Abstract
Expression of estrogen-related receptor alpha (ERRα) has recently been shown to carry negative prognostic significance in breast and ovarian cancers. The specific role of this orphan nuclear receptor in tumor growth and progression, however, is yet to be fully understood. The significant homology between estrogen receptor alpha (ERα) and ERRα initially suggested that these receptors may have similar transcriptional targets. Using the well-characterized ERα-positive MCF-7 breast cancer cell line, we sought to gain a genome-wide picture of ERα-ERRα cross-talk using an unbiased microarray approach. In addition to generating a host of novel ERRα target genes, this study yielded the surprising result that most ERRα-regulated genes are unrelated to estrogen-signaling. The relatively small number of genes regulated by both ERα and ERRα led us to expand our study to the more aggressive and less clinically treatable ERα-negative class of breast cancers. In this setting we found that ERRα expression is required for the basal level of expression of many known and novel ERRα target genes. Introduction of an siRNA directed to ERRα into the highly aggressive breast carcinoma MDA-MB-231 cell line dramatically reduced the migratory potential of these cells. Although stable knockdown of ERRα expression in MDA-MB-231 cells had no impact on in vitro cell proliferation, a significant reduction of tumor growth rate was observed when these cells were implanted as xenografts. Our results confirm a role for ERRα in breast cancer growth and highlight it as a potential therapeutic target for estrogen receptor-negative breast cancer.
Keywords: estrogen-related receptor, breast cancer, ESRRA
Introduction
Estrogen-related receptor alpha (ERRα, NR3B) is an orphan member of the nuclear receptor (NR) superfamily that shares considerable structural homology to the classical estrogen receptors ERα and ERβ. Although its primary function appears to be the activation of fatty acid oxidation and mitochondrial biogenesis in tissues that have high energy demands (e.g., cardiac and skeletal muscle), its expression has also been associated with a negative outcome in breast and ovarian cancers (1–3). This latter finding is somewhat paradoxical given that cancers are generally thought to rely predominantly on glycolytic metabolism (4). It is unclear therefore, considering the known biology of ERRα, how this receptor is involved in tumor pathophysiology.
One proposed mechanism regarding the role of ERRα in breast cancer biology stems from the observation that ERRα and ERα share a high degree of structural similarity. It has been hypothesized in breast cancer that ERRα may function as a modulator of estrogen signaling. Indeed, crosstalk between the ERα and ERRα signaling pathways has been observed at several levels (5–8).
It was initially suggested that ERRα might function as an alternate receptor for estrogens. However, it now appears that neither physiological nor synthetic ERα ligands are likely to interact in a significant manner with ERRα. Indeed, crystallographic analysis of apo-ERRα indicates that even without ligand this orphan receptor adopts an active conformation, recruiting cofactors and initiating transcription (9).
Despite the inability of ERRα to respond directly to estrogens and antiestrogens, this receptor may affect ERα signaling at the level of target gene transcription. Notably, ERα and ERRα are more highly homologous within their DNA binding domains. Not surprisingly, therefore, direct DNA-binding studies and reporter gene assays have indicated that many classical palindromic Estrogen Response Elements (EREs) can function as Estrogen-Related Receptor Response Elements (ERREs) (10). The majority of the ERREs mapped in genes involved in metabolism, however, do not interact in a significant manner with ERα. That EREs may represent a subset of ERREs supports a model in which ERRα exhibits activities both related and unrelated to ERα signaling in breast cancer. Definition of the ERRα transcriptome in breast cancer will likely help to determine the extent to which its role in this context involves cross-talk with the ERα-signaling pathway and to identify the mechanisms by which this receptor impacts tumor pathology.
Whereas a physiological role for ERRα as a regulator of estrogen signaling remains to be described, its role as a modulator of metabolic processes in muscle, heart and liver are well-characterized. In these settings, ERRα enhances β-oxidation of fatty acids and regulates flux through the tricarboxylic acid (TCA) cycle and oxidative phosphorylation (OXPHOS) by transcriptional induction of rate limiting enzymes in each process (11–14). Although certain tumors rely heavily on glycolysis, the capacity to generate ATP through OXPHOS is not always completely lost. For example, cancer cells may retain the ability to use a variety of oxidative substrates, including fatty acids, glutamine, and ketone bodies (15, 16). Recent studies have shown that the TCA cycle and OXPHOS pathway are utilized in tumors not only to generate ATP, but also to supply the biosynthetic precursors required for sustained proliferation (17). Considering the complexity of tumor metabolism, the extent to which ERRα function in breast cancer is related to either its metabolic functions or its interaction with the ERα-signaling pathway remains to be determined.
In view of this potential duality in ERRα function, we sought to define the ERα-related and ERα-unrelated transcriptional effects of ERRα activity in breast cancer cells. The significant number of genes affected by ERRα activity but not by estrogen treatment led to the hypothesis that an ERα-independent activity of ERRα may partially account for the negative correlation between ERRα expression and patient outcomes. We used a xenograft model of ERα-negative breast cancer to test the impact of ERα-independent ERRα activity on breast cancer pathology and found that ERRα knockdown significantly inhibited tumor growth. These studies provide compelling evidence that ERRα is involved in breast cancer growth and justify further investigation into the use of ERRα as a therapeutic target.
Materials and Methods
Cell culture
MDA-MB-231 cells were cultured in DMEM, AU565 and BT474 cells in RPMI, and MCF-7 cells in DMEM/F12 (Invitrogen). Stable MDA-MB-231 luciferase cells and derived cells were grown in DMEM supplemented with antibiotics.
Generation and transduction of adenoviruses
The generation of PGC-1α adenoviruses has been described (18). Cells were infected at a multiplicity of infection (MOI) of 20–100 (2 h at 20°C). For microarray experiments, MCF-7 cells were cultured in estrogen-free media (48 h) before adenoviral infection (MOI 50), treated 12 h later and harvested after an additional 12 h. In experiments with siRNA and PGC-1α infections, cells were infected with siRNA adenovirus 48 h prior to a infection with PGC-1α (11). Immunoblot analysis verified that ERRα knock-down did not alter ERα expression (Supplementary Figure 1).
RNA preparations and analysis
Total RNA was isolated using Qiagen or BioRad RNA purification columns and DNase treated (BioRad). An iScript cDNA synthesis kit (BioRad) was used (1µg RNA). Real-time PCR was performed (0.25µL cDNA, 0.4µM specific primers (Table S1), and BioRad iQ SYBRGreen supermix) and results quantified using the 2−ΔΔCt method (19).
Microarray data analysis
Probe synthesis, hybridization on Affymetrix HG133A GeneChips (Affymetrix, Santa Clara, CA) and scanning were performed at the Expression Analysis Institute (Durham, NC). Probe-level analysis was performed in dChip (20). Normalization and statistical cutoffs are described in detail in the Supplementary Materials and Methods.
