 | 122,760 | 120,596 | 111,662 | 77,295 | 13,304 |
| Rmeas, % | 11.0 (32.8) | 15.5 (40.7) | 16.1 (46.0) | 14.4 (75.9) | 16.6 (98.1) | 11.8 (186.9) |
| Rmrgd-F, % | 13.3 (38.8) | 14.9 (39.9) | 15.8 (45.4) | 16.4 (72.2) | 16.7 (77.7) | 20.0 (197.7) |
The space group is in each case F222 with a = 158 Å, b = 164 Å, and c = 596 Å. Values in parentheses refer to the highest-resolution shell.
Data Processing and Structure Solution.
Data were processed by XDS (30). Heavy-atom sites were found with SHELXD (31) and initial phases calculated with SHARP (32). Automated model building with RESOLVE (33) yielded ≈20% of the Cα trace. Manual model building was performed by using the program COOT (34). Refinement was done by Refmac (35), CNS (36), and PHENIX (37). Based on an analysis of heme-binding proteins in the Protein Data Bank (38) the distances from His-Nε2 to the heme iron were restrained to 2.2 ± 0.1 Å. The occupancy of heme in the HasA-HasR-Ile671Gly–heme complex was refined and found to be 0.9 (see Table 3).
Table 3.
Summary of refinement data
| HasA∼HasR∼heme | HasA∼HasR | HasA∼HasR-I671G∼heme | |
|---|---|---|---|
| Refinement | |||
| Resolution, Å | 49.2–2.7 (2.73–2.70) | 49.4–3.0 (3.03–3.0) | 39.2–2.8 (2.83–2.80) |
| No. of reflections | 99,334 (2,329) | 77,295 (2,431) | 92,482 (2,123) |
| Completeness, % | 95.03 (71) | 99.17 (93) | 98.1 (71) |
| Rwork, % | 23.7 (34.9) | 21.4 (37.4) | 22.6 (46.6) |
| Rfree*, % | 27.3 (38.4) | 24.3 (39.1) | 26.2 (48.3) |
| Model composition | |||
| Protein residues | 1,850 | 1,850 | 1,850 |
| Heme atoms | 86 | 0 | 86 |
| Water molecules | 58 | 19 | 13 |
| B-factors | |||
| Protein | 93.5 | 80.2 | 110.6 |
| Heme | 84.6 | — | 120.4 |
| Deviation from ideal values | |||
| Bond lengths, Å | 0.010 | 0.010 | 0.006 |
| Residues with bad bond lengths†, % | 0 | 0.05 | 0 |
| Bond angles, ° | 0.61 | 1.27 | 1.08 |
| Residues with bad bond angles†, % | 0.22 | 0.71 | 0.550 |
| Ramachandran plot† | |||
| Favored regions, % | 92.4 | 89.6 | 89.7 |
| Allowed regions, % | 99.2 | 99.5 | 99.1 |
Values in parentheses refer to the highest resolution shell.
*Calculated with 5% of the reflections.
†As determined by MOLPROBITY (41).
ITC Measurements.
Titrations were performed at 25 °C by using a MicroCal MCS titration calorimeter (MicroCal). Proteins and hemin were in 20 mM sodium phosphate at pH 7, 0.08% ZW 3-14. Samples were thoroughly degassed before use by stirring under vacuum. All injections were carried out at 3-min intervals. Because of heme adsorption, the calorimeter cell and the micro syringe used for injections were extensively washed after each experiment. The heat of dilution of the ligand was measured either by injecting the ligand into the buffer alone or by injecting more ligand into the cell after saturation. The value obtained was subtracted from the heat of reaction to give the effective heat of binding. For the titration experiment the concentration of HasA∼HasR was 1.4 μM.
The resulting titration data were analyzed and fitted by using the origin for ITC software package supplied by Microcal to obtain the stoichiometry (n), the dissociation constants (Kd) and the enthalpy (ΔH) changes of binding. For the fit any constraints on the stoichiometry and ΔH were not fixed. ITC titrations of HasA∼HasR with hemin showed 20% of variation of stoichiometry ≈1. This was because of the imprecision in the determination of hemin concentration in the presence of detergent.
Supplementary Material
Acknowledgments.
