Abstract
SWI/SNF, an evolutionarily conserved ATP-dependent chromatin-remodeling complex, has an important role in transcriptional regulation1. In Saccharomyces cerevisiae, SWI/SNF regulates the expression of ~6% of total genes through activation or repression2. Swi1, a subunit of SWI/SNF, contains an N-terminal region rich in glutamine and asparagine, a notable feature shared by all characterized yeast prions—a group of unique proteins capable of self-perpetuating changes in conformation and function3. Here we provide evidence that Swi1 can become a prion, [SWI+]. Swi1 aggregates in [SWI+] cells but not in nonprion cells. Cells bearing [SWI+] show a partial loss-of-function phenotype of SWI/SNF. [SWI+] can be eliminated by guanidine hydrochloride treatment, HSP104 deletion or loss of Swi1. Moreover, we show [SWI+] is dominantly and cytoplasmically transmitted. Our findings reveal a novel mechanism of ‘protein-only’ inheritance that results in modification of chromatin-remodeling and, ultimately, global gene regulation.
Yeast prions are epigenetic elements that are transmitted as particular protein conformations4, similarly to the mammalian prion that causes infectious diseases5. [PSI+], [URE3] and [PIN+] are three reported prions in S. cerevisiae6-9, and their protein determinants are Sup35, Ure2 and Rnq1, respectively. They all contain an intrinsic domain with high glutamine and asparagine content essential for prion formation and propagation (Fig. 1a). Yeast prions can be formed and lost at a low spontaneous rate10. Overproduction of a prion protein considerably increases de novo formation of the corresponding prion8,11,12. [PSI+] induction by Sup35 overproduction requires a [PSI+] inducible factor (Pin+), which can be an event such as overproduction of particular glutamine-, asparagine- or glutamine and asparagine-rich proteins, or another prion13-15. Notably, several members of the yeast SWI/SNF complex, including Swi1 and Snf5, also contain glutamine and asparagine-rich regions16 (Fig. 1a), and Swi1 overproduction has been implicated as a Pin+ factor9. SWI/SNF is an evolutionarily conserved, ATP-dependent chromatin-remodeling complex that has a regulatory role in gene expression17. In S. cerevisiae, the SWI/SNF complex is composed of at least 11 subunits, for a total molecular weight of ~1 MDa18. Null mutants of SWI/SNF are viable, but show phenotypes of slow growth, reduced mating-type switching, and inability to utilize nonfermentable carbon sources, such as raffinose, galactose, glycerol and sucrose19. In mammals, Ini1, a Snf5 homolog, is essential for embryo viability and tumor suppression20. Various mutations affecting the human Ini1 or Snf2 homologs, BRM and BRG-1, are associated with many types of malignant tumors21. The importance of SWI/SNF function, the notable glutamine and asparagine content of some of its components, and the Pin+ property of Swi1 led us to investigate whether SWI/SNF function can be subject to prion-like epigenetic regulation.
Figure 1.
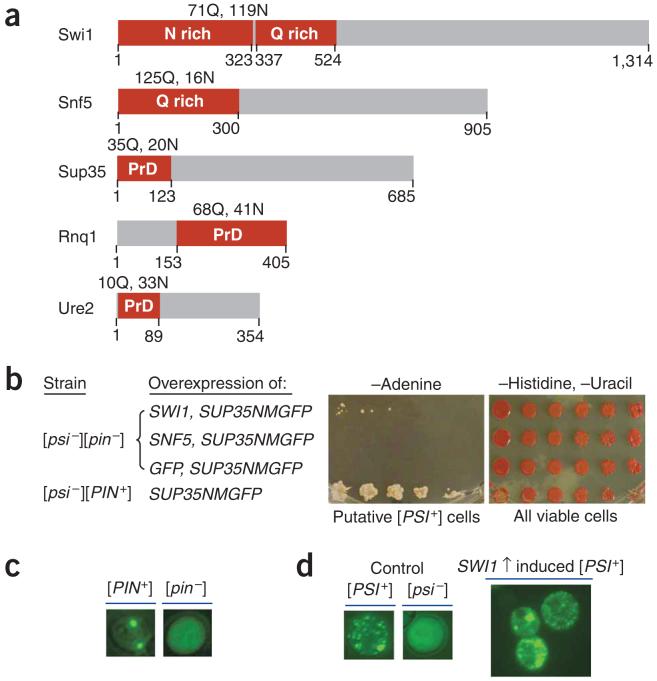
Overexpression of SWI1 but not SNF5 functions as Pin+. (a) Diagrams of Swi1 and Snf5 with their glutamine (Q) and asparagine (N) rich regions shown (red boxes). The prion domains (PrDs) of Sup35, Rnq1 and Ure2 are also indicated (red boxes). (b) Overexpression of SWI1 can substitute for [PIN+] to promote [PSI+] formation. We grew 74D-694 cells [(psi-][pin-]) containing pRS313CUP1NMGFP and p426GPDSWI1, pRS313CUP1NMGFP and p426GPDSNF5, or pRS313CUP1NMGFP and p426GPDGFP to early-log phase and induced them for NMGFP expression by addition of CuSO4 to 100 μM. After 24-h induction, cells were spotted onto the indicated plates with a fivefold serial dilution. Pictures were taken after 5-d incubation for cells on synthetic complete medium lacking histidine and uracil (-Histidine, -Uracil) or after 9 d for cells on medium lacking adenine (-Adenine). (c) Fluorescence microscopy assay of Rnq1 conformational status. Before conducting experiments shown in panel b, we confirmed the Rnq1 conformational status by transforming with p416CUP1RNQ1GFP, inducing for 4 h with 100 μM CuSO4 and analyzing by fluorescence microscopy. (d) Sup35 is aggregated in Ade+ cells obtained by SWI1 and SUP35NMGFP co-overexpression. After eliminating p426GPDSWI1 and pRS313CUP1NMGFP, we transformed Ade+ isolates from panel b with p416CUP1NMGFP and analyzed them by fluorescence microscopy to confirm their [PSI+] status. Shown here are fluorescence patterns of a representative [PSI+] isolate and isogenic [PSI+] and [psi-] controls.