Immunobloting
Protein extracts were separated by 8% SDS-PAGE and transferred onto nitrocellulose membrane. ERRα protein was detected using a mouse anti-ERRα monoclonal antibody (18), and Glyceraldehyde-3-phosphate dehydrogenase detected using goat anti-GAPDH (V-18) (Santa Cruz Biotechnology, Inc).
Retroviral constructs and gene silencing
ERRα siRNA oligonucleotides (11) and control siRNA oligonucleotides (18) were ligated into PSR-GFP/Neo (OligoEngine). Stable cell lines were constructed and sorted using standard techniques as described in Supplementary Materials and Materials. To achieve ERRα re-expression, an siRNA-resistant ERRα (ERRsiR) was constructed by introducing silent mutations into the siRNA targeted region.
Proliferation and migration assays
For proliferation assays, 5000 cells per well were plated in a 96-well plate. Cell number was assayed using FluoReporter Blue Fluorometric dsDNA Quantitation kit (Molecular Probes). For migration assays, serum starved cells migrated for 4 h, were crystal violet stained and then counted. See Supplementary Materials and Methods for details.
In vivo tumor formation
All animal handling and procedures were approved (Duke University Medical Center). 2 × 106 cells (in 50% matrigel (BD Matrigel Matrix, BD Biosciences) in PBS) were injected into the axillary mammary fat pad of 6–8 week-old female athymic nu/nu mice (10 mice per group) (21). Primary tumor volume was quantified using calipers (volume = (width2)(length)/2) and tumors removed either at a maximum of 1500 mm3 or upon ulcerated. Tumors were excised and frozen in liquid N2.
Statistical analyses
Proliferation, migration and RT-qPCR data are represented as mean ± SEM for biological replicates unless otherwise noted. Significance was evaluated by Mann-Whitney U-test for all biological data (GraphPad) and Student’s t-test as indicated.
Results
The majority of ERRα target genes in breast cancer are unrelated to ERα signaling
The relationship between the ERα and ERRα transcriptomes was evaluated in MCF-7 breast cancer cells using a classical microarray approach. Since a small molecule ligand has not been identified for ERRα, its transcriptional activity in these studies was induced using its known coactivator PGC-1α (peroxisome proliferator-activated receptor-γ coactivator-1α) as a protein ligand (22). Although complementary approaches such as knocking down ERRα were subsequently employed, PGC-1α was used as a coactivator to highlight ERRα target genes by providing a large dynamic range of regulation amenable to microarray technology. PGC-1α enhances the transcriptional activity of several nuclear receptors (NRs) and non-NR transcription factors resulting in the regulation of a variety of metabolic processes including lipid metabolism, mitochondrial biogenesis and oxidative metabolism (14, 23). To isolate the ERRα-PGC-1α complex in these studies, we utilized a customized PGC-1α that selectively binds to and activates the ERRs (ERRspPGC1). Non-NR-dependent activities of PGC-1α were controlled for using a variant PGC-1α in which the leucines within the NR-interacting domain had been mutated to alanines (L2L3M). We previously reported the construction, validation and use of ERRspPGC1 (therein termed PGC-1α 2X9) and L2L3M in the context of hepatic gene regulation and function of ERRα (18).
For the microarray study, MCF-7 cells were grown in estrogen-free conditions for 48 hours prior to adenoviral infection with wild-type (WT) PGC-1α, ERRspPGC1, L3L3M or β-galactosidase (βgal). When maximal expression of the PGC-1α construct was achieved (12 h), cells were treated with either vehicle or estradiol (10nM) for an additional 12 hours. This enabled the identification of genes regulated by ERRα or ERα in breast cancer cells and analysis of pathway overlap. We confirmed equivalent expression of each PGC-1α variant and that ERRspPGC1 activates known ERRα target genes (e.g., medium chain acyl dehydrogenase, ACADM) similarly to wild-type PGC-1α, but does not activate ERα targets (e.g., progesterone receptor, PR) (Figure 1C and Supplementary Figure S2). We also verified that canonical ERα target genes (e.g., PR) were activated by estradiol treatment under our experimental conditions (Supplementary Figure S2).
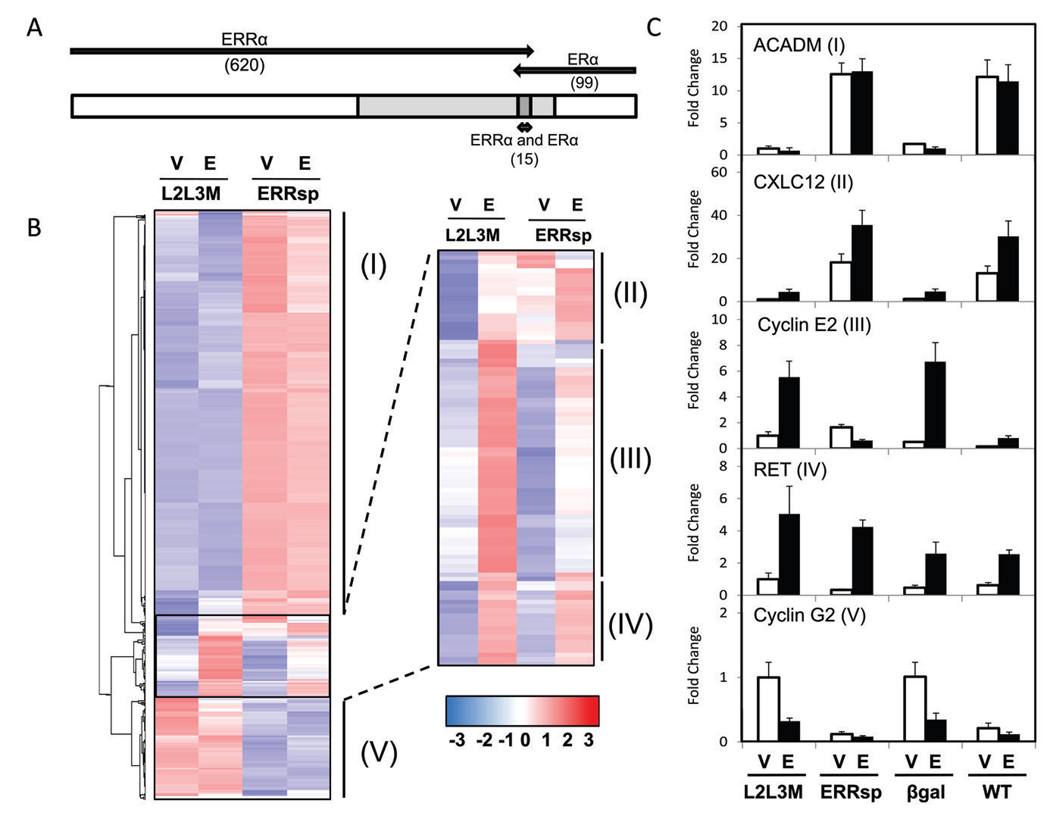
Figure 1. Definition of the overlap between ERRα and ERα signaling in MCF-7 cells.