We thank Emmanuel Frachon from the Plate-Forme 5—Production de Protéines Recombinantes et d'Anticorps at the Pasteur Institute for expert assistance in bacterial cell growth in fermentors, Julie Raimon for characterization of the HasR-I671G mutant, and the staff of the SLS and ESRF beamlines for their help with data collection. Ms. Keller has improved the English wording of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates for HasA∼HasR∼heme, HasA∼HasR, and HasA∼HasR-I671G∼heme have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3CSL, 3CSN, and 3DDR, respectively)
This article contains supporting information online at www.pnas.org/cgi/content/full/0809406106/DCSupplemental.
References
- 1.Braun V, Günter K, Hantke K. Transport of iron across the outer membrane. BioMetals. 1991;4:14–22. doi: 10.1007/BF01135552. [DOI] [PubMed] [Google Scholar]
- 2.Cornelissen CN, Sparling PF. Iron piracy: Acquisition of transferrin-bound iron by bacterial pathogens. Mol Microbiol. 1994;14:843–850. doi: 10.1111/j.1365-2958.1994.tb01320.x. [DOI] [PubMed] [Google Scholar]
- 3.Prinz T, Meyer M, Pettersson A, Tommassen J. Structural characterization of the lactoferrin receptor from Neisseria meningitidis. J Bacteriol. 1999;181:4417–4419. doi: 10.1128/jb.181.14.4417-4419.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wandersman C, Stojiljkovic I. Bacterial heme sources: The role of heme, hemoprotein receptors and hemophores. Curr Opin Microbiol. 2000;3:215–220. doi: 10.1016/s1369-5274(00)00078-3. [DOI] [PubMed] [Google Scholar]
- 5.Izadi N, et al. Purification and characterization of an extracellular heme-binding protein, HasA, involved in heme iron acquisition. Biochemistry. 1997;36:7050–7057. doi: 10.1021/bi962577s. [DOI] [PubMed] [Google Scholar]
- 6.Létoffé S, Ghigo JM, Wandersman C. Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc Natl Acad Sci USA. 1994;91:9876–9880. doi: 10.1073/pnas.91.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deniau C, et al. Thermodynamics of heme binding to the HasA(SM) hemophore: Effect of mutations at three key residues for heme uptake. Biochemistry. 2003;42:10627. doi: 10.1021/bi030015k. [DOI] [PubMed] [Google Scholar]
- 8.Arnoux P, et al. The crystal structure of HasA, a hemophore secreted by Serratia marcescens. Nat Struct Biol. 1999;6:516–520. doi: 10.1038/9281. [DOI] [PubMed] [Google Scholar]
- 9.Ghigo J-M, Létoffé S, Wandersman C. A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J Bacteriol. 1997;179:3572–3579. doi: 10.1128/jb.179.11.3572-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izadi-Pruneyre N, et al. The heme transfer from the soluble HasA hemophore to its membrane-bound receptor HasR is driven by protein–protein interaction from a high to a lower affinity binding site. J Biol Chem. 2006;281:25541–25550. doi: 10.1074/jbc.M603698200. [DOI] [PubMed] [Google Scholar]
- 11.Létoffé S, Nato F, Goldberg ME, Wandersman C. Interactions of HasA, a bacterial haemophore, with haemoglobin and with its outer membrane receptor HasR. Mol Microbiol. 1999;33:546–555. doi: 10.1046/j.1365-2958.1999.01499.x. [DOI] [PubMed] [Google Scholar]
- 12.Létoffé S, Delepelaire P, Wandersman C. Free and hemophore-bound heme acquisitions through the outer membrane receptor HasR have different requirements for the TonB-ExbB-ExbD complex. J Bacteriol. 2004;186:4067–4074. doi: 10.1128/JB.186.13.4067-4074.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paquelin A, Ghigo J-M, Bertin S, Wandersman C. Characterization of HasB, a Serratia marcescens TonB-like protein specifically involved in the haemophore-dependent haem acquisition system. Mol Microbiol. 2001;42:995–1005. doi: 10.1046/j.1365-2958.2001.02628.x. [DOI] [PubMed] [Google Scholar]
- 14.Rossi MS, Paquelin A, Ghigo J-M, Wandersman C. Haemophore-mediated signal transduction across the bacterial cell envelope in Serratia marcescens: The inducer and the transported substrate are different molecules. Mol Microbiol. 2003;48:1467–1480. doi: 10.1046/j.1365-2958.2003.03516.x. [DOI] [PubMed] [Google Scholar]