If Swi1 or Snf5 is a prion protein, it might be more likely to adopt a prion conformation when it functions as a Pin+, because prion proteins can interact with each other, and the non-[PSI+] yeast prions, [PIN+] and [URE3], can function as Pin+. Moreover, [PSI+] can be easily analyzed in strains such as 74D-694, which contains a nonsense mutation in ADE1. Whereas 74D-694 [PSI+] cells can grow in media without adenine, [psi-] cells cannot. [PSI+] colonies appear white in rich media, such as YPD, and [psi-] colonies appear red as a result of accumulation of a pigment byproduct. To test whether Swi1 and/or Snf5 are prion proteins, we conducted [PSI+]-induction experiments in 74D-694 [psi-][pin-] cells by co-overexpression of SWI1 or SNF5 and SUP35NMGFP, a fusion gene encoding the Sup35 prion domain and the green fluorescence protein (GFP). Consistent with a previous report9, co-overexpression of SWI1 and SUP35NMGFP allowed [PSI+] formation in [pin-] cells (Fig. 1b-d). Although the N-terminal region of Snf5 is also rich in glutamine (Fig. 1a), overexpression of SNF5 did not function as a Pin+ (Fig. 1b). Therefore, we next focused on examining whether Swi1 is a prion protein.
If Swi1 becomes a prion (termed [SWI+]), it might form amyloid-like structure(s), as do other identified prions22. Cells bearing [SWI+] should show compromised SWI/SNF functions, as the amyloid-form of Swi1 will recruit the nonprion Swi1 isomers and sequester them from their natural contacts. To identify [SWI+], we first obtained a large number of Ade+ colonies ([PSI+] candidates) from 74D-694 [psi-][pin-] cells by co-overexpression of SWI1 and SUP35NMGFP. We then screened for [SWI+] candidates by testing individual Ade+ isolates for phenotypes indicative of compromised SWI/SNF function, specifically, poor growth in raffinose medium (Raf-). Among ~400 tested Ade+ isolates, 32 showed the Raf- phenotype. Only two were able to grow on raffinose after 5 mM guanidine hydrochloride treatment (Fig. 2a). This guanidine hydrochloride treatment eliminates all known yeast prions, but does not usually cause nucleic acid mutations23. Like SWI/SNF mutants, both candidates grew poorly in galactose (Gal-) and glycerol (Gly-) (Fig. 2b). However, they showed only a mild defect in using sucrose (Fig. 2b). Unlike SWI/SNF mutants, they did not show any detectable sensitivity to 0.3 M LiCl or 1 M NaCl (Fig. 2b and data not shown). These results suggest that the two [SWI+] candidates show an epigenetically conferred, partial loss-of-function SWI/SNF phenotype.
Figure 2.
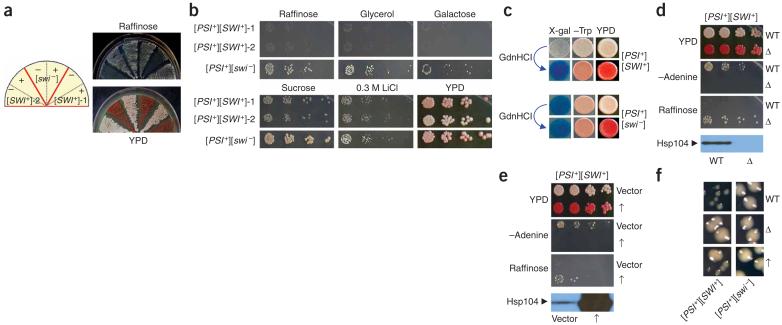
Isolation and characterization of [SWI+] candidates. (a) [SWI+]-1 and [SWI+]-2, two [SWI+] candidates isolated from [PSI+] cells upon co-overexpression of SUP35NMGFP and SWI1, showed a guanidine hydrochloride-curable Raf- phenotype. Shown are cell streaks of the candidates before (-) and after (+) guanidine hydrochloride (GdnHCl) treatment on indicated media. Note that both of the plasmids, pRS313CUP1NMGFP and p426GPD-SWI1, were eliminated before 5 mM guanidine hydrochloride treatment. (b) Phenotypic assays of the [PSI+][SWI+] candidates. [SWI+] candidates and [SWI-] control were grown in YPD to mid-log phase and then spotted to the indicated media with a fivefold serial dilution. (c) [SWI+]-2 (upper) and [SWI-] (lower) cells containing pLS7 plasmid (Trp+), a SWI/SNF lacZ reporter controlled by a chimeric promoter composed of the SWI/SNF regulatory sequence of SUC2 promoter and the core promoter of LEU2 (ref. 24), were spotted onto +sucrose -tryptophan synthetic complete media + 20 μg/ml X-gal plates (X-gal); +glucose -tryptophan synthetic complete medium plates (-Trp); and YPD (YPD) plates. (d) HSP104 deletion eliminated [PSI+] and the Raf- phenotype. [SWI+]-2 cells with (Δ) or without (WT) HSP104 disruption were assayed on indicated media. (e) The effect of HSP104 overexpression on Raf- phenotype. [SWI+]-2 cells containing p2HGHSP104 (↑) or p2HG (vector) were grown in synthetic complete liquid media lacking histidine and then spotted to the indicated media. HSP104 disruption and overexpression were confirmed by immunoblot analysis using a polyclonal Hsp104 antibody. (f) Enlarged images showing the colony sizes of [PSI+][SWI+] and [PSI+][SWI-] cells after HSP104 deletion (Δ), overexpression (↑) or no treatment (WT). A [PSI+] isolate with Raf+ phenotype obtained under identical conditions for [SWI+] candidate isolation was used as the nonprion ([SWI-]) control in all experiments described in this figure.