A, Number of genes regulated by ERRα (induced or repressed by infection of ERRspPGC1 compared to L2L3M) versus genes regulated by ERα (induced or repressed by estrogen treatment) and those that belong to both groups (15 genes). 620 ERRα-regulated and 99 ERα-regulated genes were identified using a Student’s t-test with a p-value cutoff of p<0.01 and fold change of >50% up or down. A comparison was then made to determine the subset of genes regulated by either pathway in which expression levels were significantly different (p<0.01) between MCF-7 cells expressing ERRspPGC1 (vehicle treated) and MCF-7 cells treated with estradiol (expressing control L2L3M). Genes in which this secondary analysis demonstrated no significant difference between ERRα and estrogen-induced expression are shown in light gray. B, Hierarchical clustering of genes regulated by ERα and ERRα, illustrated on a subset of MCF-7 microarray conditions. Cells were infected with adenovirus and treated twelve hours later with vehicle or estradiol (10nM). ERRα regulated genes were defined as the set of genes affected by ERRspPGC1 (I, II, III, V) and ERα targets as genes induced or repressed by addition of estradiol (predominately in Classes II, III, IV). C, Real-time PCR of representative genes from each class found in the microarray. Four different infections (negative control β-galactosidase and wild-type PGC-1α in addition to the two PGC-1α mutants) were analyzed with vehicle or estradiol treatment. 36B4 was used as internal normalization control, and a representative experiment shown for each gene. ACADM (medium chain acyl dehydrogenase) represents genes regulated only by ERRα activation (Class I) and RET (ret proto-oncogene) represents genes regulated only by ERα (Class IV). Note that in addition to the jointly regulated genes in Class II and III (e.g, CXCL12 (stromal derived factor-1) and cyclin E2), Class V was found to contain some dually regulated genes (e.g., cyclin G2).
Analysis of microarray expression data revealed that the vast majority of genes were similarly regulated by ERRspPGC1 and WT PGC-1α compared to either control condition (βgal or PGC-1α L2L3M; Supplementary Figure S3). This result agrees with our previous findings (24), and suggests that ERRα may be a major conduit of PGC-1α signaling in breast cancer cells. We did find, however, that WT PGC-1α regulated a subset of genes differently than ERRspPGC1 (particularly in estradiol treated samples), which is likely due to PGC-1α coactivation of other NRs (e.g., ERα) (25). Therefore, to determine the degree of cross-talk between ERRα and ERα, we identified genes regulated by either ERRspPGC1 (vehicle treated, PGC-1α L2L3M versus ERRspPGC1 infected) or estradiol (PGC-1α L2L3M infected, vehicle versus estradiol). Applying identical statistical thresholds for each comparison (Student’s t test p<0.01, fold change >50%), 620 ERRα-regulated and 99 ERα-regulated genes were identified (Supplementary Table S2). Initial inspection revealed that only 15 probesets were present in both groups, and we next sought to address the difference in the gene expression patterns between the samples with activated ERRα versus with activated ERα (ERRspPGC1 infected, vehicle treated samples versus L2L3M infected, estradiol treated samples). This analysis indicated that the effects of ERRα and ERα may not be as distinct as was thought based on the small number of genes regulated by both receptors in our initial analysis. Specifically, 34% of the 704 initially identified genes were not differentially regulated by ERRα versus by ERα (Figure 1A light gray region, Supplementary Table S2).
Unsupervised hierarchical clustering of the genes regulated by either ERRα or ERα revealed five major expression patterns (Figure 1B). We validated genes subject to each pattern of regulation by RT-qPCR (Figure 1C and Supplementary Figure S4). The majority of genes are induced predominately by ERRα, with little ERα contribution (Classes I). Class II represents genes that are positively regulated by ERα and ERRα, while Class III is comprised of genes upregulated by ERα and repressed by ERRα. Although direct competition for DNA binding may explain the activity of ERRα on Class III genes, it may also be due to cofactor competition, a mechanism that is likely to be relevant in cells where both receptors are activated. The reduction of basal levels of gene expression by ERRspPGC1 on some genes in this group raises the possibility that the ERRα actively represses genes through a mechanism independent of ERα. Genes in Class IV are upregulated by estradiol with little or no contribution of ERRα. Finally, the dominant feature of Class V genes is repression by ERRα. Within this class, however, resides a subset of genes repressed by both ERRα and ERα (e.g., Cyclin G2). Although these results taken together suggest some overlap between the ERα and ERRα pathways in breast cancer cells, they also demonstrate that a large portion of ERRα activity may be unrelated to ERα-signaling. This finding motivated our continued investigations into the role of ERα-independent ERRα activity in breast cancer.
Functional classes of genes regulated by ERRα in breast cancer cell lines
As a first step in defining the functional role of ERRα in breast cancer, we performed gene ontology (GO) analysis on the ERRα regulated genes identified above. This analysis yielded 12 overrepresented GO terms, the majority of which are composed of genes involved in aerobic metabolism (Supplementary Table S3). Notably, we found that the majority of enzymes of the TCA and electron transport chain are induced by ERRα. Several of these genes have been shown previously to be physiologically relevant ERRα targets in other tissues, and their potential roles in tumor pathophysiology is a matter of active research (24).