- 15.Cwerman H, Wandersman C, Biville F. Heme and a five-amino-acid hemophore region form the bipartite stimulus triggering the has signaling cascade. J Bacteriol. 2006;188:3357–3364. doi: 10.1128/JB.188.9.3357-3364.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chimento DP, Kadner RJ, Wiener MC. Comparative structural analysis of TonB-dependent outer membrane transporters: Implications for the transport cycle. Proteins. 2005;59:240–251. doi: 10.1002/prot.20416. [DOI] [PubMed] [Google Scholar]
- 17.Barjon C, Wecker K, Izadi-Pruneyre Delepelaire P. Mutagenesis and molecular modeling reveal three key extracellular loops of the membrane receptor HasR that are involved in hemophore HasA binding. J Bacteriol. 2007;189:5379–5382. doi: 10.1128/JB.00251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Létoffé S, Debarbieux L, Izadi N, Delepelaire P, Wandersman C. Ligand delivery by haem carrier proteins: The binding of Serratia marcescens haemophore to its outer membrane receptor is mediated by two distinct peptide regions. Mol Microbiol. 2003;50:77–88. doi: 10.1046/j.1365-2958.2003.03686.x. [DOI] [PubMed] [Google Scholar]
- 19.Schneider S, Marles-Wright J, Sharp KH, Paoli M. Diversity and conservation of interactions for binding in b-type heme proteins. Nat Prod Rep. 2007;24:621–630. doi: 10.1039/b604186h. [DOI] [PubMed] [Google Scholar]
- 20.Krell T, et al. Insight into the structure and function of the transferrin receptor from Neisseria meningitidis using microcalorimetric techniques. J Biol Chem. 2003;278:14712–14722. doi: 10.1074/jbc.M204461200. [DOI] [PubMed] [Google Scholar]
- 21.Wolff N, et al. Histidine pKa shifts and changes of tautomeric states induced by the binding of gallium-protoporphyrin IX in the hemophore HasASM. Protein Sci. 2002;11:757–765. doi: 10.1110/ps.3630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caillet-Saguy C, et al. Direct-detected 13C NMR to investigate the iron(III) hemophore HasA. J Am Chem Soc. 2006;128:150–158. doi: 10.1021/ja054902h. [DOI] [PubMed] [Google Scholar]
- 23.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Siderophore-mediated iron transport : Crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 24.Locher KP, et al. Transmembrane signaling across the ligand-gated FhuA receptor: Crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell. 95:771–778. doi: 10.1016/s0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- 25.Ferguson AD, et al. Structural basis of gating by the outer membrane transporter FecA. Science. 2002;295:1715–1719. doi: 10.1126/science.1067313. [DOI] [PubMed] [Google Scholar]
- 26.Chimento DP, Mohanty AK, Kadner RJ, Wiener MC. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat Struct Biol. 2003;10:394–401. doi: 10.1038/nsb914. [DOI] [PubMed] [Google Scholar]
- 27.Cobessi D, Celia H, Pattus F. Crystal structure at high resolution of ferric-pyochelin and its membrane receptor FptA from Pseudomonas aeruginosa. J Mol Biol. 200;352:893–904. doi: 10.1016/j.jmb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J. Crystal structure of a catalytic intermediate of the maltose transporter. Nature. 2007;450:515–521. doi: 10.1038/nature06264. [DOI] [PubMed] [Google Scholar]
- 29.Huché F, et al. Purification, crystallization and preliminary X-ray analysis of the outer membrane complex HasA-HasR from Serratia marcescens. Acta Crystallogr F. 2006;62:56–60. doi: 10.1107/S1744309105041394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 31.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 32.Vonrhein C, et al. Automated structure solution with autoSHARP. Methods Mol Biol. 2006;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 33.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr D. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, et al. Coot: Model building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, et al. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Brünger A, et al. Crystallography and NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 38.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolff N, et al. Comparative analysis of structural and dynamic properties of the loaded and unloaded hemophore HasA : Functional implications. J Mol Biol. 2007;376:517–525. doi: 10.1016/j.jmb.2007.11.072. [DOI] [PubMed] [Google Scholar]
- 40.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Davis, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.