To further investigate whether the observed effect is SWI/SNF specific, we developed a blue-white visual assay using a previously described lacZ reporter construct, pLS7, whose expression requires SWI/SNF24. A similar visual assay was successfully applied to test the recombinant prion element NMGR25. The candidate cells containing pLS7 appeared white on X-gal-sucrose plates but became blue after guanidine hydrochloride treatment (Fig. 2c). Isogenic [swi-] cells were blue before and after guanidine hydrochloride treatment. This result provides supporting evidence that the [SWI+] candidate cells have compromised SWI/SNF functions.
We next examined the effect of Hsp104, a member of the Clp/Hsp100 family of AAA+ ATPases26, on the[SWI+] candidates. HSP104 disruption eliminates all known yeast prions. In contrast, HSP104 overexpression abolishes [PSI+] but not [PIN+] or [URE3]10. We found that [SWI+] candidates became Ade- and Raf+ upon HSP104 disruption (Fig. 2d and Supplementary Table 1 online) and maintained these phenotypes after reintroduction of a HSP104 expression plasmid (data not shown). These results demonstrate that, as for [PSI+], a transient loss in Hsp104 production eliminates the Raf- phenotype; therefore, Hsp104 is essential for maintaining the Raf- trait. We conclude that the Raf- phenotype is protein-based and that the [SWI+] candidates actually contain two prions: [SWI+] and [PSI+].
We also examined the effect of HSP104 overexpression on [SWI+] by transforming [PSI+][SWI+] cells with p2HG-HSP104, a multicopy HSP104 plasmid with the strong and constitutive GPD (glyceraldehyde-3-phosphate dehydrogenase) promoter. All fresh transformants were mixtures of Ade+/Ade- (~36-42% Ade+) and Raf+/Raf- cells (~82-90% Raf-). After a longer time expression, most cells had lost [PSI+], yet they retained the mixed Raf+/Raf- phenotype (~68 - 87% Raf-) (Supplementary Table 2 online). Subsequent removing of p2HG-HSP104 did not alter their phenotypes. The growth phenotype of one representative [psi-] isolate obtained upon HSP104 overexpression is shown in Figure 2e,f. Although some [PSI+][SWI+] cells lost both prions upon HSP104 overexpression, all examined [PSI+] cells were Raf-. Moreover, [SWI+] was retained in most [psi-] cells (Supplementary Table 2). Thus, [SWI+] is tolerant to Hsp104 overproduction.
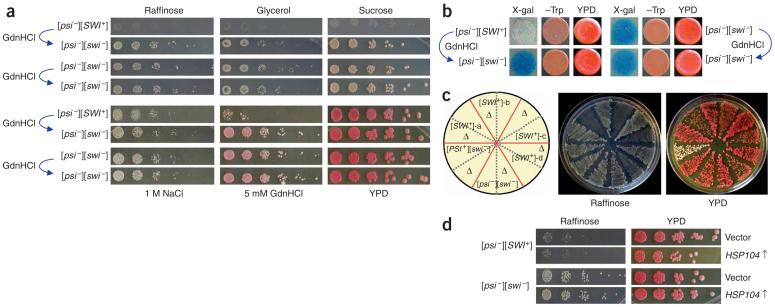
We obtained several [psi-][SWI+] isolates with stable but guanidine hydrochloride-curable Raf- phenotypes after Hsp104 overproduction (Fig. 3a and data not shown). When pLS7 was introduced into these isolates, they appeared white on X-gal-sucrose plates but blue upon guanidine hydrochloride treatment. However, the isogenic [psi-][swi-] cells were blue in both conditions (Fig. 3b). These data show that [SWI+] can exist as a prion independent of [PSI+]. Similar to [PSI+][SWI+] cells, [psi-][SWI+] cells were also Gly- and Gal-(Fig. 3a and Supplementary Fig. 1 online). [SWI+] in the [psi-] background could be eliminated by HSP104 disruption (Fig. 3c, Supplementary Table 1 and Supplementary Fig. 1) but not by Hsp104 overproduction (Fig. 3d), suggesting that the partial loss of [SWI+] in the [PSI+] background upon Hsp104 overproduction is a result of the loss of [PSI+].