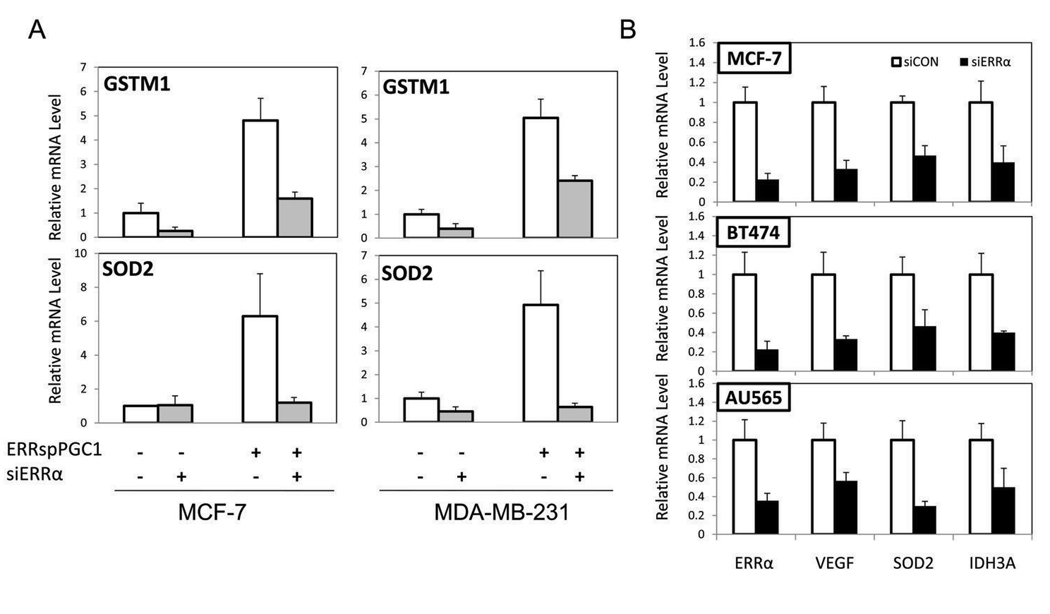
While GO analysis revealed a major impact of ERRα signaling on genes involved in aspects of energy metabolism, inspection of the list of ERRα-regulated genes also provided insight into its role in tumor biology. Considering both the intrinsic and treatment-induced metabolic stress in the tumor microenvironment, it is notable that several genes involved in the response to oxidative stress, including superoxide dismutase 2 (SOD2) and detoxification enzyme glutathione s-transferase M1 (GSTM1) were induced by ERRspPGC1 (Figure 2A). Using a knock-down approach to compliment activation of ERRα by PGC-1α, we verified that these genes were regulated by ERRspPGC1 in an ERRα-dependent manner in both ERα-positive and ERα-negative cell lines (Figure 2A). Furthermore, and of particular relevance to cancer biology, ERRspPGC1 induced expression of the highly angiogenic growth factor VEGF (vascular endothelial growth factor), which has recently been identified as a direct ERRα transcriptional target in murine skeletal muscle (26). We found that the basal level of many ERRspPGC1-induced genes, including VEGF, SOD2 and the key TCA cycle enzyme IDH3A (isocitrate dehydrogenase 3A) is dependent on ERRα expression in the ERα-positive MCF-7 and BT474 and ERα-negative AU565 and MDA-MB-231 cell lines (Figure 2B and data not shown).
Figure 2. The regulation of genes involved in the response to oxidative stress by ERRα and the dependence of basal levels of ERRα target genes on ERRα expression.
A, PGC-1α induction of SOD2 and GSTM1, ERRα target genes identified in the MCF-7 array, is dependent on ERRα expression. MCF-7 and MDA-MB-231 cells were serially infected with the siRNA to ERRα (siERRα) or scrambled control (siCON) adenovirus indicated followed by ERRspPGC1 or the control β-galactosidase virus. mRNA levels are assessed by RT-qPCR, normalized to the expression of 36B4 and represented ± SEM.B, Basal levels of ERRα target genes in breast cancer cell lines MCF-7 and BT474 (ERα-positive) and AU565 (ERα-negative) cell lines analyzed by RT-qPCR following adenoviral infection with siRNA to ERRα (siERRα) or scrambled control (siCON).
Taken together, our data suggest that ERRα is critical for expression of the detoxification genes SOD2 and GSTM1, the angiogenic gene VEGF, and that it can regulate the expression of a numerous genes involved in energy metabolism. Furthermore, ERRα plays a role in the basal expression of these genes in both ERα-positive and ERα-negative breast cancer cell lines. Encouraged that endogenous ERRα contributes to the basal expression of these potentially cancer-associated genes, we sought to determine the phenotypic effects of manipulating ERRα activity. Because of the limited cross-talk found in the MCF-7 microarray as well as the potential significance of the many ERRα-regulated genes unrelated to ERα signaling, we focused on delineating the role of ERRα in ERα-negative breast cancer.
Evaluation of the role of ERRα on proliferation and migration in a cellular model of ERα-negative breast cancer
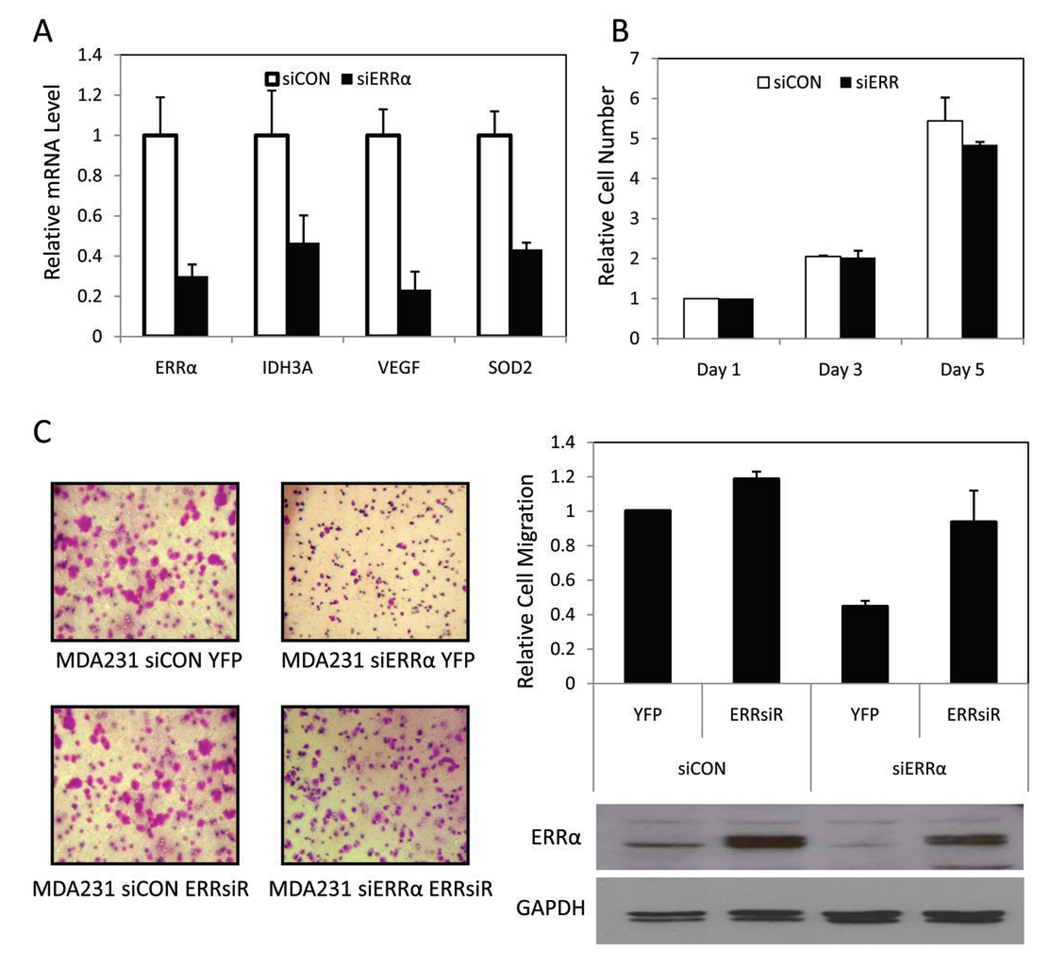
We generated a series of derivatives of the MDA-MB-231-luciferase cell line which is a well-validated model of ERα-negative breast cancer that can be studied in vitro or as a xenograft (27). Derivative stable cell lines were constructed by retroviral-mediated expression of an siRNA directed against ERRα or a scrambled control siRNA and transformants selected using a co-expressed GFP protein (11). The resultant polyclonal populations (a) MDA-MB-231-luc siERRα and (b) MDA-MB-231-luc siCON (referred to as MDA231 siERRα and MDA231 siCON) were then subjected to further characterization. We demonstrated quantitative inhibition of ERRα expression in the MDA231 siERRα cells compared to the control cell line (Figure 3A). The efficacy of the siRNA was confirmed as expression of ERRα target genes was decreased in the cells with ERRα knocked-down compared to control cells (Figure 3A).