Figure 3.
[SWI+] can exist independently from [PSI+]. (a) Phenotypic analysis of a representative [psi-][SWI+] isolate derived from [PSI+][SWI+]-2 upon Hsp104 overproduction. After eliminating the HSP104 overexpression plasmid, [psi-][SWI+] cells treated with or without 5 mM guanidine hydrochloride were spotted onto indicated plates with a fivefold serial dilution. Pictures were taken after 5-d incubation at 30 °C. (b) Isogenic [psi-][SWI+] and [psi-][SWI-] cells containing pLS7 were treated with or without 5 mM guanidine hydrochloride and were grown in -tryptophan synthetic complete liquid media to mid-log phase before spotting to +sucrose -tryptophan media + 20 μg/ml X-gal (X-gal), +glucose -tryptophan media (-Trp) and YPD (YPD) plates. (c) HSP104 disruption eliminates [SWI+] in [psi-][SWI+] isolates. Shown are cell streaks of four isolates on YPD and raffinose plates before and after HSP104 gene disruption. Δ, HSP104 disruption. (d) The [SWI+] prion is tolerant to Hsp104 overproduction. Isogenic [psi-][SWI+] and [psi-][SWI-] cells containing p2HG (vector) or p2HGHSP104 (HSP104↑) were cultured and spotted to the indicated plates.
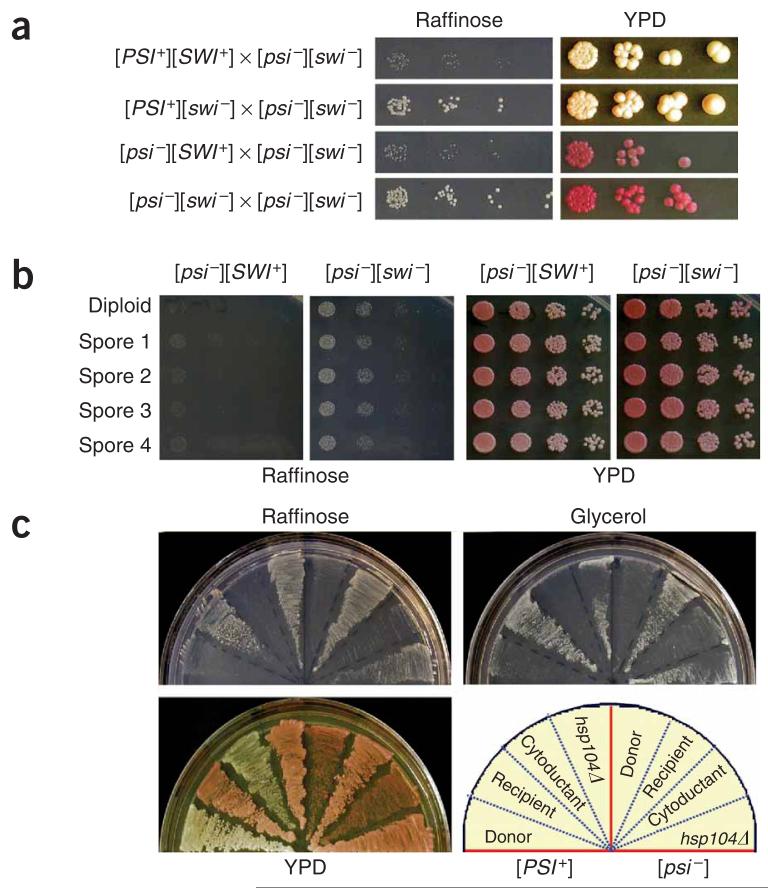
Because of their ‘infectious’ nature, all yeast prions identified to date are transmitted cytoplasmically as dominant traits. To test whether this is also the case for [SWI+], we crossed [SWI+] cells in both [PSI+] and [psi-] backgrounds with isogenic [swi-][psi-] cells. The resulting diploids showed the Raf- phenotype (Fig. 4a and Supplementary Fig. 2 online), indicating that [SWI+] is dominant. We also observed [SWI+] dominance when 74D-694 [SWI+] cells were crossed with isogenic NMΔ-SUP35 ([pin-][swi-]) and hsp104Δ cells, or outcrossed with [swi-] cells of c10B-H49 and S288C (Supplementary Fig. 2). In all cases, [SWI+] diploids could be cured by guanidine hydrochloride treatment (Supplementary Fig. 2). Homologous diploids of SWI/SNF null mutants are deficient in sporulation19. [SWI+] diploids are also poorly sporulated. After repeated attempts, we obtained full spores of seven tetrad sets from 74D-694 [SWI+] diploids, all of which were Raf-, showing that [SWI+] is transmitted as a nonmendelian element (Fig. 4b and Supplementary Fig. 3 online). We next used a kar1 mutant strain to carry out cytoduction experiments, which allow mating partners to mix their cytoplasm without nuclear material transfer27. [SWI+] in both [PSI+] and [psi-] backgrounds was successfully cytoduced from a 74D-694 strain to a c10BH49 strain containing kar1-1. The c10B-H49 cytoductants of [SWI+] grew poorly on raffinose, and this Raf- phenotype could be eliminated by guanidine hydrochloride treatment or HSP104 disruption (Fig. 4c and Supplementary Fig. 4 online). The cytoduction efficiencies of [SWI+][PSI+] and [SWI+][psi-] were about 89% and 49%, respectively (Supplementary Table 3 online). The higher cytoduction rate of [SWI+] in the [PSI+] background suggests that [PSI+] can facilitate [SWI+] transmission. From these data, we conclude that [SWI+] can be transmitted between cells through a simple cytoplasm mixing, and thus is ‘infectious’.