Figure 3. Generation and characterization of MDA-MB-231-luciferase cells expressing ERRα shRNA.
A, RT-qPCR of ERRα and target genes in MDA231 siERRα cells versus MDA231 siCON cells. Data normalized to 36B4 and shown as fraction of siCON mRNA ± SEM. B, Results of in vitro proliferation assays showing no statistically significant differences between cell lines. Data from three biological replicates plotted as fold over cells plated ± SEM. C, Down-regulation of ERRα results in significantly reduced ability to migrate while re-expression of ERRsiR restores migration. The indicated stable MDA-MB-231 cell lines were serum starved for 18 h and allowed to migrate toward serum in a Boyden Chamber. Migrated cells were stained, imaged and counted. Cells were plated in triplicate and data of biological replicates plotted as normalized ratio to cells migrated in the MDA231 siCON YFP cell line. Western blot analysis of the protein levels of ERRα in whole cell extract from each MDA-MB-231-derived cell lines with GAPDH shown as a loading control.
We next evaluated the effects of ERRα knockdown on in vitro proliferation using the MDA231 siERRα and MDA231 siCON cell lines. Under optimal growth conditions, we determined that altering ERRα activity does not affect the basal rate of cell growth in these cells (Figure 3B). Comparable results were observed in other breast cancer cell lines similarly manipulated (data not shown). We next addressed whether ERRα is required for the invasive phenotype of MDA-MB-231 cells. Using an in vitro transwell migration assay we observed that ERRα knock-down resulted in a 65% reduction in cell migration (Figure 3C). Importantly, re-expression of ERRα using an siRNA-resistant ERRα (ERRsiR) resulted in reversion to the control level of migration. Although the molecular basis for this change in migratory ability remains unknown, these results indicate that ERRα can affect in vitro migration and would suggest that ERRα may alter the in vivo correlate, tumor metastasis.
Decreased ERRα expression compromises the growth of ERα-negative tumors in athymic nude mice
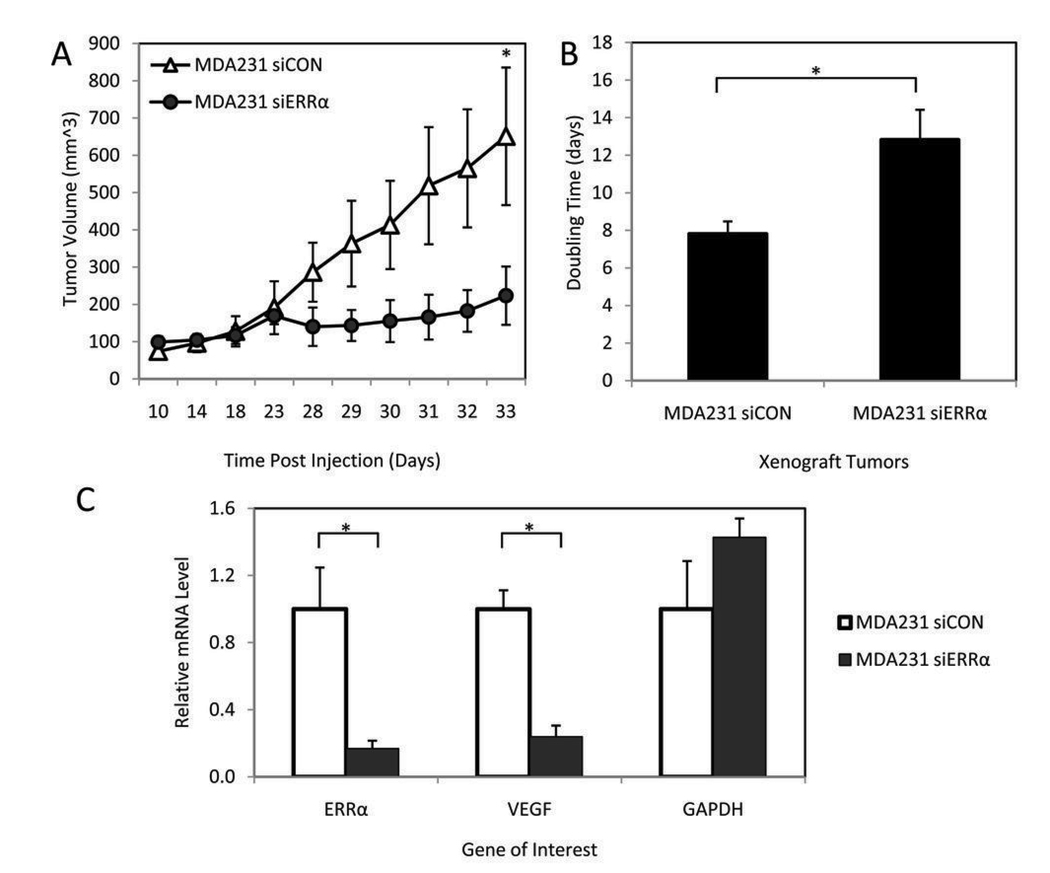
Given that the majority of genes regulated by ERRα in breast cancer cells are involved in metabolism, we hypothesized that if ERRα had a role in cell growth, it might be evident only under the metabolic pressures of in vivo tumor growth. Therefore, we proceeded to evaluate the role of ERRα in cells propagated as murine xenografts, a model that would allow us to evaluate the impact of tumor microenvironment on ERRα function. Specifically, MDA231 siERRα or MDA231 siCON cells were injected orthotopically into the mammary fat pads of athymic nude mice (10 mice per group). Although the initial take rate of each cell line was equivalent, the growth rate of the resultant tumors was significantly lower in the ERRα knockdown cells versus control (Figure 4A). Intriguingly, the MDA231 siERRα-derived tumors exhibited a lag phase prior to resumption of tumor growth such that seven out of ten tumors reached over 500 mm3 by day 46. Analysis of the growth curves for each tumor demonstrated that the doubling time of the siERRα tumors was 12.8 days versus 7.8 days for the tumors arising from control siCON cells (Figure 4B). We hypothesize that under the selective pressure of the tumor environment, the siERRα tumors eventually develop alternative mechanisms to adapt to the metabolic conditions in vivo. The development of resistance to ERα-inhibition is similarly manifest in a growth curve lag phase and increased doubling time when MCF-7 cells are grown as xenografts in the presence of fulvestrant (28).
Figure 4. Silencing of ERRα decreases the growth rate of MDA-MB-231-luciferase cells grown as xenografts.
Stable cell lines were injected into the mammary fat pad of nude mice. A, Tumor growth rate of the cell lines after orthotopic injection. Points, mean volume for 10 mice per cell line; bars, SEM (*P<0.05). B, Doubling time of each tumor was calculated by best fit exponential curve and plotted as the average ± SEM (*P<0.05). C, The expression of ERRα and VEGF are decreased in ERRα knock-down tumors compared to control tumors while the control GAPDH is not. mRNA was collected and target gene expression measured by RT-qPCR. Data are normalized to cyclophilin expression and represented ± SEM (*P<0.05).
Real time PCR analysis of the primary tumors harvested from mice revealed that ERRα remained silenced in the siERRα xenografts (Figure 4C). Given the regulation of VEGF by ERRα in vitro and the prominent role of VEGF in tumor angiogenesis, we examined VEGF expression in the xenograft tumors. Although VEGF mRNA was significantly decreased in the MDA231 siERRα-derived tumors (Figure 4C), there was no detectible difference in blood vessel density at the time of tumor excision (as indicated by CD31 staining, Supplementary Figure S5). This may be due to the excision timing as all surgeries were performed when a significant tumor burden was reached (500–1500 mm3) rather than at the same number of days post injection. We speculate that while ERRα regulation of VEGF may play a role in the delayed growth kinetics of the MDA231 siERRα xenografts, activation of compensatory pathways may obscure any effect on vessel development by the time the tumors reach maximal size. Finally, whole-animal imaging for luciferase expression after primary tumor excision did not yield evidence of metastatic spread. Although the MDA-MB-231 cell line is characterized as highly aggressive, we hypothesize that the lack of metastases in our experiment may be due to heterogeneity of MDA-MB-231 cell lines and the particular subline used. Additional models such as cardiac and tail vein injection as well as alternative breast cancer cell lines will be required to test the effects of ERRα, manifest as a decrease in migration in vitro, on tumor metastasis in vivo.
Discussion
Since the first correlative study was performed demonstrating a relationship between ERRα expression and breast cancer prognosis (1), additional associations between the expression of this receptor and patient prognosis have been observed in several type of tumors (29–31). However, a causal relationship between ERRα activity and disease outcomes has not been established. In the present study, we have investigated the role of ERRα signaling in breast cancer by identifying ERRα-regulated genes and examining the effect of altering ERRα activity in vitro and in vivo. Our initial characterization of ERRα target genes in relation to estrogen-regulated genes revealed that the majority of ERRα activity is unrelated to ERα signaling and led us to focus further phenotypic characterization on the role of ERRα in ERα-negative tumors. In vivo studies revealed that ERRα knockdown had a dramatic inhibitory effect on tumor growth in mice, a finding of pivotal importance in the validation of this NR as a therapeutic target in breast cancer.
One of the primary hypotheses we sought to address in this study was that the role of ERRα in breast cancer is that of a modulator of estrogen signaling. This hypothesis was based on the observation that ERα and ERRα exhibit similar DNA-binding site preferences and the finding that both receptors are capable of activating ERE-mediated gene transcription in transient transfection assays. It was not surprising, therefore, that our studies in MCF-7 cells revealed that a large number of ERα-regulated genes were modified by ERRα activity. One particularly interesting example of ERRα-ERα cross-talk is CXCL12, a gene previously identified as an ERα target, that is thought to be involved in mediating the mitogenic actions of estrogen and that has also been implicated in the metastasis of breast cancer to bone (32, 33). Unlike CXCL12, the majority of the genes co-regulated by ERRα and ERα are positively regulated by estradiol and negatively regulated by ERRα following its activation by PGC-1α. This is consistent with reporter gene assays in which ERRα expression led to decreased estrogen-responsiveness of several promoters in MCF-7 cells (34). Previous studies have attributed this finding to competition for DNA binding by the two receptors or to transcriptional squelching subsequent to the sequestration of limiting coactivators (7, 10). Whole genome studies, such as ChIP, will enable an evaluation of the extent to which ERα and ERRα binding sequences overlap in endogenous genes and the relative importance of this mechanism of receptor cross-talk in breast tumor pathology.
Although potential points of ERRα-ERα cross-talk were identified, our microarray analysis demonstrated that a large portion of ERRα activity in breast cancer may be completely unrelated to estrogen signaling. It should be noted, however, that in contrast with previously reported MCF-7 microarray data, the majority of genes significantly regulated by ERα in this study were induced rather than repressed by estrogen treatment (35, 36). We hypothesize that is due to a combination of the specific experimental conditions and the parameters used for microarray analysis (applied uniformly to select ERRα and ERα regulated genes). In total, we may have under-estimated the ERα-ERRα cross-talk, particularly with respect to genes down-regulated by estradiol. Despite this potential limitation, we identified genes co-regulated by these receptors in different patterns, which likely represent distinct mechanisms of receptor crosstalk. Furthermore, under the conditions of our analysis, the predominant regulation on the largest class of genes was due to ERRα activation. As assessed by gene ontology, PGC-1α coactivation of ERRα had the largest impact on genes classified as regulators of metabolism, few of which were affected by ERα. This result led us to investigate the ERα-independent activity of ERRα with the hypothesis that this activity may contribute towards breast cancer pathology. We observed that ERRα knock-down in the ERα-negative MDA-MB-231 cell line significantly attenuated the growth rate of these cells as murine xenografts, suggesting that ERRα may be a viable therapeutic target for patients with ERα-negative tumors. Although the present studies are restricted to the ERα/PR/ERBB2 negative MDA-MB-231 cell line, a model for a particularly aggressive class of breast cancer, our preliminary and ongoing experiments using other cells suggest that the role of ERRα is not restricted to this type of breast cancer. Specifically, the observation that ERRα expression supports target gene expression in multiple cell lines lends credence to the hypothesis that ERRα has a general role in breast cancer biology (Figure 2B). Additional in vivo experiments will be necessary to determine whether the similarities in ERRα function at the level of target gene regulation translate to the level of cell phenotype and tumor growth.