Figure 4.
[SWI+] is dominantly and cytoplasmically inherited. (a) [SWI+] diploids show Raf- phenotype. Diploid cells derived from the indicated crosses were grown in YPD to mid-log phase and spotted onto a raffinose or YPD plate with a fivefold serial dilution. (b) All meiotic progenies of [SWI+] show the Raf- phenotype. Shown are representative phenotypic assays of a full set of spores derived from isogenic [SWI+] or [SWI-] diploids. (c) [SWI+] cytoductants of c10B-H49 showed the Raf- phenotype that could be eliminated by hsp104Δ. Shown are donors, 74D-694 [PSI+][SWI+] (left) or [psi-][SWI+] (right); recipient, c10B-H49 [psi-][SWI-]; cytoductants, [PSI+][SWI+] (left) or [psi-][SWI+] (right); and the hsp104Δ derivatives of the corresponding cytoductants.
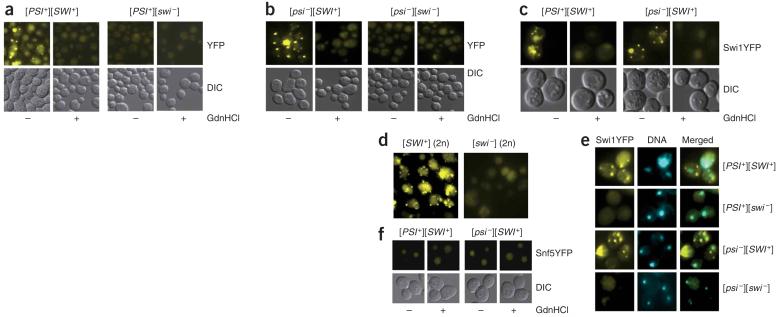
We next tested whether Swi1 had undergone conformational changes in [SWI+] cells. We transformed [PSI+][SWI+] and [PSI+][swi-] cells with p416TEFSWI1YFP, a centromere-based plasmid expressing the Swi1-YFP fusion under a weak and constitutive TEF (translation elongation factor 1α) promoter. We observed that [PSI+][SWI+] cells showed numerous sharp fluorescent foci, indicative of aggregates, but isogenic [PSI+][swi-] cells did not (Fig. 5a). Moreover, [PSI+][SWI+] cells treated with guanidine hydrochloride did not show detectable fluorescent foci (Fig. 5a). Similarly, numerous fluorescent foci were observed in [psi-][SWI+] cells but not in [psi-][swi-] cells or in [psi-][SWI+] cells treated with guanidine hydrochloride (Fig. 5b).
Figure 5.
Swi1 but not Snf5 exists in distinct conformational states in [SWI+] and [swi-] cells. (a,b) Swi1-YFP was aggregated in [PSI+][SWI+] (a) and [psi-][SWI+] (b) but not in isogenic [swi-] cells or [SWI+] cells after guanidine hydrochloride treatment. Cells as indicated were treated with (+) or without (-) 5 mM guanidine hydrochloride. (c) c10B-H49 cytoductants of [SWI+] showed guanidine hydrochloride-curable fluorescent foci of Swi1-YFP. (d) Swi1-YFP was aggregated in the 74D-694 [psi-][SWI+] diploids but not in [psi-][swi-] diploids. (e) Swi1-YFP aggregates were mainly localized in cytoplasm. We stained 74D-694 [SWI+] cells in both [PSI+] and [psi-] backgrounds with Hoechst dye and analyzed them as described in Methods. (f) Endogenous Snf5-YFP is not aggregated in [SWI+] or [swi-] cells. Cells were treated with (+) or without (-) 5 mM guanidine hydrochloride and analyzed under a fluorescence microscope. All cells were transformed with p416TEFSWI1YFP and analyzed by fluorescence microscopy assays after growth in synthetic complete medium lacking uracil to mid-log phase.
We also examined the fluorescence patterns of Swi1-YFP in c10BH49 cytoductants of [PSI+][SWI+] and [psi-][SWI+]. Like their 74D-694 [SWI+] donors, [SWI+] cytoductants containing p416TEFSWI1YFP showed fluorescent foci of Swi1-YFP, but isogenic [swi-] cells or [SWI+] cells after guanidine hydrochloride treatment or HSP104 disruption did not (Fig. 5c and data not shown). Moreover, Swi1-YFP aggregates were also observed in [SWI+] diploids containing p416TEF-SWI1YFP but not in the isogenic [swi-] diploids (Fig. 5d). Our data thus demonstrate that Swi1 exists in altered and aggregated conformational states in [SWI+] cells. The Swi11-YFP foci seemed to be mainly localized in the cytoplasm, as the signals of Swi1-YFP and Hoechst dye (a DNA-binding dye) generally did not overlap (Fig. 5e). As the 74D-964 [SWI+] strain described in this study carries a YFP tag inframe with the chromosomal SNF5 (see Methods), we examined the fluorescence pattern of Snf5-YFP in both [SWI+][PSI+] and [SWI+][psi-] cells containing no plasmids. We did not detect Snf5-YFP fluorescent foci in any of these cells (Fig. 5f).