One of our most interesting findings was that ERRα knockdown did not impact the proliferation rate of MDA-MB-231 cells in vitro whereas the same cells exhibited compromised growth propagated as xenografts. This is not completely unexpected given that the physiological roles of ERRα have been primarily described in tissues with high metabolic demand. We hypothesize that the microenvironment of a rapidly growing tumor recapitulates these conditions and thus highlights the functional significance of ERRα. Indeed, an examination of the MCF-7 array data revealed that all of the TCA enzymes and at least one subunit of each complex of the mitochondrial respiratory chain were positively regulated by ERRα. Building on experiments performed in skeletal muscle, our preliminary data (not shown) indicates that oxygen consumption is increased in MCF-7 breast cancer cells with activated ERRα (13). However, whether or not increased OXPHOS provides tumor cells with a growth advantage is a matter of considerable debate. Although an increased rate of OXPHOS has been associated with production of reactive oxygen species (ROS) and subsequent caspase-mediated apoptosis (37), recent evidence suggests that ERRα may enhance ROS detoxification. Specifically, breast cancer brain metastases exhibit increased levels of ERRα and PGC-1α and exhibit enhanced protection against the deleterious effects of oxidative stress (38). Interestingly, we observed in several breast cancer cell lines that ERRα increases the expression of SOD2, which inactivates superoxide anions, and GSTM1, an enzyme that buffers cells from ROS (Figure 2A). These anti-oxidant defenses may allow cancer cells to capitalize on the ability of ERRα to increase OXPHOS activity. This hypothesis is also supported by compelling studies that have defined a role for PGC-1α coactivation of ERRα in the response to oxidative stress in pathologic processes such as neurodegeneration (39). In addition to providing substrates for further aerobic metabolism, the TCA cycle may contribute to tumor growth in an OXPHOS-independent manner. Indeed, it has been shown recently that a primary function of the TCA cycle is to provide the intermediates needed for the production of non-essential amino acids, a rate-limiting process in rapidly growing cells (17). Clearly, the strong link between ERRα, metabolism and the growth and progression of breast tumors warrants further investigation.
Taken together our findings provide significant validation for ERRα as a therapeutic target in ERα-negative breast tumors. Pharmaceutical interest in ERRα as a drug target was initially stimulated by studies that highlighted the potential utility of agonists as treatments for metabolic disorders such as diabetes and obesity. However, despite tremendous effort, ERRα has proven to be an intractable target with respect to the development of agonists (40). In contrast, several small molecules have been developed that function as antagonists or inverse agonists by altering ERRα in such a manner as to block coactivator interactions (41). Although there are no published studies that examine the in vivo activities of these compounds, the mild phenotype observed in the ERRα knockout mouse suggests that systemic administration of this class of ERRα antagonists may exhibit minimal significant side effects (42). A priority for future studies, therefore, will be the analysis in vivo of appropriate ERRα inverse agonists and antagonists for their ability to recapitulate the effects on tumor growth observed subsequent to siRNA-mediated knockdown of ERRα. In conjunction with the growing body of literature defining the role of ERRα in tumor pathology, our work highlights the activity of ERRα in cellular models of ERα/PR/ERBB2 triple negative breast cancer, and suggests that ERRα may be an attractive therapeutic target in this aggressive and treatment-resistant type of breast cancer.
Supplementary Material
Acknowledgments
Grant support: NIH grant DK074652 and DK48807 (D.P. McDonnell), DOD grant W81XWH-06-1-0045 (R.A. Stein), Duke University Breast Cancer SPORE (M.W. Dewhirst)
We thank McDonnell lab members for helpful discussions and Drs. Chris Counter, Sally Kornbluth, Patrick Casey and Patrick Kelly for technical advice and reagents. This manuscript is dedicated to the memory of our colleague Alexander Ney for sharing with us his intellectual creativity and scientific insight.
References
- 1.Suzuki T, Miki Y, Moriya T, et al. Estrogen-related receptor alpha in human breast carcinoma as a potent prognostic factor. Cancer Research. 2004;64:4670–4676. doi: 10.1158/0008-5472.CAN-04-0250. [DOI] [PubMed] [Google Scholar]
- 2.Ariazi EA, Clark GM, Mertz JE. Estrogen-related receptor alpha and estrogen-related receptor gamma associate with unfavorable and favorable biomarkers, respectively, in human breast cancer. Cancer Research. 2002;62:6510–6518. [PubMed] [Google Scholar]
- 3.Fujimoto J, Alam SM, Jahan I, Sato E, Sakaguchi H, Tamaya T. Clinical implication of estrogen-related receptor (ERR) expression in ovarian cancers. J Steroid Biochem Mol Biol. 2007;104:301–304. doi: 10.1016/j.jsbmb.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Garber K. Energy boost: the Warburg effect returns in a new theory of cancer. J Natl Cancer Inst. 2004;96:1805–1806. doi: 10.1093/jnci/96.24.1805. [DOI] [PubMed] [Google Scholar]
- 5.Bonnelye E, Aubin JE. Estrogen Receptor-Related Receptor alpha: A mediator of estrogen response in bone. J Clin Endocrinol Metab. 2005;90:3115–3121. doi: 10.1210/jc.2004-2168. [DOI] [PubMed] [Google Scholar]
- 6.Vanacker JM, Pettersson K, Gustafsson JA, Laudet V. Transcriptional targets shared by estrogen receptor-related receptors (ERRs) and estrogen receptor (ER) alpha, but not by ER beta. EMBO Journal. 1999;18:4270–4279. doi: 10.1093/emboj/18.15.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang ZP, Teng CT. Estrogen receptor alpha and estrogen receptor-related receptor alpha 1 compete for binding and coactivator. Molecular And Cellular Endocrinology. 2001;172:223–233. doi: 10.1016/s0303-7207(00)00372-5. [DOI] [PubMed] [Google Scholar]
- 8.Yang NY, Shigeta H, Shi HP, Teng CT. Estrogen-related receptor, hERR1, modulates estrogen receptor-mediated response of human lactoferrin gene promoter. Journal of Biological Chemistry. 1996;271:5795–5804. doi: 10.1074/jbc.271.10.5795. [DOI] [PubMed] [Google Scholar]