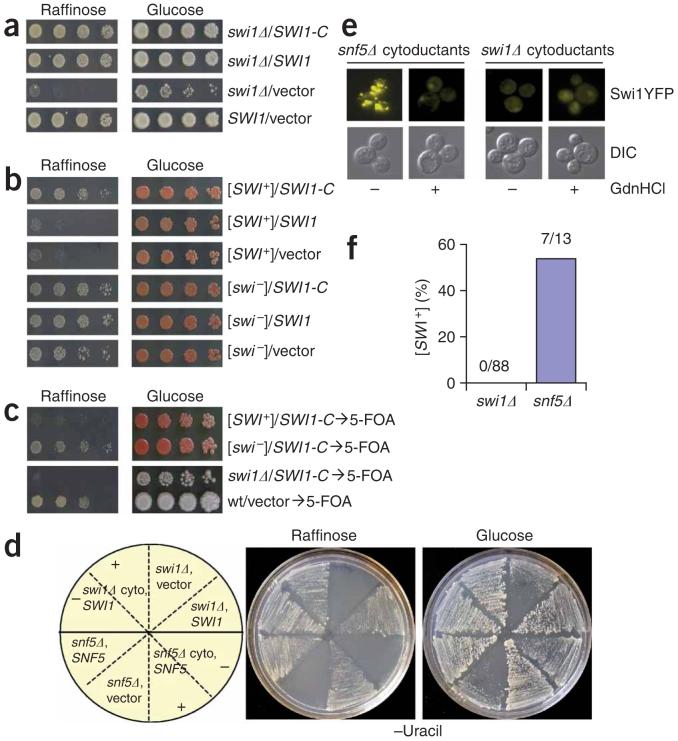
It has been shown that some nonprion aggregation-prone proteins, such as poly-Q peptides, can form aggregates in yeast15,28. Thus, Swi1-YFP aggregation in [SWI+] cells suggests but does not prove that Swi1 is the protein determinant of [SWI+]. To obtain further proof, we created a truncated Swi1 (Swi1-C) protein that lacks the first 535 amino acids containing the glutamine and asparagine-rich regions (Fig. 1a). Ectopic expression of both SWI1-C and the full-length SWI1 were able to fully complement SWI1 deletion in S288C (Fig. 6a, Supplementary Fig. 5 online). This result demonstrates that Swi1-C is sufficient for Swi1 function. Of note, Swi1-C could suppress the Raf- phenotype of [SWI+], whereas the full-length SWI1 could not (Fig. 6b). Both swi1Δ and [SWI+] strains restored the Raf- phenotype upon elimination of the SWI1-C and SWI1 plasmids (Fig. 6c). These results indicate that the ectopically expressed full-length Swi1 was recruited by [SWI+] aggregates, whereas Swi1-C was not. The fact that Swi1-C is capable of masking the [SWI+] phenotype strongly supports the notion that Swi1 is the determinant of [SWI+].
Figure 6.
Swi1 is the protein determinant of [SWI+]. (a) The C domain is sufficient for Swi1 function. Shown are S288C swi1Δ cells containing p416TEFSWI1-C (SWI1-C), p416TEFSWI1 (SWI1) or p416TEF (vector). Wild-type S288C with p416TEF was included as a control. (b) SWI1-C, but not the full-length SWI1, was able to ‘complement’ the Raf- phenotype of [SWI+]. Cells of 74D-694 [PSI+][SWI+] and [PSI+][swi-] containing p416TEFSWI-C (SWI1-C), p416TEFSWI1 (SWI1) or p416TEF (vector) were examined for growth on the indicated plates. (c) Swi1-C was able to phenotypically ‘mask’ but not cure [SWI+]. After plasmid removal, [SWI+] and swi1Δ cells were re-assayed as described in a and b. (d) Swi1 is required for [SWI+] propagation. [SWI+] was cytoduced to isogenic swi1Δ and snf5Δ cells, and the resulting cytoductants were transformed with a SWI1 expression plasmid for swi1Δ cells (swi1Δ cyto, SWI1) or SNF5 for snf5Δ cells (snf5Δ cyto, SNF5). Isogenic swi1Δ and snf5Δ cells were also included as controls. Cell streaks of each representative transformant before (-) or after (+) 5 mM guanidine hydrochloride treatment are shown on the indicated plates. (e) Swi1-YFP is aggregated in the snf5Δ but not in the swi1Δ cytoductants after [SWI+] cytoduction. Cytoductants of swi1Δ and snf5Δ were treated with (+) or without (-) 5 mM guanidine hydrochloride followed by transformation of p416TEF-SWI1YFP and analysis by fluorescence microscopy. (f) Summary of the [SWI+] cytoduction experiments. Shown are the results of three independent experiments.