- 9.Xie W, Hong H, Yang NN, et al. Constitutive activation of transcription and binding of coactivator by estrogen-related receptors 1 and 2. Molecular Endocrinology. 1999;13:2151–2162. doi: 10.1210/mend.13.12.0381. [DOI] [PubMed] [Google Scholar]
- 10.Johnston SD, Liu XD, Zuo FG, et al. Estrogen-related receptor alpha 1 functionally binds as a monomer to extended half-site sequences including ones contained within estrogen-response elements. Molecular Endocrinology. 1997;11:342–352. doi: 10.1210/mend.11.3.9897. [DOI] [PubMed] [Google Scholar]
- 11.Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERR alpha) Journal Of Biological Chemistry. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 12.Huss JM, Torra IP, Staels B, Giguere V, Kelly DP. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor at signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Molecular And Cellular Biology. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mootha VK, Handschin C, Arlow D, et al. ERR alpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huss JM, Imahashi K, Dufour CR, et al. The nuclear receptor ERRalpha is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 2007;6:25–37. doi: 10.1016/j.cmet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Kallinowski F, Schlenger KH, Runkel S, et al. Blood flow, metabolism, cellular microenvironment, and growth rate of human tumor xenografts. Cancer Res. 1989;49:3759–3764. [PubMed] [Google Scholar]
- 16.Mazurek S, Zwerschke W, Jansen-Durr P, Eigenbrodt E. Metabolic cooperation between different oncogenes during cell transformation: interaction between activated ras and HPV-16 E7. Oncogene. 2001;20:6891–6898. doi: 10.1038/sj.onc.1204792. [DOI] [PubMed] [Google Scholar]
- 17.DeBerardinis RJ, Mancuso A, Daikhin E, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaillard S, Dwyer MA, McDonnell DP. Definition of the molecular basis for estrogen receptor-related receptor-alpha-cofactor interactions. Mol Endocrinol. 2007;21:62–76. doi: 10.1210/me.2006-0179. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasper G, Reule M, Tschirschmann M, et al. Stromelysin-3 over-expression enhances tumourigenesis in MCF-7 and MDA-MB-231 breast cancer cell lines: involvement of the IGF-1 signalling pathway. BMC Cancer. 2007;7:12. doi: 10.1186/1471-2407-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schreiber SN, Emter R, Hock MB, et al. The estrogen-related receptor alpha (ERR alpha) functions in PPAR gamma coactivator 1 alpha (PGC-1 alpha)-induced mitochondrial biogenesis. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin J, Wu P-H, Tarr PT, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Gaillard S, Grasfeder LL, Haeffele CL, et al. Receptor-selective coactivators as tools to define the biology of specific receptor-coactivator pairs. Mol Cell. 2006;24:797–803. doi: 10.1016/j.molcel.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Arany Z, Foo SY, Ma Y, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature. 2008;451:1008–1012. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- 27.Mukhopadhyay RTR, Price JE. Increased levels of alpha6 integrins are associated with the metastatic phenotype of human breast cancer cells. Clin Exp Metastasis. 1999;17:325–332. doi: 10.1023/a:1006659230585. [DOI] [PubMed] [Google Scholar]
- 28.Macedo LF, Sabnis GJ, Goloubeva OG, Brodie A. Combination of anastrozole with fulvestrant in the intratumoral aromatase xenograft model. Cancer Res. 2008;68:3516–3522. doi: 10.1158/0008-5472.CAN-07-6807. [DOI] [PubMed] [Google Scholar]
- 29.Sun P, Sehouli J, Denkert C, et al. Expression of estrogen receptor-related receptors, a subfamily of orphan nuclear receptors, as new tumor biomarkers in ovarian cancer cells. Journal of Molecular Medicine. 2005;26:1699–1706. doi: 10.1007/s00109-005-0639-3. [DOI] [PubMed] [Google Scholar]
- 30.Cavallini A, Notarnicola M, Giannini R, et al. Oestrogen receptor-related receptor alpha (ERR alpha) and oestrogen receptors (ER alpha and ER beta) exhibit different gene expression in human colorectal tumour progression. European Journal of Cancer. 2005;41:1487. doi: 10.1016/j.ejca.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 31.Cheung CP, Yu S, Wong KB, et al. Expression and functional study of estrogen receptor-related receptors in human prostatic cells and tissues. J Clin Endocrinol Metab. 2005;90:1830–1844. doi: 10.1210/jc.2004-1421. [DOI] [PubMed] [Google Scholar]
- 32.Hall JM, Korach KS. Stromal cell-derived factor 1, a novel target of estrogen receptor action, mediates the mitogenic effects of estradiol in ovarian and breast cancer cells. Mol Endocrinol. 2003;17:792–803. doi: 10.1210/me.2002-0438. [DOI] [PubMed] [Google Scholar]
- 33.Muller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 34.Kraus RJ, Ariazi EA, Farrell ML, Mertz JE. Estrogen-related receptor alpha 1 actively antagonizes estrogen receptor-regulated transcription in MCF-7 mammary cells. Journal Of Biological Chemistry. 2002;277:24826–24834. doi: 10.1074/jbc.M202952200. [DOI] [PubMed] [Google Scholar]
- 35.Frasor J, Danes JM, Komm B, Chang KCN, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: Insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 36.Carroll JS, Meyer CA, Song J, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 37.Hervouet E, Simonnet H, Godinot C. Mitochondria and reactive oxygen species in renal cancer. Biochimie. 2007;89:1080–1088. doi: 10.1016/j.biochi.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 38.Chen EI, Hewel J, Krueger JS, et al. Adaptation of Energy Metabolism in Breast Cancer Brain Metastases. Cancer Res. 2007;67:1472–1486. doi: 10.1158/0008-5472.CAN-06-3137. [DOI] [PubMed] [Google Scholar]
- 39.St-Pierre J, Drori S, Uldry M, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 40.Hyatt SM, Lockamy EL, Stein RA, et al. On the Intractability of Estrogen-Related Receptor alpha as a Target for Activation by Small Molecules. J Med Chem. 2007;50:6722–6724. doi: 10.1021/jm7012387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Busch BB, Stevens WC, Jr, Martin R, et al. Identification of a selective inverse agonist for the orphan nuclear receptor estrogen-related receptor alpha. J Med Chem. 2004;47:5593–5596. doi: 10.1021/jm049334f. [DOI] [PubMed] [Google Scholar]
- 42.Luo JM, Sladek R, Carrier J, Bader JA, Richard D, Giguere V. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha. Molecular And Cellular Biology. 2003;23:7947–7956. doi: 10.1128/MCB.23.22.7947-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.