Prion propagation requires a continuous supply of substrate, such that even transient loss of a prion protein results in loss of the corresponding prion. To test whether constitutive expression of SWI1 is required for [SWI+] propagation, we cytoduced [SWI+] to swi1Δ cells ([psi-][pin-]) and then reintroduced a SWI1 expression plasmid. This transient loss of Swi1 production caused the loss of [SWI+] (Fig. 6d-f); however, transient loss of Snf5 production did not affect [SWI+] propagation. The snf5Δ cytoducants (containing a SNF5 plasmid) showed a guanidine hydrochloride-curable Raf- phenotype, but the isogenic swi1Δ cytoductants (containing a SWI1 plasmid) did not (Fig. 6d). Moreover, we observed that Swi1-YFP foci were only in snf5Δ cytoductants (Fig. 6e). Among 88 examined swi1Δ cytoductants, none retained [SWI+]. However, [SWI+] existed in 7 of 13 examined snf5Δ cytoductants (Fig. 6f). These results demonstrate that Swi1 is the protein determinant of [SWI+].
We have identified a novel prion that epigenetically modifies the function of the chromatin-remodeling complex SWI/SNF. Swi1 can adopt altered and heritable conformations without SWI1 overexpression (Supplementary Fig. 6 online), suggesting that [SWI+] can occur naturally. Although the frequency of [SWI+] occurrence remains to be determined, the fact that Swi1 is able to spontaneously switch between alternative conformations strongly suggests that prion-mediated ‘protein inheritance’ can be a regulatory mechanism contributing to global gene expression.
METHODS
Yeast strains
The [SWI+] prion was initially established in a 74D-694 strain whose chromosomal SNF5 was fused inframe with YFP (MATa ade1-14 trp1-289 his3-200 ura3-52 leu2-3, 112 SNF5YFP::kanMX4 [psi-][pin-]). To test [SWI+] dominance, we used a nontagged 74D-694 strain (MATα ade1-14 trp1-289 his3-200 ura3-52 leu2-3, 112 [psi-][pin-]) for mating to generate diploids and spores. Strain c10B-H49 (MATα ade2-1 SUQ5 lys1-1 his3-11,15 leu2-3, 112 kar1-1 ura3::KanMX4 [psi-] [pin-]) was used for cytoductions. We used a S288C strain (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 gpa1Δ [psi-] [pin-]) in mating and for SWI/SNF phenotypic assays. A swi1Δ strain (MATa his3Δ1 lys1 leu2Δ0 met15Δ0 ura3Δ0 swi1:: KanMX4 [psi-][pin-]) obtained from sporulating a S288C diploid (SWI1/swi1Δ, ATCC number: 4022812) was used in SWI1 complementation analysis and cytoduction. A S288C snf5Δ strain (MATa his3Δ1 lys1 leu2Δ0 met15Δ0 ura3Δ0 snf5:: KanMX4 [psi-][pin-], ATCC number: 4007175) was used in cytoduction. Diploids from outcrossing of 74D-694 to c10B-H49 or c10B-H49 to S288C were formed occasionally in cytoduction and partially characterized.
Yeast culture and general manipulations were done as previously described29. For testing SWI/SNF phenotypes, 1 μg/ml antimycin A from Streptomyces sp. (Sigma) was supplemented to media containing 2% of the following nonfermentable carbon sources: raffinose, sucrose, or galactose.
Plasmids
The SNF5 ORF was amplified by PCR using plasmid pLY14 (ref. 30) as a template. The 3.5-kb PCR product was initially cloned into p416GPD then subcloned into the BamHI and XhoI sites in p426GPD to obtain p426GDPSNF5. The SWI1 ORF was amplified by PCR from 74D-694 genomic DNA. The 4.0-kb PCR fragment was first ligated to the SpeI and XhoI sites in p416TEF then subcloned into the same sites in p426GPD to obtain p426GPDSWI1. We created plasmid p416TEFSWI1YFP by fusing the 4.0-kb SWI1 PCR product inframe to the 5′ terminus of the YFP gene using the SpeI and SmaI sites in p416TEFYFP. To create p426GPDGFP, we cloned the 0.75-kb GFP fragment into p416TEF through NheI and SacI and then subcloned it into p426GPD through SpeI and XhoI sites. To create the SWI1-C allele, we amplified the 2.3-kb fragment of SWI1 lacking the 5′ region encoding the first 535 amino acids from p416TEFSWI1YFP by PCR. The PCR products were ligated to p416TEF through SpeI and XhoI sites to produce the plasmid p416TEFSWI1-C. All constructs were confirmed by DNA sequencing.
The reporter plasmid pLS7 (ref. 24) was provided by M. Carlson. Plasmids p416CUP1RNQ1GFP, pRS313CUP1NMGFP, p2HGHSP104 and p2HG have been described previously29. The HSP104 disruption plasmid was a gift from S. Lindquist.
Cytoduction
We carried out cytoductions by co-incubating donor and recipient cells overnight at 30 °C and then spreading the incubation mixture onto selective media for cytoductants. Individual isolates that had recipient mating types and that lacked donor's chromosomal markers and contained recipient auxotrophic markers were scored as cytoductants. To cytoduce 74D-694 (MATa) [SWI+] in both [PSI+] and [psi-] backgrounds to c10B-H49 (MATα, kar1-1), we integrated the donors with a URA3 marker by transforming the linearized plasmid pRS306 after StuI digestion. The resulting Ura+ transformants were further transformed with p425GAL1 (Leu+) and used as donors. Isolates that were Leu+, Trp+, Ura- and MATα were scored as cytoductants. To cytoduce [SWI+] to S288C swi1Δ and snf5Δ cells, we incubated [SWI+] cells of c10B-H49 (MATα, kar1-1) containing p416TEFSWI1-C with swi1Δ and snf5Δ cells (S288C MATα) with an integrated HIS3 marker overnight at 30°C. Cytoductants were selected on synthetic complete medium that lacked uracil and histidine. Isolates that were Ura+, His+, Met-, Lys+ and MATα were scored as cytoductants. We eliminated the p416TEFSWI1-C plasmid from the cytoductants by growing them on 5-fluoroorotic acid (5-FOA) containing media before 5 mM guanidine hydrochloride treatment, then we transformed p416TEFSWI1 to swi1Δ cytoductants, pLY14 (SNF5 expression plasmid) to snf5Δ cytoductants, or p416TEFSWI1YFP to both swi1Δ and snf5Δ transformants. The resulting transformants were subsequently examined for their abilities to grow on media containing raffinose as the sole carbon source and to form Swi1-YFP aggregates. A corresponding empty vector was included as control for each transformation experiment.
Nuclear localization
We used Hoechst 33342 dye (Invitrogen) to stain DNA according to the manufacturer's instructions.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank M. Carlson (Department of Genetics and Development, Columbia University) for the gift of the pLS7 plasmid; B.C. Laurent (Department of Oncological Sciences, Mount Sinai School of Medicine) for the gift of the pLY14 plasmid; S. Lindquist (Whitehead Institute for Biomedical Research, Department of Biology, Massachusetts Institute of Technology and Howard Hughes Medical Institute) for the Hsp104 antibody; J. Workman for helpful discussions; E. Crow and G.E. Kim for technical assistance; R. Lawrence, C. Kowalczyk, R. Miller, T. Volpe, C. Long and E. Crow for critical comments and manuscript editing. This work was partially supported by grants from the United States Army (0850-370-R744), the Ellison Medical Foundation and the US National Institutes of Health (R01NS056086) to L.L.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
References
- 1.Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 2003;13:136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 2.Sudarsanam P, Iyer VR, Brown PO, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuite MF, Cox BS. Propagation of yeast prions. Nat. Rev. Mol. Cell Biol. 2003;4:878–890. doi: 10.1038/nrm1247. [DOI] [PubMed] [Google Scholar]
- 4.Wickner RB. [URE3] as an altered Ure2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 5.Prusiner SB. Prions. Proc. Natl. Acad. Sci. USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox B. [PSI], a cytoplasmic suppressor of super-suppression in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- 7.Lacroute F. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol. 1971;206:519–522. doi: 10.1128/jb.106.2.519-522.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol. Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 9.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN+] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 10.Uptain SM, Lindquist S. Prions as protein-based genetic elements. Annu. Rev. Microbiol. 2002;56:703–741. doi: 10.1146/annurev.micro.56.013002.100603. [DOI] [PubMed] [Google Scholar]
- 11.Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr. Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- 12.Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 13.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 14.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derkatch IL, et al. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI.] prion in yeast and aggregation of Sup35 in vitro. Proc. Natl. Acad. Sci. USA. 2004;101:12934–12939. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc. Natl. Acad. Sci. USA. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 18.Smith CL, Horowitz-Scherer R, Flanagan JF, Woodcock CL, Peterson CL. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat. Struct. Biol. 2003;10:141–145. doi: 10.1038/nsb888. [DOI] [PubMed] [Google Scholar]
- 19.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guidi CJ, et al. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell. Biol. 2001;21:3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts CW, Orkin SH. The SWI/SNF complex-chromatin and cancer. Nat. Rev. Cancer. 2004;4:133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- 22.Ross ED, Minton A, Wickner RB. Prion domains: sequences, structures and interactions. Nat. Cell Biol. 2005;7:1039–1044. doi: 10.1038/ncb1105-1039. [DOI] [PubMed] [Google Scholar]
- 23.Tuite MF, Mundy CR, Cox BS. Agents that cause a high frequency of genetic change from [PSI+] to [psi-] in Saccharomyces cerevisiae. Genetics. 1981;98:691–711. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neigeborn L, Carlson M. Mutations causing constitutive invertase synthesis in yeast: genetic interactions with snf mutations. Genetics. 1987;115:247–253. doi: 10.1093/genetics/115.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Lindquist S. Creating a protein-based element of inheritance. Science. 2000;287:661–664. doi: 10.1126/science.287.5453.661. [DOI] [PubMed] [Google Scholar]
- 26.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 27.Conde J, Fink GR. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl. Acad. Sci. USA. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park KW, Hahn JS, Fan Q, Thiele DJ, Li L. De novo appearance and “strain” formation of yeast prion [PSI+] are regulated by the heat-shock transcription factor. Genetics. 2006;173:35–47. doi: 10.1534/genetics.105.054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laurent BC, Treitel MA, Carlson M. The SNF5 protein of Saccharomyces cerevisiae is a glutamine- and proline-rich transcriptional activator that affects expression of a broad spectrum of genes. Mol. Cell. Biol. 1990;10:5616–5625. doi: 10.1128/mcb.10.11.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.